Abstract
Neuron migration defects are an important aspect of human neuropathies. The underlying molecular mechanisms of such migration defects are largely unknown. Actin dynamics has been recognized as an important determinant of neuronal migration, and we recently found that the actin-binding protein profilin1 is relevant for radial migration of cerebellar granule neurons (CGN). As the exploited brain-specific mutants lacked profilin1 in both neurons and glial cells, it remained unknown whether profilin1 activity in CGN is relevant for CGN migration in vivo. To test this, we capitalized on a transgenic mouse line that expresses a tamoxifen-inducible Cre variant in CGN, but no other cerebellar cell type. In these profilin1 mutants, the cell density was elevated in the molecular layer, and ectopic CGN occurred. Moreover, 5-bromo-2′-deoxyuridine tracing experiments revealed impaired CGN radial migration. Hence, our data demonstrate the cell autonomous role of profilin1 activity in CGN for radial migration.
Keywords: actin-binding protein, actin dynamics, actin treadmilling, cerebellar granule neurons, cerebellar development, cerebellum, cerebellar cortex, neuronal migration, profilin, radial migration
Abbreviations
- βGal
β-galactosidase
- βGal+
βGal-positive
- βGal−
βGal-negative
- BrdU
5-bromo-2′-deoxyuridine
- BrdU+
BrdU-positive
- CaMKIV
Ca2+/calmodulin-dependent protein kinase IV
- CGN
cerebellar granule neurons
- EGL
external granule cell layer
- IGL
internal granule cell layer
- Math1-Cre
Math1-mediated Cre expression
- ML
molecular layer
- NeuN
neuron-specific nuclear protein
- P
postnatal day
- Pfn1
profilin1
- PI
propidium iodide
Introduction
Neuron migration defects are an important aspect in human diseases, including lissencephaly, mental retardation and epilepsy.1 However, the mechanisms that contribute to neuron migration defects are poorly understood. An increasing number of studies emphasized a crucial role of actin dynamics in neuron migration,2–4 and mutations in actin regulatory proteins can cause neuron migration defects in humans.1 We recently reported that the actin-binding protein profilin1 is important for glial cell adhesion and radial migration of cerebellar granule neurons (CGN).5–7 As a consequence of impaired radial migration, we found an aberrant organization of cerebellar cortex layers and ectopically localized CGN in the molecular layer of profilin1 mutant mice. In these studies, by exploiting conditional profilin1 mutant mice and transgenic mice expressing Cre recombinase under the control of the Nestin promoter,8,9 we deleted profilin1 in all neural cells, including neurons and glial cells. Hence, it remained unknown whether profilin1 activity must be present in CGN to enable their radial migration. To clarify this, we here made use of Math1-Cre transgenic mice expressing a tamoxifen-inducible variant of Cre recombinase in CGN, but in no other cell type of the cerebellum.10 Tamoxifen injection during early postnatal development, prior to radial migration, resulted in efficient activation of Cre recombinase in CGN. Using this strategy, we inactivated profilin1 specifically in CGN during early postnatal development. By doing so, CGN radial migration was impaired and resulted in ectopic CGN in the molecular layer. These data demonstrate the in vivo relevance of profilin1 activity in CGN for radial migration and cerebellar development.
Results
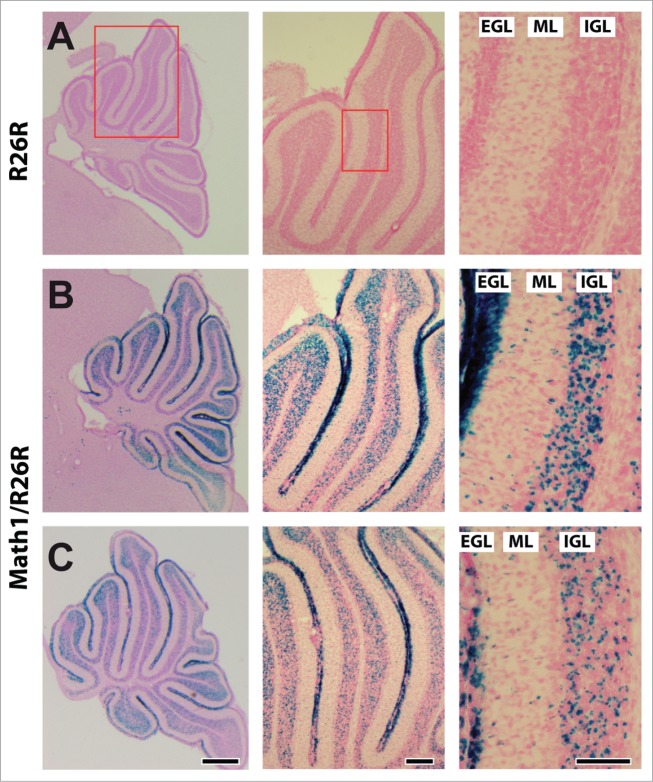
To test whether profilin1 activity in CGN is required for radial migration in vivo, we made use of conditional profilin1 mutants (Pfn1flx/flx) and Math1-Cre transgenic mice. The latter express a tamoxifen-inducible variant of Cre recombinase in hair cells of the inner ear and in CGN.8,10 To confirm efficient Cre activation in CGN, we intercrossed Math1-Cre mice with Rosa26 reporter (R26R) mice that express β-galactosidase (βGal) upon Cre activation.11 As expected, X-gal staining of 12-days-old Math1-Cre/R26R mice that received 3 tamoxifen injections during early postnatal development revealed βGal expression in all cerebellar folia, including folium 4 that was analyzed in further experiments (Fig. 1). In Math1-Cre/R26R mice, X-gal staining was evident in all 3 layers of the cerebellar cortex, consistent with an expression in CGN that, during postnatal development, migrate from the external granule cell layer (EGL) across the molecular layer (ML) into the internal granule cell layer (IGL).12 Hence, Pfn1flx/flx,Math1-Cre mice that receive tamoxifen injections during early postnatal development represent a valuable tool to clarify the in vivo relevance of profilin1 activity in CGN for the radial migration of these neurons.
Figure 1.
Tamoxifen-induced activation of Cre recombinase in CGN. (A–C) X-Gal (blue) and nuclear fast red (red) stained cerebelli of a R26R control and 2 Math1-Cre/R26R mice at P12 after 3 tamoxifen injections during early postnatal development. As indicated by the red boxes in A, high magnification (right micrograph) shows a section of folium 4 that was used for further analyses throughout this study. Active Cre, as indicated by blue X-Gal staining, is evident in all 3 layers of the cerebellar cortex, namely the external granule cell layer (EGL), the molecular layer (ML) and the internal granule cell layer (IGL). Scale bars in C correspond to 250 μm (left), 100 μm (middle) and 100 μm (right).
We first investigated the architecture of the cerebellar cortex in Pfn1flx/flx,Math1-Cre mice at postnatal day (P) 12 by exploiting propidium iodide, a fluorescent intercalating dye that non-specifically labels the somata of all cells. By doing so, we found all cerebellar folia being present in Pfn1flx/flx,Math1-Cre mice, and the cerebellar cortex layering appeared grossly normal (Fig. 2A, B), thereby excluding severe developmental defects. However, we noted a roughly 16% reduction in the IGL width in Pfn1flx/flx,Math1-Cre mice, with unchanged EGL and ML widths (Fig. 2C). Additionally, the ML cell density was increased by 19% in Pfn1flx/flx,Math1-Cre mice (Fig. 2D–F; Pfn1flx/flx: 4,424 ± 159 cells/mm2, n=18 images from 4 mice; Pfn1flx/flx,Math1-Cre: 5,741 ± 174 cells/mm2, n=17/4; P < 0.001). No changes in IGL width or ML cell density were noted in controls (Pfn1flx/flx,Math1-Cre mice that received corn oil injections; Fig. 2C and F).
Figure 2.
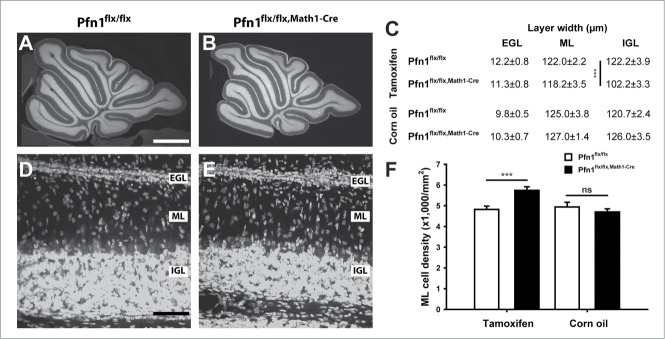
Elevated cell density in the molecular layer of P12 Pfn1flx/flx,Math1-Cre mice. (A and B) Propidium iodide-stained cerebellar sections of a Pfn1flx/flx control and a Pfn1flx/flx,Math1-Cre mouse at P12. Scale bar in A corresponds to 750 μm. (C) Layer thickness of the cerebellar cortex in tamoxifen- and corn oil-treated mice. IGL width, but not ML or EGL width, was reduced in tamoxifen-treated Pfn1flx/flx,Math1-Cre mice. No changes were observed in corn oil-treated Pfn1flx/flx,Math1-Cre controls. (D and E) Propidium iodide-stained folia 4 at high magnification. Scale bar in D corresponds to 40 μm. (F) ML cell density was increased in tamoxifen-injected Pfn1flx/flx mice at P12, but not in corn oil-treated Pfn1flx/flx,Math1-Cre controls. ns: not significant, ***: P < 0.001.
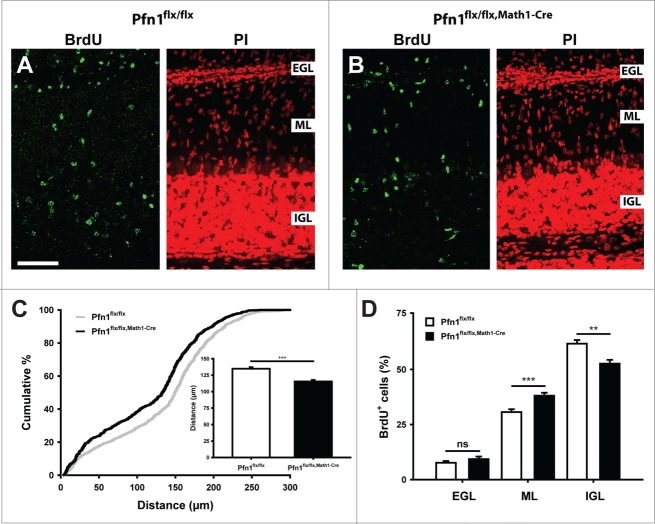
An increased ML cell density in Pfn1flx/flx,Math1-Cre mice would be in agreement with defective CGN radial migration. We therefore investigated radial migration in 5-bromo-2-deoxyuridine (BrdU) tracing experiments. To do so, we injected BrdU at P8, sacrificed the mice 96 h later, visualized BrdU-positive (BrdU+) cells in brain sections by immunohistochemistry (Fig. 3A and B) and quantified i) the distance of BrdU+ cells to the pial surface and ii) the distribution of BrdU+ cells within the cerebellar cortex. Compared to Pfn1flx/flx controls, the mean migration distance of BrdU+ cells was decreased by 15% in Pfn1flx/flx,Math1-cre mice (Fig. 3C; Pfn1flx/flx: 134.7 ± 2.3 μm, n=863 cells/14 images/3 mice; Pfn1flx/flx,Math1-Cre: 115.5 ± 2.3 μm, n=852/10/3; P < 0.001), and the distribution of BrdU+ cells within the cerebellar cortex was altered (Fig. 3D). While the relative number of BrdU+ cells in the EGL was similar between mutants and controls (Pfn1flx/flx: 7.7 ± 0.8%, Pfn1flx/flx,Math1-Cre: 9.3 ± 1.2%, P=0.270), it was increased in the ML and reduced in the IGL (ML: Pfn1flx/flx: 30.6 ± 1.3%, Pfn1flx/flx,Math1-Cre: 38.1 ± 1.3%, P < 0.001; IGL: Pfn1flx/flx: 61.7 ± 1.7%, Pfn1flx/flx,Math1-Cre: 52.6 ± 1.7%, P < 0.01). No changes were noted in the migration distance or distribution of BrdU+ cells in corn oil-injected Pfn1flx/flx,Math1-cre controls (data not shown). Hence, our data demonstrate that profilin1 activity in CGN is relevant for CGN radial migration in vivo.
Figure 3.
Impaired CGN radial migration in Pfn1flx/flx,Math1-Cre mice. (A and B) BrdU-positive cells (green) in folia 4 of a Pfn1flx/flx control and a Pfn1flx/flx, Math1-Cre mouse 96 h after a BrdU injection at P8. Sections were counterstained with propidium iodide (PtdIns, red). Scale bar in A corresponds to 50 μm. (C) Compared to Pfn1flx/flx controls, the migration distance of BrdU-positive cells is decreased in Pfn1flx/flx,Math1-Cre mice, as it is obvious from the right shift of the cumulative curve and the reduction in the mean migration distance (inset). (D) Distribution of BrdU-positive cells within the cerebellar cortex. Equal numbers were found in the EGL, whereas the relative number of BrdU-positive cells was increased in the ML and decreased in the IGL of Pfn1flx/flx,Math1-Cre mice. ns: not significant, **: P < 0.01, ***: P < 0.001.
We next investigated the cerebellar cortex architecture in Pfn1flx/flx,Math1-cre mice at P30, when cerebellum development is accomplished.13 At this stage, the cerebellum appeared grossly normal (Fig. 4A and B), and calbindin and GFAP immunolabeling revealed normal development and organization of Purkinje cells and Bergmann glia, respectively (Fig. 4C–F). However, the ML cell density was increased by 32% in Pfn1flx/flx,Math1-cre mice, as judged from propidium iodide-stained sections (Fig. 5A–C; Pfn1flx/flx: 2,296 ± 138 cells/mm2, n=12/4; Pfn1flx/flx,Math1-Cre: 3,027 ± 150 cells/mm2, n=12/4; P < 0.01), suggesting the presence of ectopic CGN. To test whether the increased ML cell density was indeed caused by ectopic CGN, we used an antibody against the neuron-specific nuclear protein (NeuN) that, within the cerebellar cortex, is solely expressed in CGN.14 Compared to Pfn1flx/flx controls, the density of NeuN-positive (NeuN+) cells was drastically increased by 169% in Pfn1flx/flx,Math1-cre mice (Fig. 5A, B, and D; Pfn1flx/flx: 350 ± 38 cells/mm2; Pfn1flx/flx,Math1-Cre: 941 ± 17 cells/mm2, n=9/3, P<0.01), while the density of NeuN-negative (NeuN-) cells was unchanged (Pfn1flx/flx: 1,616 ± 118 cells/mm2; Pfn1flx/flx,Math1-Cre: 1,859 ± 122 cells/mm2, n=9/3, P > 0.05). The presence of ectopic CGN in Pfn1flx/flx,Math1-Cre mice was confirmed by antibody staining against Ca2+/calmodulin-dependent protein kinase IV (CaMKIV), a marker for mature, postmigratory CGN15 (Fig. 5E and F). No increase was noted in the density of NeuN+ cells (Fig. 5D; Pfn1flx/flx: 224 ± 37 cells/mm2; Pfn1flx/flx,Math1-Cre: 183 ± 20 cells/mm2, n=4/2, P > 0.05) or the presence of CaMKIV+ cells in the ML of corn oil-injected Pfn1flx/flx,Math1-Cre controls (data not shown). Taken together, our data reveal ectopic CGN in the ML of Math1-Cre-Pfn1flx/flx mice and suggest that these cells appear to differentiate normally.
Figure 4.
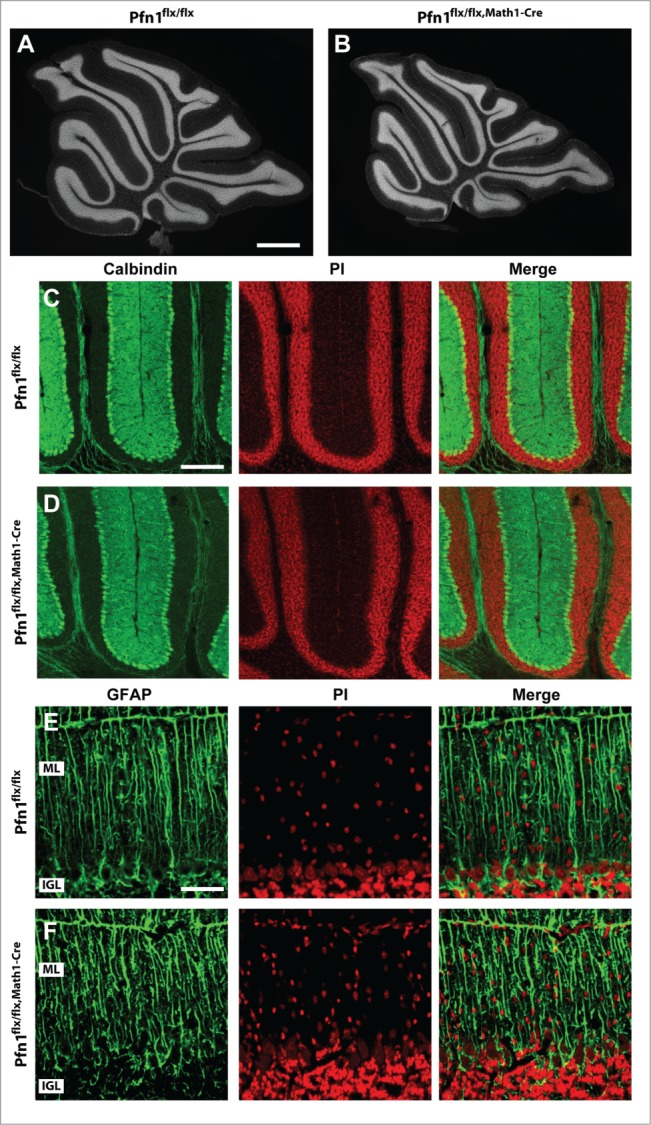
Normal organization of Purkinje cells and Bergmann glia in Pfn1flx/flx,Math1-Cre mice. (A and B) Propidium iodide-stained cerebellar sections of a Pfn1flx/flx control and a Pfn1flx/flx,Math1-Cre mouse at P30. Scale bar in A corresponds to 1 mm. (C and D) Calbindin immunoreactivity (green) revealed a normal density and organization of Purkinje cells in Pfn1flx/flx,Math1-Cre mice at P30. Sections were counterstained with propidium iodide (PI, red). Scale bar in C corresponds to 100 μm. (E and F) Likewise, the density and organization of Bergmann glia appeared normal in Pfn1flx/flx,Math1-Cre mice, as judged from GFAP immunoreactivity (green). Scale bar in E corresponds to 50 μm.
Figure 5.
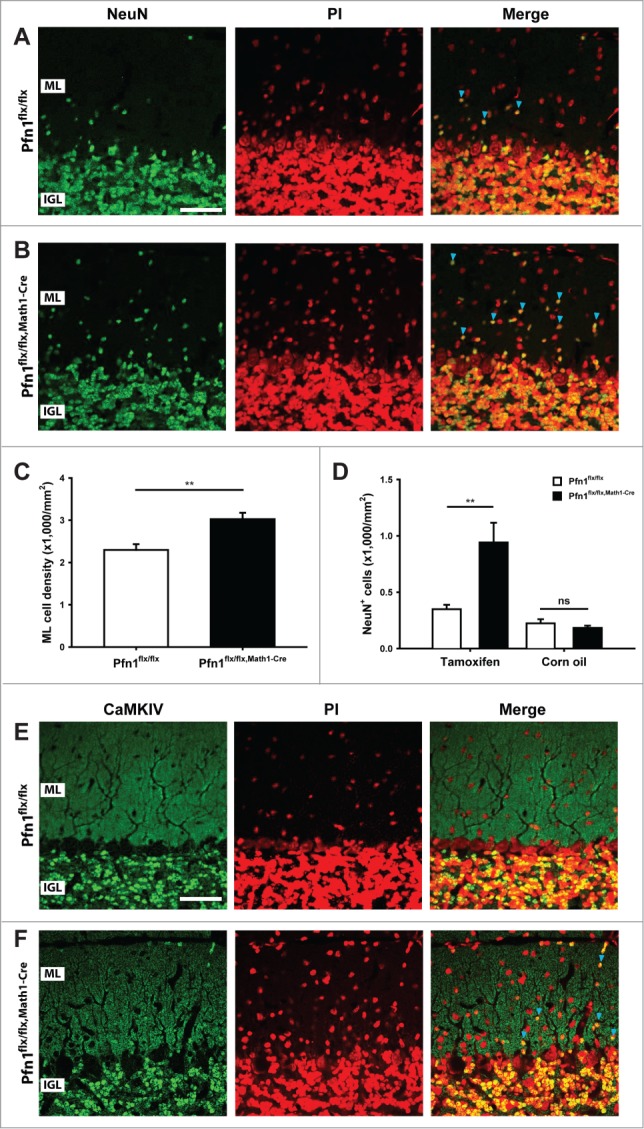
Ectopic CGN in the molecular layer of Pfn1flx/flx,Math1-Cre mice. (A and B) NeuN immunoreactivity (green) revealed ectopic CGN in the ML of Pfn1flx/flx,Math1-Cre mice at P30. Sections were counterstained with propidium iodide (PtdIns, red). Scale bar in A corresponds to 40 μm. Blue arrowheads mark NeuN-positive cells in the ML. (C) ML cell density was increased in P30 Pfn1flx/flx,Math1-Cre mice. (D) Likewise, the density of NeuN-positive cells was increased in the ML of tamoxifen-injected, but not of corn oil-injected Pfn1flx/flx,Math1-Cre mice. (E and F) CaMKIV immunoreactivity (green) confirmed the presence of ectopic CGN in the ML of Pfn1flx/flx,Math1-Cre mice (blue arrowheads). CaMKIV-positive cells were not observed in the ML of Pfn1flx/flx controls. Sections were counterstained with propidium iodide (PI, red). Scale bar in E corresponds to 40 μm. ns: not significant, **: P < 0.01.
Discussion
In the present study, we specifically deleted the actin-binding protein profilin1 in CGN, prior to their migration along radial processes of Bergmann glia. Deletion was accomplished by exploiting conditional profilin1 mutants and Math1-Cre transgenic mice. These CGN-specific profilin1 mutants displayed an elevated cell density in the ML and ectopic CGN. Moreover, an impaired CGN radial migration was demonstrated by BrdU tracing experiments. Hence, we conclude that profilin1 activity is relevant in CGN for their radial migration in vivo. This is in agreement with our previous in vitro studies that revealed a reduced velocity of isolated, profilin1-deficient CGN while migrating along processes of control glia cells.5 In the CGN-specific mutants, the impairments in cerebellar cortex cytoarchitecture and CGN radial migration are less pronounced than in mutants, in which profilin1 is removed from all brain cells, including CGN and Bergmann glia.5 For example, the migration distance of BrdU+ cells (96 h after BrdU injection at P8) was reduced by 35% in the brain-specific mutants, but only by 15% in the CGN-specific mutants. Furthermore, the number of NeuN+ cells was increased 5-fold in brain-specific mutants, but only 2-fold in CGN-specific mutants. These discrepancies can be explained by the mosaic Cre activation upon tamoxifen injection that we observed in our experiments (Fig. 1) and that was described for Math1-Cre mice.10,16 However, as suggested from our cell culture experiments with control CGN that showed a reduced velocity while migrating along processes of profilin1-deficient glia cells,5 profilin1 activity in Bergmann glia may be relevant for CGN radial migration, too, and the loss of profilin1 activity in Bergmann glia could contribute to the more profound defects in the brain-specific profilin1 mutants. Future analysis of glia cell-specific profilin1 mutants will clarify the relevance of profilin1 activity in glia cells for CGN radial migration. In summary, we here demonstrate the relevance of profilin1 activity in CGN for radial migration in vivo. Moreover, our study highlights the usefulness of Math1-Cre mice for studying CGN migration and function.
Material and Methods
Mice
Pfn1flx/flx,Math1-Cre mice were generated by intercrossing conditional profilin1 (Pfn1) mutants with transgenic mice expressing a tamoxifen-inducible variant of Cre recombinase, driven by the Math1 promoter.8,10 Deletion of Pfn1 was achieved by 2 to 3 intra-peritoneal injections of tamoxifen (0.1 mg/g body weight), diluted in corn oil, at P1 and P2 (2 injections) or P1, P2, and P3 (3 injections), respectively. If not otherwise stated, age-matched, tamoxifen-injected Pfn1flx/flx mice were used as controls throughout the study. In some experiments, corn oil-injected Pfn1flx/flx,Math1-Cre mice were used as additional controls. To label Cre expressing CGN, Math1-Cre mice were also intercrossed with Rosa26 reporter mice.11 Treatment of mice was in accordance with the German law for conducting animal experiments and followed the NIH guide for the care and use of laboratory animals.
X-gal staining
Twelve-day-old mice were perfused with 4% paraformaldehyde. Brains were post-fixed in the same fixative for 6 h and, thereafter, incubated o/n in 30% sucrose/PBS for cryoprotection. 50 μm-thick coronal sections were generated by using an HM 400R sliding microtome (Microm, Germany) and collected in 15% sucrose/PBS. X-Gal staining: Sections were fixed for 10 min in a PBS-based fixation solution containing (in %) 0.1 sodium desoxycholate, 0.2 NP-40, 1 paraformaldehyde, and 0.2 glutaraldehyde. Thereafter, sections were stained at 30°C for at least 2 h in a PBS-based staining solution containing 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.1% sodium deoxycholate, 0.2% NP-40, and 1 mg/ml X-gal in PBS. After washing, sections were mounted on glass slides, counterstained with Nuclear Fast Red (Vector Laboratories, USA), dehydrated in ethanol and xylol, and embedded in Entellan (Merck, Germany). Images were generated using an Axioskop2 microscope (Zeiss, Jena, Germany), a DP20 CCD camera system (Olympus), and CellP software (Olympus).
Antibodies used were as follows: BrdU (1:500, clone BU33, #B2531, Sigma-Aldrich), calbindin (1:1,000, clone CL-300, #C8666, Sigma-Aldrich), NeuN (1:500, clone A60, #MAB377, Millipore), and GFAP (1:500, Z0334, DakoCytomation). Antibody against CaMKIV was described before.15
Immunohistochemistry
50-μm-thick parasagittal brain slices were generated as mentioned above and blocked for 1 hour in 2% BSA, 3% normal goat serum, 0.5% NP-40/PBS at room temperature. Sections were incubated overnight with primary antibodies in blocking solution at 4°C followed by 3 washing steps of 5 min each in PBS. After an additional 30 min blocking step, sections were incubated for 1 h with secondary antibodies (Alexa Fluor 488- or 647-conjugated goat anti-mouse or goat anti-rabbit, Invitrogen) in 2% BSA, 0.5% NP-40/PBS. Propidium iodide (1:1,000, #P4170, Sigma Aldrich) was used for nuclear counterstaining in some experiments. Sections were mounted using a homemade medium based on glycerol, polyvinylalcohol, and DABCO after washing in PBS. Fluorescence micrographs of whole parasagittal cerebellum slices were acquired with an Axioskop2 microscope, 2.5x/0.12 objective and F-View II FW camera system and manually assembled. All other fluorescence micrographs were acquired on a Zeiss LSM 510 confocal microscope using 20x/0.8 and 40x/1.3 objectives and processed as described before (Herde et al., 2010). Folium 4 was chosen for counting NeuN+ and CaMKIV+ in the ML at P30.
BrdU experiments
P8 mice received a single BrdU injection (0.1 mg BrdU/g body weight). Tissue preparation and processing was performed 96 h later as described above. Prior to BrdU immunostaining, DNA was denaturated by 30 min incubation in 4 M HCl, followed by neutralization for 10 min in 0.1 M sodium borate at pH 8.5. Two 5-min washing steps were performed prior to the immunohistochemical procedure. Folium 4 was chosen for image acquisition. For quantifying CGN migration, distances of all BrdU+ cells in an image stack (optical thickness 10 × 3 μm) were measured using ImageJ software.
Statistics
Data are presented as mean values ± standard error of the mean. The unpaired 2-tailed Student´s t-test was used when comparing 2 sets of data with normal distribution. *P < 0.05, **P < 0.01, ***<0.001.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank K Ociepka and T Kehrwald for excellent technical assistance, Dr. W Witke for critical discussions on the manuscript, Dr. SJ Baker for Math1-Cre mice, Dr. P Soriano for Rosa26 reporter mice and Dr. H Sakagami for CaMKIV antibodies.
References
- 1. Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell 2007; 128:29-43; PMID:17218253; http://dx.doi.org/ 10.1016/j.cell.2006.12.021 [DOI] [PubMed] [Google Scholar]
- 2. Rivas RJ, Hatten ME. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J Neurosci 1995; 15:981-9; PMID:7869123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bellenchi GC, Gurniak CB, Perlas E, Middei S, Ammassari-Teule M, Witke W. N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev 2007; 21:2347-57; PMID:17875668; http://dx.doi.org/ 10.1101/gad.434307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron 2009; 63:63-80; PMID:19607793; http://dx.doi.org/ 10.1016/j.neuron.2009.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kullmann JA, Neumeyer A, Gurniak CB, Friauf E, Witke W, Rust MB. Profilin1 is required for glial cell adhesion and radial migration of cerebellar granule neurons. EMBO Rep 2012; 13:75-82; PMID:22081137; http://dx.doi.org/ 10.1038/embor.2011.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rust MB, Kullmann JA, Witke W. Role of the actin-binding protein profilin1 in radial migration and glial cell adhesion of granule neurons in the cerebellum. Cell Adh Migr 2012; 6:1-5; PMID:22647934; http://dx.doi.org/ 10.4161/cam.19845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kullmann JA, Neumeyer A, Wickertsheim I, Bottcher RT, Costell M, Deitmer JW, Witke W, Friauf E, Rust MB. Purkinje cell loss and motor coordination defects in profilin1 mutant mice. Neuroscience 2012; 223:355-64; PMID:22864186; http://dx.doi.org/ 10.1016/j.neuroscience.2012.07.055 [DOI] [PubMed] [Google Scholar]
- 8. Böttcher RT, Wiesner S, Braun A, Wimmer R, Berna A, Elad N, Medalia O, Pfeifer A, Aszódi A, Costell M, Fässler R. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J 2009; 28:1157-69; PMID:19262563; http://dx.doi.org/ 10.1038/emboj.2009.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nature Genet 1999; 23:99-103; PMID:10471508; http://dx.doi.org/ 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- 10. Chow LM, Tian Y, Weber T, Corbett M, Zuo J, Baker SJ. Inducible Cre recombinase activity in mouse cerebellar granule cell precursors and inner ear hair cells. Dev Dyn 2006; 235:2991-8; PMID:16958097; http://dx.doi.org/ 10.1002/dvdy.20948 [DOI] [PubMed] [Google Scholar]
- 11. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999; 21:70-1; PMID:9916792 [DOI] [PubMed] [Google Scholar]
- 12. Chedotal A. Should I stay or should I go? Becoming a granule cell. Trends Neurosci 2010; 33(4):163-72; PMID:20138673; http://dx.doi.org/ 10.1016/j.tins.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 13. Carletti B, Rossi F. Neurogenesis in the Cerebellum. Neuroscientist 2008; 14:91-100; PMID:17911211; http://dx.doi.org/ 10.1177/1073858407304629 [DOI] [PubMed] [Google Scholar]
- 14. Weyer A, Schilling K. Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J Neurosci Res 2003; 73:400-9; PMID:12868073; http://dx.doi.org/ 10.1002/jnr.10655 [DOI] [PubMed] [Google Scholar]
- 15. Sakagami H, Umemiya M, Kobayashi T, Saito S, Kondo H. Immunological evidence that the beta isoform of Ca2+/calmodulin-dependent protein kinase IV is a cerebellar granule cell-specific product of the CaM kinase IV gene. Eur J Neurosci 1999; 11:2531-6; PMID:10383642; http://dx.doi.org/ 10.1046/j.1460-9568.1999.00675.x [DOI] [PubMed] [Google Scholar]
- 16. Ruiz de Almodovar C, Coulon C, Salin PA, Knevels E, Chounlamountri N, Poesen K, Hermans K, Lambrechts D, Van Geyte K, Dhondt J, et al . Matrix-binding vascular endothelial growth factor (VEGF) isoforms guide granule cell migration in the cerebellum via VEGF receptor Flk1. J Neurosci 2010; 30:15052-66; PMID:21068311; http://dx.doi.org/ 10.1523/JNEUROSCI.0477-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]