Abstract
Disruption of epigenetic gene control mechanisms involving histone acetylation in the brain causes cognitive impairment, a debilitating hallmark of most neurodegenerative disorders. Histone acetylation regulates cognitive gene expression via chromatin packaging control in neurons. Unfortunately, the histone acetyltransferases (HATs) that generate such neural epigenetic signatures and their mechanisms of action remain unclear. Our recent findings provide insight into this question by demonstrating that Tip60 HAT action is critical for morphology and function of the mushroom body (MB), the learning and memory center in the Drosophila brain. We show that Tip60 is robustly produced in MB Kenyon cells and extending axonal lobes and that targeted MB Tip60 HAT loss results in axonal outgrowth disruption. Functional consequences of loss and gain of Tip60 HAT levels in the MB are evidenced by defects in memory. Tip60 ChIP-Seq analysis reveals enrichment for genes that function in cognitive processes and accordingly, key genes representing these pathways are misregulated in the Tip60 HAT mutant fly brain. Remarkably, increasing levels of Tip60 in the MB rescues learning and memory deficits resulting from Alzheimer's disease associated amyloid precursor protein (APP) induced neurodegeneration. Our studies highlight the potential of HAT activators as a therapeutic option for cognitive disorders.
Keywords: amyloid precursor protein (APP), axonal outgrowth, cognitive function, Epigenetics, histone acetyltransferases (HAT), mushroom body, Tip60
Environmental stimuli provide neurons in the brain with instructive information that shapes synaptic connections, which in turn, impacts cognitive function.1-5 Such flexibility in neuronal response to a constantly changing environment relies on precise regulation of dynamic gene expression profiles that promote neuroadaptation.6-13 One of the most important of such experience-driven behavioral changes is learning and memory formation as it directly impacts cognitive ability.2,4,14,15 Epigenetic gene control has recently emerged as a fundamental mechanism by which activity dependent cognitive genes transcriptionally respond to external cues in post-mitotic neurons.5 One of the best characterized epigenetic mark crucial for learning and memory is histone acetylation, that regulates cognitive gene expression by controlling chromatin packaging in neurons.3,5,7-9,15-17 Histone acetylation is regulated by the antagonistic activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs).7,18 HATs catalyze the transfer of an acetyl group from acetyl-CoA to the ε-amino group of specific lysine residues within the N-terminal tails of nucleosomal histones. Importantly, HATs also exhibit specific substrate preference for certain histone, lysine, and gene targets and thus generate different acetylation patterns within the neural epigenome.19,20 These HAT generated epigenetic signatures, in conjunction with additional DNA and histone modifications, serve as docking sites for the recruitment of distinct chromatin regulatory complexes that drive gene expression profiles critical for learning and memory.
Cognitive decline involving memory loss is a typical part of the aging process and has been associated with aberrant changes in gene expression in the hippocampus and frontal lobe of the brain.16 An emerging hypothesis is that age related accumulation of aberrant histone acetylation marks in chromatin in the adult brain cause gene misregulation that drives cognitive impairment. Indeed, over the past decade, numerous studies have reported reduced histone acetylation levels in the brains of animal models for multiple types of neurodegenerative diseases, including models for Alzheimer's, Parkinson's and Huntington's disease.8,21,22 Such acetylation loss in a p25-induced neurodegenerative mouse model causes an epigenetic blockade of learning and memory gene transcription with concomitant cognitive impairment.8 Accordingly, pharmacological treatments aimed at increasing global acetylation levels through the use of non-selective pan-HDAC inhibitors have shown promising effects in reversing cognitive deficits in a variety of neurodegenerative animal models23 making this a pow-erful therapeutic strategy.24-27 However, many currently used HDAC inhibitors (HDACi) lack target specificity18,28-33 and act by increasing global acetylation levels in the brain with potential detrimental effects, raising concerns about their applicability. Unlike some HDACs,18,29 select HATs have non-redundant neural functions that may not only restore general acetylation balance, but also modulate particular gene expression programs that work together to promote neuroprotection.7,10,34-39 Unfortu-nately, little is known about the select HATs that modify the neural epigenome, and the corresponding gene expression programs they control. Thus, a detailed analysis of the molecular mechanisms underlying neural HAT action and their gene target specificity in animals will likely provide safer and more selective ways to promote histone acetylation mediated cognitive enhancement benefits in clinical settings.
Insight into HAT based mechanisms underlying cognition and neuropathology was facilitated by our studies on Tip60 neural HAT function.7,17,30,34,40-44 While Tip60 was shown to be the second highest expressed HAT of the 18 HATs in mammalian adult brain, its involvement in neural function was unknown.45 We previously demonstrated that Tip60 HAT action is critical for nervous system development and cognitive processes such as synaptic plasticity, axonal outgrowth and transport via its transcriptional regulation of genes enriched for a variety of specific neuronal processes.40 Consistent with a role for Tip60 in nervous system function, our laboratory 7,17,30,34,40-44,46 and others 7,30,47-50 have shown that the HAT Tip60 is implicated in Alzheimer's disease (AD) based on its role in epigenetic neuronal gene control via its formation of a transcriptionally active complex with the processed C- terminal amyloid precursor protein (APP) intracellular domain (AICD).30,43,47,48,51-57 Loss of Tip60 HAT activity and/or improper recruitment of this complex to specific gene promoters causes epigenetic misregulation of a variety of genes causatively associated with neurodegeneratation.30,43,47,50,51
During the past several years, my laboratory has published a compendium of studies characterizing a functional interaction between Tip60 and the APP-C terminus (AICD) in mediating multiple cognitive neuronal processes using Tip60;APP transgenic Drosophila we generated as a robust model system.7,17,30,34,40-43 To develop this system, we utilized well characterized APP Drosophila lines that express equivalent and moderate levels of either GAL 4 responsive full length human APP (hAPP695) or APP lacking the AICD domain (APP-dCT)43,58,59 and adapted them to harbor our GAL4 responsive Drosophila Tip60 wild-type (Tip60WT) or dominant negative HAT mutant (Tip60E431Q) transgenes.40,43,46 This Tip60;APP Drosophila system enables us to modulate controlled Tip60 HAT levels in specific neural circuits in the fly of choice under APP induced neurodegenerative conditions, in vivo.40,43,46 Using this model, we demonstrated that a number of cognition linked processes known to be impaired during early AD neuropathology that include neuronal apoptosis, axonal outgrowth and transport are mediated via a functional interaction between Tip60 and AICD. We also made the exciting discovery that increasing in vivo Tip60 HAT levels in the Drosophila nervous system under APP induced neurodegenerative conditions rescues AD associated neuronal impairments such as apoptotic neurodegeneration in the central nervous system (CNS),43 axonal outgrowth 41,42 and synaptic vesicle transport in motor neurons.30 Excess Tip60 also restores associated disrupted complex functional abilities impaired in AD that include sleep cycles41,42 and locomotor function30 with concomitant induction of genes critical for the function of these neural processes.30,43 In direct contrast, loss of Tip60 HAT function in the fly nervous system causes gene misregulation and exacerbates such AD associated impaired phenoytpes.17,30,41-43 Our results highlight a novel functional interaction between Tip60 and AICD in neuronal processes associated with AD and support model in which Tip60 HAT action plays a neuroprotective role in early neurodegenerative progression via epigenetic reprogramming of gene sets that act together to promote neuroprotection.
While our previous findings highlighted a critical role for Tip60 in cognitive neuronal processes, the question of whether Tip60 HAT action is directly required for mediating gene expression changes that underlie learning and memory formation remained to be elucidated. To explore these questions, we chose to use the mushroom body (MB) in the Drosophila brain as a well-characterized model to study cognitive function in vivo. The MB is ideal for studying the transcriptional regulation of cognitive interneuronal development because it is highly plastic and forms discrete and stereotypical axonal projections that are easily visualized and tractable (Fig. 1). Moreover, MB neurons function to control multiple experience driven behavioral and cognition linked functions such as olfactory learning, decision making under uncertain conditions and courtship conditio-ning.60-64 The MB is comprised of the neuronal Kenyon cells (KC) that undergo ordered differentiation to generate 3 types of neurons, the α/α′, β/β′ and γ neurons.65 Each neuron projects dendrites that comprise the large dendritic field termed the calyx and an axon that travels anteroventrally to generate the α/α′ lobes and medially to form the β/β′ and γ lobes. During fly development, the α/β, α′/β′ and γ neurons undergo considerable structural reorganization.66,67 During metamorphosis, the γ neurons undergo a stereotypical process of axon destruction where the axons are dramatically pruned back to the peduncle and subsequently re-extend medially during pupal remodeling.68,69 These remodeling events rely on activity dependent refinement of neural circuits that lay the foundation for sustained synaptic plasticity5 in mature animals. This process is critical for organisms to learn from changing environmental stimuli and to remember critical concepts learned. One such complex behavioral learning paradigm that the Drosophila MB functions in is termed courtship conditioning. This cognitive process requires multimodal external sensory input, involving chemosensory, mechanosensory, visual and olfactory pathways and is thus well suited to study experience dependent synaptic plasticity involved in learning and memory.70-73 The ease and reproducibility of the courtship conditioning learning and memory assay, and the vast array of genetic tools that enable targeted gene expression manipulation to specific subregions of the MB74-76 make the MB a powerful model system for molecular dissection of the morphological and functional circuitry underlying experience driven cognitive plasticity.
Figure 1.
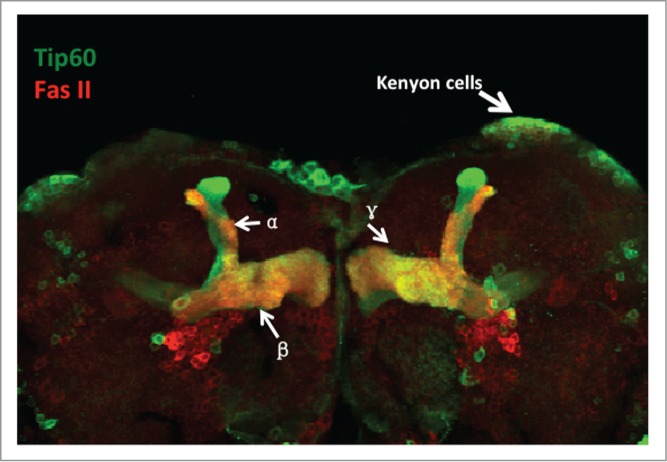
Drosophila mushroom body (MB) neurons co-immunostained with antibodies that label Tip60 shown in green and Fasciclin II (Fas II) shown in red. Fas II is a cell adhesion molecule that is expressed strongly in the MB α/β lobes and weakly in the γ lobe in the MB of the adult fly brain. Appropriate levels of Tip60 are required for axon outgrowth in the adult Drosophila MB.
In this study by Xu et al., we set out to test the hypothesis that Tip60 HAT action plays an epigenetic-based transcriptional regulatory role in cognitive function using the MB as a well-characterized cognitive model system. We found that Tip60 is widely produced throughout the adult brain, including the mushroom body (MB) lobes. Robust levels of Tip60 are localized within the nucleus of the Kenyon cells with reduced levels found localized within the cytoplasm. In addition to the Kenyon cells, Tip60 is also detected in the α/α′, β/β′ and γ lobes. Furthermore, while expression of a wild-type version of Tip60 (Tip60WT) in the MB using the MB specific OK107 driver reveals no defects in adult MB morphology, targeted MB expression of dominant negative HAT mutant Tip60 (Tip60E431Q) causes significantly thinner and shorter α/α′, β/β’ and γ lobes, indicative of axonal outgrowth impairment. The defects in the α and β lobes are observed in both sides of the brain in the dTip60E431Q mutants, indicating that the axonal defects are common to both the brain hemispheres. Importantly, no defects are observed in third instar larvae MB expressing either HAT mutant Tip60E431Q or Tip60WT, suggesting that Tip60 HAT activity is critical exclusively for adult MB development. We next tested whether Tip60 is required for MB function in learning and memory using the conditioned courtship suppression assay. Expression of either HAT mutant Tip60E431Q or wild-type Tip60WT in the MB does not impact the ability of flies to learn. In direct contrast, both loss and gain of Tip60 HAT levels in the MB result in impairment of immediate recall memory, indicating that appropriate levels of Tip60 are required for memory formation (Table 1). Consistent with the experience dependent memory formation deficits we observed, our ChIP-Seq analysis reveals that Tip60 target genes are enriched for functions in activity dependent cognitive processes, and that key genes representing these pathways are downregulated in the Tip60 HAT mutant fly brain with concomitant loss of Tip60 binding and Tip60 mediated histone acetylation marks at these specific gene loci (our unpublished results). Based on these data, we propose that misregulation of Tip60 mediated acetylation in the adult fly MB leads to aberrant changes in the chromatin landscape, causing epigenetic misregulation of genes that are induced following patterned synaptic stimulation, such as behavioral experiences. Such genes are thereby unable to carry out their function in converting the activity in neural circuits into shaping synaptic connections in the brain that allow for accessible memories. Interestingly, while we do observe that Tip60 HAT loss in the fly MB causes defects in adult but not larval MB axonal outgrowth, excess wild-type Tip60 HAT activity in the fly MB shows no obvious general developmental defects in both larval and adult stages. However, despite the lack of developmental defects, we find the memory in the Tip60WT flies is impaired as dramatically as in the HAT mutant Tip60E431Q flies. We speculate that such memory impairment is due to disruption of Tip60 mediated cellular acetylation homeostasis with subsequent negative consequences on MB synaptic connections, a model that our laboratory is currently exploring using higher resolution assays. Of note, our ChIP-Seq analysis also revealed a fraction of Tip60 target genes that function in general neural development. Moreover, while we observe no defects in third instar larval MB in response to loss or gain of Tip60, we do observe axonal outgrowth defects in the Tip60 HAT mutant adult MB. Therefore, we do not rule out the possibility that loss of Tip60 also causes transcriptional defects in genes required for general adult MB development, thereby compromising MB neuron function in memory.
Histone acetylation plays a critical role in cognitive function,7,34,48,77 and accordingly, reduced histone acetylation levels are found in the brains of animal models for multiple types of neurodegenerative diseases, including Alzheimer's disease (AD).8 Thus, we hypothesized that increasing Tip60 HAT levels in the brain would rescue AD associated cognitive deficits potentially resulting from APP expression in the MB. To test this hypothesis, we again used the conditioned courtship suppression assay to assess learning and memory, this time using our Tip60;APP Drosophila model that co-expresses equivalent levels of dTip60WT or dTip60E431Q with either APP or APP-dCT (APP lacking the C terminus that forms the transcriptional regulatory Tip60/AICD complex).78 We find that flies exhibit both learning and short-term memory defects when APP expression is targeted to the fly MB using GAL4 driver OK107. In direct contrast, equivalent levels of MB targeted APP-dCT expression causes no impairment in either learning or memory, indicating that APP induced cognitive deficits are dependent upon the presence of the Tip60 interacting C-terminus of APP. Of note, we find that targeted wild-type MB Tip60WT expression alone causes no learning deficits but does cause memory impairment and accordingly, these same phenotypes (Table 1) are recapitulated when APP-dCT that exhibits no phenotype on its own, is co-expressed with Tip60WT. Importantly, our studies reveal that targeted expression of wild-type Tip60WT in the fly MB rescues APP induced learning and memory deficits while MB expression of HAT mutant Tip60 does not rescue these APP induced deficits. Together, these findings indicate that Tip60 rescue of APP induced learning and memory defects is dependent upon a functioning Tip60 HAT domain.59,79 Our results support a neuroprotective role for Tip60 HAT activity that protects and/or promotes cognitive function under APP induced neurodegenerative conditions.
An important question to be addressed from our findings is how does Tip60 rescue both the learning and memory deficits caused by targeted APP expression in the MB? A clue to answering this question is the recent observations that histone acetylation is significantly reduced in the brains of animal models for a variety of neurodegenerative disorders.80 Accordingly, our recent findings reveal that histone acetylation is significantly reduced in the brains of APP expressing flies (our unpublished results) with concomitant loss of cognitive gene expression and induction of apoptotic cell death genes.30,43 Moreover, increasing Tip60 levels under APP conditions rescues these gene expression profiles and their Tip60 mediated histone acetylation marks. Consistent with our findings, recent studies have demonstrated that Tip60 not only functions as a transcriptional co-activator but also as a co-repressor. While the exact mechanism for how Tip60 represses gene expression remains unclear, in some instances it has been shown to occur via direct recruitment and interaction of Tip60 with transcriptional silencers and/or histone deacetylases.81,82 Based on these findings, we propose a model by which increased levels of Tip60 in the APP fly brain “resets” the histone acetylation chromatin landscape to mediate epigenetic reprogramming of gene expression programs via their activation or repression that in turn, protect and/or promote MB function in learning and memory (Fig. 2). Tip60 might exert such gene reprogramming either by itself or by forming complexes with other peptides like AICD for its recruitment to select genes via promoter bound TFs such as those we identified in our ChIP-Seq analysis. Indeed, other HATs have been shown to exert neuroprotective effects under neurodegenerative conditions in a similar fashion. For example, CBP was shown to ameliorate learning and memory deficits in a mouse AD model by increasing brain derived neurotropic factor (BDNF) 38 while p300 but not HDAC inhibitors was found to promote axonal regeneration by inducing axonal outgrowth genes.83 It is important to consider that modulation of specific HAT levels and/or activity might epigenetically reprogram multiple specific genes that function together to produce a neuroprotective effect, as suggested by our ChIP-Seq analysis for Tip60.7,34 Therefore, it will be critical to identity the full array of these genes to further dissect their neuroprotective nature for more effective design of specific HAT based therapeutic strategies.
Figure 2.
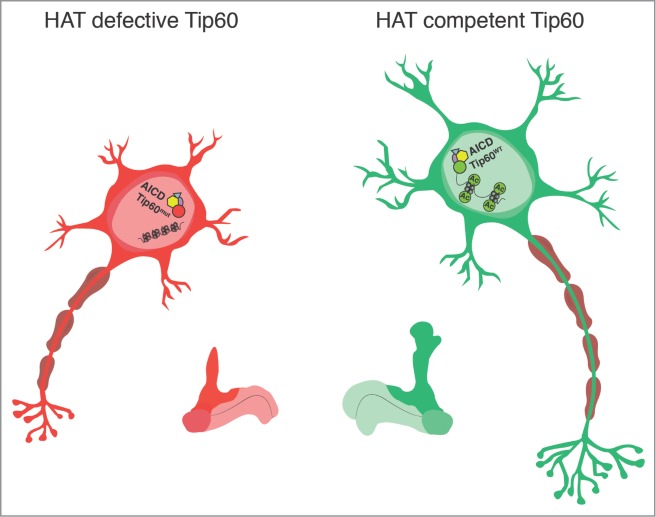
Model for the molecular events underlying rescue of APP learning and memory deficits by Tip60 HAT action under APP induced neurodegenerative conditions. Loss of Tip60 HAT activity (Tip60MUT) disrupts neural epigenetic histone acetylation signatures in the adult Drosophila MB Kenyon cells. Such changes in the chromatin landscape result in repression of cognitive genes, causing impairment of MB axonal outgrowth and synaptic connections with concomitant learning and memory deficits. Increasing cellular levels and/or enzymatic activity of specific HATs like Tip60 under APP neurodegenerative conditions might epigenetically reprogram cognitive gene cassettes that together, function to promote neuroprotection. As illustrated here, under APP induced neurodegenerative conditions, HAT competent Tip60 (Tip60WT) exerts neuroprotective effects either alone, or by complexing with AICD to epigenetically reprogram gene expression profiles via targeted histone acetylation that in turn, protects and/or promotes MB function. Graphic design and illustration of model figure by Cameryn S. Richards.
Another important question to consider is how does Tip60 respond to external cues to mediate a transcriptional response in neurons? Neural activity has been shown to modulate chromatin acetylation in hippocampal neurons in part, by controlling shuttling of certain HDACs in and out of the nucleus that influences their activity in gene control.3,84 Intriguingly, we observe both cytoplasmic and nuclear localization for the HAT Tip60 in activity dependent fly neuronal circuits that include the NMJ synaptic boutons44 and MB Kenyon cells. Thus, it is possible that external stimuli that is read as synaptic input might induce Tip60 shuttling into the nucleus that in turn, influences the neural epigenetic acetylation landscape and gene activity, a model that unpublished data from our laboratory supports. Future investigations into elucidating the mechanisms underlying Tip60 HAT function in experience driven cognitive function should serve as the foundation when exploring the utility of specific HAT activators that could potentially compliment existing non-invasive behavioral strategies for early therapeutic intervention of cognitive disorders.
Table 1.
Tip60 induced defects on MB axonal outgrowth and/or learning and memory function Drosophila adult flies
| Genotype | Effect on MB | Learning defect | Memory defect |
|---|---|---|---|
| Tip60 HAT mut | Partial defect | No | Yes |
| APP | Partial defect | Yes | Yes |
| APP; Tip60HAT mut | Complete loss | Yes | Yes |
| Tip60OE | No effect | No | Yes |
| APP; Tip60OE | Minor defect | No | No |
MB directed expression of HAT defective mutant Tip60 (Tip60HAT mut) causes MB axonal outgrowth defects and memory deficits. APP overexpression flies display a similar level of MB structural and functional defects in both learning and memory. These impairments are exacerbated in flies that co-express Tip60 HAT mutant with APP (APP;Tip60HAT mut) resulting in more severe MB morphological defects and learning and memory dysfunction. Tip60 overexpression results in memory defects with no significant effects on MB axonal structure, indicating appropriate levels of Tip60 are critical for memory formation. MB overexpression of Tip60 in conjunction with APP (APP;Tip60OE) flies rescues both learning and memory deficits and MB morphological defects possibly via epigenetic reprogramming of cognition linked expression profiles.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Cameryn S. Richards for graphic design and illustration of the model figure. We also thank Priyalakshmi Panikker and Rona Wilf for discussion of their scientific data.
Funding
This work was supported by National Institutes of Health (R01HD057939 to FE).
References
- 1.Puckett RE, Lubin FD. Epigenetic mechanisms in experience-driven memory formation and behavior. Epigenomics 2011; 3:649-64; PMID:22126252; http://dx.doi.org/ 10.2217/epi.11.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carulli D, Foscarin S, Rossi F. Activity-dependent plasticity and gene expression modifications in the adult CNS. Front Mol Neurosci 2011; 4:1-11; PMID:21441980; http://dx.doi.org/ 10.3389/fnmol.2011.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riccio A. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat Neurosci 2010; 13:1330-7; PMID:20975757; http://dx.doi.org/ 10.1038/nn.2671 [DOI] [PubMed] [Google Scholar]
- 4.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol 2011; 1:1-21; PMID:21555405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron 2008; 60:961-74; PMID:19109904; http://dx.doi.org/ 10.1016/j.neuron.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peixoto L, Abel T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 2013; 38:62-76; PMID:22669172; http://dx.doi.org/ 10.1038/npp.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirooznia K, Elefant F. Targeting specific HATs for neurodegenerative disease treatment: translating basic biology to therapeutic possibilities. Front Cell Neurosci 2013; 7:1-18; PMID:23355802; http://dx.doi.org/ 10.3389/fncel.2013.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M. et al.. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 2012; 483:222-6; PMID:22388814; http://dx.doi.org/ 10.1038/nature10849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci 2010; 13:1313-8; PMID:20975754; http://dx.doi.org/ 10.1038/nn1110-1313 [DOI] [PubMed] [Google Scholar]
- 10.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S. et al.. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010; 465:182-7; PMID:20393465; http://dx.doi.org/ 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron 2011; 70:813-29; PMID:21658577; http://dx.doi.org/ 10.1016/j.neuron.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res 2007; 61:58R-63R; PMID:17413844; http://dx.doi.org/ 10.1203/pdr.0b013e3180457635 [DOI] [PubMed] [Google Scholar]
- 13.Roth TL, Roth ED, Sweatt JD. Epigenetic regulation of genes in learning and memory. Essays Biochem 2010; 48:263-74; PMID:20822498; http://dx.doi.org/ 10.1042/bse0480263 [DOI] [PubMed] [Google Scholar]
- 14.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci 2005; 6:108-18; PMID:15654323; http://dx.doi.org/ 10.1038/nrn1604 [DOI] [PubMed] [Google Scholar]
- 15.Nelson ED, Monteggia LM. Epigenetics in the mature mammalian brain: Effects on behavior and synaptic transmission. Neurobiol Learn Mem 2011; 1:53-60; http://dx.doi.org/ 10.1016/j.nlm.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweatt JD. Neuroscience. Epigenetics and cognitive aging. Science 2010; 328:701-2; PMID:20448174; http://dx.doi.org/ 10.1126/science.1189968 [DOI] [PubMed] [Google Scholar]
- 17.Xu S, Wilf R, Memon T, Sarthi J, Elefant F. Epigenetic regulation of learning and memory by Tip60 HAT action. Genetics 2014; 198(4):1571-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci 2010; 31:605-17; PMID:20980063; http://dx.doi.org/ 10.1016/j.tips.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 19.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 2000; 64:435-59; PMID:10839822; http://dx.doi.org/ 10.1128/MMBR.64.2.435-459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem 2001; 70:81-120; PMID:11395403; http://dx.doi.org/ 10.1146/annurev.biochem.70.1.81 [DOI] [PubMed] [Google Scholar]
- 21.Graff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiology Reviews 2011; 91:603-49; http://dx.doi.org/ 10.1152/physrev.00012.2010 [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ. et al.. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008; 40:897-903; PMID:18552846; http://dx.doi.org/ 10.1038/ng.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazantsev AG, Thompson L. M. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov 2008; 7:854-68; PMID:18827828; http://dx.doi.org/ 10.1038/nrd2681 [DOI] [PubMed] [Google Scholar]
- 24.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA. et al.. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A 2003; 100:2041-6; PMID:12576549; http://dx.doi.org/ 10.1073/pnas.0437870100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardian G, Browne SE, Choi DK, Klivenyi P, Gregorio J, Kubilus JK, Ryu H, Langley B, Ratan RR, Ferrante RJ. et al.. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J Biol Chem 2005; 280:556-63; PMID:15494404; http://dx.doi.org/ 10.1074/jbc.M410210200 [DOI] [PubMed] [Google Scholar]
- 26.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ. et al.. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science 2007; 317:516-9; PMID:17588900; http://dx.doi.org/ 10.1126/science.1143780 [DOI] [PubMed] [Google Scholar]
- 27.Monti B, Gatta V, Piretti F, Raffaelli SS, Virgili M, Contestabile A. Valproic acid is neuroprotective in the rotenone rat model of Parkinson's disease: involvement of alpha-synuclein. Neurotox Res 2010; 17:130-41; PMID:19626387; http://dx.doi.org/ 10.1007/s12640-009-9090-5 [DOI] [PubMed] [Google Scholar]
- 28.Langley B, Gensert JM, Beal MF, Ratan RR. Remodeling chromatin and stress resistance in the central nervous system: histone deacetylase inhibitors as novel and broadly effective neuroprotective agents. Curr Drug Targets CNS Neurol Disord 2005; 4:41-50; PMID:15723612; http://dx.doi.org/ 10.2174/1568007053005091 [DOI] [PubMed] [Google Scholar]
- 29.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009; 10:32-42; PMID:19065135; http://dx.doi.org/ 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson AA, Sarthi J, Pirooznia SK, Reube W, Elefant F. Increasing Tip60 HAT Levels Rescues Axonal Transport Defects and Associated Behavioral Phenotypes in a Drosophila Alzheimer's Disease Model. J Neurosci 2013; 33:7535-47; PMID:23616558; http://dx.doi.org/ 10.1523/JNEUROSCI.3739-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Wang X, Liu L. HDAC inhibitor trichostatin A-inhibited survival of dopaminergic neuronal cells. Neurosci Lett 2009; 467:212-6; PMID:19835929; http://dx.doi.org/ 10.1016/j.neulet.2009.10.037 [DOI] [PubMed] [Google Scholar]
- 32.Rouaux C, Panteleeva I, René F, Gonzalez de Aguilar JL, Echaniz-Laguna A, Dupuis L, Menger Y, Boutillier AL, Loeffler JP. Sodium valproate exerts neuroprotective effects in vivo through CREB-binding protein-dependent mechanisms but does not improve survival in an amyotrophic lateral sclerosis mouse model. J Neurosci 2007; 27:5535-45; PMID:17522299; http://dx.doi.org/ 10.1523/JNEUROSCI.1139-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franklin TB, Mansuy EM. The prevalence of epigenetic mechanisms in the regulation of cognitive funcitons and behaviour. Curr Opin Neurobiol 2010; 20:441-9; PMID:20451368; http://dx.doi.org/ 10.1016/j.conb.2010.04.007 [DOI] [PubMed] [Google Scholar]
- 34.Pirooznia K, Elefant F. Modulating epigenetic HAT activity: A promising therapuetic option for neurological disease? J Mol Cloning Genetic Recombination 2012; 1:1-3; http://dx.doi.org/ 10.4172/2325-9787.1000e102 [DOI] [Google Scholar]
- 35.Schneider A, Chatterjee S, Bousiges O, Selvi BR, Swaminathan A, Cassel R, Blanc F, Kundu TK, Boutillier AL. Acetyltransferases (HATs) as targets for neurological therapuetics. Neurotherapeutics 2013; 10:568-88; PMID:24006237; http://dx.doi.org/ 10.1007/s13311-013-0204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvi BR, Cassel JC, Kundu TK, Boutillier AL. Tuning acetylation levels with HAT activators: therapeutic strategy in neurodegenerative diseases. Biochim Biophys Acta 2010; 1799:840-53; PMID:20833281; http://dx.doi.org/ 10.1016/j.bbagrm.2010.08.012 [DOI] [PubMed] [Google Scholar]
- 37.Marek R, Coelho CM, Sullivan RK, Baker-Andresen D, Li X, Ratnu V, Dudley KJ, Meyers D, Mukherjee C, Cole PA. et al.. Paradoxical enhancement of fear extinction memory and synaptic plasticity by inhibition of the histone acetyltransferase p300. J Neurosci 2011; 31:7486-91; PMID:21593332; http://dx.doi.org/ 10.1523/JNEUROSCI.0133-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A 2010; 107:22687-92; PMID:21149712; http://dx.doi.org/ 10.1073/pnas.1012851108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. et al.. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J 2003; 22:6537-49; PMID:14657026; http://dx.doi.org/ 10.1093/emboj/cdg615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorbeck M, Pirooznia K, Sarthi J, Zhu X, Elefant F. Microarray analysis uncovers a role for Tip60 in nervous system function and general metabolism. PLoS One 2011; 6:e18412; PMID:21494552; http://dx.doi.org/ 10.1371/journal.pone.0018412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirooznia SK, Chiu K, Chan MT, Zimmerman JE, Elefant F. Epigenetic regulation of axonal growth of Drosophila pacemaker cells by histone acetyltransferase tip60 controls sleep. Genetics 2012; 192:1327-45; PMID:22982579; http://dx.doi.org/ 10.1534/genetics.112.144667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pirooznia SK, Elefant F. A HAT for sleep?: epigenetic regulation of sleep by Tip60 in Drosophila. Fly (Austin) 2013; 7:99-104; PMID:23572111; http://dx.doi.org/ 10.4161/fly.24141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pirooznia SK, Sarthi J, Johnson AA, Toth MS, Chiu K, Koduri S, Elefant F. et al.. Tip60 HAT activity mediates APP induced lethality and apoptotic cell death in the CNS of a Drosophila Alzheimer's disease model. PLoS One 2012; 7:e41776; PMID:22848598; http://dx.doi.org/ 10.1371/journal.pone.0041776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarthi J, Elefant F. dTip60 HAT activity controls synaptic bouton expansion at the Drosophila neuromuscular junction. PLoS One 2011; 6:e26202; PMID:22046262; http://dx.doi.org/ 10.1371/journal.pone.0026202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.K-Lysine acetyltransferase 2a regulates a hippocampal gene expression network linked to memory formation. PloS one 2014; 33:1912-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu X, Singh N, Donnelly C, Boimel P, Elefant F. The cloning and characterization of the histone acetyltransferase human homolog Dmel\TIP60 in Drosophila melanogaster: Dmel\TIP60 is essential for multicellular development. Genetics 2007; 175:1229-40; PMID:17179074; http://dx.doi.org/ 10.1534/genetics.106.063685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. et al.. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 2002; 110:55-67; PMID:12150997; http://dx.doi.org/ 10.1016/S0092-8674(02)00809-7 [DOI] [PubMed] [Google Scholar]
- 48.Fischer A. Targeting histone-modifications in Alzheimer's disease. What is the evidence that this is a promising therapeutic avenue? Neuropharmacology 2014; 80:95-102; PMID:24486385; http://dx.doi.org/ 10.1016/j.neuropharm.2014.01.038 [DOI] [PubMed] [Google Scholar]
- 49.Muller T, Meyer HE, Egensperger R, Marcus K. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease. Prog Neurobiol 2008; 85:393-406; PMID:18603345; http://dx.doi.org/ 10.1016/j.pneurobio.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 50.Ghosal K, Vogt DL, Liang M, Shen Y, Lamb BT, Pimplikar SW. Alzheimer's disease-like pathological features in transgenic mice expressing the APP intracellular domain. Proc Natl Acad Sci 106:18367-72; http://dx.doi.org/ 10.1073/pnas.0907652106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belyaev N, Naliaeva NN, Makova NZ, Turner AJ. Neprilysin gene expression requires binding of the amyloid precursor protein intracellular domain to its promoter: implications for Alzhiemer disease. EMBO 2008; 10:94-100; http://dx.doi.org/ 10.1038/embor.2008.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim HS, Kim EM, Lee JP, Park CH, Kim S, Seo JH, Chang KA, Yu E, Jeong SJ, Chong YH. et al.. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J 2003; 17:1951-3; PMID:12923068 [DOI] [PubMed] [Google Scholar]
- 53.Cao X, Sudhof TC. Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem 2004; 279:24601-11; PMID:15044485; http://dx.doi.org/ 10.1074/jbc.M402248200 [DOI] [PubMed] [Google Scholar]
- 54.Cao X, Sudhof TC. A transcriptionally ; correction of transcriptively active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 2001; 293:115-20; PMID:11441186; http://dx.doi.org/ 10.1126/science.1058783 [DOI] [PubMed] [Google Scholar]
- 55.Muller T, Concannon CG, Ward MW, Walsh CM, Tirniceriu AL, Tribl F, Kögel D, Prehn JH, Egensperger R. et al.. Modulation of gene expression and cytoskeletal dynamics by the amyloid precursor protein intracellular domain (AICD). Mol Biol Cell 2007; 18:201-10; PMID:17093061; http://dx.doi.org/ 10.1091/mbc.E06-04-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hass MR, Yankner BA. A gamma-secretase-independant mechanism of signal transduction by the amyloid precursor protein. J Biol Chem 2005; 280:36895-904; PMID:16103124; http://dx.doi.org/ 10.1074/jbc.M502861200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem 2001; 276:40288-92; PMID:11544248; http://dx.doi.org/ 10.1074/jbc.C100447200 [DOI] [PubMed] [Google Scholar]
- 58.Fossgreen A, Brückner B, Czech C, Masters CL, Beyreuther K, Paro R. Transgeneic Drosphila expressing human amyloid precursor protein show gamma-secretase activity and a blistered- wing phenotype. Proc Natl Acad Sci 1998; 96:13703-8; http://dx.doi.org/ 10.1073/pnas.95.23.13703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron 2001; 32:389-401; PMID:11709151; http://dx.doi.org/ 10.1016/S0896-6273(01)00496-2 [DOI] [PubMed] [Google Scholar]
- 60.Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci 2003; 4:266-75; PMID:12671643 [DOI] [PubMed] [Google Scholar]
- 61.Guven-Azkan T, DR L. Functional neuroanatomy of Drosophila olfactory memory formation. . Learn Mem 2014; 21:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farris SM. Evolution of complex higher brain centers and behaviors: behavioral correlates of mushroom body elaboration in insects. Brain Behav Evol 2013; 82:9-18; PMID:23979452; http://dx.doi.org/ 10.1159/000352057 [DOI] [PubMed] [Google Scholar]
- 63.Akalai DB, Wilson CF, Zong L, Tanaka NK, Ito K, Davis RL. Roles for Drosophila mushroom body neurons in olfactory learning and memory. Learn Mem 2006; 13:659-68; PMID:16980542; http://dx.doi.org/ 10.1101/lm.221206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr Biol 2005; 15:R700-713; PMID:16139203; http://dx.doi.org/ 10.1016/j.cub.2005.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 1999; 126:4065-76; PMID:10457015 [DOI] [PubMed] [Google Scholar]
- 66.Scott EK, Lee T, Luo L. enok encodes a Drosophila putative histone acetyltransferase required for mushroom body neuroblast proliferation. Curr Biol 2001; 11:99-104; PMID:11231125; http://dx.doi.org/ 10.1016/S0960-9822(01)00020-3 [DOI] [PubMed] [Google Scholar]
- 67.Rybak J, Menzel R. Anatomy of the mushroom bodies in the honey bee brain: the neuronal connections of the alpha-lobe. J Comp Neurol 1993; 334:444-65; PMID:8376627; http://dx.doi.org/ 10.1002/cne.903340309 [DOI] [PubMed] [Google Scholar]
- 68.Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 2003; 38:871-85; PMID:12818174; http://dx.doi.org/ 10.1016/S0896-6273(03)00295-2 [DOI] [PubMed] [Google Scholar]
- 69.Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol 2004; 14:678-84; PMID:15084282; http://dx.doi.org/ 10.1016/j.cub.2004.03.035 [DOI] [PubMed] [Google Scholar]
- 70.Mehren JE, Ejima A, Griffith LC. Unconventional sex: fresh approaches to courtship learning. Curr Opin Neurobiol 2004; 14:745-50; PMID:15582378; http://dx.doi.org/ 10.1016/j.conb.2004.10.012 [DOI] [PubMed] [Google Scholar]
- 71.Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci 2007; 8:341-54; PMID:17453015; http://dx.doi.org/ 10.1038/nrn2098 [DOI] [PubMed] [Google Scholar]
- 72.Busto GU, Cervantes-sandoval I, Davis RL. Olfactory learning in Drosophila. Physiology 2010; 26:338-46; http://dx.doi.org/ 10.1152/physiol.00026.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dubnau J, Tully T. Functional anatomy: from molecule to memory. Curr Biol 2001; 11:R240-243; PMID:11301272; http://dx.doi.org/ 10.1016/S0960-9822(01)00115-4 [DOI] [PubMed] [Google Scholar]
- 74.Kahsai L, Zars T. Learning and memory in Drosophila: behaviour, genetics and neural systems. Int Rev Neurobiol 2011; 99:139-67; PMID:21906539; http://dx.doi.org/ 10.1016/B978-0-12-387003-2.00006-9 [DOI] [PubMed] [Google Scholar]
- 75.Fiala A. Olfaction and olfactory learning in Drosophila: recent progress. Curr Opin Neurobiol 2007; 17:720-6; PMID:18242976; http://dx.doi.org/ 10.1016/j.conb.2007.11.009 [DOI] [PubMed] [Google Scholar]
- 76.Siwicki KK, Ladewski L. Associative learning and memory in Drosophila: beyond olfactory conditioning. Behav Processes. 2003; 64:225-38; http://dx.doi.org/ 10.1016/S0376-6357(03)00137-2 [DOI] [PubMed] [Google Scholar]
- 77.Peixoto L, Abel T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 2013; 38:62-76; PMID:22669172; http://dx.doi.org/ 10.1038/npp.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siegel RW, Hall JC. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc Natl Acad Sci U S A 1979; 76:3430-4; PMID:16592682; http://dx.doi.org/ 10.1073/pnas.76.7.3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merdes G, Soba P, Loewer A, Bilic MV, Beyreuther K, Paro R. Interference of human and Drosophila APP and APP-like proteins with PNS development in Drosophila. EMBO J 2004; 23:4082-95; PMID:15385958; http://dx.doi.org/ 10.1038/sj.emboj.7600413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Graff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev 2011; 91:603-49; PMID:21527733; http://dx.doi.org/ 10.1152/physrev.00012.2010 [DOI] [PubMed] [Google Scholar]
- 81.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell 2000; 6:1195-205; PMID:11106757; http://dx.doi.org/ 10.1016/S1097-2765(00)00116-7 [DOI] [PubMed] [Google Scholar]
- 82.Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, Basu S, Riley JL, Hancock WW, Shen Y. et al.. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A 2007; 104:4571-6; PMID:17360565; http://dx.doi.org/ 10.1073/pnas.0700298104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaub P, Joshi Y, Wuttke A, Naumann U, Schnichels S, Heiduschka P, Di Giovanni S. The histone acetyltransferase p300 promotes intrinsic axonal regeneration. Brain 2011; 134:2134-48; PMID:21705428; http://dx.doi.org/ 10.1093/brain/awr142 [DOI] [PubMed] [Google Scholar]
- 84.Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependant nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem 2003; 85:151-9; http://dx.doi.org/ 10.1046/j.1471-4159.2003.01648.x [DOI] [PubMed] [Google Scholar]


