Abstract
The transcription factor GATA1 helps regulate the expression of thousands of genes involved in blood development, by binding to single or double GATA sites on DNA. An important part of gene activation is chromatin looping, the bringing together of DNA elements that lie up to many thousands of basepairs apart in the genome. It was recently suggested, based on studies of the closely related protein GATA3, that GATA-mediated looping may involve interactions of each of two zinc fingers (ZF) with distantly spaced DNA elements. Here we present a structure of the GATA1 ZF region bound to pseudopalindromic double GATA site DNA, which is structurally equivalent to a recently-solved GATA3-DNA complex. However, extensive analysis of GATA1-DNA binding indicates that although the N-terminal ZF (NF) can modulate GATA1-DNA binding, under physiological conditions the NF binds DNA so poorly that it cannot play a direct role in DNA-looping. Rather, the ability of the NF to stabilize transcriptional complexes through protein–protein interactions, and thereby recruit looping factors such as Ldb1, provides a more compelling model for GATA-mediated looping.
Keywords: GATA1, chromatin looping, DNA binding, protein–DNA structure, transcription factor complex
Introduction
GATA1 is a transcription factor that is a master regulator of red blood cell and megakaryocyte development. Mice in which GATA1 is disrupted die mid gestation with a complete absence of red blood cells.1 It is critical for the up- and down-regulation of thousands of genes, including the β-globin genes, where it is known to promote chromatin looping—bringing together enhancer elements in the locus control regions (LCR) and β-globin promoters.2 GATA1 and several of its associated proteins can contribute to long range gene regulation through DNA-looping. For example, chromatin conformation capture (3C) experiments show that each of GATA1, FOG1/ZFMP1, and LIM domain binding protein 1 (LDB1), are required to mediate looping between the β-globin locus control region (LCR) and the β-globin promoter.2,3 Other members of the GATA family have been shown to be involved in making long-range chromatin interactions, such as GATA2-associated DNA looping at the KIT1 gene,4 and GATA3 at the Th2 cytokine locus.5 Whereas the majority of transcription factors are displaced from chromatin during mitosis, GATA1 is retained at key haematopoietic regulatory genes.6 There GATA1 likely provides a “bookmarking” function for the re-recruitment of co-regulatory proteins such as TAL1/SCL1 and FOG1 as cells progress through the cell cycle.
GATA proteins contain two highly conserved C4-type ZFs, separated by a short basic linker: the more N-terminal N-finger (NF) and the more C-terminal C-finger (CF). Hereafter this whole ZF region is referred to as NC. The GATA1CF binds with high affinity and specificity to (A/T)GATA(A/G) motifs throughout the genome [e.g., Ref.7]. The GATA1NF can bind proteins, such as FOG18–10 and LIM-only protein 2 (LMO2).11 The GATA1NF has also been reported to bind independently to variant GAT(C/G) sequences,12 and modulate binding to ‘double GATA sites’, including the pseudopalindromic mPal sequence (CATCTGATA) in which GATA and GATG sites (underlined) exist on opposite strands [e.g., Ref.13]. The most commonly GATA-occupied double sites in the genome correspond to the mPal sequence.7
Structures have previously been determined for the isolated domains of GATA1 proteins,8,14–16 or for other GATA family proteins bound to DNA,17,18 including a structure of GATA3NC bound to mPal DNA.19 That structure showed one GATA3NC molecule binding to a single fragment of dsDNA. Interestingly, two additional GATA3NC–DNA complexes from the same study, involving other double GATA sites showed that GATA3NF and GATA3CF bound to separate strands of DNA, prompting the authors to suggest that GATA3 could be directly involved in DNA looping through binding to separate regions of DNA. Here we characterize the binding of GATA1NC to DNA. We present structures of two different crystal forms of GATA1NC bound to mPal DNA. Using a series of EMSAs and biophysical binding studies, we demonstrate that in the context of GATA1NC, the GATA1NF can indeed modulate binding to double GATA sites. However, the GATA1NF shows negligible independent binding to DNA at physiological concentrations of salt and is unlikely to mediate looping through contacts with DNA by the GATA1NF and GATA1CF. Rather, interactions with other proteins, including Ldb1-containing transcription factor complexes provide the likely physical basis of looping.
Results
Structure of GATA1NC bound to pseudopalindromic double-GATA site DNA
We generated GATA1NC, which includes both ZF domains of mouse GATA1. Size-exclusion chromatography with multiangle light scattering (SEC-MALLS) indicated that the protein was monomeric in solution (Supporting Information Fig. S1; MW 12.5±2.4 kDa, compared with a theoretical MW of 13 kDa). NMR chemical shift perturbation experiments using 15N-labelled GATA1NC and mPal DNA showed complete binding (peaks no longer shifted with increasing concentrations of DNA) at an apparent molar ratio of 2.2 molar equivalents of dsDNA:protein (Supporting Information Fig. S2). Using the same ratio, with 20-bp mPal-containing oligonucleotides equipped with complementary 2-bp overhangs (AA and TT), GATA1NC–mPal complexes were crystallized. Two different crystal forms gave rise to high quality diffraction data (Table1). The P21 form diffracted to a resolution of 1.98 Å, and contained a single 1:1 complex in the asymmetric unit (PDB ID: 3vd6). The P1 form diffracted to a resolution of 2.63 Å, and contained two identical copies of the 1:1 complex in the asymmetric unit (PDB ID: 3vek; r.m.s.d. over Cα of protein chains in this structure=0.08 Å).
Table 1.
| Space group | P21 | P1 |
|---|---|---|
| Unit-cell parameters (Å°) | a=54.33, b=37.40, c=65.48, α=90, β=98.57, γ=90 | a=35.20, b=62.42, c=66.05, α=72.39, β=74.57, γ=73.63 |
| Resolution (Å) | 15.00–1.98 (2.01–1.98) | 48.68–2.63 (2.70–2.63) |
| Completeness (%) | 99.5 (99.9) | 97.0 (94.3) |
| Redundancy | 3.4 (3.4) | 4.7 (4.4) |
| <I/σ(I)> | 19.1 (2.0) | 14.2 (3.4) |
| Rmerge† | 0.049 (0.58) | 0.088 (0.373) |
| Reflections in working set | 17316 (1142) | 13737 (676) |
| Reflections in test set | 887 (58) | 732 (56) |
| Rwork/Rfree | 0.218/0.247 | 0.235/0.275 |
| Contents of asymmetric unit | 1 Protein:DNA complex | 2 Protein:DNA complexes |
| Number of atoms | ||
| Protein | 756 | 1512 |
| DNA | 814 | 1628 |
| Ligand/ion | 7 | 14 |
| Water | 93 | 0 |
| B factors (Å2) | ||
| Protein | 53 | 62 |
| DNA | 60 | 69 |
| Ligand/ion | 62 | 67 |
| Water | 34 | N/A |
| R.m.s.d bond lengths (Å) | 0.005 | 0.004 |
| R.m.s.d bond angles (°) | 1.08 | 0.92 |
| Ramachandran plot | ||
| Favored (%) | 96.7 | 97.3 |
| Disallowed (%) | 0.0 | 0.0 |
| Refinement program | REFMAC5 | REFMAC5 |
| PDB code | 3VD6 | 3VEK |
Values in parentheses are for highest-resolution shell.
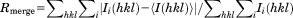 .
.
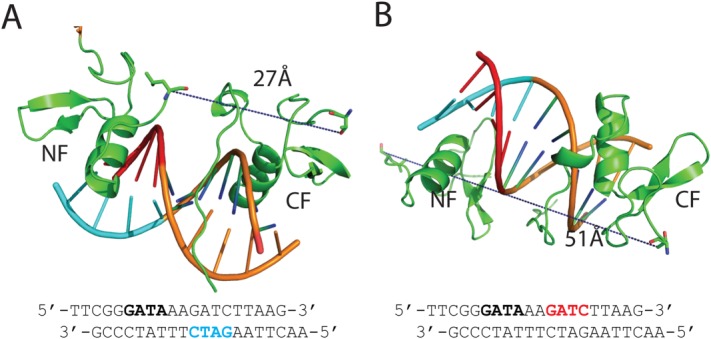
The structures of the molecules are identical in the two crystal forms [Supporting Information Fig. S3(A,B); r.m.s.d. over Cα of proteins=0.38–39 Å]. Residues A201–L241 and T256–S310 of GATA1NC are visible in the electron density maps. The missing 14 residues that link the two fingers (residues 242–255) are apparently disordered although SDS-PAGE analysis indicated that this region remained intact in the crystals. The GATA1NF and GATA1CF bind the DNA double helix on opposite sides [Fig. 1(A)]. Both fingers display a typical GATA-type structure comprising two β-hairpins followed by an α-helix. In each case a zinc ion is coordinated by four cysteine residues (C4 coordination), two each in the first β-hairpin and the α-helix. The GATA1CF contains an additional tail region (Q290–S310) that wraps around the DNA, and which includes a short helical turn (L295–Met297). Electron density exists for the complete DNA sequences, which are present as B-DNA. Base-pairing between adjacent strands [Supporting Information Fig. S3(C)] results in the DNA forming long, very straight double helical structures that lie side-by-side to form extended sheets; the relative orientations of the DNA in successive sheets varies between the two crystal forms. Subsequent analysis refers to the P21 crystal form. (PDB ID: 3vd6). With the exception of the GATA1CF tail, the GATA1NF and GATA1CF bind the pseudopalindromic DNA sequence in a generally symmetric fashion, with the main helix from each finger lying in the major groove of the DNA. The core regions of the GATA1NF and GATA1CF are 55% identical in terms of sequence and the structures are highly conserved (r.m.s.d. over the backbone atoms 0.63 Å for residues 202–241 and 256–295). Common sidechains involved in zinc coordination (C204/258, C207/261, C225/279, C228/282), hydrophobic packing (T212/266, W215/269) and DNA-binding (L214/268, R216/270, N226/280, L230/284, R239/293) all assume identical conformations [Fig. 1(B)]. In each case the protein principally makes contacts with the complementary strand of the GAT(A/G) motif, (C/T)ATC. However, the GATA1CF tail wraps around the DNA to make many additional interactions with DNA. In particular, the sidechains of residues R305 and R307 protrude deep into the minor groove. Based on the hydrogen bonds and hydrophobic contacts, sequence specific recognition is accomplished predominantly by the amino acids P213, L214, R216, N226, and L230 of the GATA1NF and the nucleobases Gua12, Cyt28, Ade29, Thy30 and Cyt31 of the GATG motif and L268, A280, L284, L288, R305 and R307 of the CF and Thy6, Thy7, Ade8, Thy9, Cyt10, Gua33, and Thy35 of the WGATAR motif [Fig. 1(C)]. In addition, Thy32, the base that separates the GATA and GATG motifs, is specifically bound by L268 and R270. Overall the GATA1CF makes many more contacts with the DNA (including both hydrogen bonds and hydrophobic interactions), than does the GATA1NF [Fig. 1(C)], which likely increases the affinity and specificity of the GATA1CF for its cognate DNA sequence.
Figure 1.
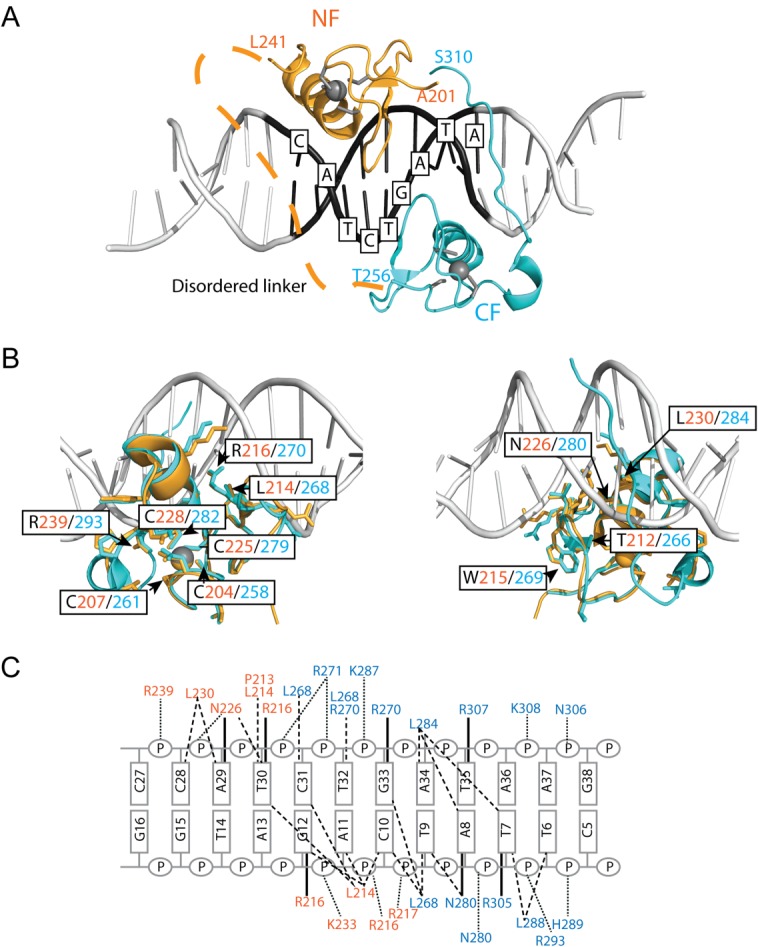
GATA1NC binds pseudopalindromic DNA in a 1:1 complex. (A) Overall structure of the GATA1NC:mPal Complex (3vd6). Ribbon diagram of GATA1NC bound to an mPal containing oligonucleotide (white and black to indicate the flanking and core mPAL sequence, respectively; the nucleotides in the core sequence are labelled). The NF is shown in orange and the CF is shown in cyan, with the zinc atoms and zinc-ligating sidechains shown as grey spheres and sticks. The linker region between the two fingers was not present in the electron density map and is represented by an orange dashed line. (B) The GATA1NF (orange) aligned with the GATA1CF (cyan) bound to DNA (white) shown from two angles. All residues that are identical in the sequences of the two domains are shown as sticks; residues discussed in the main text are labelled. (C) Schematic of DNA-protein interactions. Orange labels indicate NF and blue labels indicate CF interactions. Bold lines—hydrogen bonds between amino acids and bases; dashed lines—hydrophobic interactions; Dotted lines—contacts with sugar-phosphate backbone. An interactive view is available in the electronic version of the article.
These GATA1NC structures are consistent with the structures of other GATA proteins [Supporting Information Fig. S4(A–C)], including that of GATA3NC bound to an mPal oligonucleotide (PDB ID: 4hca).19 The sequence differences between GATA1NC and GATA3NC (most of which are homologous in nature) are largely restricted to the ends of structured regions of the domains, surface exposed residues, or residues in the NF-tail region, none of which appear to have an obvious role in DNA binding [Supporting Information Fig. S4(C,D)].
GATA1NC does not directly induce long-range contacts in DNA
The study that produced the GATA3NC-mPal structure also resulted in two related structures of GATA3NC bound to different double site oligonucleotides. In both of those structures, a molecule of GATA3NC protein spanned two adjacent stands of DNA, such that the GATA3CF contacted a GATA site on one strand of DNA and the GATA3NF contacted a GATT site on another strand of DNA (PDB IDs: 4hc7 and 4hc9; the asymmetric unit of the former contains a second molecule of GATA3NC, which apparently binds to a single molecule of DNA through both fingers as suggested previously,17 although the residues from the NF-tail are missing from the structures). The authors used an ingel FRET (Förster resonance energy transfer) assay as evidence that a single molecule of GATA3NC could facilitate looping in a oligonucleotide that contained Cy3- and Cy5-labelled ends. Each end comprises 10-bp dsDNA fragment, each containing a GATC or GATA site, separated by a 20-bp ssDNA linker.19 We attempted to use the same in-gel FRET approach to determine if GATA1NC could induce FRET in a similar experiment. In our hands 10-bp ends were not stable during electrophoresis (even at 4°C) so we redesigned the oligonucleotides to contain 30-bp ends of dsDNA with a 20-bp ssDNA polyT linker. Under these conditions we could see binding by GATA1NC, and a slightly different binding pattern for GATA1NC-R216W [Supporting Information Fig. S5(A)], a mutation which is known to inhibit binding of the GATA1NF (but not the GATA1CF) to DNA.20 These band shift patterns were very similar to those shown for GATA3NC and the equivalent GATA3NC-R276E mutation.19 However, although we saw some bleed-through of fluorescence under FRET conditions (Cy3 excitation and Cy5 emission), we saw no evidence of FRET [Supporting Information Fig. S5(B)]. FRET can be complicated by both distance and orientation of the fluorophores, so we carried out two-colour fluorescent EMSA experiments (which does not rely on FRET) in which separate dsDNA oligonucleotides containing either a Cy5-tag and a GATA site, or a Cy3-tag and a GATG site, were allowed to bind separately, or simultaneously, to GATA1NC, GATA1NC-R216W, and GATA1CF. In these experiments we saw some evidence of binding for each protein oligonucleotide pair. However, there was no significant difference between the binding patterns of the proteins to each of the isolated oligonucleotides, compared to when they were combined, suggesting that none of the proteins was able to bridge the two oligonucleotides (Fig. 2). Unexpectedly, it was noted that both GATA1NC-R216W and GATA1CF (neither of which contain a DNA-binding NF) showed evidence of binding to the GATG oligonucleotide [Fig. 2(B,C)]. This binding manifested as discrete bands at intermediate concentrations for GATA1NC-R216W and smeared rather than discrete bands for GATA1CF. Binding was apparently weaker than for the Cy5-GATA oligonucleotide, but suggested that the double banding pattern seen for ingel FRET experiments could arise from interactions of GATA1NC via the CF with each of the GATA and GATC sites, in the manner previously described for GATA3NC binding oligonucleotides that contain double GATA sites separated by 3-bp where the dsDNA cannot allow looping.17
Figure 2.
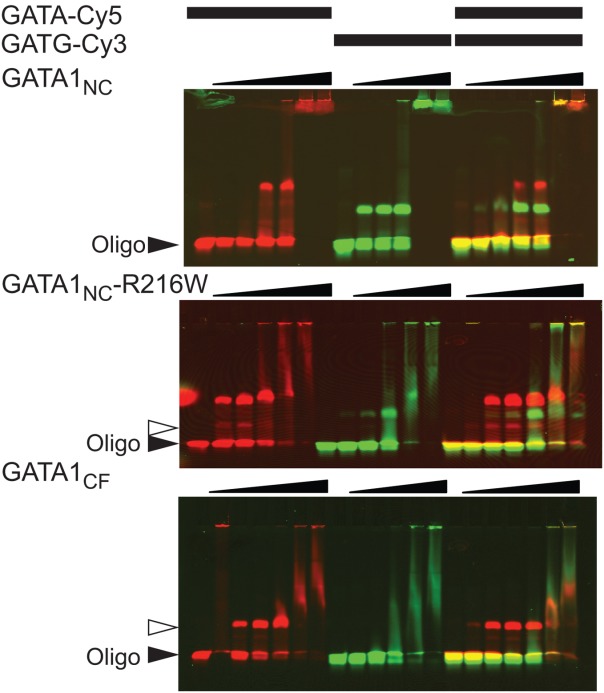
GATA1 is unable to bridge binding sites on separate DNA strands. EMSA analysis of GATA1NC, GATA1NC-R216W and GATA1CF binding to GATA-Cy5 (protein concentrations of 50, 100, 200, 400, and 600 nM protein, top and bottom panels also include 20 nM protein) and GATG-Cy3 (50, 100, 200, 400, and 600 nM protein) and both the GATA-Cy5 and GATG-Cy3 simultaneously (20, 50, 100, 200, 400, and 600 nM protein). Each oligonucleotide was at 20 nM. Images show the overlay of Cy5 (red) and Cy3 (green) fluorescence. The solid triangle indicates unbound DNA, the open triangle indicates GATA1CF bound to DNA. Other complexes are discussed in the text.
Under physiological salt concentrations GATA1CF, but not GATA1NF, can independently bind variant GATA sites
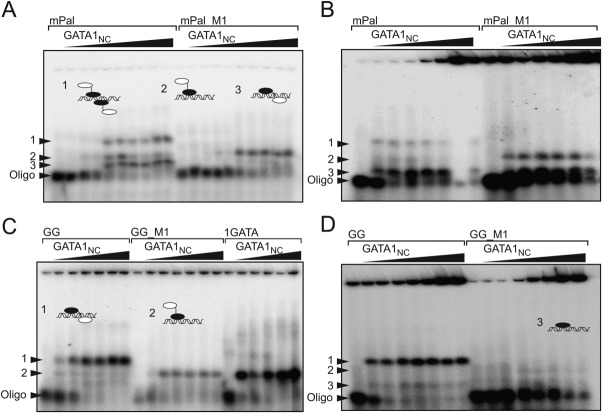
We further investigated the ability of GATA1NC, GATA1NF, and GATA1CF to bind single- and double-site dsDNA using standard radiolabelled EMSA. Many previous studies that probed GATA1-DNA binding used low concentrations of salt. All of the structural information to date indicates that the interactions have a strong electrostatic component, so we used both physiological salt (150 mM NaCl) concentrations to obtain a better understanding of GATA1-DNA interactions at the ionic strength conditions expected in the nucleus, and low salt (15 mM NaCl) for comparison with other studies. The oligonucleotides used included: mPal and GG (which contains the core sequence GGATAAAGATCT, originally shown to be conserved in the rat and mice testis promoter region21), variants of these sequences in which the GATG and GATC sites were mutated (mPal_M1 and GG_M1; Supporting Information Methods) and single site oligonucleotides as indicated. GATA1NF was only observed to bind DNA by EMSA under low salt conditions when the construct included an N-terminal GST-tag (Fig. 3), which is able to form dimers with a KD ≈ 10−6 M. 22 In contrast, untagged GATA1CF binds to both GG, mPal and mutant oligonucleotides; under low salt conditions for GG and GG_M1 we saw a single bandshift of similar migration to GG only [Fig. 3(B)], whereas for mPal and mPal_M1 we could see several additional bands that we assumed corresponded to additional molecules of GATA1CF binding to DNA [Fig. 3(C)].
Figure 3.
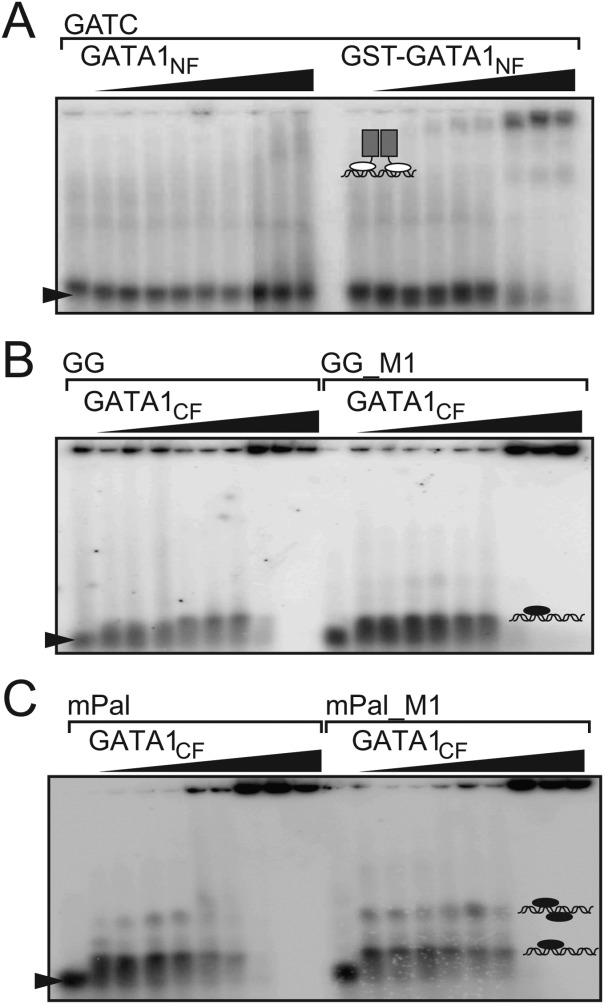
GATA1CF but not GATA1NF shows robust binding to DNA. EMSA analysis of (A) GATA1NF (lanes 2–10) or GST-GATA1NF (lanes 13–20) binding to a GATC-containing oligonucleotide at concentrations of 100, 200, 400, 600, 800, 1000, 2000, and 3000 nM at low salt (15 mM NaCl); and GATA1CF binding to (B) GG (lanes 2–10) or GG_M1 (lanes 12–20) at concentrations of 50, 100, 200, 400, 500, 800, 1000, 2000, 5000, and 7500 nM in 15 mM NaCl, or (C) to mPal (lanes 2–10) or mPal_M1 (lanes 12–20) at concentrations of 50, 100, 200, 400, 500, 800, 1000, 2000, 5000, and 7500 nM in 15 mM NaCl.
For GATA1NC binding to mPal, by EMSA we saw three shifted bands, and for mPal_M1 one shifted band that migrated to the same position as the middle-most mPal band. Under physiological salt conditions, the lowest mPal band became relatively more intense [Fig. 4(A,B)]. Based on these gel-shift patterns for both oligonucleotides we assumed that the middle band corresponded to one GATA1NC molecule binding via the GATA1CF to the GATA site, the lowest band corresponded to one GATA1NC molecule binding as shown in the crystal structure [Fig. 1(A)], and the top band probably corresponded to two molecules of GATA1NC binding to DNA. For GG, we saw one main gel-shifted band under both high and low salt conditions, with one or two less intense bands at lower positions, with the middlemost band migrating to the same position as bands seen for GG_M1 and another single site oligonucleotide (1GATA). Under high salt conditions that band was very smeared for GG_M1 [Fig. 4(C,D)]. We assumed that the middle band corresponded to one molecule of GATA1NC bound to the GATA site via the GATA1CF, and suspected that the lowest band represented small amounts of GATA1CF, originating from degraded GATA1NC binding to DNA (a similar band for the mPal oligonucleotide would be obscured by the main band). The top band was assumed to correspond to either a single molecule of GATA1NC binding to DNA via the GATA1NF and GATA1CF, or a more complex arrangement involving more than one copy of GATA1NC and/or DNA. Note that the migration position depends on the charge, shape (including the position of binding) so it is not possible to distinguish stoichiometry by EMSA alone.
Figure 4.
EMSA analysis of GATA1NC binding to double-site DNA. Binding of GATA1NC to (A) mPal (lanes 2–10) or mPal _M1 (lanes 12–20) at 10, 20, 50, 100, 200, 400, 600, 800, and 1000 nM on low salt (15 mM NaCl), (B) mPal (lanes 2–8) or mPal_M1 (lanes 10–15) at concentrations of 0.5, 1, 2, 3, 4, 5, and 6 µM in physiological salt (150 mM NaCl) (C) GG at 20, 50, 100, 200, 400, and 600 nM (lanes 2–7), GG_M1 oligonucleotide at 100, 200, 400, 600, 800, and 1000 nM (lanes 9–14) and to a single site GATA oligonucleotide (1GATA) at 50, 100, 200, 400, and 600 nM (lanes 16–20) in low salt (15 mM NaCl), (D) GG (lanes 2–8) and GG_M1 (lanes 9–16) at 0.5, 1, 2, 3, 4, 5, and 6 µM in physiological salt (150 mM NaCl). Schematics of complexes (indicated with numbered arrows) show the GATA1NF in white and GATA1CF in black. Note that the change in intensity throughout the final lane of panel C is an artefact from phosphorimaging.
We further tested binding using more quantitative equilibrium techniques, isothermal titration calorimetry (ITC) and microscale thermophoresis (MST; which monitors changes in the properties of isolated molecules and complexes moving across a thermal gradient). Under physiological salt concentrations we could see no binding of GATA1NF to GG DNA by ITC, suggesting that any binding would be weaker than KD=10−3 M [Table1; Supporting Information Fig. S6(A)]. By MST we could detect very weak binding to a GATC-containing oligonucleotide which was consistent with binding KD≥10−3 M, and no significant binding to a GATG-containing oligonucleotide [Supporting Information Fig. S6(B) and (C)]. Notably, we could measure binding of GATA1CF to the same oligonucleotides, both with a KD of ∼1 × 10−7 M, as determined by ITC [Supporting Information Fig. S6(D) and (E)]. We could also see binding by MST, but in these experiments the data was not adequately fitted by a simple 1:1 binding curve. Regardless, the inflection point in the binding curves was ∼1 × 10−6 M, indicating that at physiological concentrations of salt, GATA1CF binds to GAT(C/G) with an affinity at least three orders of magnitude higher than GATA1NF [Supporting Information Fig. S6(B) and (C)].
GATA1NF weakly modulates binding at double GATA sites
We used isothermal titration calorimetry (ITC) to estimate the binding affinities of GATA1NC and GATA1CF to mPal, mPal_M1, GG, and GG_M1 DNA (Table1). All of these interactions show evidence of biphasic binding (e.g., Supporting Information Fig. S7), consistent with NMR titration data indicating ∼2:1 binding (Supporting Information Fig. S2) rather than the 1:1 binding observed in the crystal structure. For most experiments, data could be fitted by a two-site model of binding, in which there was one stronger (KD≤∼10−8 M) and one weaker binding event (KD≥∼10−6 M). We note that because the data were fitted to a two-state model, and some binding events were very strong (KD≤∼10−8 M) or required fixing of parameters to fit the data, the absolute values of each parameter are likely to be less accurate than stated from the fits, but the trends are very consistent. For GATA1CF binding to mPal and mPal_M1, and GATA1NC binding to mPal there was a tight binding event corresponding to a KD≤∼10−8 (at the limit of detection by ITC) and a weak binding event at ∼10−6 M. For GATA1NC binding to mPal_M1, binding affinity for the tight event was reduced by ∼10-fold, but the weak event was not significantly different from the others in this series. For GG and GG_M1, binding was weaker but followed a similar pattern and was fitted by a two-site binding model (Table1). The exceptions here were GATA1NC binding to GG and GG_M1, both of which showed evidence of a weak binding event (in the final stages of the titration data was only slowly converging to zero ΔH with increasing concentrations of protein; Supporting Information Fig. S7) but the data could not be fitted by a two-site model. Rather a manually assigned baseline corresponding to a weak binding event was subtracted from the data before fitting to a single binding site model. Based on these experiments GATA1NC bound GG (KD=2 × 10−8 M) slightly more strongly than GG_M1 (KD=8.3 × 10−8 M). For the GATA1CF binding to GG and GG_M1, the tight binding events were identical within the error of the method (KD ≈ 2 × 10−8 M) but the weaker binding event was ∼5-fold weaker for GATA1CF binding to GG_M1 (KD=9 × 10−6 M) compared with GATA1CF binding to GG (KD=1.5 × 10−6 M). We interpret the tight binding events as GATA1NC binding through the GATA1CF to single GATA sites, or both the GATA1CF and GATA1NF to double GATA sites, with the weak binding event as the GATA1CF binding to GAT(C/G) sites and/or nonspecific binding. We note that weak and nonspecific binding are not distinguished by ITC analysis, but structural data exists for GATA3CF binding to a variant GATT site.17
A model for GATA1NC binding to GG DNA indicates binding on opposite strands
Given that the GATA1NF does not appear to be able to mediate an independent interaction with DNA, the ITC data and EMSA data for GATA1NC binding to GG are consistent with the main species being a tight 1:1 complex in which the GATA1CF binds the GATA site and the GATA1NF binds the GATC site. As the GATC site is palindromic, it is possible that the GATA1NF could bind in either orientation. We attempted to determine the structure of a GATA1NC-GG complex to resolve this issue. NMR titrations indicated that the interaction reached completion at the same apparent ratio as the mPal complex (Supporting Information Fig. S8), supporting the same 1:1 mode of binding, but we were unable to obtain crystals that diffracted to a sufficiently high resolution. However, molecular modelling indicates that the distances between the termini of the GATA1NF and GATA1CF are 27 Å for binding on the opposite stand and 51 Å for binding on the same strand. The linker region is not long enough to bridge this longer distance suggesting that GATA1NF binds to the opposite strand (Fig. 5).
Figure 5.
Model of GATA1NC in complex with GG DNA. (A) Model in which the NF contacts the GATC site (cyan) on the 3′ → 5′ strand. (B) Model in which the NF contacting the GATC site (red) on the 5′ → 3′ strand. The DNA sequence shown with the binding motif indicated. The minimum distances between the ends of the NF and CF are indicated.
GATA2NF and GATA3NF show negligible binding to DNA under physiological concentrations of salt
Finally, we tested the ability of the GATA2NF and GATA3NF to independently bind (GATC/G)-containing oligonucleotides using MST. Neither protein showed appreciable levels of binding to these sequences under physiological salt concentrations [Supporting Information Fig. S9(A) and (B)]. At 30 μM NaCl (the minimum salt concentration that prevented binding artefacts in the MST capillaries) all three GATANF proteins showed reasonable (∼μM) levels of binding to both sequences [Supporting Information Fig. S9(C) and (D)], but accurate estimates of binding are compromised by weaker, probably nonspecific, binding at higher concentrations of proteins.
Discussion
In contrast to earlier studies that suggested dimerisation of the GATA1NF and GATA1CF [e.g., Ref.23–24], we could see no evidence for dimerisation of GATA1NC. The purified protein behaved as a monomer in solution and none of our data are consistent with protein dimerisation. Note that the pulldown experiments used to define GATA–GATA interactions in those studies could give rise to binding artefacts.25 For example, interactions originally identified using GST-pulldown experiments between GATA1CF and another DNA-binding protein were subsequently shown to be false positives by other methods and were most likely mediated by indirect binding to nucleic acids.26
Our structural data indicate that, as might be expected for proteins that have such a high degree of sequence similarity, GATA1NC and GATA3NC bind pseudopalindromic DNA in an essentially identical manner. However, because the ability of GATA1NF to bind DNA under physiological salt concentrations is so weak, GATA1 is unlikely to directly mediate looping by making interactions with different fragments of DNA through the GATA1NF and GATA1CF of the same protein. We note that in some instances weak interactions between transcription factors and DNA are biologically important. For example, low affinity binding of homeobox transcription factor complexes were shown to bind clusters of low affinity sites in enhancers in vivo.27 In that case, the low affinity binding provides specificity, and binding appears to be enhanced through avidity effects, as multiple clustered sites were required for robust expression. In the case of GATA1, chromatin binding is dominated by WGATAR and mPal sequences7 and even if GATA1 does bind variant sites, our data indicates that the CF is a far better candidate for binding than the NF. Indeed mutations on the DNA-binding surface of the NF (R216Q and R216W; two disease-associated mutations in GATA1NF) do not significantly impair GATA1 target site occupancy in vivo.20 Thus, GATA1-associated looping is more likely to take place through interactions with other proteins, several of which are mediated through interactions with GATA1NF. In particular, GATA1 interacts with LDB1 via their common partner, LMO2,11 which is in turn recruited by TAL1/E2A complexes at GATA1-activated genes.28 LDB1, which can multimerize through an N-terminal self-association domain,29,30 was previously shown to be essential for GATA1-mediated DNA looping. 3,31. The role of this protein as a key mediator of looping was recently demonstrated by studies in which the self-association domain of LDB1 tethered to an artificial DNA-binding domain and targeted to LCR of the β-globin promoter was sufficient to induce looping and gene activation of targeted β- and γ-globin genes.32,33. For other GATA proteins we saw the same low DNA binding affinity for equivalent NF constructs (Supporting Information Fig. S9), suggesting that previous observations of direct looping by the GATA3 NF and CF could be artefacts. For example, bridging of DNA with a 3-bp spacer by GATA3NC could be crystallisation artefacts. However, it has been shown that extended GATA2NF and GATA3NF constructs that include a basic N-terminal region can have higher DNA-binding affinities.34 These sequences could allow a higher contribution of NF-binding to chromatin for those GATA factors, but are not present in GATA1.
The inability of GATA1NF to independently target DNA is supported by ChIP-seq studies showing that the main sequences bound by GATA1 in the genome are the canonical WGATAR and mPal sequences, and not GATC/G sequences.7 Our data are consistent with previous observations that the GATA1NF does modulate binding to double GATA sites [e.g., Ref.13]. For the double GATA sites we have tested, the loss of the GATA1NF or NF-binding sites tends to have a small effect on binding, from no significant difference for GATA1NC and GATA1CF binding to GG DNA, to less than an order of magnitude for GATA1NC binding to mPal or mPal_M1. Despite these small differences in binding affinity, many of our EMSA data show better defined gel shifts when the GATA1NF is available to bind, suggesting that this additional binding event changes the kinetics of binding, and the overall persistence of complexes—especially when GATA1 is targeting less highly favored sites (e.g., GATG or GGATA from the 2-color fluorescent EMSA and GG/GG_M1 sequences, rather than TGATA from mPal sequences, Supporting Information Figs. 3 and 4). Note that even though GATA1-bound mPal sites are highly represented in GATA1-activated genes, following an initial binding event the GATA1NF must be released from DNA in order to bind LMO2.11 So it is possible that the GATA1NF helps in recruiting GATA1 to DNA, or staying bound during mitosis, but is then made available to recruit other binding partners. Given that GATA1 can bind so many different target sites, and that looping can be sufficient to induce gene activation,32 it seems prudent to have extra levels of control (through recruitment of additional factors) in place to regulate looping and gene activation.
Material and Methods
SEC-MALLS
GATA1NC (residues 200–318 from murine GATA1) was generated as previously described.11 Chromatography used a Superose 12 10/30 column (GE Healthcare) at a flow rate of 0.5 mL/min in 50 mM Tris, 150 mM NaCl, 40 µM ZnSO4, 1 mM DTT, pH 7.4 with monitoring at 280, 215, and 260 nm. Light scattering and concentration data were collected by inline miniDawn Tristar laser-light scattering and Optilab DSP interferometric refractometer instruments (Wyatt Technology).
NMR spectroscopy
NMR spectra at 310 K were obtained for GATA1NC (∼0.2 mM), and GATA1NC:DNA complexes, in 20 mM 2-(N-morpholino)ethanesulfonic acid (MES), 150 mM NaCl and 1 mM DTT pH 6.5 with 5% D2O and 17 μM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS; used as an internal reference). Data were collected on Bruker AvanceIII 600 or 800 MHz spectrometers equipped with 5-mm triple resonance TCI cryoprobes, processed using Topspin (Bruker Biospin Ltd), and analyzed with Sparky (T. D. Goddard and D. G. Kneller, University of California at San Francisco).
Structure determination
Crystals were prepared by hanging-drop vapour diffusion at 293 K in which 400 nL each of protein:DNA complex (11 mg/ml in 50 mM TRIS pH 7.4, 100 mM NaCl, 40 μM ZnSO4, 1 mM DTT) and precipitant were mixed and suspended over a 75 μL reservoir of undiluted precipitant. Monoclinic crystals formed in 0.2 M ammonium acetate, 0.1 M BIS–TRIS pH 5.5, 25% PEG3350 within 1–3 days, and triclinic crystals grew in 0.01M ZnCl2, 0.1 M MES pH 6.0, 20% PEG6000 within 1–2 weeks. Crystals were cryoprotected with mother liquor supplemented with 30% ethylene glycol and flash-cooled in a cold nitrogen stream (100 K). X-ray diffraction data were collected inhouse using copper Kα X-rays produced by a Rigaku 007HF rotating-anode generator with Osmic Varimax optics and recorded on a MAR345 image plate (Marresearch GmBH). Data were indexed, integrated and scaled using HKL2000.35 The structure of the monoclinic crystal form was solved by molecular replacement using the coordinates of the C-terminal zinc finger of GATA3 bound to DNA (PDB ID 3dfx)17 as the search model in PHASER.36 The refined structure of this form was used as a search model for the triclinic crystal form. Refinement was carried out in REFMAC5.37 Manual map inspection and model building was performed in COOT.38 The quality of the model was checked using MolProbity.39
Interaction assays
EMSA using radiolabelled probes was carried out as previously described.11 For all oligonucleotide sequences see Supporting Information Methods. For two-colour fluorescent EMSA, complementary pairs of oligonucleotides in 20 mM Tris, 100 mM NaCl pH 7 were annealed by heating at 95°C for 15 min, then cooling slowly overnight to generate dsDNA with short single-stranded overhangs carrying fluorescent labels. GATA1NC, GATA1NC-R216W or GATA1CF were added to dsDNA (30 nM) containing either a GATA site and a Cy5 label, a GATG site and a Cy3 label, or both oligonucleotides simultaneously. Samples were made up in EMSA buffer (20 mM HEPES, 150 mM NaCl, 1 mM DTT, 6% Ficoll, pH 7.6), incubated on ice for 0.5 h, and run on an 8% polyacrylamide gel in 0.5× TBE buffer at 150 V for 2 h. For ingel FRET experiments equivalent amounts of ssDNA 30-mers containing either a GATA site and a Cy5 label, GATC site, and a Cy3 label were annealed to each end of a ssDNA 80-mer containing to generate 30-mer ds ends separated by a 20-nt poly(dT) spacer. GATA1NC or GATA1NC-R216W were added in EMSA buffer, and incubated on ice for 0.5 h. Samples were run on a 6% polyacrylamide gel in 0.5× TBE buffer at 150 V for 2 h at 4°C. All gels were imaged on a Typhoon FLA 9000 variable mode imager (Amersham Biosciences). For EMSA carried out with fluorescent probes, gels were imaged at an excitation wavelength of 532 nm for excitation of Cy3, or 650 nm, for excitation of Cy5. Fluorescence images were detected at emission wavelengths of 580 nm for Cy3 (false colored green) or 670 nm for Cy5 (false colored red). Where indicated, the two images were overlaid. Precipitated material is evident in the wells at the top of most EMSA gels.
For microscale thermophoresis (MST) or isothermal titration calorimetry (ITC), proteins and dsDNA oligonucleotides (with a 5′ fluorescein label for MST) were dialyzed overnight in at 4°C into 20 mM Tris, 1 mM DTT pH 7.5 with 30 or 150 mM NaCl, and passed through a 0.22 µM filter. MST experiments were performed at 25°C with a LED power of 50% and a laser power of 40%. dsDNA was held at a final concentration of 100 pM, protein concentrations are indicated. Data were analyzed using GraphPad. ITC experiments were performed at 10 or 20°C using VP-ITC or i200 microcalorimeters (MicroCal and GE healthcare). dsDNA was used at a concentration of ∼20 μM and GATA1 at ∼200 μM). Baseline data were measured by titration into buffer and subtracted from experimental data. Data were analyzed using the Origin 7.0 ITC Data analysis software package (MicroCal).
Acknowledgments
The authors have no conflicts of interest to declare.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
References
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the [beta]-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie HF, Schindler Y, Moran TB, Cheng Y, Yu DN, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CK, Simpson RJ, Kwan AH, Crofts LA, Loughlin FE, Matthews JM, Crossley M, Mackay JP. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc Natl Acad Sci USA. 2005;102:583–588. doi: 10.1073/pnas.0407511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Kowalski K, King GF, Mackay JP, Crossley M. Key residues characteristic of GATA N-fingers are recognized by FOG. J Biol Chem. 1998;273:33595–33603. doi: 10.1074/jbc.273.50.33595. [DOI] [PubMed] [Google Scholar]
- Matthews JM, Kowalski K, Liew CK, Sharpe BK, Fox AH, Crossley M, MacKay JP. A class of zinc fingers involved in protein–protein interactions biophysical characterization of CCHC fingers from fog and U-shaped. Eur J Biochem. 2000;267:1030–1038. doi: 10.1046/j.1432-1327.2000.01095.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson-White L, Gamsjaeger R, Dastmalchi S, Wienert B, Stokes PH, Crossley M, Mackay JP, Matthews JM. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci USA. 2011;108:14443–14448. doi: 10.1073/pnas.1105898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A, Mackay J, Crossley M. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem. 2001;276:35794–35801. doi: 10.1074/jbc.M106256200. [DOI] [PubMed] [Google Scholar]
- Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high- affinity interaction. Mol Cell Biol. 1996;16:2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski K, Czolij R, King GF, Crossley M, Mackay JP. The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG. J Biomol NMR. 1999;13:249–262. doi: 10.1023/a:1008309602929. [DOI] [PubMed] [Google Scholar]
- Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Biol. 1997;4:732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- Bates DL, Chen Y, Kim G, Guo L, Chen L. Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J Mol Biol. 2008;381:1292–1306. doi: 10.1016/j.jmb.2008.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich MR, Wikström M, Arst HN., Jr Clore GM, Gronenborn AM. The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors. J Mol Biol. 1998;277:605–620. doi: 10.1006/jmbi.1998.1625. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bates Darren L, Dey R, Chen P-H, Machado Ana Carolina D, Laird-Offringa Ite A, Rohs R, Chen L. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep. 2012;2:1197–1206. doi: 10.1016/j.celrep.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Wilkinson-White L, Mackay JP, Matthews JM, Blobel GA. Dissecting molecular pathways that underlie disease-causing GATA1 mutations. Blood. 2013;121:5218–5227. doi: 10.1182/blood-2013-03-488080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera K, Yomogida K, Suwabe N, Takahashi S, Muraosa Y, Hayashi N, Ito E, Gu L, Rassoulzadegan M, Engel JD, Yamamoto M. Conserved structure, regulatory elements, and transcriptional regulation from the GATA-1 gene testis promoter. J Biochem. 1997;121:251–263. doi: 10.1093/oxfordjournals.jbchem.a021581. [DOI] [PubMed] [Google Scholar]
- Riley LG, Ralston GB, Weiss AS. Multimer formation as a consequence of separate homodimerization domains: the human c-Jun leucine zipper is a transplantable dimerization module. Protein Eng. 1996;9:223–230. doi: 10.1093/protein/9.2.223. [DOI] [PubMed] [Google Scholar]
- Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay JP, Kowalski K, Fox AH, Czolij R, King GF, Crossley M. Involvement of the N-finger in the self-association of GATA-1. J Biol Chem. 1998;273:30560–30567. doi: 10.1074/jbc.273.46.30560. [DOI] [PubMed] [Google Scholar]
- Mackay JP, Sunde M, Lowry JA, Crossley M, Matthews JM. Protein interactions: is seeing believing? Trends Biochem Sci. 2007;32:530–531. doi: 10.1016/j.tibs.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Wissmueller S, Font J, Liew CW, Cram E, Schroeder T, Turner J, Crossley M, Mackay JP, Matthews JM. Protein–protein interactions: analysis of a false positive GST pulldown result. Protein. Struct Funct Gen. 2011;79:2365–2371. doi: 10.1002/prot.23068. [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, Mann RS, Stern DL. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015;160:191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Pfaff SL, Gill GN. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- Cross AJ, Jeffries CM, Trewhella J, Matthews JM. LIM domain binding proteins one and two have different oligomeric states. J Mol Biol. 2010;99:133–144. doi: 10.1016/j.jmb.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra R-J, Stevens M, Kockx C, van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory Philip D, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, Blobel GA. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. Embo J. 1997;16:2874–2882. doi: 10.1093/emboj/16.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information