Abstract
Protein-linked glycans play key roles in cell differentiation, cell–cell interactions, cell growth, adhesion and immune response. Aberrant glycosylation is a characteristic feature of tumor cells and is involved in tumor growth, escape from apoptosis, metastasis formation, and resistance to therapy. It can serve as cancer biomarker and treatment target. To enable comprehensive screening for the impact of tumor driving mutations in colorectal cancer cells we present a method for specific analysis of tumor driver-induced glycome changes. The strategy is based on a combination of three technologies, that is recombinase-mediated cassette exchange (RMCE), Click-It chemistry and mass spectrometry. The new method is exemplified by the analysis of the impact of inactivating mutations of the TGF-ß-receptor type II (TGFBR2) on sialic acid incorporation into protein-linked glycans of the colon cancer cell line HCT116. Overall, 70 proteins were found to show de novo sialic acid incorporation exclusively upon TGFBR2 expression whereas 7 proteins lost sialylation upon TGFBR2 reconstitution. Validation of detected candidate glycoproteins is demonstrated with the cell surface glycoprotein nectin-3 known to be involved in metastasis, invasion and prognosis of various cancers. Altogether, our new approach can help to systematically puzzle out the influence of tumor-specific mutations in a major signaling pathway, as exemplified by the TGFBR2 tumor suppressor, on the tumor glycome. It facilitates the identification of glycan-based tumor markers that could be used for diagnostic and therapeutic applications. In principle the outlined strategy can be adapted to any cancer cell line, tumor driver mutation and several glycan-building blocks.
Keywords: colorectal cancer, microsatellite instability, sialylation, TGFBR2, nectin-3
Introduction
Protein glycosylation represents the most common post-translational modification. There are striking examples that glycosylation can have profound effects on protein function contributing to regulation of activity and structure stabilization. Moreover, most plasma membrane-associated proteins are glycosylated with glycan chains protruding to the extracellular space thereby functioning in cell adhesion, transmembrane signaling as well as in intercellular recognition and communication.1 Consequently, a cell’s protein glycosylation state may influence its biochemical and physiological functions in a similar way as the proteome itself. In addition, aberrant glycosylation is a hallmark of various pathological conditions, as observed for example in neurological disorders,2 inflammatory diseases3 or congenital glycosylation disorders.4 Singular evidence for specific glycosylation changes during cancer development and progression was also established during the past years.5 Therefore the impact of tumor driver mutations on glycome changes has become a prominent topic in tumor biology. In particular, mutations in key players of signal transduction as epidermal growth factor receptor or transforming growth factor receptor are known to play a significant role in the growth and spread of a number of cancers.
To enable specific screening for the impact of such mutations on colorectal cancer (CRC) cells we now extended a previously established method for specific analysis of tumor driver-induced proteome changes6 to the identification of glycome changes. The method combines three existing technologies: First, recombination-mediated cassette exchange is used to establish a cell line that provides regulated target gene expression of the tumor driver. Second, an azido-sugar is metabolically incorporated into glycoproteins through the permissive nature of the oligosaccharide biosynthesis pathway which allows extraction of the metabolically labeled glycoproteins by Click-It chemistry. Third, de novo synthesized glycoproteins are identified by mass spectrometry. Because ∼30% of all colon cancers and more than 90% of microsatellite instability (MSI) colon cancers carry TGF-β-receptor type II (TGFBR2) mutations,7–9 demonstrating that it is a common target of mutational inactivation in this cancer, we chose this target to test the utility of our new approach to screen for driver gene-dependent glycome changes.
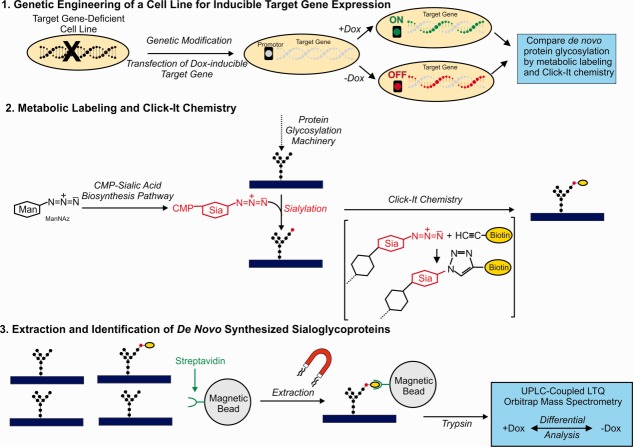
Sialic acids are a family of derivatives of neuraminic acid. The most common member of this family, N-acetyl-neuraminic acid (Neu5Ac) is synthesized from the precursor molecule N-acetyl-mannosamine (ManNAc). Neu5Ac can become substituted at specific sites by Golgi-located enzymes leading to a variety of sialic acid forms. The most common derivatives are N-acetyl-9-O-acetyl-neuraminic acid (Neu5,9Ac2) and N-glycol-neuraminic acid (Neu5Gc). The combinational diversity of specific substitutions is exemplified additionally by studies using sialoglycan microarrays.10–12 Sialic acids are typically found as terminal monosaccharides attached to cell surface glycoproteins. They play many important roles in various physiological and pathological processes. In particular, in cancer they have been shown to be involved in tumor growth, escape from apoptosis, metastasis formation, and resistance to therapy.13 Previous studies also indicated that sialylation of the cell surface of colon cancer cells is influenced by TGFBR2 expression and signaling.14 Therefore, we selected metabolic labeling for sialic acid incorporation for the present study. The complete experimental design consisting of three steps, (1) engineering of a cell line for inducible target gene expression, (2) metabolic labeling combined with click chemistry and (3) purification/extraction with magnetic streptavidin beads followed by mass spectrometric identification of the extracted proteins, is outlined in Figure 1. Comparing the results from +doxycycline (Dox) and -Dox treated cells reveals TGFBR2-dependent changes in sialylation.
Figure 1.
1. A genomic targeting approach to generate a genetically modified MSI colorectal tumor cell line enables doxycycline (Dox) -inducible and reversible expression of a target transgene in an isogenic background. A previously described recombination-mediated cassette exchange strategy was applied.14 2. Untreated controls and Dox-treated cells are metabolically labeled by adding tetraacetylated N-azido-acetyl-d-mannosamine (ManNAz) to the culture medium. ManNAz is taken up by the cells and metabolized to CMP-coupled N-acetyl-neuraminic acid and finally incorporated into nascent protein-bound glycans (for N-glycans via dolichol). After cell lysis metabolically labeled glycoproteins are tagged with biotin via a “clicking” reaction with biotin-alkyne. 3. The tag enables specific isolation of metabolically labeled sialoglycoproteins by extraction with streptavidin-coupled magnetic beads. After on bead tryptic digestion the isolated de novo sialoglycoproteome is analyzed by UPLC-coupled LTQ Orbitrap mass spectrometry. Comparing the results from untreated controls to Dox-treated cells reveals the effects of target gene expression on the sialoglycoproteome.
In addition we also performed a universally applicable double isotope labeling procedure to validate candidates obtained by the Click-It approach. In particular, this method discriminates whether altered sialylation of a protein is indeed due to changes in the protein’s glycan structure and not feigned by altered expression of the protein. In the present article we prove the utility of the whole procedure with the example of nectin-3 which is involved in mechanisms that underlie contact inhibition of cell movement and proliferation and known to contribute to the cancer phenotype.15
Results
In recent work we applied MSI-H CRC cell lines carrying frameshift mutations in the common suppressor gene activin-receptor type IIA (ACVR2A) as a model for investigating the effects of failure of expression of this gene on proteome changes.6 We now adapted this strategy to the identification of glycoproteins with altered sialic acid incorporation into their glycan chains after TGFBR2 reconstitution in a TGFBR2-deficient human CRC cell line, HCT116. Metabolic labeling of de novo synthesized sialo-oligosaccharide structures with an azido-derivative of the sialic acid precursor N-acetyl-mannosamine and subsequent extraction by Click-It chemistry turned out to cover a broad range of different proteins. Supporting Information Table SI compiles a total number of 491 (±Dox each in triplicate) metabolically labeled sialoglycoproteins, with specific occurrence of 314 candidates in TGFBR2-deficient and 434 candidates in TGFBR2-proficient cells. Non-specific binding as determined in unlabeled control cells (±Dox each in triplicate) appeared to be rather low (72 individual proteins, marked by asterisks) and was predominantly attributable to protein contamination by the procedure (various keratins) and proteins bound to the blocked beads prior to the experiments.6 To determine TGFBR2-dependent changes, results obtained with cells grown in the presence (+TGFBR2) or absence (−TGFBR2) of doxycycline were compared. Seventy proteins were found to exhibit de novo sialic acid incorporation exclusively upon TGFBR2 expression whereas seven proteins lost sialylation upon TGFBR2 reconstitution (Table1). However, the observed changes also could simply be explained by an effect of TGFBR2 reconstitution on the expression of the respective protein. Thus careful controls are required to provide proof for a specific effect on sialic acid incorporation. To exemplify the validation strategy, the target protein nectin-3, also known as poliovirus receptor-related protein 3 (PVRL3), was chosen from our list of de novo sialylated proteins. Western blotting indicated that nectin-3 expression is not affected by TGFBR2 reconstitution (Fig. 2). To confirm TGFBR2-induced sialylation of this target protein, a dual-isotope labeling strategy was applied. [35S]-L-methionine was used to metabolically label the peptide part of the glycoprotein and concomitant labeling with [3H]-N-acetyl-d-mannosamine was applied as a tracer for sialic acid incorporation into its glycan part. Immunoprecipitation with a specific antibody was performed to extract nectin-3 from the cell lysates of the parental HCT116-Tet-On cell line and two TGFBR2-inducible cell clones (HCT116-TGFBR2 #5 and #22). The precipitates were counted for [3H]- and [35S]-radioactivity in a liquid scintillation counter applying a dual label counting program. [35S]-radioactivity representing synthesis of the peptide part of nectin-3 was identical for −TGFBR2 and +TGFBR2 cells [Fig. 3(A)]. [3H]-incorporation, indicating sialylation, was virtually absent in TGFBR2-deficient cells whereas TGFBR2 expression induced a strong signal [Fig. 3(B)]. Altogether, the described strategy was successfully applied to detect and validate altered sialylation of nectin-3 as a novel function of TGFBR2 signaling in colon cancer cells.
Table 1.
Differentially Sialylated Proteins Identified by Mass Spectrometry in Absence or Presence of TGFBR2 Using HCT116-TGFBR2 #5 in at Least Two Out of Three Replicates
| Accessiona | Description | Mass (Da) | MASCOT score | Signif. peptidesb | Coverage (%)c |
|---|---|---|---|---|---|
| TGFBR2-deficient identified proteins (-Dox) | |||||
| DDX3X | ATP-dependent RNA helicase DDX3X | 73597 | 82/42 | 1/0 | 4/3 |
| DKK1 | Dickkopf-related protein 1 | 29793 | 52/36 | 1/1 | 3/3 |
| H31 | Histone H3.1 | 15509 | 97/136 | 1/1 | 34/38 |
| ITB3 | Integrin beta-3 | 90194 | 34/34 | 1/1 | 1/1 |
| NUCB1 | Nucleobindin-1 | 53846 | 79/57 | 1/1 | 11/4 |
| SNX14 | Sorting nexin-14 | 111081 | 30/30 | 0/0 | 1/1 |
| TBB2A | Tubulin beta-2A chain | 50274 | 62/80 | 1/2 | 6/6 |
| TGFBR2-proficient identified proteins (+Dox) | |||||
| 1433G | 14-3-3 protein gamma | 28456 | 201/182/93 | 2/3/1 | 21/22/11 |
| 1A69 | HLA class I histocompatibility antigen, A-69 alpha chain | 41236 | 202/147 | 3/2 | 22/20 |
| 1B35 | HLA class I histocompatibility antigen, B-35 alpha chain | 40715 | 182/171 | 2/3 | 20/17 |
| AATC | Aspartate aminotransferase, cytoplasmic | 46447 | 64/45 | 1/0 | 5/4 |
| ACTN4 | Alpha-actinin-4 | 105245 | 70/72 | 0/1 | 2/3 |
| ARF5 | ADP-ribosylation factor 5 | 20631 | 78/78 | 1/0 | 17/17 |
| AT1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 114135 | 138/189/130 | 2/3/2 | 5/5/4 |
| C1QBP | Complement component 1 Q subcomponent-binding protein, mitochondrial | 31742 | 30/48 | 1/1 | 11/11 |
| CAP1 | Adenylyl cyclase-associated protein 1 | 52325 | 33/43 | 1/0 | 4/6 |
| CD109 | CD109 antigen | 162500 | 53/81/51 | 0/1/1 | 2/3/2 |
| CLIC1 | Chloride intracellular channel protein 1 | 27248 | 90/64/63 | 1/1/1 | 17/10/10 |
| DHX9 | ATP-dependent RNA helicase A | 142181 | 130/123 | 3/1 | 4/4 |
| EFNB2 | Ephrin-B2 | 37356 | 37/32 | 1/1 | 4/4 |
| EIF3H | Eukaryotic translation initiation factor 3 subunit H | 40076 | 37/36 | 1/1 | 5/5 |
| ELAV1 | ELAV-like protein 1 | 36240 | 46/68 | 0/1 | 6/6 |
| EPHA2 | Ephrin type-A receptor 2 | 109679 | 45/35 | 1/1 | 1/1 |
| FKBP4 | Peptidyl-prolyl cis-trans isomerase FKBP4 | 52057 | 69/41 | 0/0 | 7/4 |
| FLNB | Filamin-B | 280157 | 75/121 | 1/2 | 1/1 |
| FSCN1 | Fascin | 55123 | 65/138/75 | 1/2/0 | 5/10/6 |
| G6PI | Glucose-6-phosphate isomerase | 63335 | 61/34 | 2/1 | 4/1 |
| GBB1 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 38151 | 42/92 | 1/1 | 3/7 |
| GBG5 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-5 | 7428 | 58/52 | 0/0 | 24/24 |
| HNRPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | 33707 | 86/77 | 2/1 | 12/12 |
| HNRPL | Heterogeneous nuclear ribonucleoprotein L | 64720 | 31/79 | 1/1 | 5/8 |
| IF2A | Eukaryotic translation initiation factor 2 subunit 1 | 36374 | 38/44 | 1/1 | 4/4 |
| IMDH2 | Inosine-5∼-monophosphate dehydrogenase 2 | 56226 | 64/75/36 | 1/1/1 | 5/5/2 |
| IMPA3 | Inositol monophosphatase 3 | 38828 | 40/33 | 1/1 | 3/3 |
| ITA5 | Integrin alpha-5 | 115605 | 73/96/127 | 2/2/2 | 2/4/4 |
| ITB5 | Integrin beta-5 | 91303 | 52/81/54 | 1/2/1 | 2/3/3 |
| K2C8 | Keratin, type II cytoskeletal 8 | 53671 | 145/146 | 1/2 | 7/8 |
| LAT1 | Large neutral amino acids transporter small subunit 1 | 55659 | 51/62 | 0/1 | 7/7 |
| LEG1 | Galectin-1 | 15048 | 92/45/48 | 0/0/0 | 33/15/13 |
| LPPRC | Leucine-rich PPR motif-containing protein, mitochondrial | 159003 | 31/74 | 0/1 | 1/2 |
| NDKB | Nucleoside diphosphate kinase B | 17401 | 109/103 | 3/1 | 23/22 |
| NICA | Nicastrin | 79103 | 108/88 | 0/0 | 5/5 |
| NONO | Non-POU domain-containing octamer-binding protein | 54311 | 40/71/54 | 1/2/1 | 2/7/7 |
| P5CS | Delta-1-pyrroline-5-carboxylate synthase | 87989 | 67/31 | 1/0 | 3/2 |
| PCBP1 | Poly(rC)-binding protein 1 | 37987 | 122/80/146 | 3/2/2 | 10/6/12 |
| PDIA1 | Protein disulfide-isomerase | 57480 | 141/50/52 | 3/0/1 | 13/6/2 |
| PDIA4 | Protein disulfide-isomerase A4 | 73229 | 85/34/37 | 2/1/1 | 4/2/2 |
| PLIN3 | Perilipin-3 | 47217 | 68/63 | 1/1 | 5/5 |
| PORIM | Porimin | 21575 | 51/51 | 0/0 | 7/7 |
| PPIB | Peptidyl-prolyl cis-trans isomerase B | 23785 | 153/181/82 | 3/2/2 | 24/19/13 |
| PRDX5 | Peroxiredoxin-5, mitochondrial | 22301 | 125/32/53 | 1/1/0 | 25/4/12 |
| PRDX6 | Peroxiredoxin-6 | 25133 | 88/58/35 | 1/1/1 | 13/10/4 |
| PSB5 | Proteasome subunit beta type-5 | 28633 | 36/57 | 1/1 | 4/6 |
| PVRL3 | Poliovirus receptor-related protein 3 (nectin-3) | 61477 | 52/35 | 1/1 | 2/2 |
| RAB1A | Ras-related protein Rab-1A | 22891 | 60/82 | 1/2 | 8/11 |
| RL10A | 60S ribosomal protein L10a | 24987 | 44/37 | 1/1 | 4/4 |
| RL30 | 60S ribosomal protein L30 | 12947 | 33/69 | 1/1 | 10/17 |
| RL34 | 60S ribosomal protein L34 | 13513 | 59/56 | 1/1 | 15/15 |
| RL6 | 60S ribosomal protein L6 | 32765 | 55/56/52 | 2/1/1 | 5/5/5 |
| RL7A | 60S ribosomal protein L7a | 30148 | 39/99 | 1/2 | 4/12 |
| RS11 | 40S ribosomal protein S11 | 18590 | 46/81 | 1/2 | 10/15 |
| RS26 | 40S ribosomal protein S26 | 13292 | 49/77 | 0/1 | 21/21 |
| RS27L | 40S ribosomal protein S27-like | 9813 | 34/33 | 1/1 | 10/10 |
| SCRB2 | Lysosome membrane protein 2 | 54712 | 105/68 | 1/1 | 8/4 |
| STIP1 | Stress-induced-phosphoprotein 1 | 63227 | 39/44 | 1/0 | 2/3 |
| SYNC | Asparagine–tRNA ligase, cytoplasmic | 63758 | 55/34 | 1/1 | 3/2 |
| SYNE1 | Nesprin-1 | 1017127 | 75/51/55 | 1/1/1 | 0.3/0.2/0.2 |
| SYVC | Valine–tRNA ligase | 141642 | 36/40 | 1/1 | 1/2 |
| TAGL2 | Transgelin-2 | 22548 | 62/35 | 1/1 | 15/6 |
| TM87A | Transmembrane protein 87A | 63788 | 31/59 | 0/1 | 2/3 |
| TMED4 | Transmembrane emp24 domain-containing protein 4 | 26097 | 50/55 | 0/0 | 8/9 |
| TMED9 | Transmembrane emp24 domain-containing protein 9 | 27374 | 53/63 | 1/2 | 7/7 |
| TPM3L | Putative tropomyosin alpha-3 chain-like protein | 26595 | 75/113 | 1/1 | 10/10 |
| TRFM | Melanotransferrin | 81760 | 89/65 | 2/1 | 4/3 |
| UBE2N | Ubiquitin-conjugating enzyme E2 N | 17184 | 33/49 | 1/1 | 7/7 |
| VDAC1 | Voltage-dependent anion-selective channel protein 1 | 30868 | 46/77/58 | 0/1/2 | 6/9/6 |
| XPO2 | Exportin-2 | 111145 | 57/68/43 | 1/2/1 | 3/2/2 |
Slashes correspond to individual experiments of triplicates.
Proteins identified by the SwissProt 2013_02 database with taxonomy set to human.
Significant peptides.
Sequence coverage includes all identified peptides.
Figure 2.
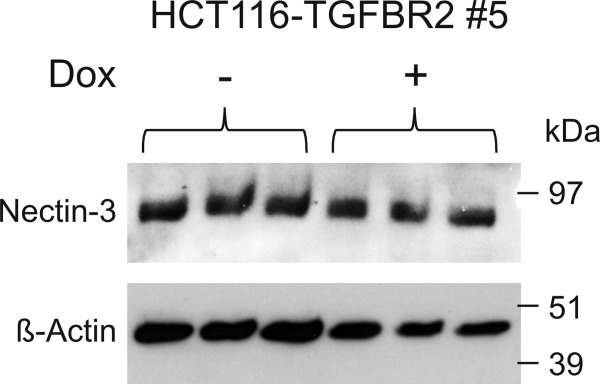
Protein expression of nectin-3 analyzed by Western blotting using 50 µg of the HCT116-TGFBR2 #5 lysates in triplicate in the absence (−) or presence (+) of 0.5 µg/ml doxycycline (Dox). β-Actin served as a loading control.
Figure 3.
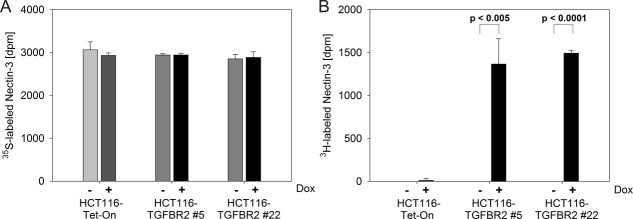
Dual radioactive labeling using [35S]-L-methionine (A) and [3H]-N-acetyl-d-mannosamine (B) of the parental HCT116-Tet-On cell line and both TGFBR2-inducible cell clones (#5 and #22), respectively in the presence (+Dox) or absence (−Dox) of TGFBR2 expression. Dpm measurements represent the mean of three independent experiments.
Discussion
Altered glycosylation is increasingly recognized as a specific hallmark of cancer cells and is considered a functional key player in cancer development and progression.16–18 Tumor specific changes of the cellular glycome have become an actual core topic of tumor biology.19 Glycomic analysis can be conducted by several methods comprising microarrays or other binding assays applying lectins or antibodies, chromatography techniques and mass spectrometry.20–23 Moreover, the targets for analysis can vary from general shifts in a cell’s whole glycome to the analysis of glycan changes in an individual glycoprotein.23,24 As a prerequisite to investigate the molecular mechanisms of specific glycosylation changes on protein function during tumor development, events that drive tumorigenesis have to be correlated with glycosylation changes on individual target proteins. Our approach enables quick and comprehensive screening for such targets.
To analyze the effects of TGFBR2 signaling, we used the human colorectal cancer cell line HCT116-TGFBR2 as a model system. These epithelial cells are characterized by MSI, carry inactivating mutations in the endogenous TGFBR2 gene and are genetically modified to allow inducible expression of the TGFBR2 transgene which has been inserted as a single copy at a defined site in the HCT116 genome, thereby ensuring expression of the wildtype protein at physiological levels.14 In this context it is important to note that results from this TGFBR2-reconstituted model reflect the inverse situation of the TGFBR2-deficiency that exists in primary colorectal tumors. Metabolic labeling with ManNAz allows the specific extraction of glycoproteins that were sialylated upon TGFBR2 induction, thereby facilitating their specific separation from the bulk of preexisting sialoglycoproteins. Comparing the results obtained from induced and uninduced cells thus enables focused screening for TGFBR2-signalling dependent protein sialylation changes.
The broad coverage of the sialoglycome by our strategy is exemplified by the extraction of 434 individual metabolically labeled sialoglycoproteins in Dox-induced cells and 314 proteins in uninduced cells. Further, confirming the experimental approach, we were able to identify well-known cell surface glycoproteins such as various clusters of differentiation (CD) antigens, integrins and mucins. The application of biotin-streptavidin interaction for targeted extraction which is based on extremely high affinity binding allows vigorous washing procedures for removing non-specifically bound proteins. Thus only 72 highly abundant proteins were found in the controls for non-specific binding. The application of a direct on-bead trypsinization step for proteomic analysis by mass spectrometry avoids possible loss of proteins by elaborate elution and extraction procedures. UPLC-coupled mass spectrometry Orbitrap provides maximum sensitivity for final analysis.
All members of the sialic acid family in humans are synthesized from ManNAc. Thus our approach in principle should cover all sialic acid derivatives. However, using ManNAz we cannot specify the type of sialic acid in the identified glycoproteins.
Application of the method to differential analysis of TGFBR2-proficient and -deficient cells revealed 70 proteins that were newly sialylated upon TGFBR2 expression and 7 proteins whose sialylation occurred in absence of the receptor. Results of the screening procedure should be validated by an independent method. This is exemplified in the present study with nectin-3 for which the described method indicated de novo sialylation upon TGFBR2 reconstitution. The observed changes in expression of sialylated nectin-3 also might be due to alterations in the expression of this protein. However, Western blotting with an antibody that is directed against the peptide part of the glycoprotein indicates no change in its overall expression level. Nevertheless, if the amount of de novo expressed nectin-3 is rather low compared to protein already present in the cells prior to TGFBR2 induction, the Western blot control might fail. Therefore, and in order to obtain an unambiguous result, a dual isotope labeling experiment should be considered. To this end, the protein part is labeled with an [35S]- or [14C]-labeled amino acid (e.g., [35S]-l-methionine) while the glycan part is labeled with a [3H]-labeled precursor of glycan building blocks (e.g., [3H]-N-acetyl-mannosamine) for metabolic radioactive sialic acid incorporation. Applying this method unambiguously demonstrates that nectin-3 sialylation exclusively occurs in the TGFBR2-proficient cells, while synthesis of the peptide part of the glycoprotein is not affected by TGFBR2 signaling.
Nectins and nectin-like proteins are immunoglobulin-like cell adhesion molecules that have been shown to be essential contributors to the formation of cell-cell adhesions and novel regulators of cellular activities, including cell polarization, differentiation, movement, proliferation and survival.15 Aberrant expression of nectins has been associated with cancer progression and metastasis.25 The underlying mechanisms are still not fully understood but an interaction with integrins, cadherins and growth factor receptors at the cell adhesion sites of contacting cells and at the leading edges of moving cells seem to be involved.15,26 In addition, modulation of the response to cytotoxic lymphocytes might play a role. The major functions of immunoglobulin superfamily receptors that bind to nectin and nectin-like proteins have recently been uncovered, and these studies have shown the in vivo and clinical importance of these receptors in mediating NK and CD8 T cell function in various models of murine and human cancer.27 Accordingly, nectin-3 has been shown to be related to metastasis, invasion and prognosis of breast and pancreatic cancer.28,29 Our study for the first time indicates a correlation between nectin-3 sialylation and the tumor suppressor TGFBR2 in colon cancer cells, thereby demonstrating the potential of our strategy to detect novel and specific protein glycosylation changes that occur as a consequence of tumor driver mutations.
In principle, the outlined strategy can be applied to any cancer cell line that can be engineered by recombinase-mediated cassette exchange. Several azido- or alkyne-derivates of glycan building blocks are available, including tetraacetylated N-azido-acetyl-galactosamine, tetraacetylated N-azido-acetyl-glucosamine and fucose alkyne. Thus our methodology can be easily adapted to screen for metabolic labeling and subsequent identification of glycoproteins with altered incorporation of N-acetyl-galactosamine, N-acetyl-glucosamine or fucose into their glycan chains.
Materials and Methods
Cell culture and metabolic labeling
All human colorectal cancer cell lines were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% heat-inactivated FBS (Life Technologies) and 100 U mL−1 penicillin and 100 µg mL−1 streptomycin (Life Technologies) using standard conditions. The genetic background and establishment of the doxycycline-inducible HCT116-TGFBR2 cell clones was detailed earlier.14 Briefly, random and single copy integration of a retroviral vector encoding two reporter genes flanked by heterospecific recombination target sites was used to identify genomic loci in independent clones, which conferred long-term and Dox-regulated expression of a marker gene cassette. In a first recombination step this marker gene cassette was then replaced by an antibiotic selection cassette resulting in a master cell line. In a second recombination step this antibiotic selection cassette was exchanged for a single copy wildtype TGFBR2 expression cassette at the original retroviral genomic integration sites resulting in the desired clones. For metabolic labeling experiments, 1–2 × 106 cells were seeded in triplicate on 10 cm dishes. The following day 10 ng mL−1 of TGF-β1 (Cell Signaling) and 40 µM of the metabolic labeling reagent tetraacetylated N-azido-acetyl-d-mannosamine (ManNAz) or for dual radioactive labeling 1.11 MBq of [3H]-N-acetyl-d-mannosamine ([3H]-ManNAc) [740 GBq mmol−1] and 0.37 MBq of [35S]-l-methionine [37 TBq mmol−1] (American Radiolabeled Chemicals) was added. The cells were either grown in absence or presence of 0.5 µg mL−1 doxycycline (Sigma) for 72 h. Cells were then washed thrice with PBS and scraped off. The cells were centrifuged for at least 10 min at 1000g at 4°C in PBS containing 1× protease inhibitor cocktail (Roche) and the pellet was either frozen at −80°C (ManNAz labeling) or directly resuspended in lysis buffer (radioactive labeling).
Tetraacetylated N-azido-acetyl-D-mannosamine (ManNAz) labeling followed by Click-iT
After labeling using ManNAz (C33366; Invitrogen), the cell pellets were lysed with 100 µl of 1% SDS in 50 mM Tris-HCl, pH 8, supplemented with protease inhibitor cocktail. Cells were sonicated for 30 s and incubated at least 30 min at 4°C while rotating. After centrifugation at 4°C for 30 min at 12,000g, protein concentration was determined by Bradford Assay. 200 µg of protein were incubated with 40 µM Biotin Alkyne in DMSO (B10185; Invitrogen) according to the manufacturer’s instructions. After methanol/chloroform precipitation, the precipitates were resolubilized in 200 µL RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium-deoxycholate, 0.1% SDS, 0.1 mM CaCl2 and 0.01 mM MgCl2), sonicated twice for 30 s and rotated overnight at 4°C. Unsolubilized material was removed by centrifugation at 4°C for 20 min at 12,000g. The supernatant, containing newly synthesized biotin-labeled sialoglycoproteins, rotated with 80 µL of prewashed streptavidin-coated magnetic bead slurry (Dynabeads MyOne Streptavidin T1, Invitrogen) using PBST (PBS/0.01% Tween 20). After 2 h at 4°C, the beads were washed thrice with 1 mL PBST containing 2% SDS followed by final washes with PBS and 40 mM ammonium bicarbonate.
Mass spectrometric analysis
Samples for mass spectrometry were prepared and analyzed as previously published.6 Briefly, sialoglycoprotein-loaded beads were reduced, alkylated and trypsinyzed at 37°C overnight. After addition of TFA, the sample was analyzed by nanoLC ESI-MS/MS. The mgf-files generated by the Xcalibur software (Thermo Scientific) were used for database searches with the MASCOT search engine (Matrix Science; version 2.4) against SwissProt database (SwissProtversion 2013_02 (539165 sequences; 191456931 residues)) with the taxonomy set to human. Candidate proteins were classified as TGFBR2-deficient or -proficient if detected in two out of three biological replicates of HCT116-TGFBR2 #5 cells and if not present in the samples without labeling reagent (ManNAz).
Dual radioactive labeling followed by Nectin-3 immunoprecipitation (IP)
After radioactive labeling, the cell pellet was resuspended in 200 µL RIPA buffer containing 2x protease inhibitor cocktail. Protein lysates were obtained as described above. For the IP, 1 mg lysate was incubated with 2 µg nectin-3 antibody (C-19; Santa Cruz). The lysate was rotated in a total volume of 300 µL containing 2× protease inhibitor cocktail overnight at 4°C. 25 µl of protein A/G agarose slurry (Oncogene) was washed thrice with RIPA buffer followed by incubation with the lysate/antibody mixture for 3 h by rotating at 4°C. The beads were washed 5 times with 1 ml RIPA buffer and eluted with 2× 200 µL 1× SDS-sample buffer (106 mM Tris-HCl, 141 mM Tris Base, 2% SDS, 10% Glycerol, 0.51 mM EDTA) at 99°C for 5 min while vortexing. The samples were then mixed with 10 ml of scintillation cocktail and counted. The dpm-measurements were conducted applying the transformed Spectral Index of the External Standard/Automatic Efficiency Control (tSIE/AEC) method.
Western blot analysis
For immunoblotting, 50 µg of extracted protein was separated on 4–12% Bis-Tris Gels (NuPAGE, Invitrogen) and electroblotted onto a nitrocellulose membrane. After blocking the membrane for 1–5 h at room temperature (RT) in 5% skim milk/TBST (20 mM Tris-HCl pH 7.5, 0.5M NaCl and 0.1% Tween-20), the following primary antibodies were used in blocking solution: goat anti-nectin-3 (C-19; Santa Cruz; 1:1000, overnight, 4°C) and mouse anti-β-Actin (MP Biomedicals; 1:10,000, RT, 30 min). After three washing steps (10 min each at RT) in TBST, the blots were incubated with secondary antibodies donkey anti-goat IgG-HRP (Santa Cruz; 1:1000) and sheep anti-mouse IgG-HRP (1:5000; GE-Healthcare) and incubated for 1 h at RT. After three washing steps, signals were detected using Western Lightning Plus ECL (PerkinElmer).
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
References
- Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Zaidi T, Iqbal K, Grundke-Iqbal I, Gong CX. Aberrant glycosylation modulates phosphorylation of tau by protein kinase A and dephosphorylation of tau by protein phosphatase 2A and 5. Neuroscience. 2002;115:829–837. doi: 10.1016/s0306-4522(02)00510-9. [DOI] [PubMed] [Google Scholar]
- Wright RD, Cooper D. Glycobiology of leukocyte trafficking in inflammation. Glycobiology. 2014;24:1242–1251. doi: 10.1093/glycob/cwu101. [DOI] [PubMed] [Google Scholar]
- Scott K, Gadomski T, Kozicz T, Morava E. Congenital disorders of glycosylation: new defects and still counting. J Inherit Metab Dis. 2014;37:609–617. doi: 10.1007/s10545-014-9720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation–potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Ballikaya S, Lee J, Warnken U, Schnölzer M, Gebert J, Kopitz J. De Novo proteome analysis of genetically modified tumor cells by a metabolic labeling/azide-alkyne cycloaddition approach. Mol Cell Proteom. 2014;13:3446–3456. doi: 10.1074/mcp.M113.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Munoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT, et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, Brattain M, Willson JKV. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Cohen M, Varki A. The sialome—far more than the sum of its parts. Omics. 2010;14:455–464. doi: 10.1089/omi.2009.0148. [DOI] [PubMed] [Google Scholar]
- Varki A. Multiple changes in sialic acid biology during human evolution. Glycoconj J. 2009;26:231–245. doi: 10.1007/s10719-008-9183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padler-Karavani V, Song X, Yu H, Hurtado-Ziola N, Huang S, Muthana S, Chokhawala HA, Cheng J, Verhagen A, Langereis MA, et al. Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. J Biol Chem. 2012;287:22593–22608. doi: 10.1074/jbc.M112.359323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull C, Stoel MA, den Brok MH, Adema GJ. Sialic acids sweeten a tumor’s life. Cancer Res. 2014;74:3199–3204. doi: 10.1158/0008-5472.CAN-14-0728. [DOI] [PubMed] [Google Scholar]
- Lee J, Ballikaya S, Schönig K, Ball CR, Glimm H, Kopitz J, Gebert J. Transforming growth factor beta receptor 2 (TGFBR2) changes sialylation in the microsatellite unstable (MSI) Colorectal cancer cell line HCT116. PLoS One. 2013;8:e57074. doi: 10.1371/journal.pone.0057074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- Taniguchi N, Kizuka Y. Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res. 2015;126:11–51. doi: 10.1016/bs.acr.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Glavey SV, Huynh D, Reagan MR, Manier S, Moschetta M, Kawano Y, Roccaro AM, Ghobrial IM, Joshi L, O’Dwyer ME. The cancer glycome: carbohydrates as mediators of metastasis. Blood Rev. 2015;29:269–279. doi: 10.1016/j.blre.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Ju T, Wang Y, Aryal RP, Lehoux SD, Ding X, Kudelka MR, Cutler C, Zeng J, Wang J, Sun X, et al. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteomics Clin Appl. 2013;7:618–631. doi: 10.1002/prca.201300024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RR. Glycosylation and cancer: moving glycomics to the forefront. Adv Cancer Res. 2015;126:1–10. doi: 10.1016/bs.acr.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Gabius HJ, Kayser K. Introduction to glycopathology: the concept, the tools and the perspectives. Diagn Pathol. 2014;9:4. doi: 10.1186/1746-1596-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst S, Wuhrer M, Rombouts Y. Glycosylation characteristics of colorectal cancer. Adv Cancer Res. 2015;126:203–256. doi: 10.1016/bs.acr.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J, Kuno A, Tateno H. Development and applications of the lectin microarray. Top Curr Chem. 2015 doi: 10.1007/128_2014_612. [DOI] [PubMed] [Google Scholar]
- Nakagawa H. Analytical Aspects: analysis of protein-bound glycans. In: Gabius H-J, editor. The sugar code: fundamentals of glycosciences. Weinheim, Germany: Wiley VCH; 2009. pp. 71–86. [Google Scholar]
- Selman MH, Hemayatkar M, Deelder AM, Wuhrer M. Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal Chem. 2011;83:2492–2499. doi: 10.1021/ac1027116. [DOI] [PubMed] [Google Scholar]
- Mandai K, Rikitake Y, Mori M, Takai Y. Nectins and nectin-like molecules in development and disease. Curr Top Dev Biol. 2015;112:197–231. doi: 10.1016/bs.ctdb.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Ogita H, Takai Y. Cross-talk among integrin, cadherin, and growth factor receptor: roles of nectin and nectin-like molecule. Int Rev Cytol. 2008;265:1–54. doi: 10.1016/S0074-7696(07)65001-3. [DOI] [PubMed] [Google Scholar]
- Chan CJ, Andrews DM, Smyth MJ. Receptors that interact with nectin and nectin-like proteins in the immunosurveillance and immunotherapy of cancer. Curr Opin Immunol. 2012;24:246–251. doi: 10.1016/j.coi.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Martin TA, Lane J, Harrison GM, Jiang WG. The expression of the Nectin complex in human breast cancer and the role of Nectin-3 in the control of tight junctions during metastasis. PLoS One. 2013;8:e82696. doi: 10.1371/journal.pone.0082696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Hirabayashi K, Nakamura N, Nakagohri T. Nectin expression in pancreatic adenocarcinoma: nectin-3 is associated with a poor prognosis. Surg Today. 2015;45:487–494. doi: 10.1007/s00595-015-1126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information