Abstract
Mutations or amplification of the MET proto-oncogene are involved in the pathogenesis of several tumours1-4, which rely on the constitutive engagement of this pathway for their growth and survival1,5. However, MET is expressed not only by cancer cells but also by tumour-associated stromal cells although its precise role in this compartment is not well characterized6-11. Here, we show that MET is required for neutrophil chemoattraction and cytotoxicity in response to its ligand HGF. Met deletion in neutrophils enhances tumour growth and metastasis. This phenotype correlates with reduced neutrophil infiltration to both primary tumour and metastatic site. Similarly, Met is necessary for neutrophil transudation during colitis, skin rash or peritonitis. Mechanistically, Met is induced by tumour-derived TNF-α or other inflammatory stimuli in both mouse and human neutrophils. This induction is instrumental for neutrophil transmigration across an activated endothelium and iNOS production upon HGF stimulation. Consequently, HGF/MET-dependent nitric oxide release by neutrophils promotes cancer cell killing, which abates tumour growth and metastasis. Following systemic administration of a MET kinase inhibitor, we prove that the therapeutic benefit of MET targeting in cancer cells is partly countered by the pro-tumoural effect rising from MET blockade in neutrophils. Our work identifies an unprecedented role of MET in neutrophils, suggests a potential “Achilles’ heel” of MET-targeted therapies in cancer, and supports the rationale for evaluating anti-MET drugs in certain inflammatory diseases.
To ensure Met deletion in the immune system only, we took advantage of the Tie2:Cre deleter that excises floxed genes in both bone-marrow (BM) and endothelial cells (EC)12 and we reconstituted lethally irradiated C57BL/6 wild-type (WT) mice with BM cells from Tie2;Metwt/wt (WT) or Tie2;Metlox/lox (KO) mice (Extended Data Fig. 2a), producing WT→WT or KO→WT mice, respectively. Compared to WT→WT controls, both growth and metastatic burden of subcutaneous LLC lung carcinomas were boosted in KO→WT mice (Fig. 1a-g), with reduced tumour apoptosis and necrosis, increased proliferation, but comparable vessel parameters and hypoxia (Extended Data Fig. 2b-r). A similar induction in tumour growth and metastasis was observed in non-irradiated KO versus WT mice (Extended Data Fig. 2s-u), but tumour growth, as well as the vascular features, were comparable in WT→WT versus WT→KO chimeras, displaying Met deletion in EC only (Extended Data Fig. 2o-r,v). Thus, Met deletion in immune cells favours cancer growth and metastasis.
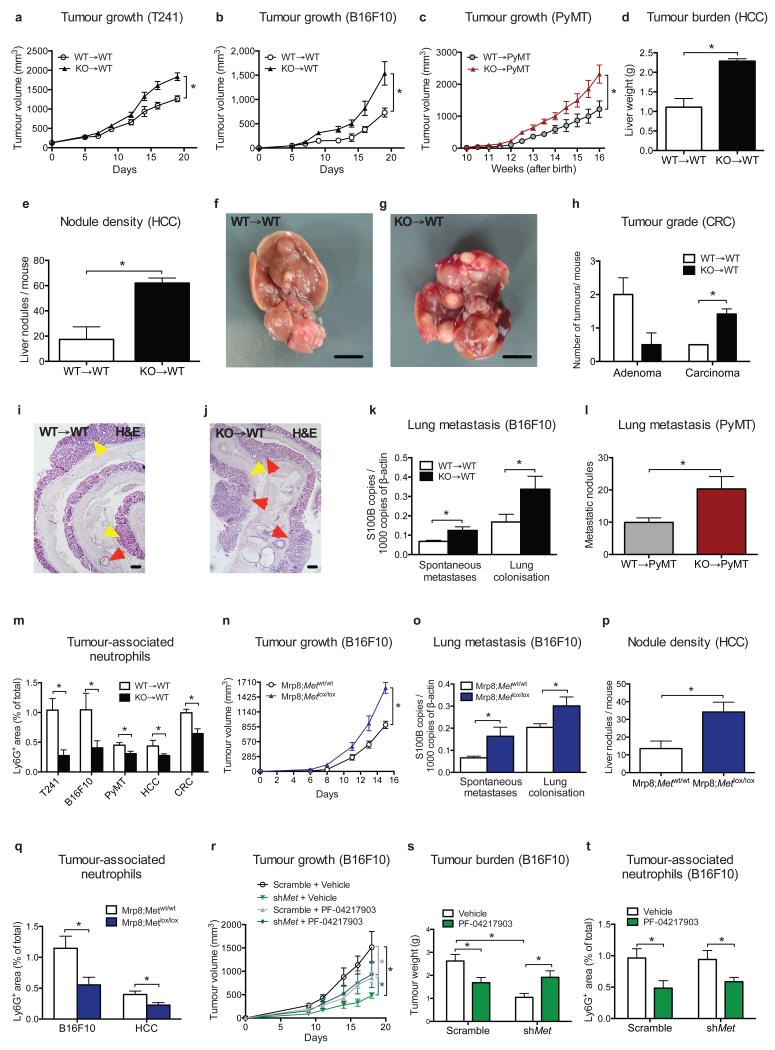
Figure 1. Met deficiency inhibits neutrophil recruitment to tumour and metastatic site.
a-g, LLC tumour growth (a), weight (b), lung macrometastases (c), metastatic area (d), representative images of H&E-stained lung sections (e,f), metastatic index (g) in WT→WT and KO→WT chimeras. Data combine 3 independent experiments; total mice: WT→WT=23, KO→WT=26. h-m, Neutrophil quantification and representative images on Ly6G-stained LLC tumour sections (h-j) or on lung sections from tumour-free and tumour-bearing mice (k) represented in (l,m). Data in (h) are representative of 4 independent experiments (6 mice/condition per experiment). Data in (k) combine 3 independent experiments; total mice: Tumour-free=10/condition, Tumour-bearing=15/condition. n-q, LLC tumour growth (n), tumour weight (o), lung macrometastases (p), TAN quantification (q) in WT→WT and KO→WT control chimeras (Mrp8:Empty) or upon neutrophil-specific Met reconstitution (Mrp8:Met). Data combine 2 independent experiments; total mice=10/condition. r-u, LLC tumour growth (r), tumour weight (s), lung macrometastases (t), TAN quantification (u) upon neutrophil-specific Met deletion (Mrp8;Metlox/lox) and controls (Mrp8;Metwt/wt). Data combine 2 independent experiments; total mice=13/condition. *, P<0.05. Scale bars: 100 μm (e,f), 50 μm (i,j,l,m). Graphs show mean ± s.e.m..
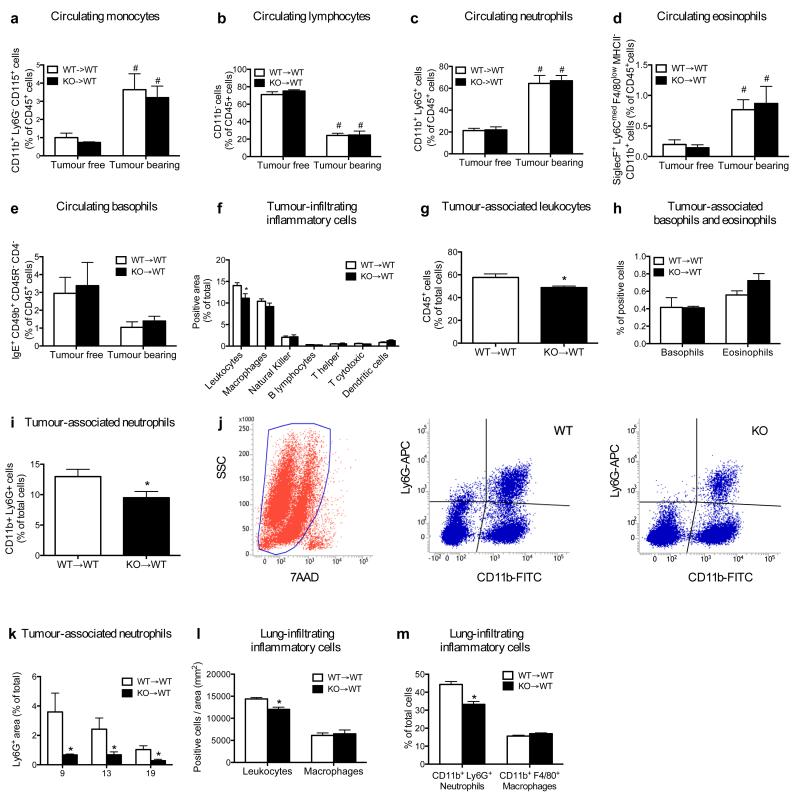
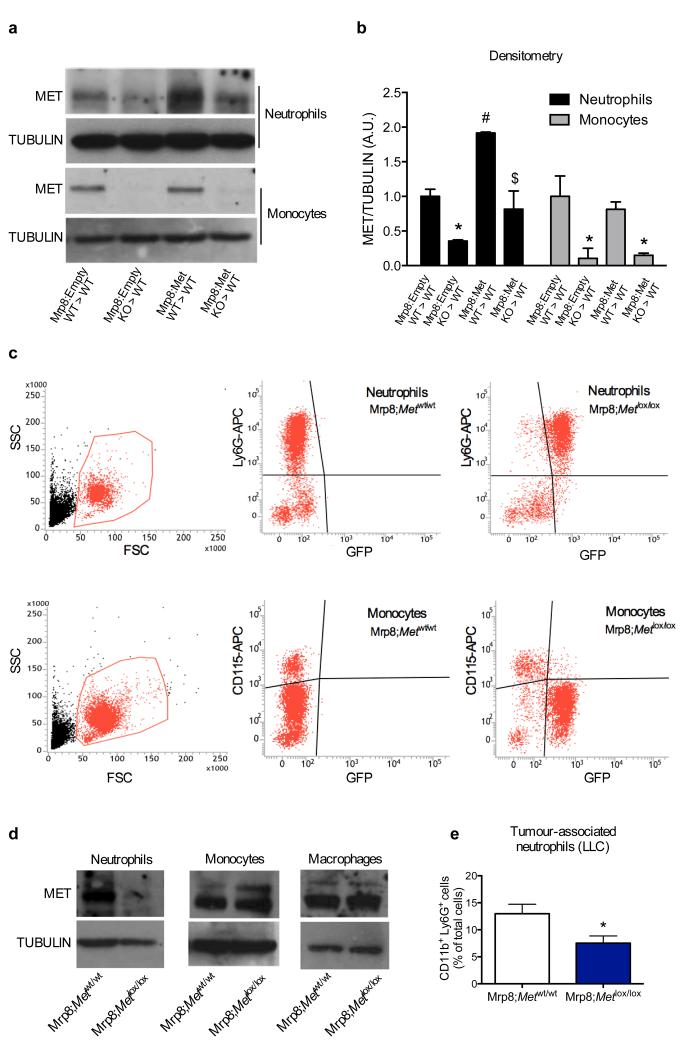
Blood counts and percentage of circulating blood cell subsets did not change in WT→WT and KO→WT mice, either at baseline or upon LLC tumour engraftment (Extended Data Fig. 3a-e; Extended Data Table 1,2). Notably, KO→WT mice displayed reduced numbers of tumour-infiltrating CD45+ leukocytes and, among all the different subpopulations, only Ly6G+ tumour-associated neutrophils (TANs) were strongly reduced in KO→WT versus WT→WT mice at any time point (Fig. 1f-j; Extended Data Fig. 3f-k). Similarly, lungs from KO→WT tumour-bearing mice contained fewer CD45+ leukocytes with decreased Ly6G+ neutrophil infiltration, while macrophages were comparable (Fig. 1k-m; Extended Data Fig. 3l,m). Furthermore, reconstitution of Met in neutrophils only13 (Extended Data Fig. 4a,b), was sufficient to rescue their recruitment and to hinder tumour growth and metastasis in KO→WT mice (Fig. 1n-q). Vice versa, restricted deletion of Met in neutrophils (Mrp8;Metlox/lox) by the neutrophil-specific Mrp8:Cre line13 (Extended Data Fig. 4c,d) led to enhanced tumour growth and dissemination, and marked TAN reduction, as in KO→WT chimeras (Fig. 1r-u; Extended Data Fig. 4e). These results indicate that MET is required for recruiting anti-tumoural neutrophils.
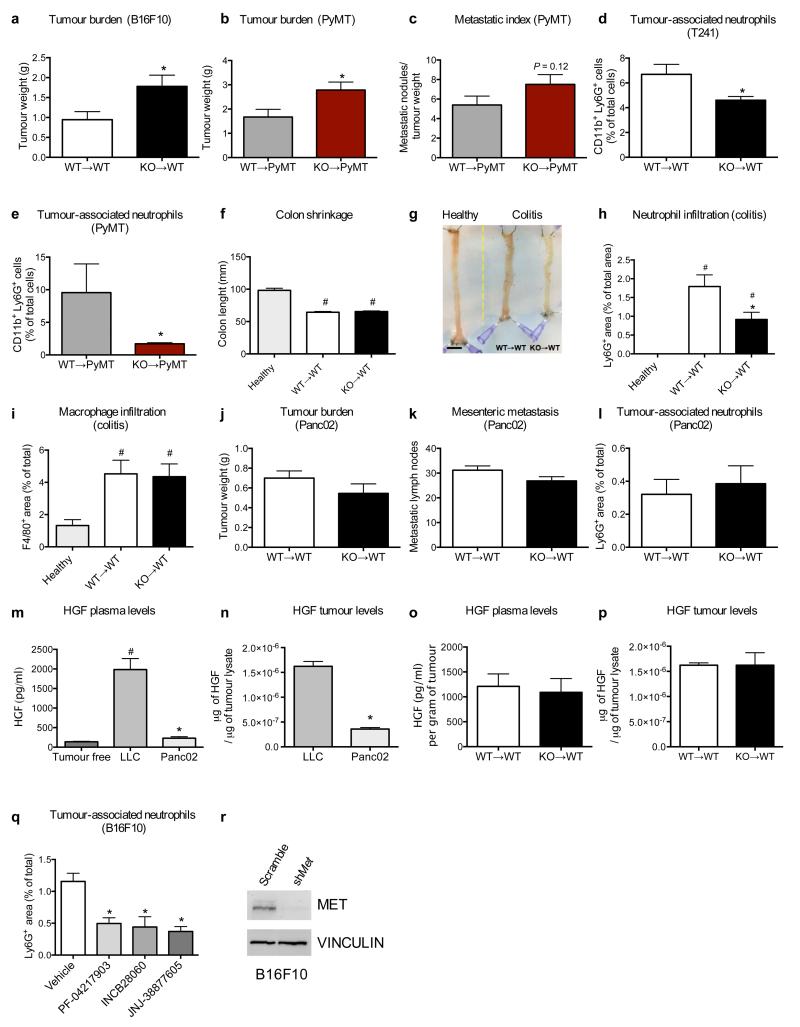
To extend our findings to other tumour types, we proved that Met deletion in the hematopoietic system increased the growth of i) orthotopic T241 fibrosarcomas and B16F10 melanomas, ii) spontaneous mammary tumours in MMTV-PyMT+ transgenic mice, iii) H-RasG12V and c-Myc-driven hepatocellular carcinomas (HCC), and iv) chemically induced colorectal cancers (CRC) (Fig. 2a-j; Extended Data Fig. 5a,b). Furthermore, lung colonisation of B16F10 melanoma cells (from either the primary tumour or after cancer cell intravenous injection) and of MMTV-PyMT+ breast tumours was boosted in Met KO chimeras (Fig. 2k,l; Extended Data 5c). In all these tumour types, Met KO TANs were fewer than WT TANs (Fig. 2m; Extended Data 5d,e). Interestingly, during chronic bowel inflammation (preceding CRC formation), neutrophil but not macrophage infiltration of the colon was also abated by hematopoietic Met deletion, but this reduction did not impinge on colitis severity (Extended Data 5f-i). B16F10 and HCC displayed enhanced tumour growth (and metastasization) as well as reduced TAN infiltration in Mrp8;Metlox/lox versus Mrp8;Metwt/wt mice (Fig. 2n-q). Conversely, orthotopic Panc02 carcinomas grew and metastasized similarly in both WT→WT and KO→WT mice, and TAN infiltration did not change (Extended Data Fig. 5j-l). However, these tumours produced little HGF compared to LLC tumours (Extended Data Fig. 5m,n). In general, plasma and intratumour HGF did not differ between genotypes (Extended Data Fig. 5o,p). In sum, Met deficiency in neutrophils promotes the progression of different (HGF-secreting) tumours.
Figure 2. Met deficiency in hematopoietic cells fosters progression of several tumour types.
a-c, Growth of T241 fibrosarcomas (a), B16F10 melanomas (b), PyMT-driven breast tumours (c). Data in (a,b) combine 2 independent experiments; total mice=14/condition (a), 8/condition (b). Data in (c) combine 3 independent experiments; total mice: WT→PyMT=13, KO→PyMT=16. d-g, Liver weight (d), nodules (e), and images (f,g) after H-RasG12V/c-Myc-driven HCC (n=4 mice/condition). h-j, Quantification (h) on H&E-stained bowel sections (i,j) of AOM/DSS-induced colon adenomas (yellow arrowheads) or carcinomas (red arrowheads). Data combine 2 independent experiments; total mice=10/condition. k, Spontaneous lung metastases from B16F10 tumours or lung colonisation after B16F10 intravenous injection. Data combine 2 independent experiments; total mice=8/condition. l, Lung macrometastases from PyMT tumours. Data combine 3 independent experiments; total mice: WT→PyMT=13, KO→PyMT=16. m, TAN quantification in T241, B16F10, PyMT, HCC, CRC tumour-tissues. Total mice: T241, CRC=10/condition, B16F10=8/condition, combining 2 experiments; WT→PyMT=13, KO→PyMT=16, combining 3 experiments; HCC=4/condition (1 experiment). n,o, B16F10 tumour growth (n), spontaneous metastases and lung colonisation (o) in Mrp8;Metwt/wt and Mrp8;Metlox/lox mice. Data combine 2 independent experiments; total mice: Mrp8;Metwt/wt=12, Mrp8;Metlox/lox=8. p, Liver nodules in Mrp8;Metwt/wt and Mrp8;Metlox/lox HCC-bearing mice. Total mice=7/condition. q, TAN quantification in B16F10 tumours (total mice=8/condition) or HCC (total mice=7/condition). r-t, Tumour growth (r), weight (s) and TANs in Met-silenced (shMet) and scramble B16F10 tumours after PF-04217903 or vehicle treatment. Data combine 2 independent experiments (total mice=11/condition for scramble, 14/condition for shMet). *, P<0.05. Scale bars: 200 μm (i,j), 0.5 cm (f,g). Graphs show mean ± s.e.m..
Systemic treatment of WT mice carrying B16F10 melanomas (which are dependent on MET14) with three different MET tyrosine-kinase inhibitors (PF-04217903, INCB28060, JNJ-38877605), strongly reduced TAN recruitment (Extended Data Fig. 5q). We then compared MET silencing in cancer cells versus systemic MET inhibition. PF-04217903 decreased weight and volume of B16F10 melanomas by 36% and 54% respectively, whereas MET knockdown in cancer cells by 58% and 75% (Fig. 2r,s; Extended Data Fig. 5r). However, combination of these two strategies was not synergic but dampened tumour inhibition to the same level as observed with PF-04217903 alone (Fig. 2r,s). TAN inhibition by PF-04217903 was comparable in both Met-silenced and scramble B16F10 melanomas (Fig. 2t). These data unveil how the therapeutic benefit of systemic MET inhibition is partly blunted by the blockade of anti-tumoural neutrophils.
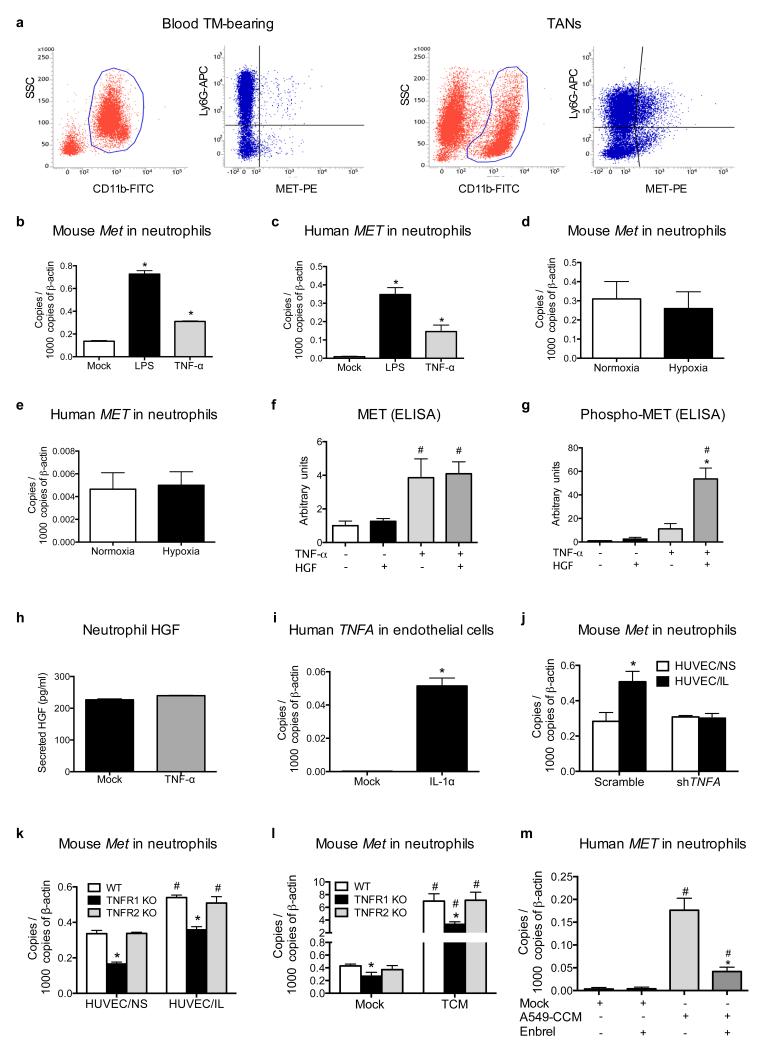
To date, MET expression in neutrophils has been poorly documented11. We thus measured MET levels in circulating or tumour-infiltrating neutrophils. Circulating Ly6G+ cells from healthy mice expressed low MET, but these levels were increased in circulating neutrophils from LLC tumour-bearing mice and even further in TANs (Fig. 3a,b; Extended Data Fig. 6a). Similarly, neutrophils isolated from non-small cell lung tumours displayed much higher MET levels than in the healthy tissue (Fig. 3c).
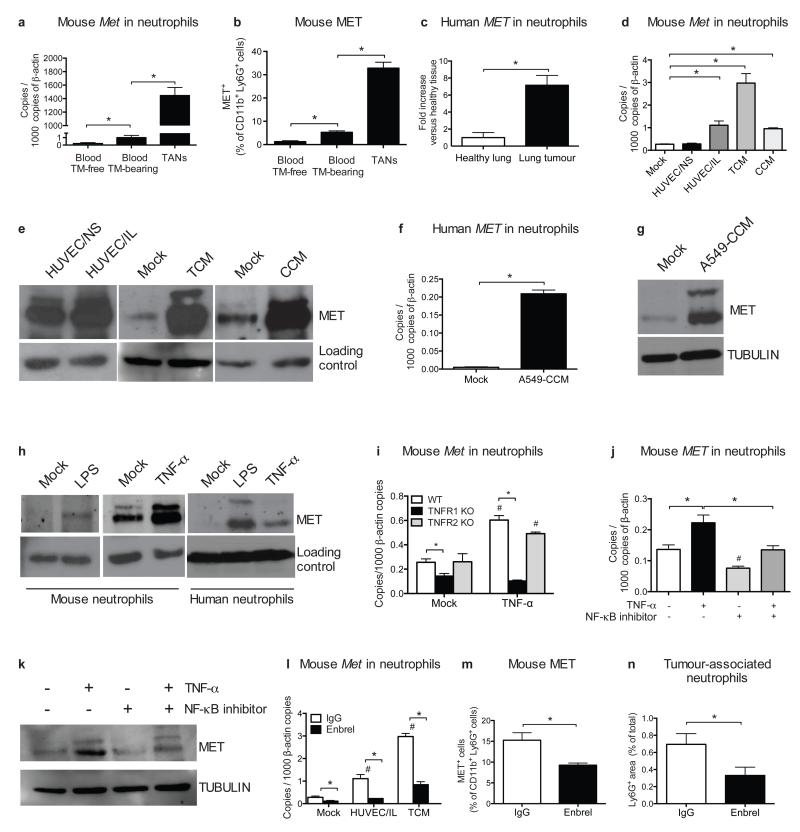
Figure 3. Met expression in neutrophils is induced by tumour-derived soluble factors.
a-b, qRT-PCR (a) and FACS (b) analysis for MET in blood neutrophils from tumour (TM)-free or LLC-tumour-bearing mice and in TANs. c, qRT-PCR for MET in human neutrophils from lung cancer versus healthy tissue. n=4 patients. d,e, MET expression by qRT-PCR (d) and Western blot (e) in circulating neutrophils from tumour-free mice after co-culture with unstimulated (HUVEC/NS) or IL-1α-pre-stimulated (HUVEC/IL) HUVEC, or after stimulation with TCM or CCM. f,g, qRT-PCR (f) or Western blot (g) for MET in circulating human neutrophils after stimulation with A549-CCM. h, Western blot for MET in mouse and human neutrophils after LPS or TNF-α stimulation. i, qRT-PCR for Met in WT, TNFR1 KO or TNFR2 KO neutrophils after TNF-α stimulation. j,k, qRT-PCR (j) and Western blot (k) for MET in mouse neutrophils after TNF-α stimulation with or without NF-κB inhibitor. l, qRT-PCR for Met in mouse neutrophils co-cultured with HUVEC/IL or stimulated with TCM in presence or absence of Enbrel. m,n, FACS for MET in TANs (m) and immunohistochemistry for Ly6G (n) in LLC tumours after Enbrel. Data combine 2 independent experiments; total mice=5/condition. All data in (a,b,d,f,i,j,l) are representative of 2 independent experiments using 4 biological replicates/condition per experiment. All Western blots were repeated 3 times on independent biological replicates. Full Western blot images are shown in Supplementary Figure 1. Loading control in (e,h) displays tubulin or actin according to Supplementary Figure 1. *, P <0.05; #, P <0.05 versus Mock (i,l), versus untreated (j). Graphs show mean ± s.e.m..
Co-culture with IL-1α pre-activated endothelium as well as stimulation with tumour- or cancer cell-conditioned medium (TCM or CCM, respectively) promoted MET expression in both mouse and human neutrophils (Fig. 3d-g). In a biased approach15-17, we found that TNF-α and LPS (but not IL-1α HGF, or hypoxia) induced MET expression in both mouse and human neutrophils (Fig. 3h; Extended Data Fig. 6b-e; not shown). TNF-α-mediated MET induction required TNFR1 and subsequent NF-κB activation (Fig. 3i-k). TNF-α alone was not able to trigger either MET phosphorylation or HGF release in neutrophils (Extended Data Fig. 6f-h).
Silencing of EC-borne TNF-α (which is 250-fold increased upon stimulation with IL-1α; Extended Data Fig. 6i), knockout of neutrophil-borne TNFR1 (but not of TNFR2), pharmacological blockade of TNF-α with the TNF-α-trap Enbrel, prevented MET induction in mouse or human neutrophils upon co-culture with activated EC or exposure to TCM/CCM (Fig. 3l; Extended Data Fig. 6j-m). Finally, systemic administration of Enbrel in LLC tumour-bearing mice diminished MET expression in neutrophils as well, resulting in their reduced recruitment to the tumour (Fig. 3m,n). Though MET is scarcely expressed in naive neutrophils, it is strongly induced by inflammatory stimuli.
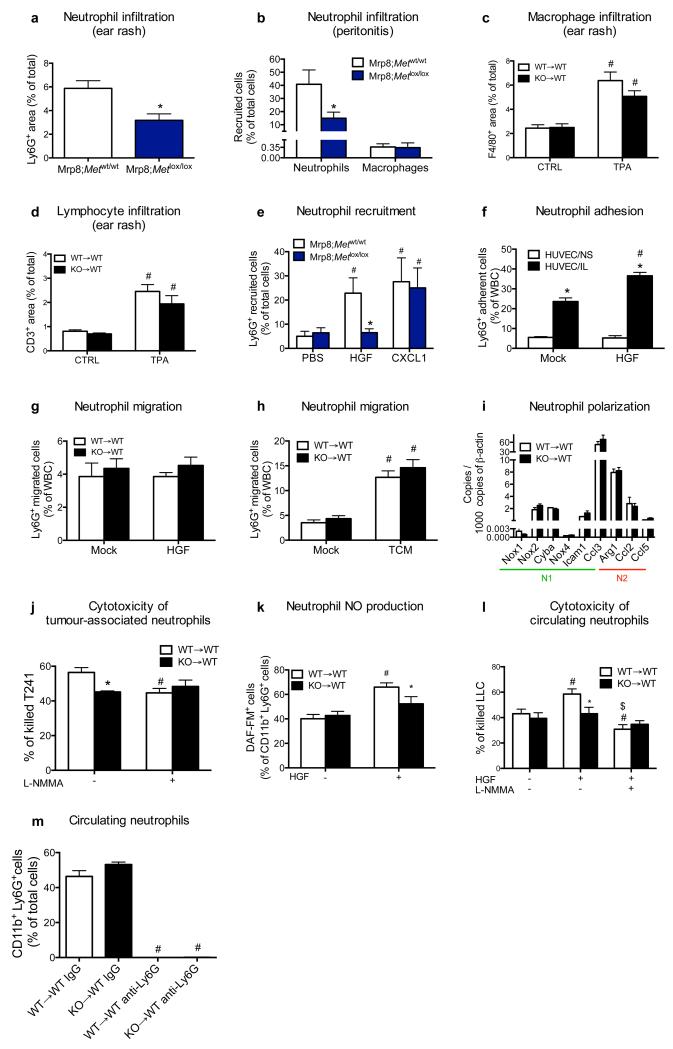
Mechanistically, impaired TAN accumulation after Met inactivation was not due to cell death as assessed in LLC tumours and in culture, both at baseline and under LPS stimulation, with or without HGF (Extended Data Fig. 7a-e), but rather to a defect in neutrophil recruitment from the blood. Indeed, in case of acute inflammation, Met KO neutrophils displayed reduced exudation to the skin or to the peritoneal cavity (Fig. 4a-d; Extended Data Fig. 8a,b). Macrophage or lymphocyte recruitment did not change (Fig. 4d; Extended Data Fig. 8c,d). Vice versa, recombinant HGF recruited WT neutrophils inside subcutaneous air pouches with similar efficacy than the neutrophil chemoattractant CXCL1 (Fig. 4e; Extended Data Fig. 8e). Instead, Met KO neutrophils did not migrate towards HGF, whilst their response to CXCL1 was preserved (Fig. 4e; Extended Data Fig. 8e). Mirroring this approach, an anti-HGF blocking antibody18 prevented neutrophil infiltration to tumours and inflamed skin (Fig. 4f).
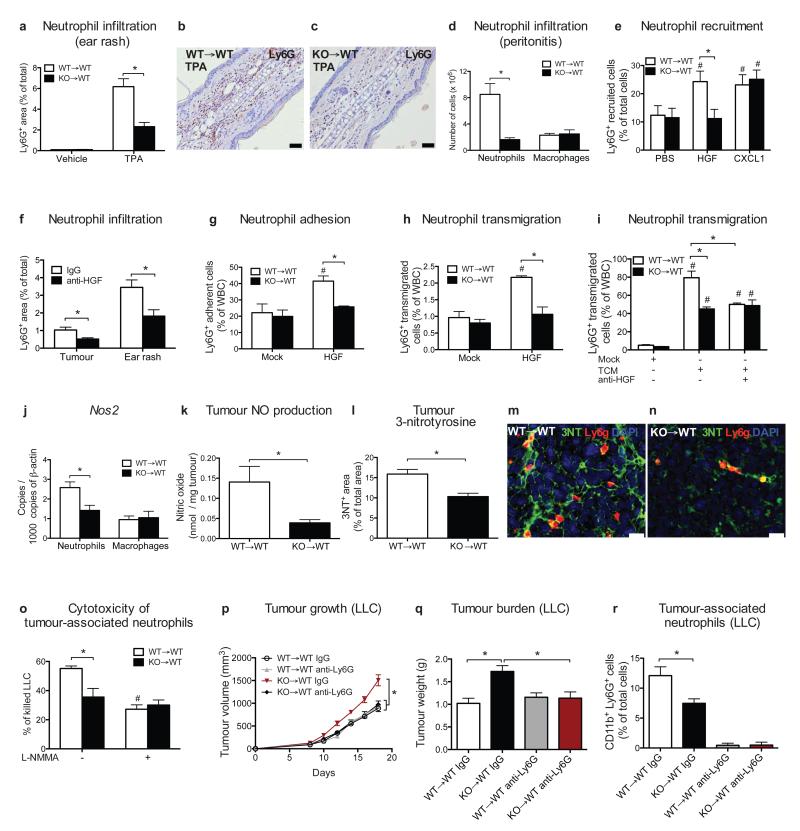
Figure 4. MET is required for neutrophil transendothelial migration and cytotoxicity.
a-c, Neutrophils quantification (a) on TPA-painted ear skin (b,c). Data combine 2 independent experiments; total mice=10/condition for vehicle, 14/condition for TPA. d, FACS analysis for Ly6G+ neutrophils or F4/80+ macrophages on peritoneal lavages after zymosan-induced peritonitis. Data are representative of 2 independent experiments using 4 mice/condition per experiment. e, FACS analysis for neutrophil recruitment towards HGF or CXCL1 in air pouch assays. Data combine 3 independent experiments; total mice=10/condition. f, Ly6G infiltration in LLC tumours or in TPA-painted ear skin after anti-HGF. Data combine 2 independent experiments; total mice: IgG=11, anti-HGF=6. g-i, Neutrophil adhesion to HUVEC/IL (g) and transendothelial migration in response to HGF (h) or TCM with or without anti-HGF (i). Data in (g-i) are representative of 3 independent experiments using 3 biological replicates/condition per experiment. j,k, qRT-PCR for Nos2 in LLC-tumour-associated neutrophils or macrophages (j) and tumour-derived NO production (k). l-n, Quantification (l) and representative images (m,n) of 3NT and Ly6G-costained LLC tumour sections. Data in (j-n) combine 2 independent experiments; total mice=8/condition. o, TAN cytotoxicity against LLC cells with or without L-NMMA. Data are representative of 4 independent experiments using 3 biological replicates/condition per experiment. p-r, LLC tumour growth (p), weight (q), TANs (r) following neutrophil-depleting anti-Ly6G treatment. Data combine 2 independent experiments; total mice=16/condition. *, P<0.05; #, P<0.05 versus PBS (e), versus Mock (g-i), versus WT→WT untreated (o). Scale bars: 100 μm (b,c), 20 μm (m,n). Graphs show mean ± s.e.m..
We then tested the relevance of MET for neutrophil migration. Stimulation of WT neutrophils with HGF promoted their adhesion and chemotaxis through an activated endothelium whereas Met KO neutrophils (displaying 85% reduction in MET protein levels compared to WT; Extended Data Fig. 2a) completely lost this response (Fig. 4g,h; Extended Data Fig. 8f). In line with this, TCM (containing 2.6 ± 0.3 ng/ml HGF) promoted transendothelial migration of WT neutrophils, but its effect was 43% lower on Met KO neutrophils (Fig. 4i). Upon HGF neutralization, WT neutrophils responded to the TCM as Met KO neutrophils (Fig. 4i). HGF or TCM did not influence neutrophil behaviour on non-activated EC or bare membranes (Extended Data Fig. 8f-h). Hence, HGF-mediated MET activation is required for neutrophil transendothelial migration to the inflammatory site.
Once migrated inside the tumour, N1 or N2 neutrophils can respectively inhibit or favour tumour progression19. Amongst the N1 and N2 genes, only the expression of the N1-marker inducible nitric oxide synthase (Nos2, otherwise iNos) was lower in Met KO versus WT TANs but similar in macrophages (Fig. 4j; Extended Data Fig. 8i). Compared to WT→WT mice, tumours harvested from KO→WT mice displayed reduced nitric oxide (NO) production and 3-nitrotyrosine (3NT) formation, a sign of NO-mediated cell damage (Fig. 4k-n). In vitro, Met KO TANs had lower cancer-cell-killing capacity than WT TANs; iNOS inhibition by L-NMMA blunted this difference (Fig. 4o; Extended Data Fig. 8j). HGF-stimulated WT but not Met KO neutrophils displayed enhanced NO release and cytotoxicity, which was abated by L-NMMA (Extended Data Fig. 8k,l).
We then hypothesized that HGF/MET pathway is key for anti-tumoural neutrophils only. Neutrophil depletion in WT→WT chimeras did not affect LLC tumour growth, implying that in this tumour model anti-tumoural and pro-tumoural neutrophils are in balance (Fig. 4p-r). The same treatment in KO→WT mice reduced tumour growth by 34% (Fig. 4p-r), indicating that Met deletion inhibits recruitment and activation of cytotoxic, but not of pro-tumoural neutrophils, which are instead blocked by the anti-Ly6G antibody (Fig. 4r; Extended Data Fig. 8m).
In sum, we demonstrate that MET is induced by inflammatory stimuli. This receptor is then required for neutrophil extravasation to inflamed tissues. Extravasated neutrophils respond to HGF by producing cytotoxic nitric oxide (Extended Data Fig. 1). All these steps restrain non-specific immune reactions to the inflammatory site, preventing damage of healthy organs.
These findings highlight a double-edged role of MET in cancer: on one side, in MET-addicted tumours, this pathway is vital for cell-cycle and survival2; on the other side, it promotes anti-tumourigenic activities in neutrophils. Thus, alternative approaches targeting MET on cancer cells only, and trials guided by new patient selection strategies will be important to maximize efficacy of MET inhibition in oncological diseases3,4,20.
Finally, given the fact that MET-inhibiting drugs are not associated to overt toxicity1, MET-targeted therapies might ameliorate the symptoms of inflammatory disorders wherein neutrophils are important for disease pathogenesis21.
METHODS
Animals
The Met floxed mice, a gift of Dr. Snorri S. Thorgeirsson (Center for Cancer Research, NCI, Bethesda, MD), were backcrossed in a C57BL/6 background. The Tie2:Cre and MMTV-PyMT transgenic lines were obtained from our mouse facility. The Mrp8:Cre mice were a gift of Dr. Clifford A. Lowell (UCSF, San Francisco, CA) and Dr. Pierre Bruhns (Pasteur Institute, Paris, FR)13,22-26. C57BL/6 mice were purchased from Harlan. TNFR1 KO mice and TNFR2 KO mice were a gift of Dr. Claude Libert (VIB, Ghent, BE). The Met floxed mice were intercrossed for at least two generations with Tie2:Cre or Mrp8:Cre in order to obtain Metlox/lox Cre negative (WT) or Metlox/lox Cre positive (KO) littermates for the specific promoter. All the experimental procedures were approved by the Institutional Animal Care and Research Advisory Committee of the KU Leuven. In all the experiments, mice were gender and age matched (within an age range between 6 and 10 weeks).
Cell lines
Murine Lewis lung carcinoma cells (LLC), melanoma B16F10, and human non-small cell lung carcinoma A549 cells were obtained from American Type Culture Collection (ATCC); the murine pancreatic tumour cell line Panc02 and the murine fibrosarcoma cell line T241 were respectively a gift from Dr. Cavallaro (IEO, Milano, IT) and Dr. Claesson-Welsh (Rudbeck Laboratory, Uppsala, SE). LLC, B16F10, A549, and T241 cells were cultured in DMEM (Gibco) supplemented with 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and containing 10% FBS (DMEM 10% FBS). Panc02 cells were cultured in RPMI (Gibco) supplemented with 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and containing 10% FBS. All these murine tumour cell lines are syngeneic in a C57BL/6 background, allowing implantation in Met conditional knockout mice or chimeras. Human Umbilical Vein Endothelial Cells (HUVEC) were isolated from human umbilical cords and maintained in M199 (Invitrogen) supplemented with 20% FBS, 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.15% Heparin, 20 μg/ml ECGS (M199 complete). 0.1% pork gelatin was used to favour the adhesion of HUVEC to the flask bottom. All cells were maintained in a humidified incubator in 5% CO2 and 95% air at 37°C. Three or four different short hairpin RNA lentiviral vectors (Sigma) were used to silence Met in LLC, B16F10 or T241 (LLC shMet, B16F10 shMet or T241 shMet; see below), or to silence TNFA in HUVEC (HUVEC shTNFA). Scramble lentiviral vectors were used as control. Transduced cells were selected with 8 μg/ml puromycin. All cancer cell lines and primary HUVEC underwent mycoplasma testing before their use. Negative mycoplasma contamination status was verified using LookOut Mycoplasma PCR Kit (Sigma) and MycoAlert Mycoplasma Detection Kit plus Assay Control (Lonza). Panc02 and T241 cells were both authenticated by Idexx Bioresearch. All cells were passaged in the laboratory for no longer than 6 months after receipt. Supplementary Table 1 lists the sequences of all the shRNA constructs used in this study.
Bone marrow transplantation and blood cell count
Recipient 6-week old female mice were lethally irradiated (9.5 Gy) and then intravenously injected with 107 bone marrow (BM) cells from WT or KO mice 16 hours later. Experiments were initiated 5 weeks after BM reconstitution. Blood cell count was determined using a hemocytometer on peripheral blood collected by retro-orbital bleeding.
Hematopoietic stem/progenitor cell (HSPCs) transduction
For MET overexpression or reconstitution in respectively WT or Met KO neutrophils, lineage negative HSPCs were enriched with the mouse hematopoietic progenitor enrichment kit (Stem cell technologies) and checked for purity by FACS according to the manufacturer’s protocol. 1×106 cells/ml were pre-stimulated for 5 hours with stem span serum-free medium (Stem Cell Technologies) supplemented with IL-3 (20 ng/ml), SCF (100 ng/ml) TPO (100 ng/ml) and FLT-3L (100 ng/ml) (Promega) and transduced with 1×108 TU/ml of a lentiviral vector expressing mouse Met under the promoter of the human gene S100A8 (Mrp8:Met), which has been engineered for neutrophil-specific transcriptional targeting, or an empty vector (Mrp8:Empty) as control. Briefly, the promoter driving Met expression in neutrophils only, corresponds to a 3.6 Kb DNA fragment encompassing the natural 5′ and 3′ regulatory regions but deleted of its exon coding sequences. Hence, Met is under the control of the 5′ and 3′ untraslated regions of the human S100A8 gene and other proximal cis regulatory sequences present in the surrogate DNA fragment13,22-26 . Ten hours after the first viral transduction, cells received a second round of their respective lentiviral vector; 7 hours later 1×106 cells were injected via tail vein in lethally irradiated C57BL/6 recipient mice. A fraction of transduced HSPCs were cultured and collected after 9 days to measure the number of integrated vector copies/cell genome (vector copy number, VCN) by qPCR using custom TaqMan assays specific for HIV-gag sequences (Applied Biosystems), as previously described27. Standard curves for HIV-gag (contained by both Mrp8:Empty and Mrp8:Met lentiviral vectors) were obtained from the corresponding plasmids. Fifty ng of genomic DNA from each sample was subjected to qPCR in quadruplicate using an ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems). VCN was determined comparing the amplification signal on the genomic DNA with the standard curve consisting of serial dilutions over a 6 log range (slope ≈ −3.3, intercept ≈ 35, efficiency % ≈ 100). Average copies per cell genome were calculated taking into account that one murine diploid genome = 5.92 pg. The results of this analysis are shown in Supplementary Table 2.
Tumour models
2×106 LLC or 1×106 B16F10 were injected subcutaneously while 2×106 T241 cells were injected intradermally in a volume of 200 μl PBS. Tumour volumes were measured 3 times a week with a calliper. At endstage, tumours were weighed and collected for histological examination or FACS analysis. MMTV-PyMT+ spontaneous breast tumours were measured 10 weeks after birth (6 weeks after BM transplantation), three times a week, and sacrificed at week 16. Lung metastases were contrasted by intratracheal injection of a 15% India ink solution, by hematoxylin eosin (H&E) staining on lung paraffin sections, or detected by qRT-PCR for the melanoma-specific gene S100B in the models involving B16F10 cells. For orthotopic pancreatic tumour growth, mice were anesthetized with isoflurane, the stomach exteriorized via abdominal midline incision, and 1×106 Panc02 tumour cells in 30 μl PBS were injected into the head of the pancreas using a 29-gauge needle. A successful intrapancreatic injection of tumour cells was identified by the appearance of a fluid bleb without intraperitoneal leakage. Mice displaying peritoneal leakage were immediately sacrificed and excluded from the analysis. At day 12, primary tumours were removed and weighed. Enlarged lymph nodes were counted under a stereoscopic microscope. For the chemically-induced colorectal cancer model, body-weight-matched mice received one intraperitoneal injection of 10 mg/kg of azoxymethane (AOM) followed by 3 cycles of 7 days of 1.5% (cycle I) or 1.7% (cycle II-III) dextran sodium sulphate (DSS) in drinking water, starting from the day of AOM injection28. After 160 days, the colon was collected and prepared for histological evaluation with the “Swiss roll” technique29. For the oncogene-driven hepatocellular carcinoma model, mice received a 1:1 molar ratio (3 μg total DNA) of piggyBac transposons encoding for c-Myc and H-RasG12V oncogenes, driven by the PGK promoter, together with the hyperactive piggyBac transposase-encoding plasmid30. DNA solutions containing transposon/transposase plasmids were diluted in 2 ml of Ringer’s solution and hydrodynamically delivered in 7 seconds through the tail vein.
Lung colonisation assay
In the experimental metastasis assays, 0.5 × 106 B16F10 cells were injected in the tail vein and lungs were collected after 12 days. To quantify pulmonary seeding, lungs were homogenized in Trizol (Ambion) and RNA was purified with the RNeasy Mini kit (Ambion) according to manufacturer’s instructions. The expression of the melanocyte specific gene S100B was measured as readout of lung colonization by qRT-PCR following reverse transcription to cDNA with the QuantiTect Reverse Transcription kit (Qiagen).
Mice treatments
To induce chronic colitis, mice received 3 cycles of 7 days of 1.5% (cycle I) or 1.7% (cycle II-III) dextran sodium sulphate (DSS) in drinking water; 2 weeks after the last DSS cycle, the colon was collected and prepared for histological evaluation as described above29. For in vivo MET inhibition, B16F10 tumour-bearing mice received 40 mg/kg PF-04217903 (AbMole Bioscience) or the corresponding vehicle (0.5% methylcellulose in saline) via oral gavage every day once a day starting from day 2 after tumour injection and twice a day from day 11 until the end of the experiment; alternatively mice were treated with 50 mg/kg INCB28060 (AbMole Bioscience) or 50 mg/kg JNJ-38877605 (Selleckchem). For TNF-α inhibition in vivo, LLC tumour-bearing mice were randomized for comparable tumour volumes at the endstage and i.p. injected with 10 mg/kg of Enbrel or human IgG in PBS, three and one day before tumour collection. For HGF inhibition in vivo, LLC tumour-bearing mice were randomized when average tumour volume was 300 mm3 and i.p. injected with 0.2 mg/mouse of anti-HGF blocking antibody (AF-2207, R&D18,31) or goat IgG in PBS. Tumours were collected 20 hours later for histological analysis; TPA ear painting was done 5 hours after antibodies administration and ears were collected 15 hours later. For neutrophil depletion, mice were treated with 50 μg/20 g body weight of rat anti-mouse Ly6G antibody (clone 1A8, BioXCell) or rat IgG every second day starting from day 4 after LLC tumour injection and every day from day 12 after tumour injection until the end of the experiment. Efficiency of neutrophil depletion was assessed by FACS in blood and tumours.
TPA model of acute skin inflammation
Phorbol ester TPA was used to induce acute skin inflammation as described before32. Briefly, TPA (2.5 μg in 20 μl acetone per mouse) was topically applied on the ear skin of anaesthetized mice. The contralateral ear was painted with acetone alone as vehicle control. Mice were sacrificed after 24 hours and ears collected in 2% PFA for histological analysis.
Zymosan-mediated acute peritonitis model
To induce acute peritonitis, zymosan A (Sigma) was prepared at 2 mg/ml in sterile PBS. Four hours after i.p. injection of 0.1 mg zymosan A per mouse, inflammatory exudates were harvested by peritoneal lavage with 2 ml PBS. Cells were counted with a Burker chamber and stained for Ly6G (1A8) and F4/80 (BM8) for FACS analysis.
Air pouch assay
To create subcutaneous air pouches, BM transplanted chimeric mice or Mrp8;Metlox/lox or Mrp8;Metwt/wt mice were injected with 3 ml of sterile air by dorsal subcutaneous injection with a butterfly 23G needle on day 0 and on day 3. On day 6, 200 ng/mouse of murine CXCL1 or HGF dissolved in 0.5 ml PBS-Heparin (15 U/ml) or PBS-Heparin (15 U/ml) as control, were injected in the newly formed dorsal camera. After 4 hours, inflammatory cells were harvested by washing the pouch with 5 ml of PBS. Cells were stained for Ly6G (1A8), washed and resuspended in PBS 0.1% BSA with unlabelled counting beads and quantified by FACS.
Mouse white blood cell (WBC) isolation
Blood was collected from the retro-orbital vein in 10% heparin. For the isolation of WBC, the blood was diluted in 1.25% dextran in saline to allow the sedimentation of red blood cells (RBC). After 30′, the supernatant was collected and washed in PBS 0.1% BSA. The remaining RBC were lysed in a hypotonic solution of 0.2% NaCl for 30 seconds and brought in isotonic condition with 1.6% NaCl and 0.1% glucose. WBC were washed in PBS 0.1% BSA, counted and resuspended accordingly with the experimental setting.
Mouse blood neutrophil isolation
Blood was collected from the retro-orbital vein in 10% heparin and diluted in an equal volume of PBS 0.5% BSA. Up to 5 ml of diluted blood was layered on top of a discontinuous gradient of Histopaque 1119 (4 ml) and Histopaque 1077 (5 ml) from Sigma. The gradient was centrifuged for 30 minutes at 700g with brake off. The neutrophil layer between the Histopaque 1077 and 1119 was collected and washed in PBS 0.5% BSA. RBC lysis was performed as above. Neutrophils were washed in PBS 0.5% BSA, counted and resuspended according to the experimental condition. Alternatively, blood was sedimented in a saline solution containing 1.25% dextran and neutrophils were negatively selected with magnetic beads33. Neutrophil purity, as assessed by the hemocytometer, was always higher than 93%.
Bone marrow neutrophil and mononuclear cell isolation
In order to reach reasonable amounts of protein, all the Western Blot analyses in mice were performed on neutrophils isolated from bone marrows. Mice were sacrificed by cervical dislocation. Femurs and tibias were collected in cold sterile Hank Balanced Salt Solution (HBSS, Invitrogen) and flushed with HBSS 0.25% BSA. Cells were layered on top of a discontinuous gradient of Percoll 81%, 62%, 55%, freshly prepared and centrifuged for 30 minutes at 2000 g with break off. Monocytes were collected at the interface between the BM cells and the layer of Percoll 55%, whereas neutrophils were collected at the interface between Percoll 55% and 62%. Cells were washed in HBSS 0.25% BSA and RBC lysis was performed as described above. Neutrophils (or monocytes) were washed again, counted and resuspended according to the experimental setting. Neutrophil (or monocyte) purity assessed by hemocytometer was higher than 87%.
FACS analysis and flow sorting of blood or tumour-associated cells
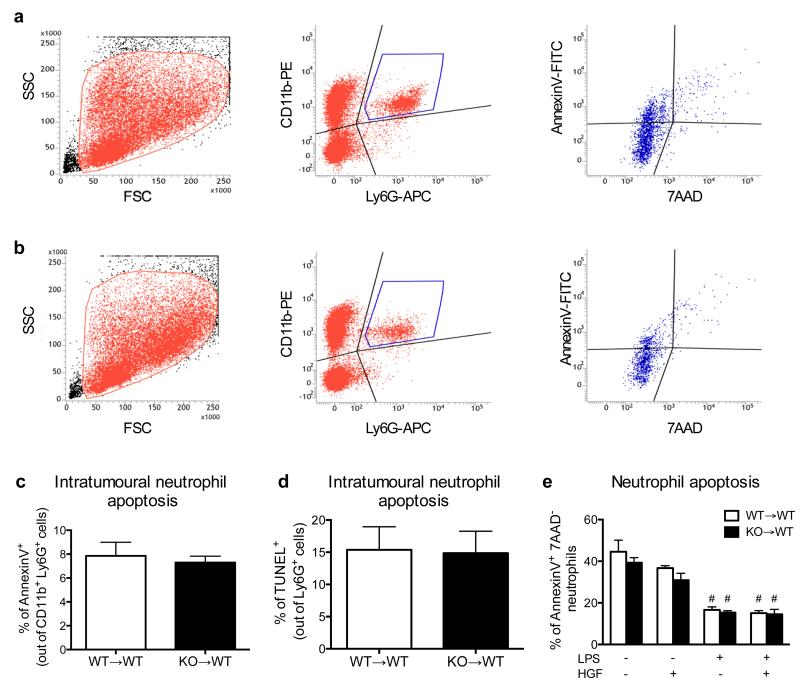
Blood was collected in 10% heparin and stained for 20 minutes at room temperature. After RBC lysis, cells were washed and resuspended in FACS buffer (PBS containing 2% FBS and 2 mM EDTA). Tumours were minced in RPMI medium containing 0.1% collagenase type I and 0.2% dispase type I (Gibco) for 30 minutes at 37°C and passed through a 70 and 40 μm cell strainer. After RBC lysis, cells were resuspended in FACS buffer and stained for 20 minutes at 4°C. Lungs were collected after 7 minutes of transcardial perfusion with saline and processed as for the tumours. Briefly, blood or single cell suspensions were incubated for 15 minutes at 4°C with Mouse BD Fc Block (2.4G2, BD Pharmingen) 1:100 in FACS buffer. The antibodies were added directly in the blocking solution in the appropriate combinations (as indicated in the figure legends). CD45 (30F-11), CD11b (M1/70), Ly6G (1A8), CD45R (RA3-6B2), CD3 (17A2), CD4 (RM4-5), IgE (R35-72), CD49b (DX5), Ly6C (AL-21) (all from BD Pharmingen), F4/80 (BM8), CD115 (AF598), MHCII (M5/114.15-12) (all from eBioscience), were used 1:200 for 2×106 in 100 μl; Siglec-F (E50-2440, BD Pharmingen) was diluted 1:750. In all the stainings 7AAD (BD Pharmingen) was used to gate out dead cells. For intra-tumour proliferation, 1 mg of BrdU was i.p. injected in each mouse 4 hours before tumour collection and cell proliferation was quantified on single cell suspensions with the FITC BrdU Flow Kit (BD Bioscience) according to manufacturer’s protocol. Tumour apoptosis was assessed by staining single cell suspension for the apoptotic marker AnnexinV (1:40, BD Bioscience), excluding 7AAD positive cells. For TAN apoptosis, tumour single cell suspensions were gated for CD11b (M1/70) and Ly6G (1A8); AnnexinV and 7ADD were used to distinguish apoptotic or dead neutrophils. In vitro neutrophil apoptosis was performed by seeding neutrophils isolated from tumour-bearing mice at a concentration of 1×106/ml and stimulating them with or without LPS (1μg/ml), alone or in combination with HGF (100 ng/ml) for 10 h at 37°C. Cells were collected, washed and stained for AnnexinV and 7AAD. The combination of CD11b (M1/70), Ly6G (1A8) and MET (eBioclone 7, eBioscience) was used to identified triple-positive MET expressing neutrophils in blood or tumour cells 7AAD-negative. Freshly stained samples were analysed by FACS Canto II (BD Bioscience). For tumour-associated neutrophil sorting, myeloid population was enriched by coating with CD11b-conjugated magnetic beads (MACS, Miltenyi Biotec) and separation through magnetic columns (MACS, Miltenyi Biotec), staining with Ly6G and sorting with a FACS Aria I (BD Bioscience). Cells were collected in RLT buffer (Qiagen) for RNA extraction or resuspended according to the experimental conditions.
Human neutrophil isolation
10 ml of venous blood from healthy volunteers were collected in citrate-coated tubes and isolated by erythrocyte sedimentation with dextran and purification with a discontinuous plasma-Percoll gradient as already described34.
Lung cancer patients
We enrolled 4 non-small cell lung carcinoma-patients with no previous history of oncological, chronic inflammatory, or autoimmune diseases within 10 years prior to this study. This patient list includes 2 males (61 and 71 years of age) and 2 females (64 and 68 years of age), of which 3 smokers (1 T1N0M0 adenocarcinoma, 1 T2N0M0 large cell carcinoma, 1 T3N0M0 squamous cell carcinoma) and 1 non-smoker (T2N0M0 adenocarcinoma). The protocol was approved by the Ethics Committee of the University Hospitals Gasthuisberg (Leuven), and all subjects consented prior to study participation.
Flow sorting of neutrophils from lung cancer patients
Lung tumour biopsies or healthy tissues were minced in RPMI medium containing 0.1% collagenase type I, 0.2% dispase type I and DNase I 100 U/ml (60 minutes at 37°C), passed through a 19 G needle and passed through a 70 and 40 μm cell strainer. After RBC lysis, cells were resuspended in FACS buffer (PBS containing 2% FBS and 2 mM EDTA) and counted. Myeloid population, enriched using CD11b-conjugated magnetic beads (MACS, Miltenyi Biotec) and separated through magnetic column (MACS, Miltenyi Biotec), was stained with anti-CD66b (G10F5, BD Pharmingen) for 20 minutes on ice and sorted with FACS Aria I (BD Bioscience). Cells were counted and resuspended in RLT buffer (Qiagen) for RNA extraction.
Endothelial cell isolation
Lungs were collected and a single cell suspension was obtained as above. Endothelial cells were obtained by performing a negative selection for CD45 (30F-11) and F4/80 (CI:A3-1) followed by a positive selection for CD31 (MEC 13.3) by using magnetic beads (Dynabeads, Invitrogen) according to the manufacturer’s protocol.
Peritoneal macrophages
5 ml of sterile PBS were injected in the peritoneum of anesthetized mice and collected after 3 minutes. Cells were centrifuged, washed and culture overnight.
Tumour-conditioned medium (TCM) and LLC (or A549)-conditioned medium (CCM) preparation
Two grams of endstage LLC tumour explanted from WT mice were chopped and incubated at 37°C in 7 ml of DMEM (supplemented with 2 mM glutamine, 100 units/ml penicillin/100 μg/ml streptomycin) FBS-free (DMEM 0% FBS). 5×104 LLC (or A549) were seeded in a 6-multiwell plate in DMEM 10% FBS and incubated at 37°C. Medium alone (DMEM 0% FBS or DMEM 10% FBS, respectively) was used to prepare mock controls. After 72 hours, the medium was filtered, supplemented with 2 mM glutamine and 20 mM HEPES and stored at −20°C. TCM and mock medium (DMEM 0% FBS) were diluted 1:5 in DMEM 10% FBS; CCM and mock medium (DMEM 10% FBS) were diluted 4:5 in DMEM FBS-free.
Quantitative RT-PCR (qRT-PCR)
For mRNA analysis, 1×105 or 3×105 mouse or human blood neutrophils, respectively, were incubated in normoxic (21% oxygen) or hypoxic (1% oxygen) conditions, or stimulated with TCM (plus 50 μg/ml Enbrel or human IgG when indicated), CCM, A549-CCM, 100 ng/ml of murine or human TNF-α, 50 ng/ml LPS, or mock medium in 96-multiwell for 4 hours at 37°C. For NF-κB inhibition, 0.18×106 neutrophils were pre-treated with 10 μM 6-amino-4-(4-phenoxyphenylethylamino) quinazoline (Calbiochem) for 1 hour at 37°C and stimulated with 100 ng/ml of murine TNF-α for 1 hours at 37°C. 2×105 HUVEC were seeded in 24-multiwell coated with 0.1% gelatin and stimulated with 5 ng/ml IL-1α in DMEM 10% FBS for 4 hours at 37°C. Cells were washed in PBS, collected in RLT buffer (Qiagen) and kept at −80°C. RNA was extracted with the RNeasy Micro kit (Qiagen) according to manufacturer’s instructions. Reverse transcription to cDNA was performed with the SuperScript® III First Strand cDNA Synthesis Kit (Life Technologies) according to manufacturer’s protocol. Pre-made assays were purchased from Applied Biosystem, except for Nos2 that was provided by IDT. cDNA, primer/probe mix and TaqMan Fast Universal PCR Master Mix were prepared in a volume of 10 μl according to manufacturer’s instructions (Applied Biosystems). Samples were loaded into an optical 96-well Fast Thermal Cycling plate (Applied Biosystems) and qRT-PCR were performed using an ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems)
ELISA
To quantify plasma, intra-tumoural, tumour-released (TCM) and neutrophil-released HGF, a murine HGF ELISA kit (R&D) was used according to manufacturer’s protocols. Blood was collected from tumour free or tumour bearing mice and plasma was prepared according to manufacturer’s instruction. TCM was prepared as above. Tumour proteins were extracted in Extraction Buffer (20M Tris HCl, 150mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM EDTA). 0.2×106 neutrophils were cultured for 20 hours in DMEM complete in presence or absence of 100 ng/ml of murine TNF-α. Medium was collected, spun down and supernatant stored at −80°C until use. Mock medium was used as negative control. For phospho-MET quantification, 6×106 mouse blood neutrophils isolated from tumour bearing mice were cultured in presence or absence of 100 ng/ml of murine TNF-α; 11 hours later, cells were stimulated or not with 100 ng/ml of mouse HGF in presence of 1 mM orthovanadate for 3 minutes at 37°C, washed in PBS supplemented with 1 mM orthovanadate and phosStop 1X (Roche) and lysed in Extraction Buffer supplemented with 1 mM orthovanadate, 2X phosStop and Complete Mini protease inhibitor cocktail (Roche) for 20 minutes at 4°C. After clearance, samples were quantified and the same amount of proteins was used for MET and phospho-MET detection using a sandwich ELISA. Briefly, 96-well microtiter plates (MaxiSorp, Nunc) were coated with 1 μg/ml of anti-mouse MET antibody (AF527, R&D Systems) overnight at 4°C and then incubated for 2 hours at room temperature (RT) in blocking buffer (PBS, 0.1% Tween-20, 6% non-fat dry milk). The same amount of proteins per sample was diluted in blocking buffer and incubated for 2 hours at RT on the ELISA plate. After 6 washes in PBS 0.1% Tween-20, samples were incubated for 2 hours at RT with the mouse anti-MET (3D4, Invitrogen) or the mouse anti-phosphotyrosine (4G10, Merck Millipore) antibodies diluted 1:500 in blocking buffer, then washed 6 times in PBS 0.1% Tween-20, and incubated with goat anti-mouse immunoglobulins conjugated to horseradish peroxidase (sc-2031, Santa Cruz Biotechnology) diluted 1:500 in blocking buffer for 2 hours at RT. Signals was developed by 15 minutes incubation with the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution (Promega). After stopping the reaction with H2SO4, absorbance was measured at 450 nm and corrected for 630 nm with a spectrophotometer.
Western blot
To assess MET deletion, BM cells, neutrophils, peritoneal macrophages, monocytes (all cultured overnight in TCM) or endothelial cells were lysed in hot Laemmli buffer (2.5% SDS, 25% Tris-HCl pH 6.8) for 10 minutes at 96°C, sonicated, cleared and quantified. Alternatively, 2×106 bone marrow derived neutrophils from WT mice were stimulated with TCM, CCM, 100 ng/ml of murine TNF-α (or mock medium 0% FBS or 10% FBS as control) for 20 hours at 37°C. For the co-culture with HUVEC, a monolayer of HUVEC was stimulated for 4 hours with 5 ng/ml IL-1α at 37°C, and washed before neutrophil seeding. After 20 hours of stimulation, neutrophils were collected using Cell Dissociation Buffer Enzyme Free PBS-Based (Gibco). Cells were washed in PBS, lysed in 15 μl of a protease inhibitor mixture and incubated for 15 minutes on ice. The protease inhibitor mixture was obtained by dissolving one tablet of Complete Mini protease inhibitor mixture (Roche) in 5 ml of PBS with 2 mM diisopropyl fluorophosphate (DFP; Acros Organics, Morris Plains, NJ). After addition of an equal amount of 2x SDS sample buffer supplemented with 4% 2-mercaptoethanol, the lysates were boiled for 15 minutes and kept at −80°C until use. NF-κB inhibition was achieved by pre-treating 7×106 neutrophils with 10 μM 6-amino-4-(4-phenoxyphenylethylamino) quinazoline (Calbiochem) for 1 hour at 37°C; cells were then stimulated with murine TNF-α (100 ng/ml) for 5 hours before lysis. Human MET was assessed by stimulating 3×107 blood neutrophils isolated from the blood of healthy volunteers with A549-CCM, 100 ng/ml human TNF-α, 50 ng/ml LPS (or mock medium 10% FBS as control) for 20 hours. Cells were incubated with 2.7 mM DFP for 15 minutes at 4°C, collected and washed in PBS supplemented with 2.7 mM DFP and Complete Mini protease inhibitor 1X, and lysed in hot Laemmli buffer at 96°C for 10 minutes. Cell lysates were sonicated, cleared and quantified. 6x loading buffer was added before loading on the gel. The following primary antibodies were used: mouse anti-mouse Met (3D4, Invitrogen), mouse anti-mouse β-actin (I-19, Santa Cruz), mouse anti-vinculin (hVIN-1, Sigma), rabbit anti-human Met (D1C2, Cell Signaling), HRP-conjugated anti-beta-tubulin (Abcam). The following secondary antibodies were used: HRP-conjugated goat anti-mouse and HRP-conjugated goat anti-rabbit (Santa Cruz). Signal was visualized by Enhanced Chemiluminescent Reagents (ECL, Invitrogen) or West Femto by Thermo Scientific according to the manufacturer’s instructions.
Adhesion assay
4×104 HUVEC were seeded in M199 20% FBS in a 96-multiwell plate, previously coated with 0.1% gelatin. After 12 hours, HUVEC were stimulated with 5 ng/ml IL-1α in DMEM 10% FBS at 37°C. After 4 hours the endothelial monolayer was thoroughly washed and 2.5×105 WBC isolated indistinctly from Tie2;Metwt/wt and Tie2;Metlox/lox mice or from WT→WT and KO→WT transplanted mice, were seeded on top of it, with or without murine HGF (50 ng/ml). After 15 minutes non-adherent cells were washed out whereas adherent cells were detached by using a Cell Dissociation Buffer Enzyme Free PBS-Based (Gibco). Cells were stained for Ly6G (clone 1A8), washed and resuspended in PBS-BSA 0.1% with unlabelled counting beads (BD Bioscience) and quantified by FACS Canto II (BD Bioscience).
Transmigration and migration assay
For the transmigration assay, 2×105 HUVEC were seeded on 3 μm polycarbonate membrane (Transwell; Costar) previously coated with 0.1% gelatin in M199 20% FBS. After 12 hours, HUVEC were stimulated for 4 hours at 37°C in DMEM 10% FBS with 5 ng/ml IL-1α and then washed. 5×105 WBC isolated indistinctly from Tie2;Metwt/wt and Tie2;Metlox/lox mice or from WT→WT and KO→WT transplanted mice were seeded on top of the endothelial monolayer, while mock medium, TCM (with or without 3 μg/ml anti-HGF antibody AF-2207, R&D18,31) or 50 ng/ml murine HGF was added in the bottom. After 2 hours at 37°C, transmigrated cells were collected from the bottom chambers and from the lower side of the filter with cold PBS 0.5% EDTA. Cells were stained and Ly6G+ cells quantified as above. In the migration assays WBC were seeded directly on top of 3 μm polycarbonate porous membranes.
Nitric oxide measurement by FACS
Neutrophils isolated from the blood of WT or KO LLC-tumour bearing mice were co-cultured for 4 hours with LLC shMet, washed and resuspended in 20 mM Hepes-PBS, and incubated for 10 minutes with 5μM DAF-FM diacetate (Molecular probes) in absence or presence of mouse HGF (100 ng/ml) at 37°C. Cells were then washed and analysed by FACS.
Cytotoxicity assay
LLC shMet or T241 shMet were transduced with a luciferase-expressing lentiviral vector (EX-hLUC-Lv114 from GeneCopoeia); 104 cells were then seeded in DMEM 10% FBS in a 96-multiwell plate. After 4 hours, 0.2×106 neutrophils purified from the blood of tumour bearing mice or directly from the tumours themselves, were co-cultured with the cancer cells in DMEM 2% FBS for 4 hours at 37°C, with or without 100 ng/ml mouse HGF or 1 mM L-NMMA (Sigma). After washing, adherent cells were lysate in 0.2% Triton, 1 mM DTT. Luciferase signal was revealed with a microplate luminometer. The use of shMet was thought to prevent any possible confounding activity of MET on cancer cell survival and thus to restrict the effect of HGF to neutrophils only.
Histology and immunostainings
To obtain serial 7-μm-thick sections, tissue samples were immediately frozen in OCT compound or fixed in 2% PFA overnight at 4°C, dehydrated and embedded in paraffin. Paraffin slides were firstly rehydrated to further proceed with antigen retrieval in citrate solution (DAKO). Cryosections were thawed in water and fixed in 100% methanol. If necessary, 0.3% H2O2 was added to methanol to block endogenous peroxidases. The sections were blocked with the appropriate serum (DAKO) and incubated overnight with the following antibodies: rat anti-CD45 (30F-11, BD Pharmingen), rat anti-Ly6G (1A8, BD Pharmingen) 1:100, rat anti-CD31 (MEC 13.3, BD Pharmingen) 1:200, rabbit anti-FITC (Serotec) 1:200, goat anti-phosphohistone H3 (Cell Signaling) 1:100, rat anti-F4/80 (CI:A3-1, Serotec) 1:100, mouse anti-NK1.1-biotin (PK136, BD Pharmingen) 1:200, rat anti-CD45R (RA3-6B2, BD Pharmingen) 1:100, rat anti-CD4 (H129.9, BD Pharmingen) 1:100, rat anti-CD8 (53-6.72, BioXCell) 1:100, hamster anti-CD11c biotin (N418, eBioscience) 1:100, mouse anti-3-nitrotyrosin (HM.11 Santa Cruz) 1:200. Appropriate secondary antibodies were used: Alexa488-or Alexa568-conjugated secondary antibodies (Molecular Probes) 1:200, HRP-labelled antibodies (DAKO) 1:100, Biotin-labelled antibodies (Jackson ImmunoResearch) 1:100. When necessary, Tyramide Signaling Amplification (Perkin Elmer, Life Sciences) was performed according to the manufacturer’s instructions. Whenever sections were stained in fluorescence, ProLong Gold mounting medium with DAPI (Invitrogen) was used. Otherwise, 3,3′-diaminobenzidine was used as detection method followed by Harris’ haematoxilin counterstaining, dehydration and mounting with DPX. Apoptotic cells were detected by the TUNEL method, using the AptoTag peroxidase in situ apoptosis detection kit (Millipore) according to the manufacturer’s instructions. For the double staining TUNEL and LY6G, TUNEL staining was performed as described above, followed by Ly6G staining by using the Vectastain ABC kit (Vector laboratories) according to the manufacturer’s instructions. Tumour necrosis and lung metastasis were evaluated by H&E staining. Necrotic area was defined as the area including necrotic cancer cells, inflammatory cells and stromal cells, compared to the total area of the field. Necrotic cells display a more glassy homogeneous appearance in the cytoplasm with increased eosinophilia, while the nuclear changes are reflected by karyolysis, pyknosis, and karyorrhexis. Alternatively, the necrotic tissue was visualized by autofluorescence as previously described35. Microscopic analysis was done with an Olympus BX41 microscope and Cell Sense imaging software or a Zeiss Axioplan microscope with KS300 image analysis software. The morphometric analysis was performed by acquiring 4-6 fields per sections on 5 independent sections (at a distance of 40 μm in depth during sectioning) from the same biological tissue sample. The values in the graphs represent the average of the means of, at least, 5 samples and the standard error indicates the variability among the different samples.
Hypoxia assessment and tumour perfusion
Tumour hypoxia was detected by injection of 60 mg/kg pimonidazole into tumour-bearing mice 1 hour before tumours harvesting. To detect the formation of pimonidazole adducts, tumour cryosections were immunostained with Hypoxyprobe-1-Mab1 (Hypoxyprobe kit, Chemicon) following the manufacturer’s instructions. Perfused tumour vessels were counted on tumour cryosections from mice injected intravenously with 0.05 mg FITC-conjugated lectin (Lycopersicon esculentum; Vector Laboratories).
Tumour-derived nitric oxide production
LLC tumours were collected 8 days after injection, cut in pieces of about 5×5 mm, weighted and incubate at 37°C in 24-multiwell with 800 μl of DMEM. After 24 hours, the media was collected, centrifuged to remove cell debris, and NO levels were measured using the Griess reagent system kit (Promega).
Statistics
Data entry and all analyses were performed in a blinded fashion. All statistical analyses were performed using GraphPad Prism software. Statistical significance was calculated by two-tailed unpaired t-test on two experimental conditions or two-way ANOVA when repeated measures were compared, with P<0.05 considered statistically significant. Data were tested for normality using the D’Agostino-Pearson omnibus test (for n>8) or the Kolmogorov-Smirnov test (for n≤8) and variation within each experimental group was assessed. Detection of mathematical outliers was performed using the Grubbs’ test in GraphPad. Sample sizes for all experiments were chosen based on previous experiences. Independent experiments were pooled and analysed together whenever possible as detailed in figure legends. All graphs show mean values ± s.e.m..
Extended Data
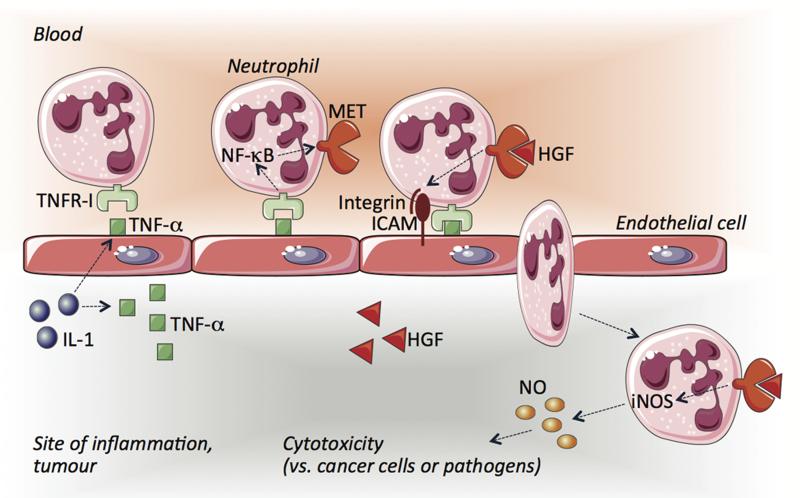
Extended Data Figure 1. Scheme illustrating the role of MET in neutrophils.
During cancer or infections, the release of cytokines such as IL-1 at the inflammatory site will promote the expression of TNF-α on the endothelium and the surrounding tissue. When circulating neutrophils will encounter the activated endothelium, TNF-α will unleash NF-κB through the binding to TNFR1, which in turn will induce MET expression on neutrophil surface. HGF, also released and proteolytically activated at the site of inflammation, will bind to MET and stimulate the firm adhesion of neutrophils to the endothelium, likely via integrin engagement, and thus neutrophil diapedesis. Once extravasated, HGF/MET pathway will still function on neutrophils by reinforcing their cytotoxic response through the induction of iNOS and NO production, ultimately favouring a bactericidal and tumouricidal neutrophil phenotype.
Extended Data Figure 2. Met deletion in immune cells, but not in EC, fosters tumour growth.
a, MET expression in total bone marrow (BM) cells, endothelial cells (EC) and neutrophils harvested from Met floxed mice intercrossed with the Tie2:Cre deleter thus generating Tie2;Metlox/lox (KO) or Tie2;Metwt/wt (WT) mice. Western blots are representative of 3 repetitions on independent biological replicates. Western blot images have been cropped for presentation. Full scan images are shown in Supplementary Figure 1. b-d, Quantification (b) and representative images of tumour sections’ TUNEL stainings (c,d) from subcutaneous endstage LLC tumours in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=10 mice/condition. e, FACS quantification of AnnexinV+ 7AAD− early apoptotic tumour cells in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=8 mice/condition. f-j, Tumour necrosis quantification in WT→WT and KO→WT mice (f), assessed by histologic evaluation of H&E stained tumour sections (g,h) and by measurement of autofluorescent tumour areas (i,j); yellow dotted lines demarcate necrosis. Data combine 2 independent experiments; total n=10 mice/condition. k-m, Quantification (k) and representative images of tumour sections stained for the proliferation marker pHH3 (l,m) from subcutaneous endstage LLC tumours in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=10 mice/condition. n, FACS quantification of BrdU+ proliferating tumour cells in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=10 mice/condition. o-r, CD31+ vessel area (o), vessel density (p), lectin perfusion (q), and hypoxic (Pimo+) area (r) in LLC subcutaneous tumours from KO→WT mice (where the hematopoietic/immune system is knocked out for Met) or WT→KO mice (where EC only are knocked out for Met) compared to control WT→WT mice. Data in o-r combine 2 independent experiments; total n: WT→WT=12, KO→WT=8, WT→KO=8. s-u, Subcutaneous LLC tumour growth (s), weight (t) and lung metastases (u) in Tie2;Metlox/lox compared to Tie2;Metwt/wt mice. Data combine 2 independent experiments; total n: Tie2;Metwt/wt=12, Tie2;Metlox/lox=10. v, Tumour growth in endothelial cell specific Met KO (WT→KO) and control (WT→WT) mice. Data combine 2 independent experiments; total n=8/condition. *, P<0.05 versus WT→WT (b,e,f,k,n), versus Tie2;Metwt/wt (s-u). Scale bars: 50 μm (c,d,l,m), 100 μm (g-j). All graphs show mean ± s.e.m.
Extended Data Figure 3. Circulating and tumour-infiltrating immune cells upon Met deletion.
a-e, FACS analysis showing percentages of circulating monocytes (a), lymphocytes (b), neutrophils (c), eosinophils (d), and basophils (e) in tumour free or in LLC-tumour bearing WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=8 mice/condition. f, Quantification of LLC-tumour sections stained for the pan-leukocyte marker CD45, the macrophage marker F4/80, the NK marker NK1.1, the B lymphocyte marker CD45R, the T helper cell marker CD4, the cytotoxic T cell marker CD8 and the dendritic cell marker CD11c (with exclusion of F4/80+ area) in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=8 mice/condition). g,h, FACS quantification for tumour-associated CD45+ leukocytes (g) or CD45+ IgE+ CD49b+ CD4− CD45R− basophils and CD45+ CD11b+ SiglecF+ Ly6Cmed F4/80low MHCII− eosinophils (h) in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=8 mice/condition. i,j, FACS quantification (i) and gating strategy (j) for tumour-associated neutrophils selected from the main tumour cell population negative for 7AAD staining; tumour-associated neutrophils were then gated as CD11b and Ly6G double positive cells. Data combine 2 independent experiments; total n: WT→WT=11, KO→WT=10. k, Ly6G+ tumour infiltration at day 9, day 13, and day 19 after LLC subcutaneous tumour injection in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=8 mice/condition. l, Morphometric quantification of leukocytes and macrophages on respectively CD45 and F480-stained lung sections from LLC tumour-bearing WT→WT or KO→WT mice. Data combine 2 independent experiments; total n=8 mice/condition. m, FACS quantification of CD11b+ Ly6G+ neutrophils and CD11b+ F4/80+ macrophages infiltrating metastatic lungs from LLC tumour-bearing WT→WT or KO→WT mice. Data combine 2 independent experiments; total n=8 mice/condition. *, P<0.05 versus WT→WT (f,g,i,k-m); #, P<0.05 versus Tumour free (a-d). All graphs show mean ± s.e.m.
Extended Data Figure 4. MET in neutrophils is required for their anti-tumour activity.
a,b, Western blot analysis (a) and relative densitometric analysis (b) for MET expression in BM neutrophils and monocytes upon reconstitution of WT recipient mice by WT or KO HSPCs transduced in vitro with an empty vector (Mrp8:Empty) or a vector expressing Met under the neutrophil-specific promoter Mrp8 (Mrp8:Met); tubulin was used as loading control. Western blots are representative of 3 repetitions on independent biological samples where each sample is the pool of neutrophils or monocytes isolated from 3 mice. Densitometric analysis has been performed on these 3 Western blots. c, FACS analysis for GFP in circulating Ly6G+ neutrophils or CD115+ monocytes, harvested from the neutrophil-specific Mrp8:Cre line carrying separate expression of GFP because of an Internal Ribosome Entry Site (IRES) downstream the Mrp8-driven Cre gene. Data combine 2 independent experiments; total n=10 mice/condition. d, MET expression in neutrophils, monocytes, and macrophages harvested from Mrp8;Metwt/wt or Mrp8;Metlox/lox mice. Western blots are representative of 3 repetitions on independent biological replicates. e, FACS analysis for CD11b+ Ly6G+ neutrophils in subcutaneous LLC tumours from Mrp8;Metwt/wt or Mrp8;Metlox/lox. Data combine 2 independent experiments; total n: Mrp8;Metwt/wt=10, Mrp8;Metlox/lox=11. Western blot images in (a,d) have been cropped for presentation. Full scan images are shown in Supplementary Figure 1. *, P<0.05 versus Mrp8:Empty WT→WT (b), versus Mrp8;Metwt/wt (e); #, P<0.05 versus Mrp8:Empty WT→WT; $, P<0.05 versus Mrp8:Empty KO→WT. All graphs show mean ± s.e.m.
Extended Data Figure 5. Pharmacologic and genetic inhibition of MET prevents the recruitment of anti-tumoural neutrophils to several neoplastic tissues and inflammatory sites.
a, Tumour weight of subcutaneous B16F10 melanomas in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n: WT→WT=8, KO→WT=9. b,c, Total tumour weight (b) and metastatic index (c) in MMTV-PyMT mice reconstituted with WT or Met KO BM cells before tumour appearance (WT→PyMT and KO→PyMT mice, respectively). Data combine 3 independent experiments; total n: WT→PyMT=13, KO→PyMT=16. d,e, FACS quantification for CD11b+ Ly6G+ neutrophils in T241 tumours harvested from WT→WT or KO→WT mice (d) or in in breast tumours spontaneously grown in WT→PyMT and KO→PyMT mice (e). Data combine 2 independent experiments; total n=10 mice/condition (d) or total n=8 mice/condition (e). f-i, Length measurement (f) and representative image (g) of the colon, as well as quantification of neutrophils (h) and macrophages (i) on bowel sections, from WT→WT and KO→WT mice upon induction of chronic colitis compared to healthy control. Data combine 2 independent experiments; total n: healthy=5, WT→WT=12, KO→WT=15. j,k, Tumour weight (j) and metastatic mesenteric lymph nodes (k) 12 days after orthotopic injection of pancreatic Panc02 cancer cells in WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=12/condition. l, Histological quantification of Ly6G+ infiltrates in Panc02 pancreatic tumours harvested from WT→WT and KO→WT mice. Data combine 2 independent experiments; total n=12 mice/condition. m, Quantification of plasma HGF in tumour (TM)-free mice, in subcutaneous LLC or orthotopic Panc02 tumour-bearing mice. Data combine 2 independent experiments; total n: TM free=10, LLC=10, Panc02=8 biological replicates. n, Quantification of HGF in subcutaneous LLC or orthotopic Panc02 tumours. Data combine 2 independent experiments; total n: LLC=10, Panc02=8 biological replicates. o,p, Quantification of HGF in plasma (o) or in subcutaneous LLC tumours (p) from tumour-bearing WT→WT and KO→WT mice. Data are representative of 2 independent experiments using 5 mice/condition per experiment. q, Quantification of Ly6G+ area on sections from B16F10 melanomas grown in C57BL/6 WT mice, daily treated with PF-04217903, INCB28060, JNJ-38877605, or vehicle as control. Data combine 2 independent experiments; total n: vehicle=14, PF-04217903=9, INCB28060=6, JNJ-38877605=4. r, Western blot analysis for MET in B16F10 melanoma cells after transduction with a lentiviral vector encoding scramble or mouse shMet under a constitutive promoter; vinculin was used as loading control. Western blot is representative of 3 independent repetitions. Western blot images have been cropped for presentation. Full scan images are shown in Supplementary Figure 1. *, P<0.05 versus WT→WT (a,d,h), versus WT→PyMT (b,e), versus LLC (m,n), versus Vehicle (q); #, P<0.05 versus Healthy (f,h,i), versus TM free (m). Scale bar: 10 mm (g). All graphs show mean ± s.e.m.
Extended Data Figure 6. HGF is required for MET activation upon induction by TNF-α.
a, Gating strategy related to Fig. 3b to quantify MET expression in blood neutrophils from LLC-tumour (TM)-bearing mice and in TANs, where live cells were first gated as CD11b positive cells; this population was finally gated for Ly6G and MET in order to identify MET-expressing Ly6G+ neutrophils. b,c, qRT-PCR for MET in mouse (b) and human (c) neutrophils after LPS or TNF-α stimulation. Data are representative of 3 independent experiments using 4 biological replicates/condition per experiment. d,e, qRT-PCR for MET expression in mouse (b) or human (c) neutrophils cultured in normoxia (21% O2) or hypoxia (1% O2). Data combine 2 independent experiments; total n=8 biological replicates/condition. f,g, ELISA for total MET (f) and phospho-MET (g) from mouse neutrophils stimulated for 3 minutes with mock medium or HGF after an overnight incubation with or without TNF-α. Data combine 3 independent experiments; total n=6 biological replicates/condition. h, HGF release by neutrophils stimulated with mock medium or TNF-α after 20 hours in culture. Data combine 2 independent experiments; total n=6 biological replicates/condition. i, qRT-PCR for TNFA in HUVEC upon stimulation with IL-1α compared to mock medium. Data combine 2 independent experiments; total n=4 biological replicates/condition. j, qRT-PCR for Met in mouse neutrophils co-cultured with HUVEC/NS or HUVEC/IL transduced with shTNFA or scramble as control. Data are representative of 3 independent experiments where 3 different shRNA sequences were used; total n=4 biological replicates/condition per experiment. k,l, qRT-PCR for Met in WT, TNFR1 KO, TNRF2 KO neutrophils upon co-culture with HUVEC/NS or HUVEC/IL (k), or after stimulation with conditioned medium (TCM) from LLC tumours (l). Data are representative of 2 independent experiments using 4 biological replicates/condition per experiment. m, qRT-PCR for MET in human neutrophils stimulated with A549-CCM in presence or absence of Enbrel or human IgG as control. Data are representative of 2 independent experiments using 4 biological replicates/condition per experiment. *, P<0.05 versus Mock (b,c,i), versus TNF-α alone (g), versus HUVEC/NS (j), versus WT (k,l), versus A549-CCM (m); #, P<0.05 versus untreated or HGF alone (f,g), versus HUVEC/NS (k), versus Mock (l,m). Graph shows mean ± s.e.m.
Extended Data Figure 7. Met deletion in neutrophils does not affect apoptosis.
a,b, Gating strategy of apoptotic WT (a) and Met KO (b) neutrophils in LLC tumours where single cells suspensions were firstly gated for physical parameters and then for CD11b and Ly6G in order to identify neutrophils as double positive cells; this population was finally gated for AnnexinV and 7AAD: AnnexinV+ 7AAD− cells display early apoptotic neutrophils whereas AnnexinV+ 7AAD+ cells display late apoptotic neutrophils. c, Quantification of apoptotic WT and Met KO tumour-associated neutrophils measured by FACS. Data combine 2 independent experiments; total n=7 mice/condition. d, Quantification of apoptotic WT and Met KO neutrophils on LLC tumour sections by immunohistochemistry. Data combine 2 independent experiments; total n: WT→WT=7, KO→WT=6. e, FACS analysis for AnnexinV and 7AAD of WT or KO neutrophils incubated for 10 hours in presence or absence of LPS and HGF, alone or in combination. Data combine 2 independent experiments; total n=6 biological replicates/condition. #, P<0.05 versus untreated or HGF alone. Graph shows mean ± s.e.m.
Extended Data Figure 8. MET affects neither neutrophil basal migration nor polarization but it is required for neutrophil recruitment and cytotoxicity.
a, Quantification of Ly6G staining in ear-sections upon phorbol ester (TPA)-induced cutaneous rash in Mrp8;Metwt/wt and Mrp8;Metlox/lox mice. Data combine 2 independent experiments; total n=8 mice/condition. b, FACS analysis on peritoneal lavages for Ly6G+ neutrophils or F4/80+ macrophages in Mrp8;Metwt/wt and Mrp8;Metlox/lox mice 4 hours after intra-peritoneal injection of sterile zymosan A. Data are representative of 2 independent experiments using 5 mice/condition per experiment. c,d, Quantification of F4/80 (c) and CD3 (d) stainings in ear-sections at baseline and upon TPA-induced cutaneous rash. Data combine 2 independent experiments; total n: WT→WT CTRL=22, KO→WT CTRL=15, WT→WT TPA=23, KO→WT CTRL=15 (c) or total n=8 mice/condition (d). e, FACS quantification of Mrp8;Metwt/wt and Mrp8;Metlox/lox neutrophils recruited into subcutaneous air pouches in response to HGF, CXCL1 or PBS. Data combine 2 independent experiments; total n=6 mice/condition. f, FACS quantification of WT neutrophil adhesion to quiescent HUVEC (HUVEC/NS) or activated HUVEC (HUVEC/IL) in presence or absence of HGF. Data are representative of 2 independent experiments using 4 biological replicates/condition per experiment. g,h, FACS quantification of WT and Met KO neutrophils migrated through a bare porous filter (i.e., in absence of HUVEC) towards HGF (g) or tumour conditioned medium (TCM) (h). Data are representative of 2 independent experiments using 3 biological replicates/condition per experiment. i, Gene expression profile for N1 and N2 markers in neutrophils sorted from LLC tumours grown in WT→WT or KO→WT mice. Data are representative of 3 independent experiments using 4 mice/condition per experiment. j, Cytotoxicity of WT and KO tumour-associated neutrophils against T241 cells in absence or presence of the NO synthase inhibitor L-NMMA. Data are representative of 3 independent experiments using 3 biological replicates/condition per experiment. k, FACS quantification of DAF-FM-positive circulating neutrophils after co-culture with LLC cancer cells as a readout of NO production in absence or presence of HGF. Data are representative of 4 independent experiments using 3 biological replicates/condition per experiment. l, Quantification of LLC cancer cell killing by WT and KO neutrophils (isolated from the blood of tumour-bearing mice), stimulated with HGF alone or in presence of L-NMMA. Data are representative of 2 independent experiments using n=12 biological replicates/condition per experiment. m, Blood neutrophils in WT→WT and KO→WT mice treated with neutrophil-depleting Ly6G antibody or rat IgG as control. Data combine 2 independent experiments; total n=16/condition. *, P<0.05 versus Mrp8;Metwt/wt (a,b), versus Mrp8;Metwt/wt + HGF (e), versus HUVEC/NS (f), versus WT→WT untreated (j), versus WT→WT + HGF (k,l); #, P<0.05 versus CTRL (c,d), versus PBS (e), versus Mock (f,h), versus WT→WT untreated (j-l), versus IgG (m); $, P<0.05 versus WT→WT + HGF (m). All graphs show mean ± s.e.m.
Extended Data Table 1. Blood count in Tie2;Metwt/wt or Tie2;Metlox/lox tumour free mice.
The values show the hematological parameters (mean ± s.e.m.) in tumour free Tie2;Metwt/wt and Tie2;Metlox/lox mice. Data combine 2 independent experiments; total n=10/condition. Abbreviations: white blood cell (WBC), neutrophil (NEU), lymphocyte (LYM), monocyte (MON), eosinophil (EOS), basophil (BAS), red blood cell (RBC), hematocrit (HCT), mean cell hemoglobin concentration (MCHC), platelet (PLT).
| Tumour free | Tie2;Metwt/wt | Tie2;Metlox/lox |
|---|---|---|
| WBC (k/μl) | 5.68±1.44 | 5.55±1.29 |
| NEU (%) | 23.03±5.45 | 29.67±7.88 |
| LYM (%) | 69.72±6.46 | 72.03±4.89 |
| MON (%) | 1.24±0.37 | 2.86±1.15 |
| EOS (%) | 0.12±0.05 | 0.17±0.12 |
| BAS (%) | 3.38±1.32 | 4.47±2.1 |
| RBC (M/μL) | 5.21±1.91 | 4.89±1.52 |
| HCT (%) | 71.3±3.43 | 60.2±13.38 |
| MCHC (g/dl) | 15.83±2.65 | 18.3±0.26 |
| PLT (K/μL) | 439.73±26.64 | 508±55.79 |
Extended Data Table 2. Blood count in WT→WT and KO→WT tumour free or tumour bearing mice.
The values show the hematological parameters (mean ± s.e.m.) in tumour free or in LLC-tumour bearing (21 days) WT→WT and KO→WT chimeric mice. Data combine 2 independent experiments; total n=10/condition. Abbreviations: white blood cell (WBC), neutrophil (NEU), lymphocyte (LYM), monocyte (MON), eosinophil (EOS), basophil (BAS), red blood cell (RBC), hematocrit (HCT), mean cell hemoglobin concentration (MCHC), platelet (PLT).
| Tumour free | WT→WT | KO→WT |
|---|---|---|
| WBC (k/μl) | 10.03±2.05 | 8.66±0.93 |
| NEU (%) | 9.18±2.1 | 10.3±3.07 |
| LYM (%) | 85.2±2.91 | 83.94±3.46 |
| MON (%) | 1.41±0.48 | 1.3±0.4 |
| EOS (%) | 0.44±0.17 | 0.25±0.05 |
| BAS (%) | 3.77±0.68 | 4.21±0.18 |
| RBC (M/μL) | 8.21±0.54 | 9.3±0.21 |
| HCT (%) | 59.86±3.52 | 70.16±1.67 |
| MCHC (g/dl) | 18.63±0.61 | 13.4±0.16 |
| PLT (K/μL) | 589.26±134.65 | 758.4±50.63 |
|
| ||
| Tumor bearing | WT→WT | KO→WT |
|
| ||
| WBC (k/μl) | 7.97±0.63 | 9.12±1.22 |
| NEU (%) | 44.3±0.37 | 53.71±7.23 |
| LYM (%) | 27.29±8.33 | 33.96±2.52 |
| MON (%) | 1.79±0.75 | 1.9±0.64 |
| EOS (%) | 0.26±0.03 | 0.45±0.1 |
| BAS (%) | 1.94±0.53 | 1.84±0.57 |
| RBC (M/μL) | 5.5±0.54 | 6.83±0.46 |
| HCT (%) | 42.13±2.93 | 49.43±3.47 |
| MCHC (g/dl) | 18.27±0.5 | 19.24±0.05 |
| PLT (K/μL) | 566.17±109.48 | 805.5±88.19 |
Supplementary Material
ACKNOWLEDGMENT
The authors thank: Rocco Stirparo, Marco Mambretti and Yannick Jönsson for technical assistance; Dr. Guido Serini (IRCC, Candiolo, IT), Dr. Livio Trusolino (IRCC, Candiolo, IT), Dr. Pierre Bruhns (Pasteur Institute, Paris, FR) for comments; Dr. Enrico Radaelli (VIB, KU Leuven, BE) for his valuable advice on histological analyses. VF and GDC were granted by FWO, AC by the Fondazione Umberto Veronesi. SRW is supported by a Wellcome Trust Senior Clinical Fellowship Award. MM is supported by an ERC starting-grant.
Footnotes
Competing Financial Interests
No competing financial interests to declare.
REFERENCES
- 1.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 2.Bertotti A, et al. Only a subset of Met-activated pathways are required to sustain oncogene addiction. Sci Signal. 2009;2:er11. doi: 10.1126/scisignal.2102er11. [DOI] [PubMed] [Google Scholar]
- 3.Lennerz JK, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29:4803–4810. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choueiri TK, et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol. 2013;31:181–186. doi: 10.1200/JCO.2012.43.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–516. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 6.Bussolino F, et al. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J Cell Biol. 1992;119:629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, et al. Hepatocyte growth factor and c-Met expression in pericytes: implications for atherosclerotic plaque development. J Pathol. 2007;212:12–19. doi: 10.1002/path.2155. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, DeFrances MC, Zarnegar R. Induction of met proto-oncogene (hepatocyte growth factor receptor) expression during human monocyte-macrophage differentiation. Cell Growth Differ. 1996;7:821–832. [PubMed] [Google Scholar]
- 9.Baek JH, Birchmeier C, Zenke M, Hieronymus T. The HGF receptor/Met tyrosine kinase is a key regulator of dendritic cell migration in skin immunity. J Immunol. 2012;189:1699–1707. doi: 10.4049/jimmunol.1200729. [DOI] [PubMed] [Google Scholar]
- 10.Adams DH, et al. Hepatocyte growth factor and macrophage inflammatory protein 1 beta: structurally distinct cytokines that induce rapid cytoskeletal changes and subset-preferential migration in T cells. Proc Natl Acad Sci U S A. 1994;91:7144–7148. doi: 10.1073/pnas.91.15.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tesio M, et al. Enhanced c-Met activity promotes G-CSF-induced mobilization of hematopoietic progenitor cells via ROS signaling. Blood. 2011;117:419–428. doi: 10.1182/blood-2009-06-230359. [DOI] [PubMed] [Google Scholar]
- 12.Takeda Y, et al. Macrophage skewing by Phd2 haplodeficiency prevents ischaemia by inducing arteriogenesis. Nature. 2011;479:122–126. doi: 10.1038/nature10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott ER, et al. Deletion of Syk in neutrophils prevents immune complex arthritis. J Immunol. 2011;187:4319–4330. doi: 10.4049/jimmunol.1100341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishi Y, et al. Systemic NK4 gene therapy inhibits tumor growth and metastasis of melanoma and lung carcinoma in syngeneic mouse tumor models. Cancer Sci. 2009;100:1351–1358. doi: 10.1111/j.1349-7006.2009.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pennacchietti S, et al. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 16.Moghul A, et al. Modulation of c-MET proto-oncogene (HGF receptor) mRNA abundance by cytokines and hormones: evidence for rapid decay of the 8 kb c-MET transcript. Oncogene. 1994;9:2045–2052. [PubMed] [Google Scholar]
- 17.Dai JY, DeFrances MC, Zou C, Johnson CJ, Zarnegar R. The Met protooncogene is a transcriptional target of NF kappaB: implications for cell survival. J Cell Biochem. 2009;107:1222–1236. doi: 10.1002/jcb.22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suga H, et al. IFATS collection: Fibroblast growth factor-2-induced hepatocyte growth factor secretion by adipose-derived stromal cells inhibits postinjury fibrogenesis through a c-Jun N-terminal kinase-dependent mechanism. Stem Cells. 2009;27:238–249. doi: 10.1634/stemcells.2008-0261. [DOI] [PubMed] [Google Scholar]
- 19.Fridlender ZG, Albelda SM. Tumor-associated neutrophils: friend or foe? Carcinogenesis. 2012;33:949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- 20.Garber K. MET inhibitors start on road to recovery. Nat Rev Drug Discov. 2014;13:563–565. doi: 10.1038/nrd4406. [DOI] [PubMed] [Google Scholar]
- 21.Wright HL, Moots RJ, Bucknall RC, Edwards SW. Neutrophil function in inflammation and inflammatory diseases. Rheumatology. 2010;49:1618–1631. doi: 10.1093/rheumatology/keq045. [DOI] [PubMed] [Google Scholar]
- 22.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Van Ziffle JA, Lowell CA. Neutrophil-specific deletion of Syk kinase results in reduced host defense to bacterial infection. Blood. 2009;114:4871–4882. doi: 10.1182/blood-2009-05-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abram CL, Roberge GL, Pao LI, Neel BG, Lowell CA. Distinct roles for neutrophils and dendritic cells in inflammation and autoimmunity in motheaten mice. Immunity. 2013;38:489–501. doi: 10.1016/j.immuni.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albanesi M, et al. Neutrophils mediate antibody-induced antitumor effects in mice. Blood. 2013;122:3160–3164. doi: 10.1182/blood-2013-04-497446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagasse E, Clerc RG. Cloning and expression of two human genes encoding calcium-binding proteins that are regulated during myeloid differentiation. Mol Cell Biol. 1988;8:2402–2410. doi: 10.1128/mcb.8.6.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamm A, et al. PHD2 regulates arteriogenic macrophages through TIE2 signalling. EMBO Mol Med. 2013;5:843–857. doi: 10.1002/emmm.201302695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 29.Moolenbeek C, Ruitenberg EJ. The “Swiss roll”: a simple technique for histological studies of the rodent intestine. Lab Anim. 1981;15:57–59. doi: 10.1258/002367781780958577. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol. 2014;184:912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schira J, et al. Significant clinical, neuropathological and behavioural recovery from acute spinal cord trauma by transplantation of a well-defined somatic stem cell from human umbilical cord blood. Brain. 2012;135:431–446. doi: 10.1093/brain/awr222. [DOI] [PubMed] [Google Scholar]
- 32.Cramer T, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotter MJ, Norman KE, Hellewell PG, Ridger VC. A novel method for isolation of neutrophils from murine blood using negative immunomagnetic separation. Am J Pathol. 2001;159:473–481. doi: 10.1016/S0002-9440(10)61719-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haslett C, Guthrie LA, Kopaniak MM, Johnston RB, Jr., Henson PM. Modulation of multiple neutrophil functions by preparative methods or trace concentrations of bacterial lipopolysaccharide. Am J Pathol. 1985;119:101–110. [PMC free article] [PubMed] [Google Scholar]
- 35.Shaked Y, et al. Rapid chemotherapy-induced acute endothelial progenitor cell mobilization: implications for antiangiogenic drugs as chemosensitizing agents. Cancer Cell. 2008;14:263–273. doi: 10.1016/j.ccr.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.