Abstract
Wnt signalling proteins regulate many aspects of animal development. We have investigated the function of mouse Wnt8b during forebrain development. Wnt8b is expressed in a highly restricted pattern including the prospective hippocampus and hypothalamus. Mutant mice lacking Wnt8b are viable and healthy. The size and morphology of the hippocampus appeared normal in mutant embryos and adults and we found no evidence of hypothalamic defects in mutants. Wnt8b is also expressed in the neurogenic region of the adult dentate gyrus, however cell proliferation was unchanged in Wnt8b−/− mutants. Mutant embryos did, however, display altered levels of expression of other Wnt genes normally expressed in forebrain. The spatial expression patterns of other Wnt genes and the overall level of canonical Wnt activity were indistinguishable from wild types. Thus, loss of Wnt8b does not give rise to an overt morphological phenotype, but does affect expression levels of other Wnts in developing forebrain.
Keywords: Wnt, gene-targeting, telencephalon, cortical hem, hippocampus
Introduction
The Wnt gene family encodes a set of highly evolutionarily conserved secretory glycoproteins, implicated in the regulation of a diverse set of developmental processes (reviewed by Logan and Nusse, 2004). There are nineteen Wnt genes in both mice and humans. Wnt signaling is mediated through several intracellular pathways, the best known of which is the canonical or β-catenin pathway. Activation of the canonical pathway causes accumulation of β-catenin and its subsequent translocation to the nucleus where, in combination with TCF/LEF transcription factors, it drives the expression of downstream target genes. Knockout mouse studies have uncovered important functions for the majority of Wnts – many null Wnt mutants have severe phenotypes, some resulting in embryonic lethality (reviewed by van Amerongen and Berns, 2006). For example, Wnt1 knockout mice lack a cerebellum and much of the midbrain and die perinatally (McMahon and Bradley, 1990) while Wnt3a knockout mice die at midgestation, lacking a number of structures including the hippocampus (Lee et al., 2000) and caudal somites (Takada et al., 1994).
Wnts have important regulatory roles at many different stages of development of the nervous system, from early patterning through to synapse formation (reviewed by Ciani and Salinas, 2005). They are commonly involved in regulating the balance between proliferation and differentiation of neural precursor cells. In the embryonic forebrain, multiple Wnts, including Wnt8b, are expressed in a part of the dorsomedial wall of the telencephalon known as the cortical hem. The cortical hem is adjacent to the hippocampal anlage and acts as a signalling centre, controlling development of the hippocampus (Grove et al., 1998; Mangale et al., 2008). Wnt3a null mutant mice lack a hippocampus (Lee et al., 2000), while mutant mice lacking either Lef1 (an effector of Wnt signaling) or Lrp6 (a Wnt co-receptor) and mice that ectopically express Dkk1 (an inhibitor of canonical Wnt signalling) have an abnormally small dentate gyrus as a result of decreased proliferation of progenitor cells (Galceran et al., 2000; Zhou et al., 2004; Solberg et al., 2008). Conversely, ectopic activation of β-catenin signaling can promote hippocampal fate (Machon et al., 2007). Mutant mice that lack the Wnt receptor Frizzled9 display subtle abnormalities of hippocampus development, including decreased cell density in the adult dentate gyrus coupled with visuospatial learning deficits (Zhao et al., 2005). Despite this clear evidence that Wnt signaling is important for the regulation of developmental processes in dorsal forebrain, the roles of most individual Wnts in these processes are not yet well understood.
In humans, chick and zebrafish, Wnt8b is also expressed in the developing hypothalamus (Lako et al., 1998; Garda et al., 2002; Lee et al., 2006). In the zebrafish, blocking wnt8b function by morpholino injection leads to the loss of proneural gene expression and blocks neurogenesis in the posterior hypothalamus (Lee et al., 2006). Here, we show that mouse Wnt8b is also expressed in the prospective hypothalamus. As yet, no roles have been described for Wnt signaling during hypothalamus development in the mouse.
To investigate the role played by Wnt8b in development of the mouse forebrain, we generated a null Wnt8b allele by gene targeting in ES cells. Surprisingly, mice lacking Wnt8b were viable and fertile and displayed no obvious morphological abnormalities. We found that rates of proliferation of neural precursor cells in mutant embryos were comparable to wild types, and saw no differences in expression of markers of specific cell types in either hippocampus or hypothalamus. The gross morphology and volume of the hippocampus in adult Wnt8b mutants were indistinguishable from those in wild types and we found no difference in cell proliferation in the adult dentate gyrus, which also expresses Wnt8b. Expression of the Wnt target gene Axin2 in Wnt8b−/− dorsomedial telencephalon was comparable to that in wild types during development, indicating that there is no overall decrease in canonical Wnt activity in the mutants. As noted above, a number of Wnt genes are expressed in patterns that overlap Wnt8b’s expression domain in the embryonic dorsomedial telencephalon. We found that the dorsomedial telencephalic expression domains of Wnts −2b, −3a, −5a, −7a and −7b were unchanged in Wnt8b−/− mutants. However, the levels of expression of Wnt3a and Wnt5a were increased in mutant telencephalon, while the levels of Wnt2b and Wnt7a were decreased and the level of Wnt7b was unchanged.
Results
Expression of Wnt8b in embryonic forebrain
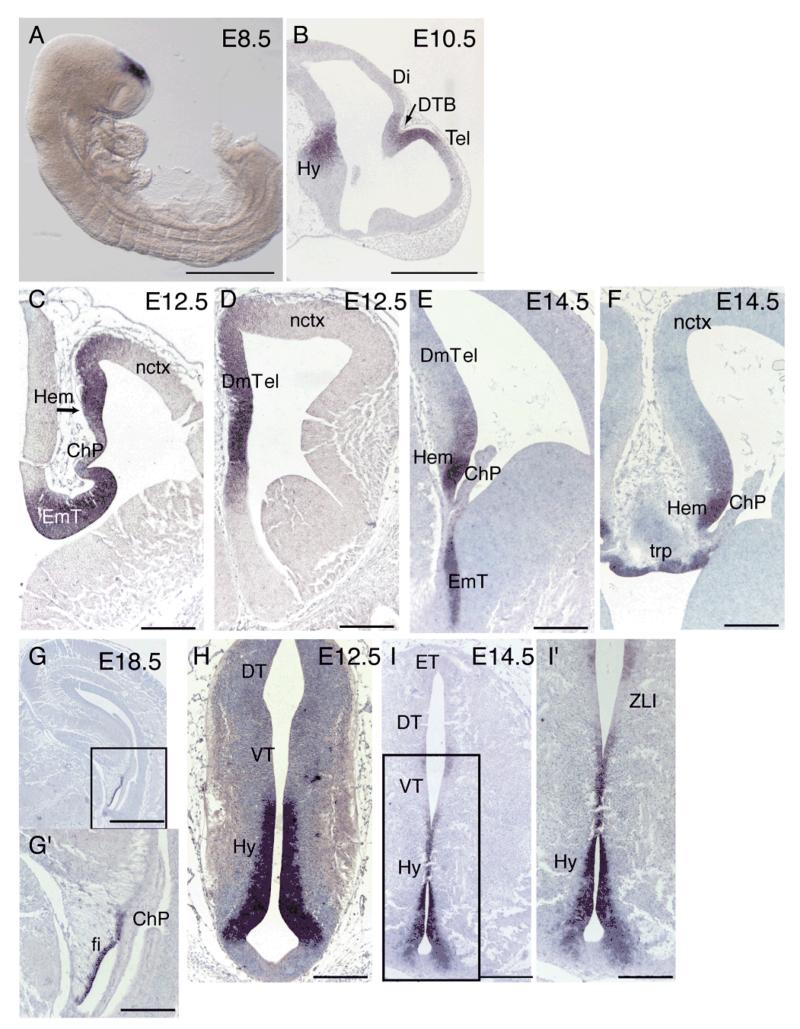
Wnt8b has been used as a marker of dorsomedial telencephalon in several studies (Tole et al., 2000; Theil et al., 2002; Kimura et al., 2005) but its full embryonic forebrain expression pattern has been described only in the chick and in human. We therefore performed a detailed expression pattern analysis of mouse Wnt8b by in situ hybridization. At E8.5, Wnt8b is strongly expressed in the dorsal prosencephalon (Fig. 1A). At E10.5 the division between the telencephalon and diencephalon has become clear and Wnt8b expression is observed at the diencephalic-telencephalic boundary (DTB) and the future dorsomedial telencephalon (Fig. 1B). These results are in agreement with previously published findings (Richardson et al., 1999). In the telencephalon at E12.5, Wnt8b expression was mainly found in the dorsomedial telencephalon, with very low amounts at the most dorso-lateral tip of the neocortex (Fig.1C,D). By E14.5, the relative extent of expression in the dorsomedial telencephalon was greatly reduced and the highest levels of expression were observed in the cortical hem (Fig. 1E,F). Expression was also found in the telencephalic roof plate (Fig. 1F). At E18.5, expression was restricted to the lateral margin of the hippocampal fimbria (Fig. 1G,G’), the remnant of the cortical hem at this stage (Bulchand et al., 2001). In the developing diencephalon, Wnt8b is expressed strongly in the presumptive hypothalamus at E10.5 (Fig 1B). Strong Wnt8b expression was observed in the eminentia thalami at both E12.5 and E14.5 (Fig. 1C and E respectively). At E12.5, Wnt8b was also found at high levels in the neuroepithelium along the ventricular zone of the developing hypothalamus, except for the base of the ventral hypothalamus (Fig. 1H). Hypothalamic expression persisted at E14.5 (Fig. 1I,I’). By E18.5, expression of Wnt8b in the hypothalamus was restricted to a very thin domain surrounding the ventricular zone (not shown). At E14.5, a small patch of Wnt8b expression was observed at the ventricular neuroepithelium of the thalamus near the zona limitans intrathalamica (Fig. 1I,I’).
Figure 1. Wnt8b expression in embryonic forebrain.
At E8.5 Wnt8b is expressed at the dorsal prosencephalic vesicle (A). At E10.5 expression is observed at the dorsal diencephalic-telencephalic boundary (DTB) (arrow in B), the dorsal-most part of the telencephalon (Tel), and the hypothalamus (Hy) (B). Wnt8b expression is seen in the E12.5 and E14.5 dorsomedial telencephalon (DmTel), with stronger levels of expression in the cortical hem (hem) (C-F). No expression is observed in the choroid plexus (ChP) (C,E,F). At E14.5 Wnt8b expression is also detected in the telencephalic roof plate (trp) (F). By E18.5, telencephalic expression of Wnt8b is restricted to the lateral margin of the hippocampal fimbria (fi) (G,G’). In the developing diencephalon, Wnt8b is strongly expressed in the E12.5 and E14.5 eminentia thalami (EmT) (C,E). Expression is also abundant in the ventricular zone of the E12.5 hypothalamus, (H). A similar, but more restricted hypothalamic expression domain is observed at E14.5 (I,I’). In addition, a small expression domain is observed between the dorsal and ventral thalamus (DT, VT) at the level of the zona limitans intrathalamica (ZLI) (I’). Panels G’ and I’ show higher magnifications of the boxed areas in G and I respectively. Scale bars: A, 500 μm; B, 50 μm; C-F, G’, H, I’, 200 μm; G, I, 400 μm. Abbreviations: Hy: hypothalamus; ET, epithalamus.
Targeted disruption of the Wnt8b gene
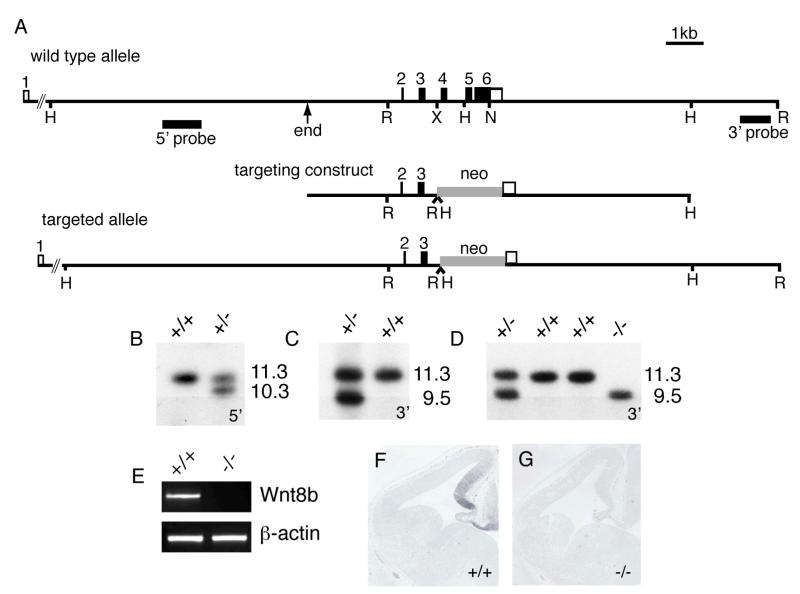
To determine Wnt8b’s role during forebrain development, we generated a null Wnt8b allele by gene targeting in ES cells (Fig. 2). Targeting resulted in the deletion of exons 4 and 5 and almost all of the coding region of exon 6. Mouse Wnt8b contains 329 amino acids and this targeting event causes the deletion of sequences encoding amino acids 81-316. Two correctly targeted clones, identified by Southern blotting (Fig. 2B,C), were injected into blastocysts to generate chimeras. Following germline transmission, Wnt8b+/− animals were back-crossed onto a C57Bl/6 genetic background. Intercrosses of Wnt8b+/− animals gave rise to expected Mendelian ratios of wild type, heterozygous and homozygous mutant offspring. Wnt8b−/− animals are viable and fertile. The absence of Wnt8b expression in Wnt8b−/− embryos was confirmed by RT-PCR and in situ hybridisation (Fig. 2E-G).
Figure 2. Generation of Wnt8b-deficient mice.
(A) Targeting construct, structures of the wild type and targeted Wnt8b alleles and probes used in Southern blotting to detect correctly-targeted clones. ‘End’ indicates the 5′ end of the 5′ homology arm and corresponds to the end of the λ clone from which the targeting vector was derived. H, HindIII; R, EcoRI; X, XbaI; N, NotI. (B,C) Southern blots using 5′ and 3′ flanking probes, showing correctly targeted ES cell clones. The 5′ probe detects an 11.3kb HindIII fragment in wild type DNA and a 10.3Kb fragment in the targeted locus. The 3′ probe detects an 11.3kb EcoRI fragment in wild type DNA and a 9.5kb fragment in the targeted allele. (D) Genotyping of mice carrying targeted allele (E) RT-PCR showing absence of Wnt8b mRNA in mutant telencephalon. (F,G) Absence of Wnt8b expression in dorsomedial telencephalon of E12.5 mutant embryo as judged by in situ hybridisation.
The dorsomedial telencephalon develops normally in Wnt8b−/− mutants
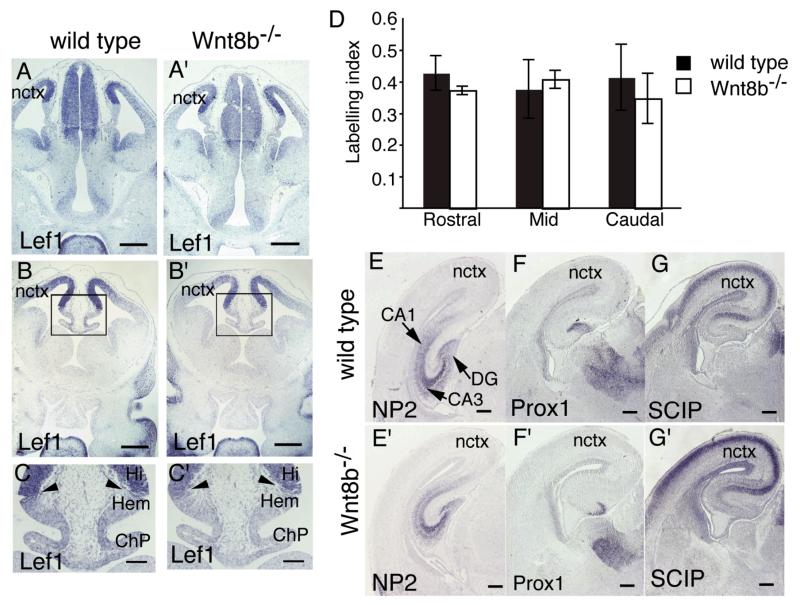
As Wnt8b is expressed in dorsomedial telencephalon and Wnt signaling is known to be important for the normal development of this region (Lee et al., 2000; Galceran et al., 2000; Zhou et al., 2004; Zhao et al., 2005; Machon et al., 2007; Solberg et al., 2008), we examined the expression pattern of the dorsomedial telencephalic marker gene Lef1 in wild type and Wnt8b−/− embryos at E12.5. In wild type embryos, Lef1 was expressed in a gradient in the cortex, with highest expression immediately adjacent to the cortical hem and decreasing expression towards the lateral cortex (Fig. 3A,C). There was no clear difference in the size or morphology of the dorsomedial telencephalon in mutant embryos (Fig. 3A’-C’) and the expression of Lef1 appeared identical between wild type and Wnt8b−/−mutants (compare Fig. 3A-C with 3A’-C’).
Figure 3. The hippocampus develops normally in Wnt8b−/− mutants.
(A-C’) In situ hybridisation for Lef1 in E12.5 wild type (A,B,C) and Wnt8b−/− mice (A’,B’,C’). The areas boxed in B and B’ are magnified in C and C’, respectively. The expression pattern of Lef1 is indistinguishable between wild types and mutants. Arrowheads in C and C’ indicate lack of Lef1 expression in the cortical hem (Hem) and choroid plexus (ChP). (D) Bar graph showing BrdU labelling indices in the dorsomedial telencephalon of Wnt8b−/− mutants and wild type controls at E14.5. n=3 animals of each genotype, error bars indicate SEM. (E-G’) In situ hybridisation on coronal sections of E18.5 wild type (E,F,G) and Wnt8b−/− (E’,F’,G’). NP2 marks the entire hippocampus (E,E’), while Prox1 (F,F’) and SCIP (G,G’) mark the dentate gyrus (DG) and CA1 field of the hippocampus respectively. Scale bars: A-B’, 200 μm; C-C’, 100 μm E-G’, 250 μm. Abbreviations: Hi: hippocampus; nctx: neocortex.
Wnt signalling is required for normal proliferation of precursor cells in the medial wall of the dorsal telencephalon (Lee et al., 2000; Galceran et al., 2000). To determine whether the loss of Wnt8b affects proliferation of these cells, we measured the BrdU labelling index in dorsomedial telencephalon at E14.5 in mutant embryos and wild type controls (Fig. 3D). Labelling indices were measured at rostral, mid and caudal levels, to check for possible regional effects of Wnt8b, but no difference was found between Wnt8b−/− mutants and their wild type littermates at any position (n=3).
The highly distinctive architecture of the hippocampal formation is readily observed at later stages of embryonic development, around E18.5. By this time, region-specific expression of several genes allows the identification of specific subdomains of the hippocampus. For example, Neuropilin2 (NP2) is expressed throughout the hippocampus (Sheng et al., 1997; Galceran et al., 2000); Prox1 is expressed in the dentate gyrus (Oliver et al., 1993; Galceran et al., 2000; Shinozaki et al, 2004) and SCIP is expressed in the neocortex, subiculum and CA1 field (Frantz et al, 1994; Tole et al., 1997). We used in situ hybridisation to compare the expression patterns of each of these markers in wild type and Wnt8b−/− mutant embryos at E18.5 (Fig. 3D-G’). Again, we found no difference in either the overall appearance of the hippocampus or in the expression domains of these marker genes in mutant embryos.
Normal patterning of the diencephalon in Wnt8b−/− mutant embryos
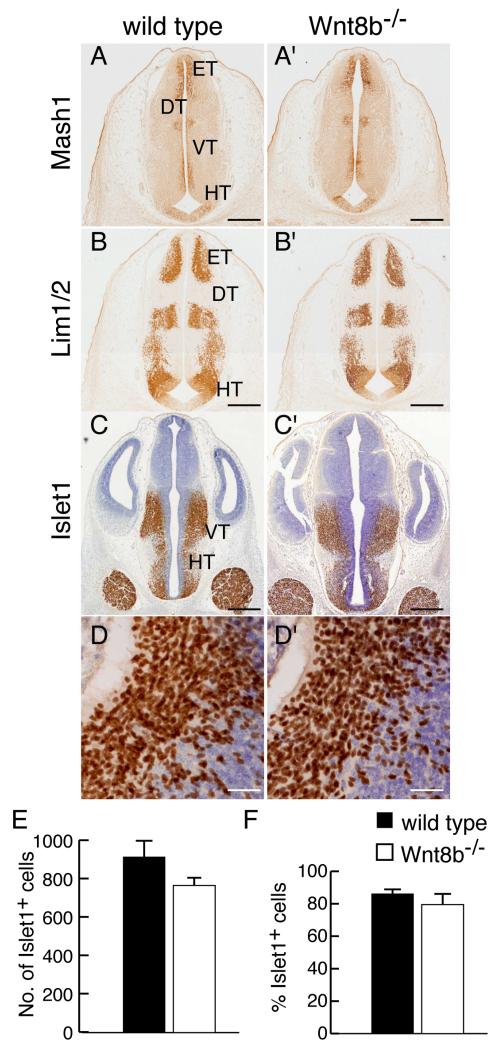
Wnt8b is also expressed in the embryonic diencephalon, primarily in the posterior hypothalamus (Fig. 1). Loss of function studies in zebrafish indicate a role for Wnt8B during diencephalic development in promoting neural differentiation in the posterior hypothalamus (Lee et al., 2006). To determine whether loss of Wnt8b affects diencephalic development in the mouse, we examined the expression of markers of diencephalic fate in mutant embryos. At E12.5 Mash1, Lim1 and Islet1 are each expressed in a specific, characteristic, subset of domains of the diencephalon (Fig. 4A-D). We found no difference in the expression patterns of any of these proteins in Wnt8b−/− mutants (Fig. 4A’-D’). To look for possible differences in cell number, we counted Islet1-expressing neurons throughout the rostro-caudal extent of the hypothalamus in mutant and wild type embryos at E12.5. We found no significant difference in either the number of Islet1-positive cells or the proportion of hypothalamic cells that expressed Islet1 (Fig. 4E,F).
Figure 4. Normal expression of hypothalamus markers in Wnt8b−/− mutants.
(A-D’) Immunoreactivity for Mash1 (A,A’), Lim1 (B,B’) and Islet1 (CD’) in coronal sections of diencephalon in E12.5 wild type (A-D) and Wnt8b−/−embryos (A’-D’). D,D’ show high magnification views of Islet1 immunoreactivity in the hypothalamus, as used for cell counting. (E,F) Graphs showing number (E) and percentage (F) of Islet1 expressing cells in the hypothalamic region of E12.5 embryos, as estimated by counting in 212.5 × 212.5 μm areas of representative sections. Bars represent mean ± SEM; n=3 per genotype. Scale bars: A-C’ 400 μm; D,D’ 50 μm. Abbreviations: DT: dorsal thalamus; ET: epithalamus; HT: hypothalamus; VT: ventral thalamus.
The adult hippocampus appears normal in Wnt8b−/− mutants
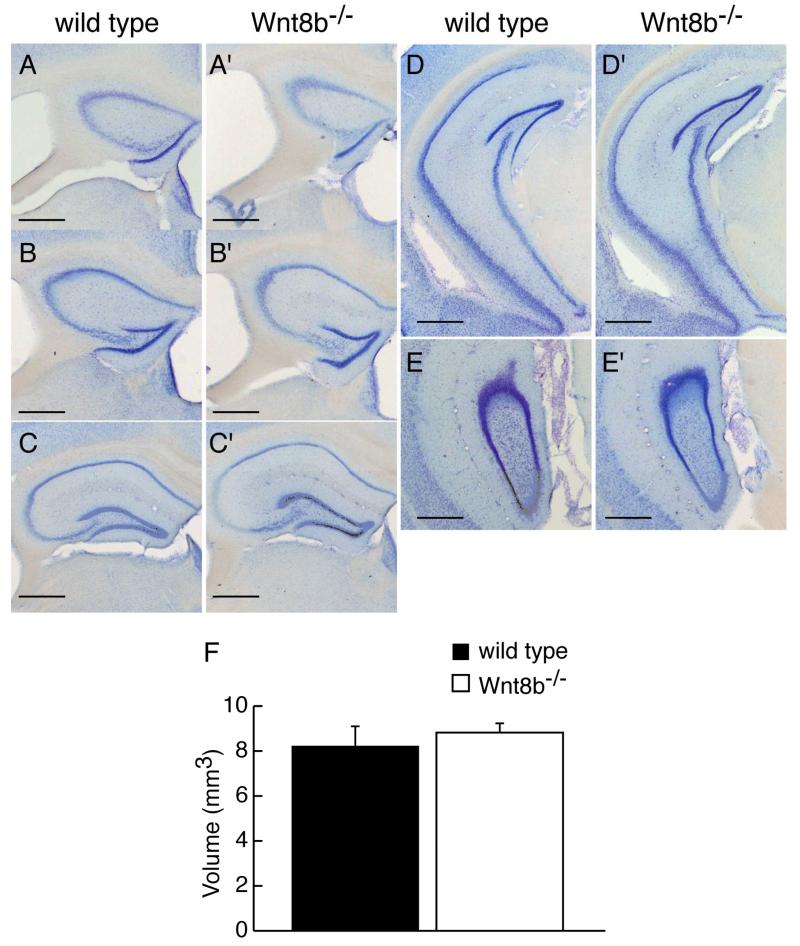
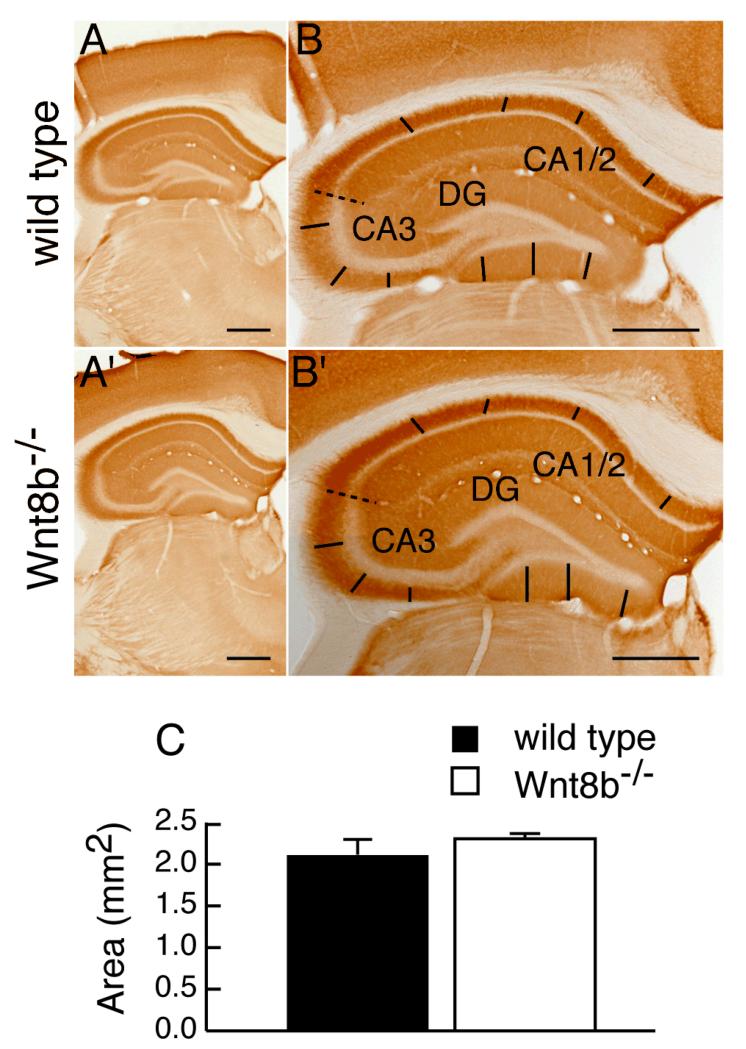
To determine whether lack of Wnt8b leads to abnormalities in the adult hippocampus, we carefully examined hippocampus morphology in mutants and age-matched wild-type controls. Fig. 5A-E’ show representative cresyl violet stained coronal sections along the rostro-caudal axis of adult wild type and mutant animals. No gross alterations in the mutant hippocampus were observed (compare Fig. 5A-E with A’-E’). We measured the area occupied by the hippocampus in serial sections of adult Wnt8b−/− mutant and control brains and used these values to estimate the volume of the adult hippocampus (Fig 5F, see materials and methods for details). These measurements revealed a slight increase in the overall volume of the hippocampus in adult mutants, but this was not statistically significant (n=3, Student’s t-test, p=0.649). Although we did not find any significant difference in the total volume of the adult hippocampus in Wnt8b−/− mutants, it remained possible that the relative sizes of the different hippocampal fields may be altered. To test this possibility, we performed a series of measurements in middle hippocampal sections. We used sections immunolabelled for mGluR5 to delineate the boundaries of the CA1/2 and CA3 cell layers and measured the length of each layer (CA1/2, CA3 and DG). mGluR5 labels the hippocampal dendritic fields but not the pyramidal cells of areas CA1-CA3, nor the granular cells of the dentate gyrus (DG). The characteristic expression pattern of mGluR5, which is reduced in the CA3 field allowed us to distinguish clearly between areas CA1/2 and CA3. The unlabelled granular cell layer of the dentate gyrus was also easily identified (Fig. 6A-B’). Although in all cases mutants showed an increase in the length of the all hippocampal layers compared to wild types, the difference was not significant (Table 1). In addition, measurements of the thickness of the basal dendritic fields of CA1/2 and CA3, as well as that of the molecular layer of he DG (Fig. 6A-B’) did not reveal any differences between wild types and mutants (Table 2). Finally, the overall area occupied by the hippocampus in these mid-level sections showed no difference in the mutants (n=3; Student’s t-test; p= 0.347) (Fig. 6C).
Figure 5. Morphology and volume of the adult hippocampus is similar in wild type and Wnt8b−/− mutant mice.
Representative coronal sections stained with cresyl violet showing morphology of the hippocampus from rostral to caudal levels in wild type (A-E) and mutant mice (A’-E’) A,A’ are most rostral, E,E’ are most caudal. (F) Volume of the hippocampus in adult wild type and Wnt8b−/− mutants. Bars represent mean ± SEM; n=3 animals per genotype. Scale bars: 100 μm.
Figure 6. Areal measurements of middle hippocampal sections are similar between wild types and Wnt8b−/− mutants.
mGluR5 immunohistochemistry on coronal wild type (A-B) and mutant (A’-B’) hippocampal sections taken at the midpoint of the rostrocaudal axis. The dashed line indicates the boundary between the CA1/2 and CA3 fields. The straight lines represent those used to measure the dendritic hippocampal fields of each layer. (C) Area of the hippocampus at the midpoint of the rostrocaudal axis in adult wild type and Wnt8b−/− mutants. Bars represent mean ± SEM; n=3 mice per genotype. Scale bars: 500 μm.
Table 1. Hippocampal cell layer lengths in adult Wnt8b+/+ and Wnt8b−/− mice.
| Hippocampal layer |
Cell layer length (mm)a (mean ± SD) |
||
|---|---|---|---|
| Wnt8b+/+ | Wnt8b−/− | pb | |
| CA1/2 | 2.178 ± 0.157 | 2.364 ± 0.127 | 0.19 |
| CA3 | 1.387 ± 0.096 | 1.427 ± 0.103 | 0.40 |
| DG | 1.952 ± 0.195 | 2.043 ± 0.168 | 0.57 |
Mean layer length from four to eight different sections for each group (n=3) of mice.
For CA1/2 and DG by Student’s t-test; for CA3 by Mann-Whitney test.
Table 2. Hippocampal layer thickness in adult Wnt8b+/+ and Wnt8b−/− mice.
| Hippocampal layer |
Dendrite Layer Thickness (mm)a (mean ± SD) |
||
|---|---|---|---|
| Wnt8b+/+ | Wnt8b−/− | pb | |
| CA1/2 | 0.110 ± 0.012 | 0.110 ± 0.005 | 0.95 |
| CA3 | 0.172 ± 0.006 | 0.175 ± 0.007 | 0.69 |
| DG | 0.160 ± 0.009 | 0.174 ± 0.012 | 0.16 |
Mean thickness of dendritic layer (basal dendritic field for CA1-CA3; molecular layer dendritic field for DG) from an average of twenty four to forty eight measurements for each layer and for each group (n=3) of mice.
Student’s t-test.
Proliferation rates in the adult dentate gyrus are unchanged in Wnt8b−/− mutants
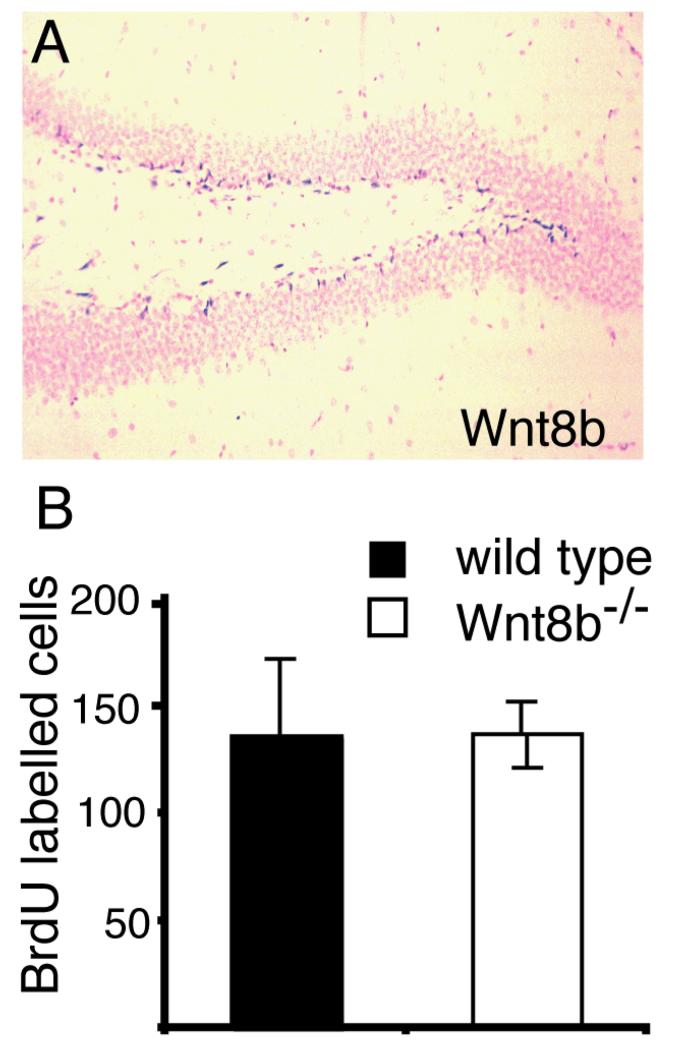
We found that expression of Wnt8b persists into adult stages in the dentate gyrus (Fig. 7A). Wnt8b is expressed in the subgranular layer of the dentate gyrus, one of the two known sites of adult neurogenesis in rodents (reviewed by Li et al., 2009). Wnt signalling has been shown to regulate proliferation of progenitor cells in this region (Lie et al., 2005). To test whether loss of Wnt8b affected cell proliferation in the adult hippocampus, we injected mutant and wild type animals with BrdU over a 12-hour period and counted the number of BrdU-labelled cells in the hippocampus at the end of the labelling period. We found no difference in the mutant animals (Fig. 7B).
Figure 7. Levels of BrdU incorporation in the adult hippocampus are not altered in Wnt8b−/− mutants.
(A) In situ hybridisation showing Wnt8b expression in the subgranular layer of the adult dentate gyrus. (B) Number of BrdU labelled cells in the hippocampus of animals after 12 hours continuous BrdU labelling. BrdU labelled cells in the entire hippocampus were counted on every tenth section throughout the full rostrocaudal extent of the hippocampus and the graph shows the total number of labelled cells counted. n=3 animals of each genotype, error bars indicate SEM.
Canonical Wnt signalling is not perturbed but expression levels of several Wnt genes are altered in the dorsomedial telencephalon of Wnt8b−/− embryos
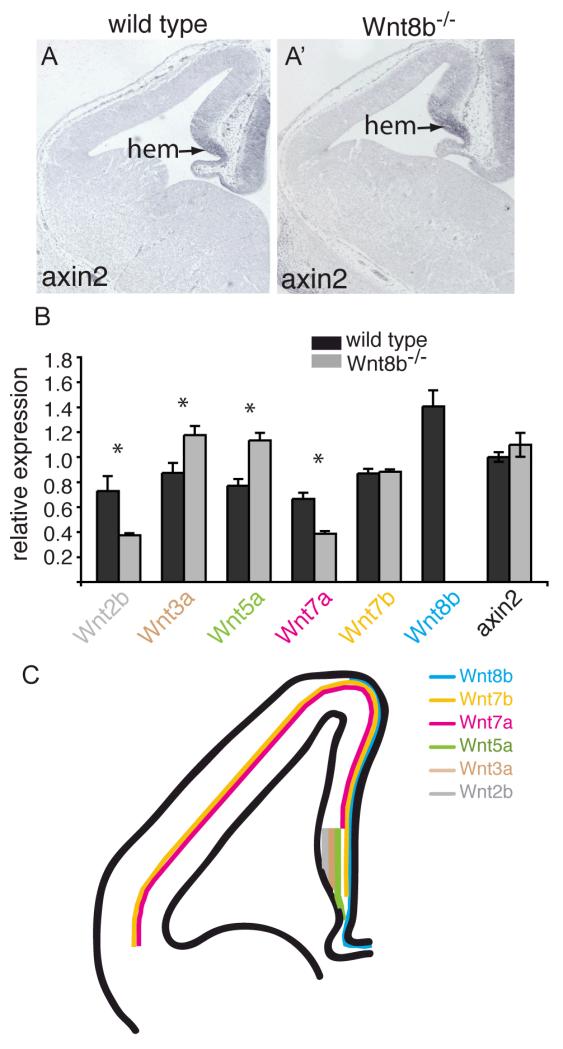
Several Wnt pathway mutations have been described that adversely affect hippocampus development (Lee et al., 2000; Galceran et al., 2000; Zhou et al., 2004; Machon et al., 2007) so it seemed surprising that we were unable to detect any abnormalities in this region of Wnt8b−/− mutants, either in embryos or adults. We therefore sought to determine whether the overall pattern of canonical Wnt signaling is altered in the forebrains of Wnt8b−/− mutant embryos. We examined the expression of the well-characterised Wnt target gene Axin2 (Jho et al., 2002) in mutant dorsomedial telencephalon both by in situ hybridisation and by qRTPCR. No obvious difference was found in either the extent (Fig 8A,A’) or the level (Fig 8B) of Axin2 expression, indicating that canonical Wnt signalling is not altered in this region of mutant embryos.
Figure 8. Canonical Wnt signalling appears normal and expression of Wnts 3a and 5a is increased in Wnt8b−/− mutants.
Axin2 in situ hybridisation on coronal sections of telencephalon in E12.5 wild type (A) and Wnt8b−/− mutant embryos (A’). The arrow points to the cortical hem (hem). (B) Quantitative RT-PCR showing the expression levels of Axin2 and several Wnt genes in the E12.5 telencephalon relative to GAPDH. Expression of Wnt3a and Wnt5a is increased in Wnt8b−/− embryos while expression of Wnt2b and Wnt7a is lower. Wnt8b expression is completely lost, as expected. Axin2 levels are unchanged. n=3 embryos of each genotype, * p<0.05, Student’s t-test. (C) Schematic summary of Wnt gene expression in E12.5 dorsal telencephalon. Note that the lines indicate only the extent of expression along the medial-lateral axis – each of these Wnts is expressed throughout the full thickness of the tissue at this stage, except Wnt7b which is expressed throughout the hem but whose cortical expression is restricted to the outermost cells (pial edge) in the cortex.
This suggested the possibility that the lack of a detectable phenotype in Wnt8b−/−mutants may be due to compensatory changes in the expression of other Wnt genes. Wnts −2b, −3a, −5a, −7a and −7b are all expressed in patterns that overlap, at least in part, that of Wnt8b in the dorsomedial telencephalon (Fig 8C; Grove et al., 1998; Theil 2005). We sought to determine whether the spatial pattern of expression of any of these five Wnt genes was altered in the dorsomedial telencephalon of Wnt8b−/− mutants. Using in situ hybridisation, we compared the expression of Wnts −2b, −3a, −5a, −7a and −7b in E12.5 mutant and control embryos, but found no differences (supplementary Fig. 1). As in situ hybridisation is not a quantitative technique, it remained possible that the levels of expression of one or more of the other Wnt genes may be altered, although the patterns were unchanged. We therefore carried out quantitative RT-PCR on mRNA isolated from E12.5 telencephalon to determine the expression levels of Wnts 2b, 3a, 5a, 7a and 7b in mutant and wild type embryos. Expression levels were calculated relative to the level of GADPH mRNA and the results are shown in Figure 8B. We found that expression of Wnt3a and Wnt5a was increased in the Wnt8b−/− mutants whereas expression of Wnt2b and Wnt7a was decreased. There was no significant difference in the level of Wnt7b expression.
Discussion
We have shown that expression of Wnt8b is restricted to three parts of the developing mouse forebrain, the hippocampus, hypothalamus, and eminentia thalami. This expression pattern is very similar to that described for human WNT8B (Lako et al., 1998) and agrees with earlier, less complete descriptions of the mouse Wnt8b expression pattern (Grove et al., 1998; Lee et al., 2000; Richardson et al., 1999). This evolutionarily conserved pattern of expression, coupled with findings that Wnt signalling is required for normal development of the hippocampus in mouse (Lee et al., 2000; Galceran et al., 2000; Zhou et al., 2004; Zhou et al., 2005; Solberg et al., 2008) and the hypothalamus in zebrafish (Lee et al., 2006) strongly suggested that Wnt8b may be important for normal development of the mouse forebrain. However, we found no overt anatomical abnormalities of forebrain development in our Wnt8b−/− mice.
We also found that Wnt8b is expressed in the subgranular zone of the adult dentate gyrus, one of the areas in which neurogenesis persists into adulthood. It has been shown that Wnt3, which is also expressed in this region, is important for regulating neurogenesis from adult hippocampal progenitor cells (Lie et al., 2005). We did not find any difference in BrdU incorporation in this region in Wnt8b−/− adults. This could indicate that the functions of Wnt3 and Wnt8b are redundant here. Alternatively, as adult hippocampal neurogenesis is regulated physiologically, for example in response to stress, it may be that Wnt8b regulates aspects of neurogenesis when mice are exposed to altered conditions.
Although we have been unable to detect a phenotype at the cellular or tissue level, we cannot exclude the possibility that there are subtle alterations in the mutants that we have failed to detect. Our finding stands in marked contrast to studies on many other mouse Wnts, as loss of function mutations in many Wnt genes lead to severe phenotypes, commonly resulting in embryonic or perinatal lethality (reviewed in van Amerongen and Berns, 2006). The Wnt8b mutant allele described here is likely to be a true null. A large part of the coding sequence is deleted; comparable to, or greater than, that deleted in targeted alleles of other Wnt genes (van Amerongen and Berns, 2006). The deleted region includes 19 of the 24 cysteine residues found in Wnt8b; these cysteine residues are a particularly highly conserved feature of Wnt proteins, and several of them are essential for Wnt activity (Mason et al., 1992). Further, no Wnt8b transcripts were detected in mutant embryos, either by in situ hybridisation or by RT-PCR.
Wnt signalling is required for normal development of the dorsomedial telencephalon in mouse. The hippocampus of Wnt3a−/− mutants is much reduced in size, as a result of decreased proliferation of precursor cells. The CA1 field and dentate gyrus are completely absent in these mutants and only a very small remnant of CA3 remains (Lee et al., 2000). A mutant allele of Lef1, which produces a Lef1-lacZ fusion that blocks transcriptional activation by Lef/Tcf proteins leads to complete absence of the hippocampus, also as a consequence of decreased proliferation of precursor cells (Galceran et al., 2000). Conditional inactivation of β-catenin in dorsal telencephalon (but excluding the cortical hem) leads to a loss of hippocampal fields CA1 and CA2 and a reduction in CA3 and the dentate gyrus (Machon et al., 2003). Ectopic expression of dkk1 (an extracellular antagonist of canonical Wnt signalling) in the dorsomedial telencephalon causes abnormal development of the dentate gyrus (Solberg et al., 2008). In contrast, we found no anatomical abnormalities or changes in proliferation in the hippocampus of Wnt8b−/− mutants. It remains possible that, although the Wnt8b mutants appear essentially normal, there may be subtle phenotypes that have not been detected by our analysis. A subtle phenotype has been reported in mice lacking the Wnt receptor Fzd9, indicating that Wnt signalling regulates highly localised developmental processes in the hippocampus. In Fzd9 mutants, the overall organisation of the dentate gyrus is normal, but mutants have slightly fewer dentate granule cells and more hilar mossy cells. These minor changes are associated with deficits in spatial learning (Zhao et al., 2005).
The lack of an obvious anatomical phenotype in Wnt8b−/− mutants may well be due, at least in part, to functional redundancy between Wnt genes. At least six Wnt family members are expressed in the embryonic dorsomedial telencephalon, along with multiple receptors and secreted inhibitors of Wnt signalling (Grove et al., 1998; Kim et al., 2001; Fischer et al., 2007). Indeed, the finding that so many Wnts are expressed in the cortical hem led to this area being described as ‘Wnt rich’ (Grove et al., 1998). Although there is substantial overlap between expression domains, each Wnt is expressed in a slightly different pattern at E12.5 – Wnt2b and Wnt3a are restricted to the cortical hem; Wnt5a is expressed in the hem and adjacent mesenchyme; Wnt7a is expressed in the majority of the cerebral cortex but is not expressed in the hem; Wnt7b is expressed throughout dorsal telencephalon, including the hem and Wnt8b is expressed throughout the dorsomedial telencephalon, including the hem. These patterns of expression are summarised in Figure 8C. The diversity of these expression patterns, coupled with the diversity of frizzled receptor expression in the same region (Kim et al., 2001; Fischer et al., 2007) suggests that different Wnts may be required for different processes that contribute to the normal development of this complex region of the emerging forebrain.
There is precedence for compensatory functions between Wnts during CNS development. Wnt1 and Wnt3a are expressed in partially overlapping patterns in the dorsal CNS, including the roof plate of the spinal cord. Wnt1−/− mutants lack a cerebellum and part of the midbrain (McMahon and Bradley, 1990), while Wnt3a−/− mutants lack a hippocampus (Lee et al., 2000). Neither single mutant has a dorsal neural tube phenotype. However, Wnt1−/−;Wnt3a−/− double mutants have a massively-decreased population of neural crest cells, due to a failure of progenitor cells in the dorsal neural tube to proliferate normally (Ikeya et al., 1997), indicating that each of these Wnt genes is able to compensate for some of the lost function of the other. Therefore, it is conceivable that increased expression of Wnt3a and Wnt5a observed in Wnt8b−/− mutants may compensate for the loss of Wnt8b. This would suggest the presence of a feedback mechanism in the embryonic forebrain that can react to altered levels of Wnt ligands by increasing the expression of other family members. However, we also found that the expression of Wnt2b and Wnt7a decreases in Wnt8b−/− mutants, indicating that the detailed operation of any feedback mechanism is likely to be complex.
Historically, Wnts have been catalogued as ‘canonical’ or ‘non-canonical’ based upon classical assays of Wnt activity (transformation of mammary epithelial cells and induction of a new antero-posterior axis in Xenopus). Wnt3a and Wnt8b fall into the ‘canonical’ group, while Wnt5a has often been described as a ‘non-canonical’ Wnt, acting through pathways other than the canonical Wnt pathway (Shimizu et al., 1997; Wong et al., 1994; Olson and Papkoff, 1994; Du et al., 1995; Slusarski et al., 1997a,b). As canonical Wnt signalling is clearly implicated in dorsomedial telencephalon development (Lee et al., 2000; Galceran et al., 2000), it may seem surprising to suggest that Wnt5a may compensate, at least in part, for the lack of Wnt8b. However, a recently published study, using purified Wnt5a protein, showed that Wnt5a promotes nuclear accumulation of β-catenin, in a context dependent manner (Mikels and Nusse, 2006). Thus, the classification of Wnts into ‘canonical’ and ‘non-canonical’ classes likely represents an oversimplification.
Wnt8b is also expressed in the presumptive hypothalamus. Using specific morpholinos that block the function of wnt8b in zebrafish, others have shown that wnt8b, acting through Lef1, is required for neuronal differentiation in the posterior hypothalamus (Lee et al., 2006). In our Wnt8b−/− mutant mice, we did not detect any difference in the size or morphology of the hypothalamus or in the expression of several specific markers, including Islet1, whose expression is lost in zebrafish Wnt8b morphants (Lee et al., 2006). Further, both the number and the proportion of Isl1 expressing cells were unchanged in E12.5 mutant hypothalamus. This difference may reflect the different methods of gene inactivation (translation-blocking morpholino as against gene-targeting) or could be due to species differences either in the function of Wnt8b, or in the ability of related genes to compensate for the loss of Wnt8b.
Our finding that expression levels of several Wnt genes are altered in Wnt8b−/− mutants indicates that Wnt8b is required, directly or indirectly, to regulate the expression of other Wnt genes. Thus, at least at the molecular level, Wnt8b activity is not completely redundant in the embryonic dorsomedial telencephalon. The molecular mechanisms governing the embryonic development of the forebrain are highly complex, and sophisticated approaches are likely to be needed to unravel the roles played by individual Wnt genes in this process.
Materials and Methods
Mice
A null mutation of Wnt8b was generated by replacing most of the coding sequence with a neomycin resistance cassette driven by a β-actin promoter (Fig. 2A). Briefly, genomic 129/Ola clones containing the Wnt8b coding sequence were isolated from a bacteriophage lambda library (gift from Andrew Smith, University of Edinburgh). Homology arms were subcloned from the isogenic lambda clones. The 5′ homology arm comprised a 3.5kb fragment extending from the XbaI site in intron 3 to an SfiI site that was introduced at the end of the lambda clone. The 3′ homology arm comprised a 5.1kb fragment extending from the NotI site at the very end of the coding sequence in exon 6 to a HindIII site 5kb downstream of the end of the Wnt8b coding sequence. Targeting leads to loss of exons 4 and 5 and almost all of the coding part of exon 6. Correctly targeted clones were identified by Southern blotting using 5′ and 3′ flanking probes (Fig. 2B,C). Two separate clones were injected into C57Bl/6 blastocysts to generate chimaeric mice. Chimaeric males successfully transmitted the targeted allele through the germline. The mutant allele (designated Wnt8btm1Jmas using Mouse Genome Informatics nomenclature) is referred to as Wnt8b− throughout this paper. Wnt8b+/− adult mice were intercrossed to yield Wnt8b−/− homozygotes. Embryonic day (E) 0.5 was defined as midday on the day of plug detection. Animal care was in accordance with institutional guidelines and UK Home Office regulations.
Genotype analysis
Genotyping was either by Southern blot using a radiolabelled 1.1kb PstI fragment subcloned from the 3′ flanking sequence (Fig. 2D) or by multiplex PCR using four primers, β-actinF: 5′-GGCCAACGCCAAAACTCTCC-3′, β-actinR: 5′-GGCCGCTCGAGCCATAAAAG-3′, (211bp product); Wnt8bF: 5′-GGTTGCAACTGCCAGAGTTC-3′ and Wnt8bR: 5-GAGGCGGGAGGGATAGATAC-3′ (494bp product).
In situ hybridisation
Embryos were fixed in 4% paraformaldehyde in phosphate buffer (0.1 M, pH 7.4) at 4°C overnight, dehydrated in an ascending ethanol series, cleared in xylene, embedded in paraffin and 10μm sections were cut using a microtome. In situ hybridisation was performed as previously described (Yu et al., 2009). In situ probes were as described previously: Wnt8b (Richardson et al., 1999); Lef1 (Galceran et al., 2000); NP2 (Sheng et al., 1997); Prox1 (Oliver et al., 1993); SCIP (Frantz et al., 1994) and Axin2 (Theil, 2005; Machon et al., 2007). RNA antisense probes were labelled using the digoxigenin RNA labeling kit (Roche, Welwyn Garden City, UK) according to the instructions of the manufacturer.
Immunohistochemistry
Immunohistochemistry on paraffin sections was performed as previously described (Fotaki et al., 2006). Mouse monoclonal antibodies were against BrdU (1:200; BD Biosciences, Oxford, UK), Mash1 (1:100; BD Biosciences), Islet1 (1:50; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) and Lim1/2 (1:200; Developmental Studies Hybridoma Bank). Binding of appropriate secondary antibodies (1:200; Dako) was revealed with the avidin-biotin-peroxidase system (Vector Laboratories) and diaminobenzidine (Sigma).
Islet1-positive cell counts in E12.5 hypothalamus
Islet1+ cells and cresyl violet cells were manually counted from 3-4 representative sections along the rostrocaudal hypothalamic axis within a square of 212.5 × 212.5 μm using ImageTool 3.0 software. Islet1+ cell density was calculated as a percentage of Islet1+ cells to total number of cells.
Bromodeoxyuridine (BrdU) analysis
For analysis of embryos, a 30 min pulse of BrdU (70 μg /g body weight, i.p.) was administered to pregnant dams, and E14.5 embryos were collected. Sections were cut serially at 10 μm and reacted as described previously (McLaughlin et al., 2003). Labelling indices (LI: labelled cells as proportion of total cells) were determined in three Wnt8b−/− mutants and three wild type littermates. For analysis of adults, mice were injected with BrdU at two-hour intervals over a 12-hour period and sacrificed two hours after the last injection. The total number of BrdU-labelled cells present in the complete hippocampal formation was counted on every tenth section throughout the full rostrocaudal extent of the hippocampus in three Wnt8b−/− mutants and three wild type littermates. All cell counting was performed blind in embryos and adults. Means were compared statistically using Student’s t-test.
RNA extraction, RT-PCR and qRT-PCR
RNA was extracted from E12.5 telencephalon of wild-type and Wnt8b−/− embryos (n=3, respectively) using an RNeasy mini kit (Qiagen, Chatsworth, CA), following the manufacturer’s protocol. cDNA was prepared by adding 100 ng of random hexanucleotide primers (Promega, Madison, WI) to 1.0 μg of extracted RNA in nuclease-free water in a total volume of 15 μL at 70°C for 10 min and briefly chilling on ice. The synthesis was continued in a total volume of 25 μL consisting of the RNA/random primers mix, 1μL RT-buffer (Promega), 1.0 mM of the four deoxyribonucleoside 5′-triphosphates (dNTPs, Promega), 1 unit of RNase inhibitor (Promega), and 200 units of MMLV reverse transcriptase (Promega) at 42°C for 1 h. RT-PCR was performed in a total volume 20 μL containing 1μL of synthesized cDNA, 1.5mM MgCl2, 0.2 mM of dNTPs, 7.0 pmol of both forward and reverse primers, and 1 unit of Taq polymerase (Promega). Quantitative RT-PCR was carried out using Quantitect SYBR Green PCR master mix (Qiagen) in an Opticon DNA Engine (MJ Research), following the manufacturer’s recommendations. Primer sequences and reaction parameters were exactly as described in Nordin et al., 2008, except for Wnt8b which was quantitated using primers 5′-AACGTGGGCTTCGGAGAGGC-3′ (forward) and 5′-GCCCGCGCCGTGCAGGT-3′ (reverse).
Morphometric analysis of adult hippocampus
Adult mice (n=3 per genotype) were anaesthetised with a terminal dose of Pentobarbital and perfused intracardially with PBS (pH 7.4) followed by 4% PFA in 0.1M Phosphate buffer, pH 7.4. Brains were removed and immersed in the same fixative overnight at 4°C then incubated in 30% sucrose in PBS overnight at 4°C. Brains were cut serially (4 series) at a coronal plane of 48 μm using a freezing microtome and kept in cryoprotective solution (40% PBS, 30% glycerol, 30% ethylene glycol) at −20°C until use.
i. Volumetric measurements of the adult hippocampus
One set of serial sections was mounted on poly-L-lysine slides and stained with cresyl violet (0.2% in H20). The most rostral section analysed was defined by the presence of CA and DG and coincided with the dorsal hippocampal commissure, approximately −0.94 mm from Bregma (Franklin and Paxinos, 1997). All sections in which the hippocampus was clearly visible were included in our analysis. The most caudal hippocampal section analysed corresponded to approximately −3.88 mm from Bregma (Franklin and Paxinos, 1997). The hippocampus was manually outlined on photographed sections using Adobe Photoshop 7.0.1. The outline of the hippocampus was drawn at the grey/white matter border with the fimbria/corpus callosum. For each animal, the areas of each of the two hippocampi were calculated using ImageJ software. To calculate the hippocampal volume, hippocampal areas were summed, multiplied by four and by the thickness of the section. Volumes are expressed as mm3 and correspond to the average of the two hippocampi in each animal.
ii. Areal, length and thickness measurements of the adult hippocampal fields
Four 48 μm-thick sections taken from the rostrocaudal midpoint of the hippocampus were used to measure the length of the hippocampal fields. Free-floating sections were immunostained as previously described (Fotaki et al., 2002) with a polyclonal rabbit antibody to mGluR5 (Millipore, 1:10,000). The cellular layers of CA1/2, CA3 and DG were manually traced and the length of each was calculated using the ImageJ software. To calculate the dendritic field thickness, the average of twenty four different measurements for each hemisphere were used. Finally, the total hippocampal area for these middle sections was measured, following the methodology described above for the cresyl violet area measurements.
The observer was blind to the mouse genotype during all measurements. Statistical analysis was performed using SigmaPlot11 software. Student’s t-test was applied for all measurements except when the equal variance test was not passed (CA3 length measurements) in which case the Mann-Whitney non-parametric test was used. p values <0.05 were considered significant.
Supplementary Material
Supplementary Figure 1. Expression patterns of Wnts −2b, 3a, −5a, 7a and 7b in Wnt8b−/− embryos and wild type controls. In situ hybridisation for each of the Wnt genes whose expression overlaps that of Wnt8b in the dorsomedial telencephalon of E12.5 embryos. Panels A,C,E,G,I show wild type embryos and panels B,D,F,H,J show equivalent sections of Wnt8b−/− embryos.
Acknowledgements
We thank Melville Richardson, Jan Ure and Christine Morrison for expert technical assistance and Biomedical Research Resources staff for animal husbandry. We are grateful to Sophie Thompson for her contribution to the quantitative analysis and Sam Gibbons-Frendo for his contribution to the in situ hybridisation analysis. We thank Andrew Smith for the 129Ola genomic DNA library, Juan Galceran Shinichi Aizawa and Ondrej Machon for supplying in situ probes and Peter Kind for the antibody to mGluR5. Antibodies to Lim1/2 and Islet1 were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa (Department of Biological Sciences, Iowa City, IA). S.J.Z was supported by a fellowship from the China Scholarship Council (CSC). Work in the authors’ laboratories is supported by the BBSRC, MRC and the Wellcome Trust.
References
- Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 2001;100:165–75. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–62. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Du SJ, Purcell SM, Christian JL, McGrew LL, Moon RT. Identification of distinct classes and functional domains of Wnts through expression of wild-type and chimeric proteins in Xenopus embryos. Mol Cell Biol. 1995;15:2625–34. doi: 10.1128/mcb.15.5.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Guimera J, Wurst W, Prakash N. Distinct but redundant expression of the Frizzled Wnt receptor genes at signaling centers of the developing mouse brain. Neuroscience. 2007;147:693–711. doi: 10.1016/j.neuroscience.2007.04.060. [DOI] [PubMed] [Google Scholar]
- Fotaki V, Dierssen M, Alcántara S, Martínez S, Martí E, Casas C, Visa J, Soriano E, Estivill X, Arbonés ML. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol. 2002;18:6636–47. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotaki V, Yu T, Zaki PA, Mason JO, Price DJ. Abnormal positioning of diencephalic cell types in neocortical tissue in the dorsal telencephalon of mice lacking functional Gli3. J. Neurosci. 2006;26:9282–9292. doi: 10.1523/JNEUROSCI.2673-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. 2nd edition Academic Press; San Diego: 1997. p. 186. [Google Scholar]
- Frantz GD, Bohner AP, Akers RM, McConnell SK. Regulation of the POU domain gene SCIP during cerebral cortical development. J. Neurosci. 1994;14:472–85. doi: 10.1523/JNEUROSCI.14-02-00472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–82. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- Garda AL, Puelles L, Rubenstein JL, Medina L. Expression patterns of Wnt8b and Wnt7b in the chicken embryonic brain suggest a correlation with forebrain patterning centers and morphogenesis. Neuroscience. 2002;113:689–98. doi: 10.1016/s0306-4522(02)00171-9. [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–25. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–70. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–83. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AS, Lowenstein DH, Pleasure SJ. Wnt receptors and Wnt inhibitors are expressed in gradients in the developing telencephalon. Mech Dev. 2001;103:167–172. doi: 10.1016/s0925-4773(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Kimura J, Suda Y, Kurokawa D, Hossain ZM, Nakamura M, Takahashi M, Hara A, Aizawa S. Emx2 and Pax6 function in cooperation with Otx2 and Otx1 to develop caudal forebrain primordium that includes future archipallium. J Neurosci. 2005;25:5097–108. doi: 10.1523/JNEUROSCI.0239-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lako MS, Lindsay S, Bullen P, Wilson DI, Robson SC, Strachan T. A novel mammalian wnt gene, WNT8B, shows brain-restricted expression in early development, with sharply delimited expression boundaries in the developing forebrain. Hum Mol Genet. 1998;7:813–22. doi: 10.1093/hmg/7.5.813. [DOI] [PubMed] [Google Scholar]
- Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–67. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- Lee JE, Wu SF, Goering LM, Dorsky RI. Canonical Wnt signaling through Lef1 is required for hypothalamic neurogenesis. Development. 2006;133:4451–61. doi: 10.1242/dev.02613. [DOI] [PubMed] [Google Scholar]
- Li Y, Mu Y, Gage FH. Development of neural circuits in the adult hippocampus. Curr Top Dev Biol. 2009;87:149–74. doi: 10.1016/S0070-2153(09)01205-8. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, Gage FH. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–5. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Machon O, van den Bout CJ, Backman M, Kemler R, Krauss S. Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience. 2003;122(1):129–43. doi: 10.1016/s0306-4522(03)00519-0. 2003. [DOI] [PubMed] [Google Scholar]
- Machon O, Backman M, Machonova O, Kozmik Z, Vacik T, Andersen L, Krauss S. A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev Biol. 2007;311:223–37. doi: 10.1016/j.ydbio.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, Li Y, Flanagan LA, Tole S, Monuki ES. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–9. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JO, Kitajewski J, Varmus HE. Mutational analysis of mouse Wnt-1 identifies two temperature-sensitive alleles and attributes of Wnt-1 protein essential for transformation of a mammary cell line. Mol Biol Cell. 1992;3:521–33. doi: 10.1091/mbc.3.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin D, Karlsson F, Tian N, Pratt T, Bullock SL, Wilson VA, Price DJ, Mason JO. Specific modification of heparan sulphate is required for normal cerebral cortical development. Mech Dev. 2003;120:1481–8. doi: 10.1016/j.mod.2003.08.008. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–85. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin N, Li M, Mason JO. Expression profiles of Wnt genes during neural differentiation of mouse embryonic stem cells. Cloning and Stem Cells. 2008;10:37–48. doi: 10.1089/clo.2007.0060. [DOI] [PubMed] [Google Scholar]
- Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- Olson DJ, Papkoff J. Regulated expression of Wnt family members during proliferation of C57mg mammary cells. Mech Dev. 1994;44:3–16. [PubMed] [Google Scholar]
- Richardson M, Redmond D, Watson CJ, Mason JO. Mouse Wnt8b is expressed in the developing forebrain and maps to chromosome 19. Mammalian Genome. 1999;10:923–925. doi: 10.1007/s003359901115. [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Bertuzzi S, Chiang C, Shawlot W, Taira M, Dawid I, Westphal H. Expression of murine Lhx5 suggests a role in specifying the forebrain. Dev Dyn. 1997;208:266–77. doi: 10.1002/(SICI)1097-0177(199702)208:2<266::AID-AJA13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Julius MA, Giarré M, Zheng Z, Brown AM, Kitajewski J. Transformation by Wnt family proteins correlates with regulation of beta-catenin. Cell Growth Differ. 1997;8:1349–58. [PubMed] [Google Scholar]
- Shinozaki K, Yoshida M, Nakamura M, Aizawa S, Suda Y. Emx1 and Emx2 cooperate in initial phase of archipallium development. Mech Dev. 2004;121:475–89. doi: 10.1016/j.mod.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Slusarski DC, Corces VG, Moon RT. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997a;390:410–3. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- Slusarski DC, Yang-Snyder J, Busa WB, Moon RT. Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev Biol. 1997b;182:114–20. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- Solberg N, Machon O, Krauss S. Effect of canonical Wnt inhibition in the neurogenic cortex, hippocampus, and premigratory dentate gyrus progenitor pool. Dev Dyn. 2008;237:1799–81. doi: 10.1002/dvdy.21586. [DOI] [PubMed] [Google Scholar]
- Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–89. doi: 10.1101/gad.8.2.174. 1994. [DOI] [PubMed] [Google Scholar]
- Theil T. Gli3 is required for the specification and differentiation of preplate neurons. Dev Biol. 2005;286:559–71. doi: 10.1016/j.ydbio.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Theil T, Aydin S, Koch S, Grotewold L, Rüther U. Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development. 2002;129:3045–54. doi: 10.1242/dev.129.13.3045. [DOI] [PubMed] [Google Scholar]
- Tole S, Christian C, Grove EA. Early specification and autonomous development of cortical fields in the mouse hippocampus. Development. 1997;124:4959–70. doi: 10.1242/dev.124.24.4959. [DOI] [PubMed] [Google Scholar]
- Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J) Dev Biol. 2000;217:254–65. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Berns A. Knockout mouse models to study Wnt signal transduction. Trends Genet. 2006;221:678–89. doi: 10.1016/j.tig.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Wong GT, Gavin BJ, McMahon AP. Differential transformation of mammary epithelial cells by Wnt genes. Mol. Cell. Biol. 1994;14:6278–86. doi: 10.1128/mcb.14.9.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Fotaki V, Mason JO, Price DJ. Analysis of early telencephalic defects in mice lacking functional Gli3 protein. J Comp Neurol. 2009;512(5):613–27. doi: 10.1002/cne.21918. [DOI] [PubMed] [Google Scholar]
- Zhao C, Avilés C, Abel RA, Almli CR, McQuillen P, Pleasure SJ. Hippocampal and visuospatial learning defects in mice with a deletion of frizzled 9, a gene in the Williams syndrome deletion interval. Development. 2005;132:2917–27. doi: 10.1242/dev.01871. [DOI] [PubMed] [Google Scholar]
- Zhou CJ, Zhao C, Pleasure SJ. Wnt signaling mutants have decreased dentate granule cell production and radial glial scaffolding abnormalities. J Neurosci. 2004;24:121–6. doi: 10.1523/JNEUROSCI.4071-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Expression patterns of Wnts −2b, 3a, −5a, 7a and 7b in Wnt8b−/− embryos and wild type controls. In situ hybridisation for each of the Wnt genes whose expression overlaps that of Wnt8b in the dorsomedial telencephalon of E12.5 embryos. Panels A,C,E,G,I show wild type embryos and panels B,D,F,H,J show equivalent sections of Wnt8b−/− embryos.