Abstract
Interpretation of mass spectra is challenging because they report a ratio of two physical quantities, mass and charge, which may each have multiple components that overlap in m/z. Previous approaches to disentangling the two have focused on peak assignment or fitting. However, the former struggle with complex spectra, and the latter are generally computationally intensive and may require substantial manual intervention. We propose a new data analysis approach that employs a Bayesian framework to separate the mass and charge dimensions. Based on this approach, we developed UniDec (Universal Deconvolution), software that provides a rapid, robust, and flexible deconvolution of mass spectra and ion mobility-mass spectra with minimal user intervention. Incorporation of the charge-state distribution in the Bayesian prior probabilities provides separation of the m/z spectrum into its physical mass and charge components. We have evaluated our approach using systems of increasing complexity, enabling us to deduce lipid binding to membrane proteins, to probe the dynamics of subunit exchange reactions, and to characterize polydispersity in both protein assemblies and lipoprotein Nanodiscs. The general utility of our approach will greatly facilitate analysis of ion mobility and mass spectra.
Introduction
Mass spectrometry (MS) is a powerful approach for structural and physical characterization of biomacromolecules.1-7 However, for MS to revolutionize these fields as it has proteomics, data analysis techniques are needed to rapidly and reliably extract quantitative information from spectra. Interpretation of mass spectra is challenging because MS detects a ratio of mass to charge, two correlated physical quantities.
In electrospray ionization (ESI) MS, separation of mass and charge is possible because ESI produces multiply charged ions for a single molecular species. If peaks can be assigned to a single charge series, their masses and charges can be determined by solving a system of equations.8 However, assignment becomes difficult when several charge state distributions overlap in m/z. As experimental approaches become more sophisticated and ever more heterogeneous systems are analyzed, rapid deconvolution of complex data has become a priority for MS-based structural biology.9-12
Deconvolution in the context of MS often refers to separating the interwoven charge and mass dimensions.13 MS deconvolution approaches generally fall into three broad categories: peak assignment, isotopic, and simulation based algorithms.14,15
Peak assignment algorithms extract a list of peaks from the spectrum and assign a charge to each.8,14-18 Because of the data reduction achieved by peak selection, these algorithms are fast and produce simple outputs. However, these algorithms often struggle with complex spectra and do not generally produce quantitative results.
Isotopic algorithms require high-resolution data with resolved isotopologues.13,19 Because the mass difference between peaks is known from the atomic masses of the isotopes, the charge can be inferred directly from difference in m/z. However, these approaches are not generally applicable, as many spectra do not display isotopic resolution, and they are not generally quantitative.
Simulation algorithms address the deficiencies of peak assignment algorithms for complex systems.10,11,20-22 Multiple hypothetical mass and charge distributions are generated from which a spectrum is simulated. The simulated spectrum that fits the data best based on a metric of minimum chi-squared11 or maximum entropy16,23,24 is then taken to be the most correct. Although this approach yields quantitative fits, current implementations tend to be computationally intensive and require substantial user guidance.
Here, we present a novel Bayesian deconvolution algorithm, which represents a means for rapid and quantitative interpretation of both mass and ion mobility (IM) spectra. We reframe the problem into a mathematical deconvolution, which is the process of retrieving the latent signal from a recorded signal that has been convolved with a given point-spread function. The approach is a special case of the Richardson-Lucy mathematical deconvolution algorithm.25,26 A similar concept was previously applied to MS as a method of deconvolving extracted intensities from a peak list.9 We have generalized and modified the algorithm to develop a new approach to deconvolving both mass spectra and ion mobility-mass spectra (IM-MS) with no reduction of the data.
The algorithm, UniDec (Universal Deconvolution), performs quantitative deconvolution and charge separation of complex spectra. It is fast, robust, and easily generalized to any number of dimensions or peak shapes. Remarkably, it does not require substantial user intervention or prior knowledge. Here, we describe the UniDec approach for studying intact protein complexes with both native MS and two-dimensional IM-MS. We explore its utility for studying the biophysics of heterogeneous systems such as membrane proteins, small heat-shock protein assemblies, and Nanodisc lipoprotein complexes. Overall, we show that UniDec provides a solution to analyzing challenging biochemical systems.
Methods
Overview of Algorithm
Our approach assumes the spectrum can be described as a convolution between a peak shape and a set of weighted delta functions (referred to here as a “delta matrix”), which describe the contribution of specific molecular ions to the spectrum. A schematic description of the UniDec approach is shown in Figure S-1, and a detailed description of the algorithm is provided in the Supplemental Methods. For MS data, we implement a two dimensional delta matrix containing m/z in one dimension and charge in the other (Figure 1B). Initially, we assume an equal probability for all charge states. The algorithm then proceeds by sequential iteration of three steps to determine the effective contribution of each m/z and charge to the overall spectrum.
Figure 1.
Description of UniDec outputs. The experimental IM-MS spectrum for AqpZ with bound POPC at 100V collision voltage is shown in D. Summing along the arrival time axis produces the mass spectrum shown in A. UniDec deconvolution of A separates the charge dimensions into a m/z vs. charge matrix and a mass vs. charge matrix as shown in B and C respectively. UniDec deconvolution of the IM-MS spectrum produces a 3D matrix of m/z vs. arrival time vs. charge, which is transformed into a matrix of mass vs. CCS vs. charge. The projections of each 3D matrix along each axis are shown on the face of cubes in E and F respectively.
First, a filter is applied to smooth the charge distributions in the delta matrix. This step modifies the Bayesian priors to include information from neighboring species. Physically, this approach assumes that a charge state series of an ion will be smooth, but no a priori assumptions are placed on the precise form of the charge state distribution. Thus, for a given m/z value, the probability of that intensity being assigned to a given charge state is related to the probability of the same m value appearing at m/(z+1) and m/(z−1). A similar filter can be applied for systems where known mass differences may be taken into account, an approach very suitable for oligomeric assemblies.
Second, the filtered delta matrix is summed along the charge axis to produce a one-dimensional m/z spectrum of weighted delta functions. The summation of delta functions is then convolved with the peak shape defined by the user to produce an m/z spectrum, which has the same dimensions as the data.
Finally, each element in the filtered delta matrix is multiplied by the ratio of the data to the projected spectrum at its specific m/z value. If the projected spectrum has a higher intensity than the data at a given m/z, which may be caused by overlapping peaks, the ratio will be less than unity, and the probability of each charge state at that m/z will be decreased. If the data is more intense than the projected spectrum, the ratio will be greater than unity, and the probability will be increased to compensate. There is no change if the data and projected spectrum are the same. It follows that the algorithm converges when the projected spectrum matches the data.
The result of the successive iteration of these three steps is a deconvolved matrix of delta functions. To better represent the data, each charge column of the matrix is convolved with the peak shape function. Because the m/z and charge are defined for each point in the delta matrix, the convolved delta matrix is then transformed into a matrix of mass vs. charge (Figure 1C). An analogous approach is used for IM-MS (Figure 1D-F) as detailed in the Supplemental Methods.
The only parameters required by the algorithm are the peak width and the range of charge states to consider. Additional restraints on allowed masses may be provided to accelerate the deconvolution and to mitigate any overfitting errors. In general, the algorithm speed scales with the size of the m/z vs. charge matrix and to a lesser degree with the size of the transformed mass vs. charge matrix. The speed is not significantly affected by heterogeneity, allowing the algorithm to scale effectively to very complex systems. Uncertainties in the data, resulting from poor resolution, high noise, or untenable complexity, will provide a practical limit on the applicability of the algorithm. A detailed discussion of potential artifacts is provided in the Supplemental Methods.
Deconvolution Parameters
We implemented UniDec in C and developed a user interface in Python to control the software and generate graphical outputs. A copy of the program is available on request or for download at unidec.chem.ox.ac.uk. Details of the implementation are provided in the Supplemental Methods, together with a detailed description of the experimental methods and deconvolution parameters. Briefly, the key parameters used for each system are as follows. All spectra were deconvolved using a charge smooth filter with a width of one elementary charge except for those from Nanodiscs. An appropriate peak shape function was determined by fitting an isolated peak to either Gaussian, Lorentzian, or split Gaussian/Lorentzian peak shapes. The split Gaussian/Lorentzian peak shape is defined as a Lorentzian on the high m/z side and a Gaussian on the other such that the FWHM is symmetrical about the maximum. The split Gaussian/Lorentzian distribution is a good model of native MS peak shapes when a long tail is observed on the high m/z side due to bound adduct species.20
Specific Applications
We describe here application of the UniDec approach to problems of increasing complexity: membrane protein AqpZ; small heat shock proteins HSP17.7, HSP16.5, and αB-crystallin; and lipoprotein Nanodiscs.
MS and IM-MS data of aquaporin Z (AqpZ) with bound 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) obtained at 100 V accelerating potential into a dedicated collision cell was analyzed using UniDec by limiting the mass range to between 95 and 105 kDa.27 An example of how the algorithm performs without mass limitations is shown in Figure S-2. Data was smoothed in MassLynx 4.1 software (Waters Corp.) before analysis with Transform and MaxEnt, which used the same mass limitation.
Deconvolution of subunit exchange data from HSP17.7 was performed by limiting the allowed mass range to between 211 kDa and 222 kDa. Tandem MS spectra of the isolated +47 charge state of HSP16.5 24-mers were summed across multiple collision voltages to compile an aggregate spectrum.28 Deconvolution was performed by limiting the charge state between 10 and 49 and manually defining the +47 charge state, which was necessary because only one charge state was isolated in the MS/MS experiment. Collision induced dissociation (CID) spectra of αB-crystallin were obtained similarly. Masses were limited to within 3000 Da of a wide range of potential oligomer complexes ranging from 1 to 74 subunits of a 20085 Da monomer. Charge was limited to between 5 and 84. In addition to the charge-smooth filter, a mass-smooth filter was applied to smooth the distribution of dimer units.
Nanodiscs with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and POPC were analyzed with a linear drift cell Waters Synapt G1 ion mobility-mass spectrometer.29 Data was deconvolved without a charge filter but by using a mass filter to smooth the distribution of lipids. Masses were limited to between 100 kDa and 175 kDa. Conversion from arrival time to collision cross section (CCS) was performed using the Mason-Schamp equation as described previously,27,29 using t0 values calibrated from alcohol dehydrogenase analyzed under the same instrumental conditions.
Results and Discussion
Deconvolution of Mass Spectra
To assess the accuracy of UniDec, we first describe the application to membrane protein AqpZ with bound POPC lipid. The spectrum contains multiple lipid adducts, yet the individual charge states are easily discerned (Figure 1A-C).27 We compared the UniDec result with the MaxEnt and Transform algorithms included in MassLynx software (Waters Corp.) as shown in Figure S-2. Similar to UniDec, MaxEnt provides a deconvolved zero-charge mass spectrum with few user-defined starting parameters.16,23,24 As seen in Figure S-2C, the raw output from UniDec agrees closely with the MaxEnt result for the same input data and restraints. However, the MaxEnt algorithm generally takes around two orders of magnitude longer to arrive at the result (0.15 s for UniDec vs. ~30 s for MaxEnt). The Transform algorithm relies on a user-defined mass window to simply sum up isolated charge states with no deconvolution. The UniDec reconvolved output agrees closely with the Transform result as seen in Figure S-2E. These results demonstrate that UniDec provides accurate, rapid, and quantitative deconvolution and transformation of spectra with well-defined charge states with minimal user input.
Deconvolution of Subunit Exchange Experiments
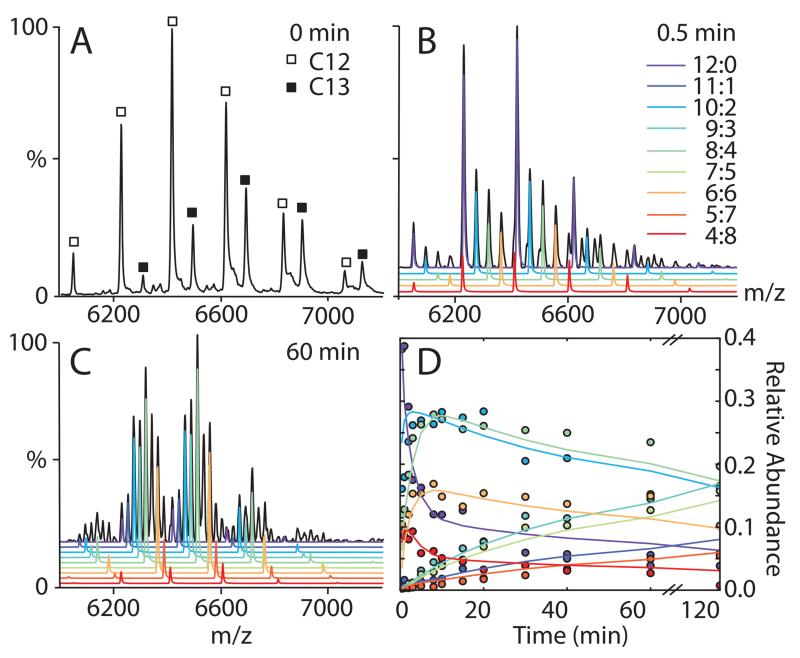
Turning to more complex spectra, we explored the utility of UniDec for quantifying the quaternary dynamics of proteins in a case where charge and mass distributions are overlapping. We examined HSP17.7, a dodecameric small heat-shock protein that is known to exchange subunits at equilibrium (Figure 2). When HSP17.7 labeled metabolically with 13C (heavy) is mixed with its unlabeled (light) equivalent, two distinct charge series are observed, corresponding to homo-dodecamers (Figure 2A).
Figure 2.
UniDec analysis of subunit exchange data for HSP17.7. The annotated mass spectrum is shown in panel A for the zero time point, B at 0.5 minutes, and C at 60 minutes. Extracted intensities for species annotated in B are shown in panel D as a function of equilibration time along with kinetic fits.
Over time, continuous dissociation and reassociation of monomers and dimers results in the gradual appearance of peaks corresponding to hetero-oligomers comprising different numbers of light and heavy subunits.30 At early time-points, dodecamers composed predominately of an even number of both heavy and light subunits are observed (Figure 2B). However, after 60 min of incubation, 12-mers comprising odd numbers of both types of subunit are observed (Figure 2C). While subunit exchange is clearly occurring, overlapping peaks make it challenging to quantify directly the time-dependent evolution of the intensities of these various species, even in these well-resolved spectra.
Using UniDec for deconvolution, we quantified the relative abundances of all the species in the evolving mixture at each time point. We fit the extracted intensities using a previously described kinetic exchange model31,32 that describes the sequential dissociation and reassociation based on a separate rates for dissociation of dimers and monomers from the dodecamers (Figure 2D). These fits allow the extraction of kinetic constants, returning koff = 0.0026 ± 0.0002 s−1 and 0.44 ± 0.07 s−1 for monomer and dimer respectively. These results provide quantitative insight into how the dynamic oligomers interconvert, confirming that the monomer exchange is much slower than the dimer exchange. Such an analysis would not be possible without a reliable method to extract relative contributions of the various oligomers involved, which is provided here by UniDec.
Although analysis of overlapping spectra like this may be possible using simulation algorithms, UniDec streamlines the pipeline from raw data to kinetic fits. Because the peak shape is relatively constant, a single set of parameters is used for all of the fits, and the deconvolution does not need to be seeded with a starting distribution. Moreover, each deconvolution takes around 0.1 s, so an entire time series can be converted to relative concentrations in a matter of seconds. As experimental approaches continue to expand in both dimension and scale, UniDec enables analysis of large data-sets, for example taking triplicate measurements of kinetics as a function of temperature and pH, without additional computational overhead or extensive manual manipulation.
Deconvolution of CID Spectra
In addition to monitoring dynamic processes that occur in solution such as subunit exchange, there is an increasing demand to quantify dynamic processes that happen in the mass spectrometer as a result of collision induced unfolding and dissociation.27,33,34 Collision-induced dissociation (CID) is used to study polydisperse ensembles because the loss of a highly charged monomer leaves behind a charge-stripped oligomer. The lower average charge increases the spacing between oligomers and reduces overlap between different species in the spectrum.28,34 Mass spectra from CID experiments pose a particular challenge for quantitative deconvolution algorithms because reconstructing the underlying distribution of oligomers requires not only deconvolution of the mass components but also knowledge of the number of subunits lost to produce each stripped complex. Although this is often trivial for CID spectra of monodisperse protein complexes, it is challenging for polydisperse proteins where oligomeric distributions overlap between native and stripped complexes.
We employed the charge and mass separation afforded by UniDec to analyze CID experiments. Figure 3A shows a combined CID spectrum of the isolated 47+ charge state of the monodisperse native HSP16.5 24-mer (N). Successive dissociation steps, which proceed by removal of monomers, lead to the appearance of 23 (N-1), 22 (N-2), and 21-mers (N-3) at higher m/z as annotated in Figure 3C.
Figure 3.
UniDec analysis of MjHSP16.5 (A, C, and E) and αB-crystallin (B, D, and F) CID spectra. Spectra combined from a number of collision voltages are shown in A and B. Results in m/z vs. charge dimensions are presented in C and D and in mass vs. charge dimensions in E and F. Lines plotted correspond to an offset from the predicted average native charge as a function of mass.
UniDec deconvolution of the combined spectrum shows lower average charge for each dissociation product as shown in Figure 3E. To assign the CID state of a particular stoichiometry, we fit the deconvolution results to an empirical relationship between mass and zA, the average charge for a complex in units of elementary charge: zA = 0.0467m0.533+F where m is the mass in Da, and F is an offset from the native state.11,12 For the HSP16.5 CID spectrum (Figure 3E), the +47 charge state has an offset of +2, indicating that the isolated state has a native charge. The dissociation products are well-separated with charge offsets of −10, −21, and −27, an effect caused by monomer units taking charges from the complex as they dissociate. Comparing the intensity of each CID state extracted as a function of collision voltage (Figure S-3A) with manually extracted results28 demonstrates that UniDec accurately extracts both charge distribution and intensities on monodisperse CID spectra.
Separation of the charge dimension now provides a novel avenue for exploring CID spectra from highly polydisperse proteins that cannot be routinely analyzed manually. Figure 3B shows a combined CID spectrum of polydisperse αB-crystallin. The m/z regions of each dissociation step were previously identified34 but overlap in m/z (Figure 3D and 4A). However, deconvolution reveals distinct regions for native oligomers and the two successive dissociation steps as shown in Figure 3F. Using the relationship described above, we plotted the mass vs. average charge relationship in Figure 3F with offset terms of +2, −17, and −26 for the native (N), singly stripped (N-1), and doubly stripped (N-2) oligomers respectively. Summing intensities along these charge curves for each dissociation state (plus or minus one charge state) yields the relative intensity of each isolated CID state (Figure S-3B). Despite the large charge and m/z range, each spectrum deconvolved in 2-3 s.
Figure 4.
Deconvolution of αB-crystallin CID spectra. Composite mass spectrum (black, Figure 3B) over a range of CID voltages is presented in panel A with the separated oligomeric and charge states colored beneath. Oligomeric states range linearly from 12 at the top (purple) to 66 at the bottom (red). Extracted distributions, corrected for subunit loss, are presented in B for the native state (N) at 140 V (red), the first dissociation (N-1) at 160 V (blue), and the second dissociation (N-2) at 200 V (black), showing that the dissociated distributions are shifted towards smaller complexes. The overall oligomeric distribution is reconstructed at different collision energies by correcting for the loss of one monomer unit per dissociation step (C).
The ability to define the CID state in mass vs. charge space allows us to interrogate how the extracted distribution depends on the collision voltage. This is a key question because differing CID efficiencies between large and small oligomers could bias the extracted distribution in the CID products towards smaller oligomers. Using UniDec, we extracted the oligomeric distribution of αB-crystallin separately from the N, N-1, and N-2 oligomers at 140, 160, and 200 V respectively (Figure 4B). This reveals that native oligomers have a slightly larger average number of subunits compared to the stripped oligomers. Because the overall distribution and thus the transmission efficiency is constant, the selective depletion of the smaller oligomeric complexes confirms that the larger complexes are more stable to CID, likely due to more internal degrees of freedom over which to distribute collisional energy.35
By correcting the stoichiometries to account for the monomer units lost by dissociation, we can fully reconstruct the initial distribution of oligomer sizes. Comparing the combined distribution across all CID states (Figure 4C), we observe that the extracted distribution is relatively constant from 140 to 200 V, despite transitioning from nearly fully N to almost completely populating the N-2 state (Figures S-3B). By separating the mass and charge dimensions, UniDec enables the monitoring of CID in highly complex and overlapping spectra that is not possible through conventional data analysis methods.
Ion Mobility Mass Spectra
IM-MS provides an additional dimension to conventional MS by recording not only the m/z ratio but also the time taken to traverse a region of inert gas under the influence of a weak electric field. As with MS, the arrival time measured in IM is related to a ratio of two physical quantities, in this case the collision cross section (CCS) of the ion divided by its charge.29,36 However, the experiment does not report on the CCS/z ratio directly. Arrival times must be converted into CCS using both experimental parameters and the charge on each ion. Charges in IM-MS spectra are usually assigned using the m/z dimension, but this is challenging in complex and overlapping spectra, where more sophisticated experimental approaches are required.37
UniDec provides a unique approach to interpreting IM-MS data by performing a simultaneous deconvolution of the m/z and arrival time dimensions. Examining the AqpZ spectrum with bound POPC presented in Figure 1D, we used UniDec to separate the IM-MS data into its constituent components. Here, the two-dimensional m/z vs. arrival time data (Figure 1D) deconvolves into a three-dimensional m/z vs. arrival time vs. charge output (Figure 1E and Figure S-4A). Because charge is known for each point in the matrix, the experimental observables of m/z and arrival time can be transformed into a three-dimensional matrix of physical quantities, mass vs. CCS vs. charge (Figure 1F).29 Figure S-3B shows the projection along the mass axis into a charge vs. CCS matrix. Here, the lower charge states remain in their native conformation while the higher charge states are unfolded as previously observed.27,38 UniDec provides a rapid (around 1 s) deconvolution and transformation of the data with little user intervention.
Deconvolution of Nanodisc Ion Mobility Spectra
To test UniDec against complex IM-MS data with overlapping mass, charge, and CCS distributions, we explored the deconvolution of IM-MS spectra of Nanodiscs, which are nanoscale discoidal lipoprotein complexes similar to high-density lipoprotein particles.39 Each Nanodisc complex contains two copies of the membrane scaffold protein belt and a number of lipids. Although the complexes are relatively monodisperse, there is an intrinsic distribution in the number of lipids per complex greater than ±5 lipids.9 Overlap between this mass distribution and the charge state distribution makes spectral assignment of Nanodiscs challenging, especially because there is resonant overlap at defined m/z values.9 An example IM-MS spectrum is shown in Figure 5A, and representative spectra as a function of collision voltage are provided in Supplementary Movies S-1 and S-2 respectively. These spectra demonstrate the same constructive overlap pattern observed in conventional MS.9
Figure 5.
IM-MS deconvolution of Nanodiscs. The IM-MS spectrum of MSP1D1(−) DMPC Nanodiscs at 120 V CID is shown in A, where the color bar indicates intensity. D shows the separation of the spectrum into its individual charge states. Here, color indicates the charge state and transparency indicates the intensity. Total CCS summed across masses and charge states is presented as a function of collision voltage for the average of three replicates for DMPC and POPC Nanodiscs in panels B and E respectively. Dissociation of lipids is shown for DMPC (blue) and POPC (red) Nanodiscs in C, and the corresponding CCS collapse is presented in F. Error bars are shown as the standard deviation of the mean of three replicate measurements. Dashed lines in F indicate the predicted CCS for a globular protein with masses from C.
Applying UniDec to simultaneously deconvolve the m/z and arrival time dimensions, we discovered that UniDec is able to separate the underlying mass, charge, and CCS distributions (Figure 5D).
Despite the complexity and dimensionality, deconvolution only takes around 1-3 seconds per spectrum. A series of spectra were analyzed with increasing collision voltage to determine mass and CCS distributions as a function of collision voltage. The total CCS distribution summed across all charge and mass states is shown for DMPC and POPC Nanodiscs (Figures 5B and 5E, respectively) with the average mass and CCS from three replicate measurements plotted in Figures 5C and 5F. Error bars report the standard deviation of the mean, demonstrating that the distributions are reproducible.
Knowledge of the mass and CCS distributions allowed us to examine the gas-phase behavior of Nanodiscs. The initial mass distributions agree with solution studies, showing 164 DMPC molecules and 131 POPC molecules in addition to the two membrane scaffold protein belts.40,41 Using a scaled projection approximation method,42 we calculated predicted CCS values based on a molecular dynamics model43 and small angle X-ray scattering data of Nanodisc complexes in solution (see Supplemental Methods for more detail).44,45 We discovered that the predicted CCS for DMPC Nanodiscs of 8560 Å2 agrees with the experimental value of 8920 Å2 with a peak width (one standard deviation) of 800 Å2 at a collision voltage of 50 V. POPC Nanodiscs have a predicted CCS of 8300 Å2, which agrees closely with experimental values of 8250±770 Å2 at 50 V. These data suggest that Nanodiscs at low collision energy have not only masses that agree with the solution values but also CCS values that agree with a discoidal shape.
As the collision voltage increases, both POPC and DMPC Nanodiscs undergo CID and show both a progressive loss of lipids9 and a corresponding decrease in CCS. To examine whether the decrease in CCS results from a structural rearrangement or is simply a consequence of the reduced mass, we calculated predicted CCS values for globular proteins of equivalent masses,29,42 shown as dashed lines in Figure 5F. Nanodiscs begin with a structure larger than an isobaric globular protein. This is expected because lipids show a larger average CCS at a given mass than peptides.46 Moreover, an idealized discoidal ellipsoid will have a larger CCS than an idealized sphere of the same volume.47
Although the loss of lipids contributes partially to the decrease in CCS, as evidenced by the downward slope in the dashed lines in Figure 5F, the mass loss alone is not sufficient to account for the CCS change. This suggests that a structural rearrangement produces collapse of the Nanodisc into a compact structure.
In contrast, most proteins show an increase in CCS with increased collision voltage due to protein unfolding, although certain topologies may show a slight collapse prior to unfolding.38 The solution structure of Nanodiscs, however, contains no empty spaces to collapse into, suggesting this collapse is more likely the result of a decreasing shape factor. Given the changes observed in the shape factors of geometric solids,47 we expect that this compact structure is more spherical than the initial discoidal ellipsoid.
Because UniDec enables deconvolution of both the m/z and arrival time dimensions, we are able to extract the CCS as a function of collision voltage and observe the structural change from a native-like to collapsed shape that is unseen in the mass spectra and very difficult to infer from the arrival times alone. This unusual behavior can only be revealed in systems as heterogeneous and complex as Nanodiscs if the charge is known for all relevant peaks, demonstrating the utility of UniDec to deconvolute complex IM-MS spectra.
Conclusion
UniDec is a fast, robust, and flexible approach to (IM-)MS data analysis. We have demonstrated its utility for systems of varying complexity, including membrane proteins, subunit exchange intermediates, and collision induced dissociation of polydisperse protein and lipoprotein complexes. UniDec robustly handles overlapping charge state distributions as well as multidimensional IM-MS data. The universal nature of the algorithm will show immediate utility in a wide range of MS experiments, including ligand binding, subunit exchange, and “top-down” experiments. As the complexity of systems expands, we anticipate that UniDec will become indispensable for analysis of multidimensional mass and ion mobility spectra.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Eamonn Reading and Andrew Aquilina for help in acquiring data for AqpZ and αB-crystallin respectively, and Eman Basha and Elizabeth Vierling for providing HSP17.7. The authors thank Tim Allison for helpful discussions and code contributions. AJB is a BBSRC David Phillip’s fellow. EGM is supported by Carl Trygger’s Foundation for Scientific Research. GKAH is supported by an EPSRC studentship. JLPB holds a University Research Fellowship of the Royal Society. MTM is funded by program grant G1000819 from the Medical Research Council. CVR is funded by European Research Council Investigator Award (IMPRESSS, grant no. 268851).
Footnotes
ASSOCIATED CONTENT
Theory describing algorithm, details on computational implementation, experimental methods, appendix describing Bayesian derivation of algorithm, supplemental figures, and movies of Nanodisc ion mobility-mass spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Sharon M, Robinson CV. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- (2).Marcoux J, Robinson Carol V. Structure. 2013;21:1541–1550. doi: 10.1016/j.str.2013.08.002. [DOI] [PubMed] [Google Scholar]
- (3).Heck AJR. Nat. Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- (4).Lanucara F, Holman SW, Gray CJ, Eyers CE. Nat. Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- (5).Heuvel R. H. H. v. d., Heck AJR. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- (6).Loo JA. Int. J. Mass Spectrom. 2000;200:175–186. [Google Scholar]
- (7).van Duijn E. J. Am. Soc. Mass. Spectrom. 2010;21:971–978. doi: 10.1016/j.jasms.2009.12.010. [DOI] [PubMed] [Google Scholar]
- (8).Mann M, Meng CK, Fenn JB. Anal. Chem. 1989;61:1702–1708. [Google Scholar]
- (9).Marty MT, Zhang H, Cui WD, Gross ML, Sligar SG. J. Am. Soc. Mass. Spectrom. 2014;25:269–277. doi: 10.1007/s13361-013-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Morgner N, Robinson CV. Anal. Chem. 2012;84:2939–2948. doi: 10.1021/ac300056a. [DOI] [PubMed] [Google Scholar]
- (11).Stengel F, Baldwin Andrew J., Bush Matthew F., Hilton Gillian R., Lioe H, Basha E, Jaya N, Vierling E, Benesch Justin L. P. Chem. Biol. 2012;19:599–607. doi: 10.1016/j.chembiol.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, Vierling E, Robinson CV, Benesch JLP. Proc. Natl. Acad. Sci. 2010 doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Horn DM, Zubarev RA, McLafferty FW. J. Am. Soc. Mass. Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- (14).Tseng Y-H, Uetrecht C, Yang S-C, Barendregt A, Heck AJR, Peng W-P. Anal. Chem. 2013;85:11275–11283. doi: 10.1021/ac401940e. [DOI] [PubMed] [Google Scholar]
- (15).Zheng H, Ojha PC, McClean S, Black ND, Hughes JG, Shaw C. Rapid Commun. Mass Spectrom. 2003;17:429–436. doi: 10.1002/rcm.927. [DOI] [PubMed] [Google Scholar]
- (16).Reinhold BB, Reinhold VN. J. Am. Soc. Mass. Spectrom. 1992;3:207–215. doi: 10.1016/1044-0305(92)87004-I. [DOI] [PubMed] [Google Scholar]
- (17).Hagen JJ, Monnig CA. Anal. Chem. 1994;66:1877–1883. [Google Scholar]
- (18).Zhang Z, Marshall AG. J. Am. Soc. Mass. Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- (19).Fernandez-de-Cossio Diaz J, Fernandez-de-Cossio J. Anal. Chem. 2012;84:7052–7056. doi: 10.1021/ac301296a. [DOI] [PubMed] [Google Scholar]
- (20).Sivalingam GN, Yan J, Sahota H, Thalassinos K. Int. J. Mass Spectrom. 2013;345-347:54–62. doi: 10.1016/j.ijms.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).van Breukelen B, Barendregt A, Heck AJR, van den Heuvel RHH. Rapid Commun. Mass Spectrom. 2006;20:2490–2496. doi: 10.1002/rcm.2620. [DOI] [PubMed] [Google Scholar]
- (22).Hilton GR, Hochberg GK, Laganowsky A, McGinnigle SI, Baldwin AJ, Benesch JL. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20110405. doi: 10.1098/rstb.2011.0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ferrige AG, Seddon MJ, Green BN, Jarvis SA, Skilling J, Staunton J. Rapid Commun. Mass Spectrom. 1992;6:707–711. [Google Scholar]
- (24).Ferrige AG, Seddon MJ, Jarvis S, Skilling J, Aplin R. Rapid Commun. Mass Spectrom. 1991;5:374–377. [Google Scholar]
- (25).Lucy LB. Astron. J. 1974;79:745. [Google Scholar]
- (26).Richardson WH. J. Opt. Soc. Am. 1972;62:55–59. [Google Scholar]
- (27).Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Benesch JLP, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Chem. Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- (29).Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal. Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- (30).Painter AJ, Jaya N, Basha E, Vierling E, Robinson CV, Benesch JLP. Chem. Biol. 2008;15:246–253. doi: 10.1016/j.chembiol.2008.01.009. [DOI] [PubMed] [Google Scholar]
- (31).Benesch JLP, Aquilina JA, Baldwin AJ, Rekas A, Stengel F, Lindner RA, Basha E, Devlin GL, Horwitz J, Vierling E, Carver JA, Robinson CV. Chem. Biol. 2010;17:1008–1017. doi: 10.1016/j.chembiol.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Baldwin AJ, Lioe H, Robinson CV, Kay LE, Benesch JLP. J. Mol. Biol. 2011;413:297–309. doi: 10.1016/j.jmb.2011.07.016. [DOI] [PubMed] [Google Scholar]
- (33).Zhong Y, Han L, Ruotolo BT. Angew. Chem. Int. Ed. 2014;53:9209–9212. doi: 10.1002/anie.201403784. [DOI] [PubMed] [Google Scholar]
- (34).Aquilina JA, Benesch JLP, Bateman OA, Slingsby C, Robinson CV. Proc. Natl. Acad. Sci. 2003;100:10611–10616. doi: 10.1073/pnas.1932958100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Benesch JP. J. Am. Soc. Mass. Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- (36).Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Nat. Protocols. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- (37).Shepherd DA, Marty MT, Giles K, Baldwin AJ, Benesch JLP. Int. J. Mass Spectrom [Google Scholar]
- (38).Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. J. Am. Chem. Soc. 2012;134:3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- (39).Bayburt TH, Grinkova YV, Sligar SG. Nano Lett. 2002;2:853–856. [Google Scholar]
- (40).Marty MT, Zhang H, Cui WD, Blankenship RE, Gross ML, Sligar SG. Anal. Chem. 2012;84:8957–8960. doi: 10.1021/ac302663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Bayburt TH, Sligar SG. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Marklund EG, Degiacomi MT, Robinson CV, Baldwin AJ, Benesch JLP. Structure. 2015 doi: 10.1016/j.str.2015.02.010. DOI: 10.1016/j.str.2015.02.010. [DOI] [PubMed] [Google Scholar]
- (43).Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Biophys. J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- (45).Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. J. Phys. Chem. B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).McLean JA. J. Am. Soc. Mass. Spectrom. 2009;20:1775–1781. doi: 10.1016/j.jasms.2009.06.016. [DOI] [PubMed] [Google Scholar]
- (47).Hewitt D, Marklund E, Scott DJ, Robinson CV, Borysik AJ. J. Phys. Chem. B. 2014;118:8489–8495. doi: 10.1021/jp501950d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.