Summary
Autophagy is a conserved membrane transport pathway used to destroy pathogenic microbes that access the cytosol of cells. The intracellular pathogen Legionella pneumophila interferes with autophagy by delivering an effector protein, RavZ, into the host cytosol. RavZ acts by cleaving membrane-conjugated Atg8/LC3 proteins from preautophagosomal structures. Its remarkable efficiency allows minute quantities of RavZ to block autophagy throughout the cell. To understand how RavZ targets preautophagosomes and specifically acts only on membrane-associated Atg8 proteins, we elucidated its structure. Revealed is a catalytic domain related in fold to Ulp-family deubiquitinase-like enzymes and a C-terminal PI3P-binding module. RavZ targets the autophagosome via the PI3P-binding module and a catalytic domain helix and preferentially binds high-curvature membranes, intimating localization to highly curved domains in autophagosome intermediate membranes. RavZ-membrane interactions enhance substrate affinity, providing a mechanism for interfacial activation that may also be used by host autophagy proteins engaging only lipidated Atg8 proteins.
Introduction
Macroautophagy is the process in eukaryotic cells whereby unnecessary or damaged intracellular proteins and organelles are sequestered within a membrane-bound compartment, the autophagosome, and then destroyed when the autophagosome matures and fuses with lysosomes (Kuballa et al., 2012; Xie and Klionsky, 2007). Autophagosome biogenesis starts with a cup-shaped precursor membrane, the phagophore, which gradually expands to either selectively or non-selectively enclose cytoplasmic material (Rogov et al., 2014). Proteins in the Atg8/LC3 family of ubiquitin-like proteins (Ubls) are involved in all stages of autophagosome maturation. Their coupling to phosphatidylethanolamine (PE) in the autophagosome membrane is a critical early step in phagophore biogenesis (Ichimura et al., 2000), and completion of autophagosome formation requires their deconjugation from PE by the papain-like Ubl-deconjugating enzyme Atg4 (Kirisako et al., 2000). Although the protein machinery that orchestrates autophagy has been largely identified, the process remains poorly understood. It is not well-established, for example, from where the phagophore membrane is derived; nor is it clear how the autophagy (atg) proteins are recruited and assembled at the phagophore/autophagosome.
Autophagy is also used as a cell-autonomous defense pathway that recognizes intracellular microbes as cargo to be enveloped in an autophagosome and targeted for destruction in an autolysosome (Huang and Brumell, 2014). The bacterium Legionella pneumophila, which causes the severe form of pneumonia known as Legionnaire’s disease, is able to evade detection by the autophagy pathway through the action of the protein RavZ (Choy et al., 2012), one of ~300 effectors that it secretes into the host cell cytoplasm during infection (Huang et al., 2011; Segal, 2013). Like Atg4, RavZ is a protease targeting the Atg8/LC3 protein family, but unlike traditional Ubl-deconjugating enzymes, RavZ does not cleave the peptide bond that was originally formed by the forward Ubl-like reaction (which in this case is the amide bond linking the terminal glycine of Atg8/LC3 and PE) (Choy et al., 2012). Instead, RavZ cleaves the amide bond between the PE-conjugated terminal glycine and the aromatic residue preceding it, thereby rendering Atg8/LC3 resistant to reconjugation (Choy et al., 2012) and thus inhibiting autophagosome maturation. RavZ also differs from Atg4 in targeting exclusively lipidated Atg8 proteins (Choy et al., 2012); as a result it does not interfere with early stages of autophagic membrane growth, when targeting and covalent association of Atg8 family proteins with the growing phagophore are taking place. How RavZ targets and ultimately proteolyzes LC3-PE is not known. Further, how a limited pool of LC3-PE can be recognized in a potential sea of soluble cytoplasmic LC3 is an important general question. A number of host cell autophagy proteins also preferentially engage lipidated LC3, but how this selectivity is achieved is not clear (Behrends et al., 2010; Birgisdottir et al., 2013). Thus, the autophagy inhibitor RavZ is an intriguing system through which to understand autophagosome maturation, including protein targeting to this compartment.
The crystal structure of a fragment of RavZ, which we present here, includes several membrane-interaction motifs beyond simple targeting of LC3-PE itself. The crystallized fragment consists of an N-terminal catalytic and a C-terminal domain, which each contribute to autophagosome membrane association. The C-terminal domain includes a phosphatidylinositol 3-phosphate (PI3P) binding site. PI3P is a phosphoinositide enriched at the autophagosome (Dall’Armi et al., 2013) and residues in the binding site are key in RavZ localization to the autophagosome. The N-terminal domain has a fold similar to the Ubl-specific protease (Ulp) family of cysteine proteases, and in addition to harboring the cysteine protease active site, it features an extended loop with a helix important for membrane association. We find further that RavZ binds more efficiently to highly curved membranes, suggesting that the protein may be preferentially targeted to the rim of the autophagophore cup. Our studies yield clues as to how RavZ is interfacially activated to function only on membranes, thus necessarily limiting its substrate pool to LC3-PE.
Results and Discussion
Overview of RavZ structure
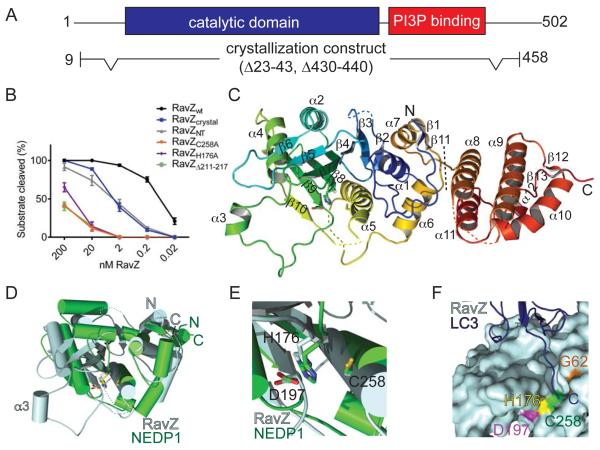
We used the single anomalous wavelength dispersion technique with selenomethionine substituted crystals (Hendrickson, 1991) to obtain the crystal structure of RavZ at 3.0 Å resolution. The crystallization construct RavZcrystal is active in an Atg8/LC3 delipidation assay ((Choy et al., 2012), Figure 1A-B) and comprises residues 10-458. It lacks sequences predicted to be disordered, including the N- and C-termini (residues 1-9, 459-502) and two loops (residues 23-43, 430-440). The structure was refined with Rwork/Rfree = 21.4/25.0 and good stereochemistry (Table S1). The final model contains residues 49-428, a barium ion, and one water molecule coordinated by it. The N- and C-terminal most residues and four flexible regions (residues 92-95, 249-253, 278-287, 347-356) of RavZcrystal lacked defined electron density and were not modeled.
Figure 1. RavZ N-terminal domain is structurally homologous to the Ulp family of Ubl deconjugating enzymes.
(A) RavZ domain organization and crystallization construct. (B) RavZ deconjugation activity assay. GL-1-PE (10 μM) on 25 nm liposomes (DOPE:POPC:blPI in a 55:35:10 molar ratio) was incubated for 1 h at room temperature with RavZWT or RavZ mutants (0.02 to 200 nM). The amount of deconjugated GL-1 relative to total GL-1 was quantified. Depicted are mean and standard deviation of three independent experiments. (C) Crystal structure of RavZ (residues 10-458, Δ23-43, Δ430-440) in ribbon representation colored blue to red from N- to C-terminus. The dotted line indicates boundary between N and C-terminal domains. (D) Secondary structure based superimposition (rmsd: 3.06 Å) of RavZ N-terminal domain (light blue) and protease domain of Ulp-family protein NEDP1 (green, PDB 2BKR). (E) RavZ (light blue) active site superposed with the NEDP1 (green, PDB 2BKR) active site. Residues numbered according to RavZ. (F) Model of the LC3 C-terminus (dark blue, ribbon representation, PDB 2Z0D) bound to the RavZ (light blue, surface representation) substrate-binding groove. RavZ/LC3 complex was generated by superimposing RavZ and LC3 onto the NEDP1/NEDD8 complex structure (PDB 2BKR). See also Figure S1.
RavZcrystal has two domains (Figure 1C). A five-helix bundle forms a C-terminal domain (α8-α12, residues 329-423), whereas the N-terminal portion folds into a 7-stranded beta sheet (β2-β4 and β7-β10) sandwiched between seven helices, four (α1 and α5-α7) on the side adjacent to the C-terminal domain and three (α2-α4) on the other side. A beta hairpin (β5 and β6) continues the sheet at the C-terminal end of strand β7 and above strand β4.
The RavZ N-terminal domain harbors its protease activity
Unexpectedly, since RavZ shares no significant sequence homology to any previously characterized protein, the fold of its N-terminal domain is closely related to cysteine proteases in the Ubl-specific protease (Ulp) family (Figure 1D). Ulp proteins both process full-length Ubl’s to their mature form prior to conjugation to their target proteins and deconjugate Ubl’s from modified proteins (Chosed et al., 2007; Mossessova and Lima, 2000; Shen et al., 2005). The RavZ N-terminal domain and Ubl-specific proteases have in common a core consisting of β4 and β7-β10 (RavZ numbering) in the central β-sheet and helices α1 and α4-α6 (Figure 1D). There are structural differences in an insertion between β4 and β7, which includes β5-β6 and α2 in the case of RavZ instead of the two helices as in the Ulp proteins, and RavZ has a long loop, connecting β9 and α4, not present in the Ulp family. This loop includes the short helix α3 and is hereafter referred to as the α3-loop. Importantly, the catalytic triad identified for the Ulp proteins (Chosed et al., 2007; Mossessova and Lima, 2000; Shen et al., 2005) appears to be conserved, and in RavZ corresponds to residues Cys258, His176 and Asp197 (close to the N-terminus of α5, and at the N-termini of β8 and β9, respectively) (Figure 1E).
To confirm that its N-terminal domain harbors RavZ cysteine protease activity and to assess the influence of the C-terminal portions on activity, we expressed and isolated the C- and N-terminal domains of RavZ for use in an Atg8/LC3 delipidation assay. As a substrate we used GABARAP-L1 (GL-1), a human ATG8/LC3 homolog, conjugated to PE (GL-1-PE). We first tested all RavZ constructs at high concentration (200 nM) (Figure S1A). Then, for those constructs that showed activity under these conditions, to approximate the degree by which delipidation activity is reduced relative to the wild-type protein, we determined the lowest concentration with measurable activity. The C-terminal domain of RavZ (RavZCT; residues 327-502) alone shows no detectable deconjugation activity even at the highest concentration while RavZcrystal and the N-terminal domain RavZNT (residues 1-326) are active, although with at least ten-fold reduced activity as compared to the full-length protein RavZWT (Figure 1B). A likely explanation for the activity reduction is that portions of RavZWT absent in RavZNT and RavZcrystal are involved in interactions with Atg8/LC3, the membrane to which it is attached, or both.
To assess whether the Cys258, His176, and Asp197 triad is important for catalytic activity, as predicted based on the structural similarity of RavZNT and the Ulp family proteins, we assayed the delipidation activities of four constructs in which these residues were mutated to alanine (RavZC258A, RavZH176A, RavZD197A, and RavZC258A,H176A). RavZD197A and RavZC258A,H176A showed no detectable deconjugation activity even at the highest concentration (Figure S1A), equivalent to at least a five-order of magnitude reduction in activity, whereas RavZC258A and RavZH176A were ~1000-fold less active than RavZWT (Figure 1B). Thus, the Cys258-His176-Asp197 triad is key for RavZ catalytic activity. In the Ulp proteins, the catalytic triad is within a groove that accommodates the Ubl C-terminus (Chosed et al., 2007; Mossessova and Lima, 2000; Shen et al., 2005), and it is likely that the analogous groove in the RavZ proteins binds the lipidated C-terminus of Atg8/LC3 substrates in a similar way (Figure 1F).
The RavZ C-terminal domain binds PI3P
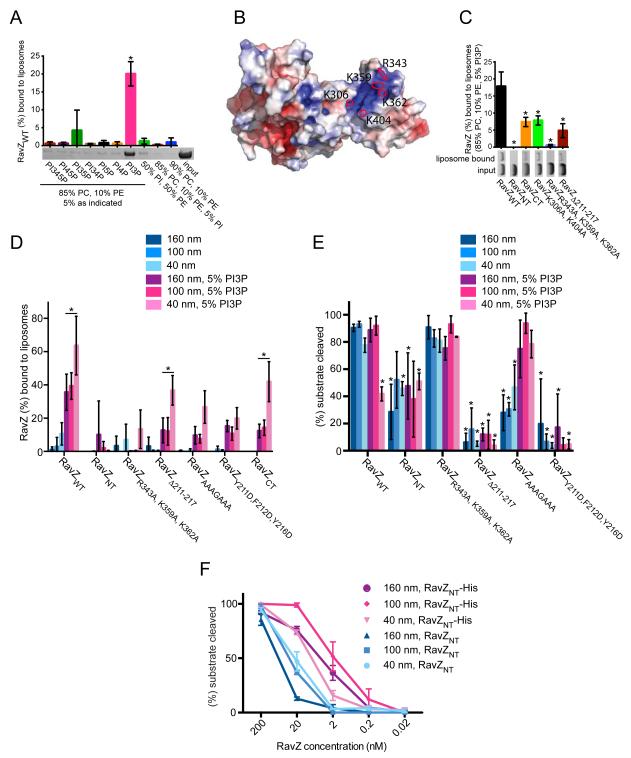
That RavZ proteolyses membrane-attached but not soluble Atg8/LC3 substrates suggested that it may interact with membranes directly. We used liposome binding assays to test for a direct interaction, discovering a specific interaction with liposomes enriched in PI3P (Figure 2A). PI3P is enriched in the autophagosome (Dall’Armi et al., 2013), so that PI3P recognition by RavZ may represent one way for targeting to the correct compartment.
Figure 2. The RavZ C-terminal domain includes a PI3P binding module responsible for RavZ autophagosomal localization.
(A, C and D) Liposome flotation assays. The amount of liposome-bound RavZ relative to input RavZ was quantified based on SDS-PAGE analysis of fractions. Mean and standard deviation (SD) of three independent experiments are indicated. In representative SDS-PAGE gels the input lane contains 5% of total input protein and sample lanes contain 12.5% of total liposome bound protein. (A) RavZWT binding to liposomes (160 nm diameter) of indicated composition. Mean and SD are indicated; *P < 0.0001 compared to control (90% PC, 10% PE). For full statistical analysis see Table S2, tab 1. (B) Electrostatic surface potential of RavZ (± 5 kBT/e). Basic residues in putative PI3P binding site are indicated. (C) Binding of RavZ mutants to 160 nm diameter liposomes of indicated composition. *P < 0.001 as compared to RavZWT. For full statistical analysis see Table S2, tab 2. (D) Binding of RavZ mutants to liposomes of different diameters with 5% PI3P (DOPE:POPC:PI3P in a 30:65:5 molar ratio) and without (DOPE:POPC:blPI in a 30:60:10 molar ratio). Selected significant results are marked; *P < 0.01. For full statistical analysis see Table S2, tab 3. (E) RavZ deconjugation activity assay. Liposomes containing GL-1-PE (10 μM) were incubated for 1 h at room temperature with RavZWT or RavZ mutants (2 nM). Liposome composition was identical to (D) to ensure comparability. The amount of deconjugated GL-1 relative to total GL-1 was quantified. Mean and standard deviation (SD) of three independent experiments are indicated. *P < 0.01; compared to RavZWT on 100nm liposomes without PI3P. For full statistical analysis see Table S2, tab 4. (F) Delipidation assay as in (E), but with liposomes containing Ni-NTA lipid (65% POPC, 30%, DOPE, 5% DGS-NTA). Activity of C-terminally hexa-histidine tagged RavZ constructs tethered to the liposomes is compared to soluble, untagged versions of same constructs. The amount of deconjugated GL-1 relative to total GL-1 was quantified, and mean and SD of three independent experiments are shown. See also Figures S2 and S3.
To begin understanding how RavZ might interact with membranes, we further inspected the crystal structure. We identified a pocket in the C-terminal domain of RavZ, which is lined with basic residues (Figure 2B), such as could interact with an acidic phospholipid, as a potential PI3P binding site. Consistent with a PI3P binding site in the C-terminal domain of RavZ, RavZCT but not RavZNT interacts with PI3P-positive liposomes in liposome binding assays (Figure 2C). To test the importance of the basic pocket for PI3P binding, we made constructs of RavZ in which basic residues in the pocket were replaced by alanine (RavZK306A,K404A, RavZR343A,K359A,K362A). RavZK306A,K404A bound PI3P-containing liposomes with lower affinity than RavZWT, and binding in the case of RavZR343A,K359A,K362A was almost entirely abrogated, indicating a role for basic pocket residues in PI3P binding and membrane association.
RavZ senses high curvature membranes
We also assayed whether the RavZ interaction is curvature dependent, finding that RavZWT as well as RavZCT associate more efficiently with smaller liposomes, which have more highly curved membranes, than larger ones (Figure 2D). Curvature sensing is PI3P-independent since the same trend is apparent in the presence and absence of PI3P (Figure 2D). High affinity membrane binding appears to be due to the C-terminal domain of RavZ as RavZNT does not exhibit any significant membrane binding even to highly curved membranes (Figure 2C, 2D). As the autophagosome is cup-shaped, with highly curved surfaces at the cup rim (Hayashi-Nishino et al., 2009), our results suggest that RavZ may preferentially localize to these regions, or possibly to highly curved small vesicles that may support local LC3 lipidation (Ge et al., 2014).
α3, a helix in the protease domain, is important for membrane binding but not curvature sensing
While RavZNT alone does not stably interact with liposomes in our non-equilibrium floatation assay, sequences in RavZNT may nevertheless engage the membrane during catalysis and possibly modulate membrane interactions mediated primarily by RavZCT. Indeed, RavZCT binds liposomes more weakly than the full-length protein (Figure 2C). As noted previously, the α3-loop insertion is one of the major differences between RavZNT and the Ulp proteases. Interestingly, α3 and its flanking regions contain predominantly basic and aromatic residues with the sequence Y211FKGKYR217. Basic residues can interact with the membrane by electrostatic interactions with negatively charged lipid head groups, and the interaction of aromatic residues is mediated by the insertion of their side chain into the lipid bilayer. Thus, consistent with a role for α3 in membrane binding, deleting residues 211-217 (RavZΔ211-217) significantly reduces RavZ association with liposomes (Figure 2C and 2D). Similarly, mutating the aromatic residues to aspartate (RavZY211D,F212D,Y217D), whose negative charge would be expected to inhibit interactions with acidic membranes, or mutating both the aromatic and basic residues in α3 to alanine (RavZAAAGAAA) reduces membrane affinity (Figure 2D). However, RavZΔ211-217 still shows increased binding to highly curved membranes (Figure 2D), suggesting curvature sensing is primarily due to the C-terminal domain.
RavZ protease activity requires α3 but not high affinity membrane interactions
As shown, RavZ associates with membrane via the C-terminal domain, which contains the PI3P-binding module and curvature sensing components, as well as helix α3 in the N-terminal domain. To test the effects of these elements on RavZ protease activity, we performed RavZ activity assays using GL-1 conjugated to liposomes of the same composition as in the RavZ curvature sensing/membrane binding studies (Figure 2D). RavZNT is not as active as the wild-type protein (Figure 2E and Figure S2), suggesting that membrane targeting mediated by the C-terminal domain may enhance activity, and consistent with this, we find that artificially tethering RavZNT to liposomes is activating (Figure 2F). Nevertheless, the increase in membrane affinity of RavZWT either in the presence of PI3P or higher membrane curvature (Figure 2D) does not correlate with higher protease activity (Figure 2E and Figure S2). Nor does the PI3P-binding deficient mutant RavZR343A,K359A,K362A, which binds only weakly to all liposome types tested (Figure 2D), have reduced activity as compared to RavZWT (Figure 2E and Figure S2). Taken together, these findings indicate that while membrane association enhances activity, as in the case of RavZNT, a weak interaction is sufficient for robust catalytic activity in vitro. Curiously, the activity of RavZWT at low concentrations (2 nM and 0.2 nM) is in fact significantly reduced on PI3P-positive, highly curved as compared to other liposomes despite a stronger interaction with membrane. A likely explanation for the apparent inhibition is that RavZWT is tightly bound to these liposomes, and under conditions where the liposome concentrations vastly exceeds the RavZ concentration (at constant total lipid, there are about 100 nM of the 40 nm liposomes, but only 6 nM of the much larger 160 nm liposomes), dissociation from the liposomes becomes the rate-limiting factor. Accordingly, the same inhibition is not observed for mutant versions of RavZ with lower affinity for membrane.
While high affinity membrane binding by the C-terminal domain of RavZ is not required for efficient delipidation, deletion (RavZΔ211-217) or mutation (RavZY211D,F212D,Y217D and RavZAAAGAAA) of α3 in the N-terminal domain leads to a significant reduction in protease activity on liposomes containing no PI3P (Figure 2E and Figure S2). In the case of RavZAAAGAAA, where α3 may still retain some reduced ability to associate with membrane via the hydrophobic alanine side chains, activity can be rescued by the presence of 5% PI3P in the liposomes. This is not true for RavZΔ211-217 or RavZY211D, F212D, Y217D, where α3 is not present or else would electrostatically repel the acidic membrane. This finding indicates that membrane binding by α3, while only contributing a small fraction of the overall membrane affinity of RavZ, is critical for RavZ protease activity.
Both RavZ C-terminal portions and α3 are required for autophagosome localization and activity in vivo
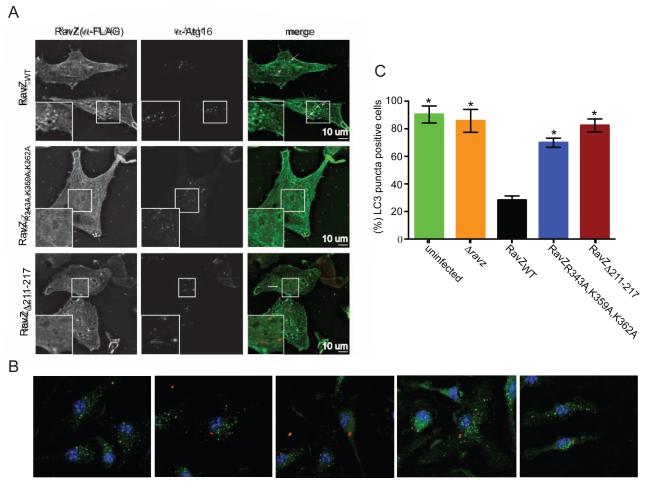
To discover whether RavZ targeting to the autophagosome depends on either the C-terminal PI3P-binding domain or α3 in the N-terminal domain, we compared localization of RavZWT, RavZR343A,K359A,K362A and RavZΔ211-217 in vivo, by overexpressing 3xFlag-tagged forms of these proteins in HeLa cells that were then starved to induce autophagy. RavZWT localizes to puncta that are positive for the autophagosome marker Atg16 (Figure 3A and (Choy et al., 2012)), whereas both RavZR343A,K359A,K362A, the construct lacking a functional PI3P binding pocket, as well as RavZΔ211-217 mislocalize primarily to the cytosol and to a lesser degree to the ER (Figure 3A). The PI3P binding pocket as well as helix α3 are therefore critical for localization to the autophagosome.
Figure 3. RavZ requires both the PI3P binding domain and α3 for autophagosome localization and activity in vivo.
(A) In vivo localization of RavZWT, RavZR343A,K359A,K362A and RavZΔ211-217 in HeLa cells, demonstrating that the residues in the putative PI3P-binding pocket and α3 are important for RavZ autophagosomal targeting. Localization of RavZ and the Atg16 autophagosome marker are shown in the first and second column, respectively. The third column shows the extent of their co-localization. (B) Images show LC3 (green) distribution in mouse bone marrow-derived macrophages infected with the indicated Legionella strains (red). (C) Percentage of LC3-puncta positive cells calculated from three independent assays, where a total of 100 cells were scored in each assay. Presented are the mean and SD; *P < 0.0001 compared to RavZWT. For full statistical analysis see Table S3.
In vivo studies were conducted to determine if α3 in the N-terminal domain or the PI3P binding module in RavZ contribute to the ability of this effector to modulate autophagy when delivered into host cells by L. pneumophila during infection. Mouse bone marrow-derived macrophages were infected with isogenic ΔravZ strains that were complemented with plasmid-encoded ravZ alleles and endogenous levels of LC3 were visualized by immunofluorescence microscopy. As expected, LC3-positive puncta formation was inhibited in macrophages infected with L. pneumophila producing the wild type RavZ protein (Figure 3B). By contrast, robust puncta formation was observed in cells infected with L. pneumophila producing the PI3P binding-mutant RavZR343A,K359A,K362A or the α3 mutant RavZΔ211-217 (Figure 3B). When puncta formation was quantified in the infected cells, a significant increase in LC3 puncta were observed in cells infected with strains producing RavZR343A,K359A,K362A or RavZΔ211-217 compared to cells producing the wild type RavZ protein (Figure 3C). The catalytically impaired RavZΔ211-217 strain was unable to delipidate LC3 in vivo, which was consistent with in vitro data. Importantly, the PI3P-binding deficient mutant RavZR343A,K359A,K362A protein, which displayed full enzymatic activity in the in vitro delipidation assay, had a severe defect in delipidation activity in the physiologically relevant infection assay. This confirms the importance of the PI3P-binding domain in targeting a limited amount of translocated RavZ to preautophagosomal structures to mediate efficient LC3 delipidation in vivo.
A model for RavZ substrate and membrane binding
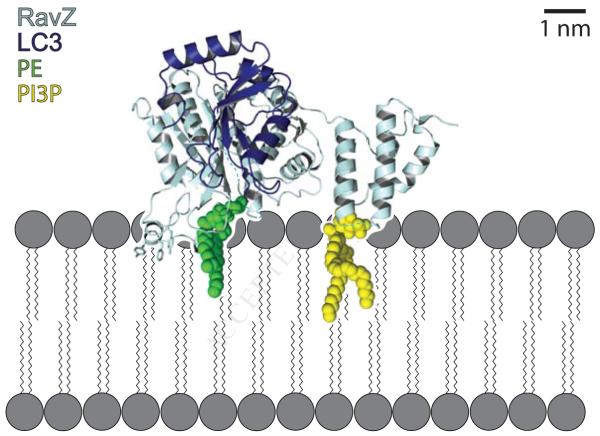
Identification of structural elements in RavZ that mediate membrane interactions suggests a model for RavZ membrane association during the deconjugation reaction (Figure 4). The electrostatic pocket in the C-terminal domain implicated in PI3P binding and the helix within the α3–loop of the N-terminal domain are on the same face of the protein. Further, the position of the catalytic triad and the substrate groove suggest that the lipidated tail of LC3-PE would also be found on this surface of RavZ prior to its cleavage. Thus RavZ is likely to engage the autophagosome through at least three sites, each relying on distinct physico-chemical properties of the local membrane environment.
Figure 4. Model of RavZ dependent Atg8/LC3 delipidation at the autophagosomal membrane.
RavZ (dark blue) is recruited to the autophagosomal membrane (grey) by interaction of its C-terminal domain with PI3P (yellow). Helix 3 in the N-terminal protease domain likely interacts with the membrane by insertion of aromatic side chains between the lipid acyl-chains and by electrostatic interactions between its basic side chains and the negatively charged lipid headgroups. The RavZ N-terminal domain recognizes Atg8-PE/LC3-PE (Atg8/LC3: light blue, PE: green) at the membrane and catalyzes its deconjugation. The interaction of Helix 3 of the RavZ N-terminal domain with membrane is critical for efficient catalysis, possibly as a “lid” clamping the C-terminus of Atg/LC3 between RavZ and the membrane to restrict LC3 mobility. RavZ/LC3 complex was generated by superimposing RavZ and LC3 (PDB 2Z0D) onto the NEDP1/NEDD8 complex structure (PDB 2BKR).
A major question regarding RavZ is why it cleaves lipidated but not soluble Atg8/LC3 substrates. The findings that RavZ constructs with mutations in α3 are impaired in their ability to associate with membranes, show significantly lower proteolytic activity, and fail to localize to the autophagosome in vivo (Figure 2D, 2E and Figure 3) suggest that the interaction between α3 and the membrane is critical for RavZ activity. In several Ulp family proteins including each of the mammalian and yeast homologues of Atg4, the flexible C-terminus of their respective substrates is retained in the active site groove through the action of a “lid”, a highly conserved tryptophan residue located above the active site. Removal of this lid residue severely reduces the proteolytic activity of these enzymes (Shen et al., 2005; Sugawara et al., 2005). The “lid” tryptophan is not present in RavZ, where this residue is a glycine (G62) (Figure 1F). However, interaction of helix α3 with the membrane would trap the lipidated Atg8/LC3 C-terminus between the substrate groove in the N-terminal domain of RavZ and the membrane (as in Figure 4). We propose that the membrane may effectively act as a lid when it is engaged by α3 in the N-terminal domain of RavZ, thus promoting Atg8/LC3 proteolysis.
In the absence of a RavZ/LC3 complex structure, we do not know whether RavZ binds only the lipidated C-terminus of its Atg8/LC3 substrate or whether, and if so how, it also binds the globular N-terminal portion. Thus, the orientation of RavZ with respect to the Atg8/LC3 N-terminal domain as depicted in the model (Figure 4) is speculative. Further, we do not know if interactions with the membrane induce conformational changes in RavZ that promote its catalytic activity. This type of interfacial activation has been best studied in lipases, where membrane association typically results in large rearrangements that allow substrate to enter a previously inaccessible active site (Casas-Godoy et al., 2012). This particular scenario does not seem to apply to RavZ, where the active site groove is solvent accessible in the crystal (i.e., in the absence of membrane). We cannot exclude, however, that structural elements that normally block access to the RavZ active site groove when RavZ is soluble are displaced due to crystal contacts and that RavZ in the crystal is, in fact, in an activated conformation. Alternatively, the membrane could induce other rearrangements that might increase affinity for the substrate or subtle conformational changes that correctly position active site residues for catalysis. We expect that further biochemical and structural studies, in particular of a RavZ-substrate complex, will provide additional insights as to the mechanisms underlying the interfacial activation of RavZ.
Concluding Remarks
Thus, we have found that RavZ is targeted to the autophagosome by targeting PI3P-enriched, high curvature membranes via a C-terminal PIP3-binding module, RavZCT, and a membrane-interacting helix in the catalytic domain RavZNT. Further, although more work is required to understand in detail how RavZ is interfacially activated, our studies provide clues, suggesting, for example, that by incorporating multiple autophagosome-targeting motifs, RavZ becomes both highly efficient and strictly specific for the processing of lipidated forms of LC3. These insights obtained for RavZ targeting and activation may apply more broadly to host cell proteins that act in autophagy. Because autophagosomes are largely devoid of transmembrane proteins (Feng et al., 2014), targeting to these organelles must rely upon other membrane cues. The best described cues are the locally enriched population of PI3P, the specific accumulation of lipidated LC3 proteins and increasingly the presence of highly curved or poorly packed membranes (Dancourt and Melia, 2014; Fan et al., 2011; Nath et al., 2014; Ragusa et al., 2012). However, neither high curvature nor PI3P are unique to autophagosomes, and while lipidated LC3 is strongly associated with autophagic processes, there is no evidence that this protein is structurally distinct from the vast pool of cytoplasmic LC3 (Ma et al., 2010); thus how proteins identify autophagosomes remains an important and intriguing question. RavZ presents one very satisfying model, that inclusion of multiple perhaps low-affinity and only moderately specific membrane recognition modules can collectively drive very specific and effective local targeting or activity of the protein. It will be interesting to explore whether other host autophagy proteins target LC3-PE through similar collections of interactions.
Experimental Procedures
For RavZ structure determination, native and selenomethionine-substituted RavZ (residues 10-458, Δ23-43, Δ430-440) were recombinantly produced as GST-fusion proteins in E. coli and purified by affinity and size exclusion chromatography. Crystals were grown at 4°C by the hanging drop method against a reservoir solution consisting of 0.1 M MES [pH 5.1], 12% PEG 3350, and 0.2M BaCl2. The crystals were transferred into mother liquor supplemented with 20% glycerol and then flash-frozen in liquid nitrogen prior to data collection. Data were collected at beamline NE-CAT 24-ID-C at Argonne National Laboratories (APS), and the structure was determined to 3.28 Å using selenomethionine substituted crystals in the single wavelength anomalous dispersion (SAD) method (Hendrickson, 1991). The structure was subsequently refined to 2.98 Å against data collected from native crystals using Phenix software (Adams et al., 2010).
RavZ constructs for biochemical assays were produced in the same way but omitting the size exclusion step. The liposome binding and delipidation activity assays were carried out as described in Nath et al., 2014, and Choy et al., 2012, respectively, with small modifications. The Legionella strains for in vivo activity assays, which harbor mutant versions of the RavZ gene, were constructed and infected into macrophages as described in Choy et al., 2012. For imaging, cells were fixed and permeabilized prior to immunostaining.
To test whether a set of experimental groups differ from each other in a statistically significant way we performed either a two-way (Figure 2D and 2E) or one-way (else) analysis of variance (ANOVA). If a significant difference was found, we compared the mean of each experimental group with the mean of each other experimental group using Tukey’s test to discover which experimental groups differ from each other. 95% confidence intervals (CI) and multiplicity-adjusted p-values for all comparisons were calculated and are reported in supplementary tables as indicated in the respective figure legends. Selected significant results are marked in the figures.
All protocols are described in full in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgements
We are grateful to the staff at NE-CAT beamline 24ID-C for assistance with data collection. This work was supported by the NIH (R01 GM080616 and R03 AG046941 to KMR, R01 GM100930 to TJM, R01 AI041699 to CRR, and T32 GM007223 for graduate student support).
Footnotes
Author contributions: FH carried out all experiments unless otherwise noted. KJK carried out the flotation assays; LK performed in vivo localizations, with participation by VS; RKS performed in vivo activity assays; KPK prepared several of the RavZ constructs for crystallization trials. FH, CRR, TJM, and KMR designed the experiments, analyzed the data, and wrote the manuscript with input from other authors.
Coordinates and structure factors for RavZ have been submitted to the PDB (accession number 5CQC).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. Journal of cell science. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- Casas-Godoy L, Duquesne S, Bordes F, Sandoval G, Marty A. Lipases: an overview. Methods in molecular biology. 2012;861:3–30. doi: 10.1007/978-1-61779-600-5_1. [DOI] [PubMed] [Google Scholar]
- Chosed R, Tomchick DR, Brautigam CA, Mukherjee S, Negi VS, Machius M, Orth K. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. The Journal of biological chemistry. 2007;282:6773–6782. doi: 10.1074/jbc.M608730200. [DOI] [PubMed] [Google Scholar]
- Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Current biology : CB. 2013;23:R33–45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J, Melia TJ. Lipidation of the autophagy proteins LC3 and GABARAP is a membrane-curvature dependent process. Autophagy. 2014;10:1470–1471. doi: 10.4161/auto.29468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell research. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 2014;3 doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature cell biology. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nature reviews Microbiology. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O’Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. The E Block motif is associated with Legionella pneumophila translocated substrates. Cellular microbiology. 2011;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. The Journal of cell biology. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annual review of immunology. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- Ma P, Mohrluder J, Schwarten M, Stoldt M, Singh SK, Hartmann R, Pacheco V, Willbold D. Preparation of a functional GABARAP-lipid conjugate in nanodiscs and its investigation by solution NMR spectroscopy. Chembiochem : a European journal of chemical biology. 2010;11:1967–1970. doi: 10.1002/cbic.201000354. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Molecular cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nature cell biology. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Molecular cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Segal G. Identification of legionella effectors using bioinformatic approaches. Methods in molecular biology. 2013;954:595–602. doi: 10.1007/978-1-62703-161-5_37. [DOI] [PubMed] [Google Scholar]
- Shen LN, Liu H, Dong C, Xirodimas D, Naismith JH, Hay RT. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. The EMBO journal. 2005;24:1341–1351. doi: 10.1038/sj.emboj.7600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. The Journal of biological chemistry. 2005;280:40058–40065. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nature cell biology. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.