Abstract
G protein–coupled receptors (GPCRs) are essential mediators of signal transduction, neurotransmission, ion channel regulation, and other cellular events. GPCRs are activated by diverse stimuli, including light, enzymatic processing of their N-termini, and binding of proteins, peptides, or small molecules such as neurotransmitters. GPCR dysfunction caused by receptor mutations and environmental challenges contributes to many neurological diseases. Moreover, modern genetic technology has helped identify a rich array of mono- and multigenic defects in humans and animal models that connect such receptor dysfunction with disease affecting neuronal function. The visual system is especially suited to investigate GPCR structure and function because advanced imaging techniques permit structural studies of photoreceptor neurons at both macro and molecular levels that, together with biochemical and physiological assessment in animal models, provide a more complete understanding of GPCR signaling.
Keywords: GPCRs, signal transduction, membrane biology, transmembrane receptors, receptor pharmacology, allosteric regulation, rhodopsin, crystal structure
G PROTEIN–MEDIATED SIGNALING: A BRIEF HISTORICAL PERSPECTIVE
Although it is difficult to identify a single event that initiates a new scientific field, the discovery of cyclic adenosine monophosphate (cAMP) is generally perceived to have initiated the study of G protein–mediated signaling in earnest. In the late 1950s, Earl W. Sutherland, Jr., Chair of Pharmacology at Western Reserve University (later renamed Case Western Reserve University), and his postdoctoral fellow, Theodore W. Rall (a subsequent Chair of the same department), discovered this distinctive nucleotide (Rall & Sutherland 1958, Sutherland & Rall 1958). Trained by Carl F. Cori (Washington University in St. Louis), a Czech emigrant biochemist/pharmacologist and 1947 Nobel laureate, Sutherland linked the formation and destruction of cyclic AMP (the secondary messenger) with the effects of epinephrine, glucagon, and insulin upon glycogen metabolism (Robison et al. 1968). Sutherland was the sole winner of the 1971 Nobel Prize in Physiology or Medicine “for his discoveries concerning the mechanisms of the action of hormones” (http://www.Nobelprize.org). Not only was Sutherland an incredibly insightful scientist, but he also initiated one of the first MD/PhD programs in the country and personally recruited Alfred G. Gilman to Cleveland to join that program. Gilman, a doctoral student of Rall (Gilman & Rall 1968a,b) who was intrigued by the role of cAMP, later discovered and characterized G proteins, molecular switches that link cellular receptors to various responses (Gilman 1987).
That hormone action is mediated via receptors had been known since the beginning of the twentieth century, but it was Rodbell and colleagues who showed that these receptors and associated proteins are organized at the cellular plasma membrane as large multiprotein complexes composed of receptors, G proteins, and effector enzymes (Schlegel et al. 1979). Both Gilman and Martin Rodbell were awarded the Nobel Prize in Physiology or Medicine in 1994 for these seminal contributions.
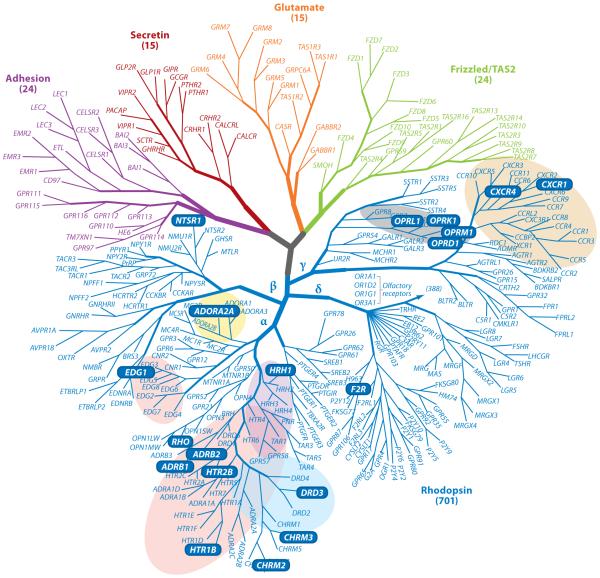
Today we know that GPCRs [also known as guanine-nucleotide-binding protein-coupled membrane receptors, seven-transmembrane (7-TM) receptors, or heptahelical receptors] are evolutionarily highly conserved (Strotmann et al. 2011) and are widely expressed in eukaryotic organisms where they control a vast range of biological processes. These receptors are classified into various subtypes on the basis of their extracellular domain topology, sequence similarity, activating ligands, type of G protein they activate, function, and other criteria (see, e.g., Park et al. 2008b) (Figure 1). All GPCRs are predicted to share a common 7-TM α-helical structure and are localized within cellular membranes. These receptors respond to a diverse array of physical and chemical entities that include photons, proteolytic events, soluble small molecules and ions, peptides, and large proteins. More than a decade ago, the first crystal structure of a native GPCR (rhodopsin) was solved (Palczewski et al. 2000), and on the basis of a vast amount of biochemical data, investigators predicted that the general features in the TM α-helical region are conserved across all GPCRs (Filipek et al. 2003b).
Figure 1.
Current phylogenetic tree for GPCRs. GPCR gene names are located on branches on the basis of their primary sequence similarity. Solved structures are represented using gene names on blue backgrounds. Colored backgrounds indicate groups of GPCRs that share sequence homology with determined X-ray structures that are 35% or higher. Major branches such as the adhesion, secretin, glutamate, Frizzled/TAS2, and rhodopsin are labeled, as well. Figure adapted from Katritch et al. (2012).
In neuronal systems, GPCRs fine-tune cellular functions by activating a heterotrimeric G protein–dependent cascade of signals (Gainetdinov et al. 2004). These fine-tuning events further regulate a variety of cellular events including synaptic modulation and neurotransmission, Ca2+ and cyclic nucleotide signaling, cellular plasticity, electrical activity, neurite outgrowth, exocytosis, gene expression, the unfolded protein response, and neuronal development/differentiation, among others. For example, a putative octopamine GPCR in sensory neurons actively suppresses innate immune responses by downregulating the expression of noncanonical unfolded-protein response genes to control stress-response pathways (Sun et al. 2011). GPCRs expressed by adult neural stem cells have also emerged as potential modulators of adult neurogenesis that are critical for self-repair of neuropathologic conditions (Doze & Perez 2012).
GPCR activities may influence higher functions such as behavior but are also involved in pathologies such as neurodegeneration, excitotoxic injury, and motor dysfunction. Here, we summarize recent progress in determining the structural biology of GPCRs and understanding GPCR signaling by using visual phototransduction as a specific example. Invertebrate and vertebrate visual systems involving phototransduction are model systems that have provided molecular and functional information about GPCR signaling for decades (Yau & Hardie 2009).
ADVANCES IN STRUCTURAL STUDIES OF GPCRs AND OTHER INTERACTING PROTEINS
GPCRs Exhibit Similar Overall Topology with Diversity in Their Ligand-Binding Sites
Among all available GPCR structures solved to date (Figure 1), the large structural diversity of the extracellular half, denoted by an average root-mean-square deviation of ~2.7 Å, contrasts sharply with the relatively similar structural features of the intracellular half, with an average root-mean-square deviation of ~1.5 Å . The molecular topology shared by all GPCRs consists of the seven transmembrane (TM) domain composed of α-helices spanning the biological membrane (Figure 2a–t). These TM helices are connected by three extracellular (ECL) and three intracellular (ICL) loops denoted as ECL-I to -III and ICL-I to -III, respectively (Salon et al. 2011). The ECLs, N-termini, and/or part of the TM α-helical region facing the extracellular space are responsible for ligand recognition, whereas the ICL and C-termini recognize the specific protein partner required for signal transmission within the cell. The orthosteric ligand-binding region is frequently located within a cavity formed by the TM helices, but in some GPCRs, such as the metabotropic glutamate receptors, the ligands are bound by a separate amino terminal discrete domain (Pin et al. 2003). Although the extracellular region has evolved to recognize a multitude of diverse ligands, the intracellular region needs to recognize and discriminate between a relatively small number of G proteins. Because the amino terminal ligand-binding domains of some GPCRs fold properly and bind ligand independently of the TM α-helical region, researchers have elucidated several structures of these domains (reviewed in Park et al. 2008b, Pin et al. 2003). Common features of the TM helices have long been recognized (Figure 2u), and efficient signal transduction through the membrane is achieved by a similar mechanism for all rhodopsin-like class A GPCRs (Mirzadegan et al. 2003) and perhaps all GPCR classes. The mechanism by which the signal is propagated involves a network of water molecules, specific conserved amino acids, and small movements of the TM helices (Figure 3a). In addition to these features, other conserved structural elements such as the disulfide bond linking ECL-II to TM-III, the (D/E)RY motif in helix III (which participates in the ionic lock between helix III and VI), the WxP motif in helix VI, and the NPxxY motif in helix VII are also thought to be involved in efficient signal transduction. Several different analyses of the activation mechanism have been published but are beyond the scope of this review (Hofmann et al. 2009, Katritch et al. 2012, Park et al. 2008b, Salon et al. 2011).
Figure 2.
Gallery of selected GPCR structures determined by X-ray crystallography. Available X-ray structures are shown in the same orientation for all GPCRs together with an alignment graph of each GPCR pair obtained using flexible structure alignment by chaining aligned fragments (Ye & Godzik 2003). (a) bovine rhodopsin (PDB code: 1U19); (b) β2-adrenergic receptor/T4 lysozyme chimera (PDB code: 2RH1); (c) thermostabilized β1-adrenergic receptor (PDB code: 2Y01); (d ) squid rhodopsin (PDB code: 3AYN); (e) adenosine receptor/T4 lysozyme chimera (PDB code: 3EML); ( f ) CXCR4 chemokine/T4 lysozyme chimera (PDB code: 3ODU); ( g) dopamine D3 receptor/T4 lysozyme chimera (PDB code: 3PBL); (h) human histamine H1 receptor/T4 lysozyme chimera (PDB code: 3RZE); (i ) human M2 muscarinic acetylcholine receptor/T4 lysozyme chimera (PDB code: 3UON); ( j) κ-opioid receptor/T4 lysozyme chimera (PDB code: 4DJH); (k) lipid GPCR/T4 chimera (PDB code: 3V2W); (l ) M3 muscarinic receptor/T4 lysozyme chimera (PDB code: 4DAJ); (m) μ-opioid receptor/T4 lysozyme chimera (PDB code: 4DKL); (n) δ-opioid receptor/T4 lysozyme chimera (PDB code: 4EJ4); (o) thermostabilized apocytochrome b562 (Bril)-nociceptin/orphanin FQ receptor fusion (PDB code: 4EA3); ( p) chemokine receptor CXCR1 (PDB code: 2LNL); (q) neurotensin receptor NTS1 (PDB code: 4GRV); (r) human protease-activated receptor 1 (PAR1) (PDB code: 3VW7); (s) chimeric protein of 5-HT2B-BRIL (HTR2B) (PDB code: 4IB4); (t) chimeric protein of 5-HT1B-BRIL (HTR1B) (PDB code: 4IAQ). GPCRs are shown in a rainbow-colored scheme: N-terminal (dark blue), C-terminal (red ), and nonnative fusion protein components ( gray). (u) Alignment of fragment pairs derived for each combination of two GPCR structures presented in panels a–t. Blue circles indicate the largest differences resulting from insertion/deletions when comparing the aligned structures of the GPCRs under study.
Figure 3.
Signal transduction in GPCRs. (a) Diagrams of the seven transmembrane (TM) helices and hydrogen-bonding network in rhodopsin (PDB ID:1U19). (Left) Helices are colored according to their primary sequence: helix-I (blue); helix-II (blue-green), helix-III ( green); helix-IV (light green); helix-V ( yellow); helix-VI (orange); helix-VII (red ); helix-8 ( purple). (Right) The chromophore is shown with balls and sticks ( pink). Water molecules are displayed as spheres (light blue). The receptor is oriented such that the extracellular space is above and the G protein–interacting cytoplasmic face is below. (b) The superposed surface representations of ground-state rhodopsin (PDB code: 1u19 in red ) and photoactivated-like rhodopsin (PDB code: 4a4m in yellow) viewed from the intracellular side of a plasma membrane. Phospholipids representing the plasma membrane are shown as a gray layer. On the right, 45° rotation reveals major secondary structure elements of rhodopsin and photoactivated rhodopsin visible outside the phospholipid membrane. Locations of intracellular loops (ICLs) and the C-terminal segment are indicated by arrows. (c) An intracellular view of metarhodopsin II ( yellow) with a bound peptide ( green) derived from the primary sequence of Gt. (d ) Superposition of the structure of ground-state rhodopsin (red ) on the structure of photoactivated rhodopsin ( panel c). The overlap of the peptide derived from Gt with ground-state rhodopsin suggests that interaction of the two would be unlikely.
Palczewski et al. (2000) revealed the first atomic resolution details of a GPCR with the crystallographic structure determination of rhodopsin. Ground state rhodopsin crystals prepared under different conditions yielded structures of rhodopsin in three different conformations with significant changes in ICL-III (Li et al. 2004, Okada et al. 2002, Salom et al. 2006). Subsequent collaborative efforts led to the discovery of rhodopsin dimerization in native membranes (Fotiadis et al. 2003), and the first reported photoactivated deprotonated rhodopsin structure (Salom et al. 2006).
A new era in the GPCR field began in 2007 including studies that produced the first structures of GPCRs bound to various diffusible and covalently bound ligands (Cherezov et al. 2007, Rasmussen et al. 2007, Rosenbaum et al. 2007), the structure of squid rhodopsin (Murakami & Kouyama 2008, Shimamura et al. 2008), and a glimpse into the GPCR activation process provided by the structure of bovine opsin (Park et al. 2008a). Some of these advances were facilitated by using protein engineering techniques to increase GPCR stability—a much sought after property for crystallization trials (Tate & Schertler 2009). Often N-termini, ICL-III, and/or C-termini were shortened to improve expression and crystallization (Mustafi & Palczewski 2009). Another strategy employed the fusion of GPCRs with fast-folding proteins such as T4 lysozyme (Cherezov et al. 2007, Rosenbaum et al. 2007) or thermostabilized apocytochrome b562 (Chun et al. 2012, Thompson et al. 2012). GPCR crystal formation has also been successfully facilitated by the formation of complexes between GPCRs and antibody Fab fragments or nanobodies (Day et al. 2007). Figure 2a–t shows all currently known three-dimensional structures of GPCRs together with Figure 2u,a one-to-one comparison half matrix that illustrates the main differences between members. Inspection of Figure 2a–t indicates that GPCRs, as described above, share a high degree of structural homology except for the CXCR4 chemokine receptor, the most distant member of the cluster of known structures from the homology tree (Figure 1; Figure 2u, column f and row f ). The structural differences observed when comparing GPCR members with the CXCR4 chemokine receptor stem from the disordered nature of helix VIII in the CXCR4 chemokine receptor.
Investigators have observed specific diversity in the GPCR ligand-binding sites, which indicates that these receptors can respond to different groups of chemicals, peptides, or proteins (Granier & Kobilka 2012). It is an incredible evolutionary triumph that hundreds of different genes have produced proteins that fold into similar structures that diversify by fine-tuning their ligand-binding sites. Thus, it was not suprising that the 2012 Nobel Prize in Chemistry recognized these accomplishments in the field. Robert J. Lefkowitz of Duke University (Durham, North Carolina), and Brian K. Kobilka of Stanford University School of Medicine (Palo Alto, California) were recipients of the award for studies of G protein–coupled receptors (http://www.Nobelprize.org).
Water Molecules in the Transmembrane Segments of GPCRs
Upon photoactivation, rhodopsin, with its covalently bound ligand 11-cis-retinal, undergoes changes that involve both deprotonation of the Schiff base and protonation/deprotonation at the cytoplasmic surface, which intuitively suggests a role for internal water molecules in proton transfer (Hofmann et al. 2009). Crystallography, particularly of GPCR structures at high resolution, has unequivocally revealed the presence of water molecules bound in specific sites within the hydrophobic TM α-helical domain (Angel et al. 2009a). More recently, radiolytic footprinting analysis of bound waters revealed that internal water was redistributed when rhodopsin was activated and the rod photoreceptor–specific G protein (transducin or Gt) was subsequently bound (Angel et al. 2009b, Orban et al. 2012b). In addition to bound water molecules, sodium ions can play an important role in GPCR function as recently proposed for the highest resolution X-ray structure (1.8 Å ) of a GPCR solved to date (Liu et al. 2012). Reorganization of water molecules identified in different states of photoactivated rhodopsin (Figure 3a) and in dynamic regions noted in both photoactivated rhodopsin and Gt revealed that the receptor undergoes relaxation upon activation and then rigidifies once the G protein binds (Figure 4). Thus, water appears to mediate the conformational changes between the ligand-binding site and the G protein–coupling domain on the cytoplasmic surface of these receptors. A general characteristic of all transmembrane proteins, even those that are not water channels, could be their use of internal water to stabilize secondary elements and allow flexibility in assembly of these elements. This underappreciated strategy perhaps allows membrane proteins to behave like highly hydrated sponges, requiring only minimal energy from ligand binding to be sensed through the receptor. This hydration possibility should also caution researchers who interpret GPCR mutagenesis studies because changes in side chains within TM α-helical segments will inevitably reorganize the internal water molecules. This hydration phenomenon could cause serious problems for rational drug (ligand) design in general if investigators do not consider the precise localization of water molecules. The number of water molecules within the TM α-helical domain is highly conserved among GPCRs, suggesting that functional roles for water-mediated contacts can also be conserved across members of this receptor family (Angel et al. 2009a, Pardo et al. 2007).
Figure 4.
GPCR: G protein complex. (a) Superposed structures of photoactivated rhodopsin in complex with Gt (red ) (Jastrzebska et al. 2011b) and the β2-adrenergic receptor in complex with Gs (blue) (Chung et al. 2011) (dark gray, T4 lysozyme present in the β2-adrenergic receptor-Gs structure; gray transparent layer, the phospholipid membrane). The section highlighted in detail (large black circle) shows superposed Gt (PDB code: 1GOT, red ) and Gs (PDB code: 4A4M, blue). The averaged angle between the Gsα-AH and Gtα-AH domains evaluated from several measurements was ~95°, whereas distances between corresponding atoms of the two domains had an average value of ~40 Å (Jastrzebska et al. 2011b). Superposition of the Gsα-AH subdomain with the Gtα-AH subdomain is achieved only after a full 180° rigid body rotation around the axis shown on the left. (b) Differences in normalized hydrogen-deuterium exchange evaluated for photoactivated rhodopsin, photoactivated rhodopsin-Gt, and Gt. Heat maps (dark blue to rose) were used to evaluate differences in normalized hydrogen-deuterium exchange for free Gt and Gt in complex with photoactivated rhodopsin and for free photoactivated rhodopsin and photoactivated rhodopsin in complex with Gt (Orban et al. 2012b). Heat maps were then placed on the three-dimensional structure of the photoactivated rhodopsin-Gt complex model (Jastrzebska et al. 2011b). Negative differences in hydrogen-deuterium exchange are shown as 0–9% ( green), 10–19% (cyan), 20–29% (light blue), 30–39% (blue), and 40–50% (darker blues). Positive differences are displayed as 0–9% ( yellow), 10–19% (light orange), 20–29% (orange), 30–39% (red ), and 40–50% (magenta to purple). The lipid bilayer where photoactivated rhodopsin is embedded is presented as a transparent gray layer. Abbreviation: AH, α-helical.
GPCR ACTIVATION AND RECRUITMENT OF G PROTEINS AND OTHER PROTEINS
Mechanism of GPCR Activation: A Brief Account
The crystal structures of several GPCRs help researchers understand how GPCRs activate, stabilize their inactive conformations, and interact with partner proteins. These receptors likely share a common mode of activation (Smith 2012). One idea is that a rearrangement of TM-V and TM-VI opens a crevice at the cytoplasmic side of the receptor into which the C-terminus of the cognate G protein can bind (Hofmann et al. 2009). Another idea, derived from a nuclear magnetic resonance study of rhodopsin in membranes, proposes that the retinal ligand initiates collective helix fluctuations on a microsecond to millisecond timescale between activated GPCR conformers at equilibrium (Struts et al. 2011). Because membranes are integral to GPCR signaling (Inagaki et al. 2012, Jastrzebska et al. 2011a, Kaya et al. 2011), this idea deserves serious consideration.
Crystallography of GPCRs has accelerated over the past three years with the determination of a large number of antagonist-bound GPCR structures as well as with more recent work that has produced agonist-bound (activated) structures (Figure 2). For example, Doré et al. (2011) determined the structures of a thermostabilized adenosine A2A receptor in complex with xanthine, a xanthine amine congener, and caffeine, as well as the A2A selective inverse agonist ZM241385. This receptor was crystallized in its inactive conformation because the ionic lock in the conserved threeamino-acid DRY motif region was observed. Endorphins and opioid alkaloids bind to three members of the GPCR family: the μ-, δ-, and κ-opioid receptors. Together the structures of the μ- and κ-, the δ-opioid receptors, and the nociceptin/orphanin FQ (N/OFQ) peptide receptor structure with bound antagonists provide insight into the conserved elements of opioid ligand recognition and also reveal those features associated with ligand-subtype selectivity (Granier et al. 2012, Manglik et al. 2012, Thompson et al. 2012, Wu et al. 2012). A more complete list of crystallized GPCRs with bound agonists, antagonists, or antibody fragments is shown in Figure 2.
Constitutively active GPCR mutants with increased basal activity have been used to elucidate the structures of activated forms of rhodopsin. Two different constitutively active mutants of rhodopsin (E113Q and M257Y) with a bound C-terminal fragment of a G protein α-subunit were used to trap the activated state of rhodopsin with the agonist all-trans-retinal present in the native-binding pocket (Deupi et al. 2012, Standfuss et al. 2011). Within crystal structure uncertainty, this conformation is virtually identical to that of opsin-all-trans-retinal bound to a similar peptide (Choe et al. 2011). Investigators have also successfully probed the dynamics of GPCRs during the activation process using hydrogen-deuterium exchange (Lodowski et al. 2010, Orban et al. 2012b, West et al. 2011) and long-time scale computational simulations (Dror et al. 2011).
Conformational changes on the cytoplasmic side of the β2-adrenergic receptor following binding of the agonist BI-167107 can be described by the rigid body movement of TM-VI. This alteration of the TM-VI position probably results from the disrupted hydrogen-bonding network within the core of this GPCR, as shown in a detailed snapshot of this region in rhodopsin. The hydrogen-bonding network starting in the immediate vicinity of the ECL region maps continuously throughout the entire core of the protein all the way to the ICL. This network involves internal water molecules and amino acid residues such as Asn302, Tyr306, and even the side chain of Pro303 in one instance (Figure 3a). The network’s stability was perturbed after activation of rhodopsin and binding of Gt (Orban et al. 2012b). However, whether disruption of the hydrogen-bonding network is the cause or the effect or works in synergy with the rigid body movement of the TM helices during agonist binding/receptor activation remains uncertain.
Structure of Light-Activated Rhodopsin in Membranes
Park and colleagues (Park et al. 2008b) proposed that GPCR action can be analogous in some respects to regulatory enzymes where the ligand-binding site is discrete from the active site (Changeux & Edelstein 2005). Transmission of the signal from the ligand-binding site of a GPCR to the nucleotide-binding site in the G protein is proposed to occur by relaxing the former, allowing transient association between the two proteins. The energy of their binding then is used to open the nucleotide-binding site and release guanosine diphosphate (GDP) from the G protein, whereas the subsequent binding of guanosine-5′-triphosphate (GTP) provides the energy to break this complex and release the G protein.
Activated rhodopsin can form a stable complex with Gt when the guanyl nucleotide-binding site in the G protein α-subunit is unoccupied (Bornancin et al. 1989). This finding was expanded in more recent studies (Jastrzebska et al. 2006, 2011a), showing that communication between the chromophore and the nucleotide-binding site on a G protein can be uncoupled once the complex is formed. The chromophore can be added in either all-trans- or 11-cis-configurations, but dissociation of the G protein restores the receptor back to the inactive intermediate (Jastrzebska et al. 2009).
As noted above, changes in the ligand-binding site are transmitted to the GPCR’s cytoplasmic surface. But only the cytoplasmic surface is recognized by the G protein to initiate signal transduction (Figure 3b). The deprotonated photoactivated form of rhodopsin was successfully crystallized by Salom and colleagues (2006). Insights into rhodopsin activation were obtained from studies of opsin at low pH that was assumed to have achieved an activated conformation (Park et al. 2008a, Scheerer et al. 2008), even though its catalytic properties were known to be extremely low compared with those of photoactivated rhodopsin (Buczylko et al. 1996, Jäger et al. 1996). Opsin crystals can bind all-trans-retinal without significant conformational changes (Choe et al. 2011, Scheerer et al. 2008) and can also bind the Gtα C-terminal peptide (Choe et al. 2011) (Figure 3c,d). The 2–5 Å changes in helix V at the cytoplasmic surface end are needed to dock G protein because, in rhodopsin, this helix blocks full opening of the binding site. This simplistic docking of just the Gtα C-terminal peptide cannot be the only binding site of Gt to rhodopsin. Gt binds to dark-state rhodopsin prior to photoactivation (Hamm et al. 1987), suggesting that this nonproductive complex does not achieve the necessary conformational changes to allow nucleotide exchange.
Oligomeric Forms of GPCRs
In biochemical assays, largely of detergent-solubilized preparations, either monomeric or multimeric forms of GPCRs can activate their cognate G-proteins (Jastrzebska et al. 2006), but they assemble into homo- and heterodimers or larger oligomers in native tissues (Palczewski 2010, Park et al. 2004, Prezeau et al. 2010, Terrillon & Bouvier 2004). In the dimeric form, GPCR monomers are not functionally equivalent because only one monomer will bind to the C-terminal helix of the cognate G protein, and ligand affinities could differ between the monomer and the dimer (Fuxe et al. 2012, Jastrzebska et al. 2013, Maurice et al. 2011). Cholesterol and palmitoyl moieties present on many GPCR C-terminal regions could play a critical role in forming the dimer interface (Maeda et al. 2010, Oates & Watts 2011, Thompson et al. 2011, Zheng et al. 2012). Moreover, each monomeric unit, when occupied by a ligand, does not transmit an equivalent signal to its cognate G protein. The efficiency of activation can depend on whether one or two ligands are present in the dimer (Pellissier et al. 2011). A mathematical model was recently created to reflect this asymmetric organization and its impact on different signaling pathways (Rovira et al. 2010). On the basis of theoretical considerations, Neri et al. (2010) proposed that one monomer of rhodopsin is responsible for light detection, whereas the other serves as the G protein activator. Moreover, GPCR homodimer activity can be controlled by an allosteric ligand-binding site when different ligands bind to each receptor monomer (Zylbergold & Hébert 2009).
Visualization of GPCRs in native tissue is a key to understanding the physiological settings of these receptors. Atomic force microscopy (AFM) of rhodopsin in native rod disc membranes provides the clearest structural picture to date of the oligomeric arrangement of a GPCR (Fotiadis et al. 2003, Liang et al. 2003) (see sidebar, Imaging GPCRs in Native Settings). AFM measurements yielded a density of 30,000–55,000 rhodopsin molecules/μm2 and ~108 rhodopsin molecules per rod, partially organized in para-crystalline arrays composed of rows of rhodopsin (Fotiadis et al. 2003, Liang et al. 2003). These rows of rhodopsin dimers were used to derive spatial constraints that were used to construct a molecular model of the oligomeric structure of rhodopsin in the rod outer segment (ROS) membrane (Fotiadis et al. 2004). This model indicates that the rhodopsin dimer offers a platform complementary to the binding of a single Gt or arrestin molecule (Figure 5).
Figure 5.
Hypothetical model for assembly on a cell surface of major proteins required for effective visual signal transduction. Visual signal transduction is carried out by a multitude of proteins and messengers. (Top left) Structures of proteins involved in phototransduction. (Bottom left) A phospholipid cell membrane represented as a gray transparent layer containing a dimer and monomer of rhodopsin (red ). Various complexes of these proteins assembled on the membranes are modeled on the right. Following activation, heterotrimeric Gt ( pink) is recruited to the cytoplasmic cell surface where it binds to photoactivated rhodopsin, forming the photoactivated rhodopsin-Gt complex. Gt-bound guanosine diphosphate (GDP) is then exchanged for guanosine-5′-triphosphate (GTP). Next, phosphodiesterase 6 holoenzyme (PDE6, green) is recruited to the membrane and carries out hydrolysis of cyclic guanosine monophosphate (cGMP). The activity of Gt is, in turn, suppressed by RGS9 (orange), which promotes its inactivation. Finally, arrestin ( yellow) and GRK1 (blue) both help deactivate photoactivated rhodopsin, a step required for resetting the dark state of rhodopsin prior to another light stimulus.
IMAGING GPCRs IN NATIVE SETTINGS.
High-resolution methods are needed for two types of GPCR imaging. First is the static self-clustering and complexing of GPCRs with other proteins in native membranes. The second is real-time imaging to understand and quantify GPCR dynamics in response to cellular changes. For static imaging, atomic force microscopy, used to resolve metal orbitals on hard surfaces, has provided the highest resolution images of several membrane-embedded or reconstituted proteins in various activated states (Engel & Gaub 2008, Müller et al. 2008). Near-field scanning optical microscopy, fluorescence correlation spectroscopy, and other advanced confocal and 2-photon microscopy techniques also permit static visualization of GPCRs in membranes (Herrick-Davis et al. 2012, Ianoul et al. 2005). Dynamic changes of GPCRs are monitored predominantly by fluorescence methods. Resonance energy transfer methods together with complementation and transactivation of functional receptors provide evidence for receptor colocalization and oligomerization. These methods are also used for imaging applications (Harrison & van der Graaf 2006). Quantum dot (QD) probes can monitor movement of a single GPCR during its life cycle (Fichter et al. 2010) or follow a ligand that binds tightly to these receptors (Zhou et al. 2007).
Only limited information is available about the dynamics and stability of GPCR dimers (Lambert 2010). To this end, development of a single fluorescent-molecule imaging method allowed investigators to observe a dynamic equilibrium for the N-formyl peptide receptor in live cells, indicating that dimers rapidly fall apart and reassemble (Kasai et al. 2011). It appears that the M3 muscarinic receptor also exists in multiple dimeric/oligomeric arrangements (McMillin et al. 2011). GPCR oligomerization could be responsible for fine-tuning receptor function and signaling (Tadagaki et al. 2012).
Great advances were made using noninvasive fluorescence- and luminescence-based techniques to observe bimolecular fluorescence complementation that can result from binding fluorescent protein fragments to nonfluorescent proteins. This innovative approach allowed researchers to visualize and measure protein interactions in neuronal cells (Ciruela et al. 2010, Vidi et al. 2010). Adapted to native tissues, the same strategy also demonstrated the presence of oxytocin receptor dimers and/or oligomers in the mammary gland (Albizu et al. 2010). An elegant application of fluorescence correlation spectroscopy with photon counting from histograms led to the recent conclusion that GPCR dimers represent the basic signaling unit (Herrick-Davis et al. 2006). Some other evidence for the functional significance of heterodimerization, such as variants of the serotonin receptor or the metabotropic glutamate receptor, is questionable on the basis of pharmacological assays using the heterologous expression system (Delille et al. 2012) (see sidebar, Heterologous Expression of GPCRs). Effects of oligomerization itself must be distinguished from the integration of inputs from different receptors. Data derived from native tissues in a physiologically relevant setting are most critical. For example, Park et al. (2012) demonstrated a functional heterodimer for monomeric GPCRs known as DAF-37 and Daf-38 in live C. elegans (Park et al. 2012).
With such a large number of GPCRs, some mechanisms of oligomerization and signaling will apply to some receptors, whereas others will not. Oligomerization or parallel signaling could have dramatic effects on medically relevant treatments. For disorders such as schizophrenia and dementia, metabotropic glutamate 2 receptor/serotonin 2A receptor–mediated changes in Gi and Gq activity could affect the psychoactive behavioral effects of a variety of pharmacological compounds (Fribourg et al. 2011).
Complexes Between GPCRs and G Proteins
The only available structural model for the photoactivated rhodopsin-Gt complex is a recently described low-resolution electron microscopic reconstruction by single particle analysis (Jastrzebska et al. 2011b). After purifying and analyzing photoactivated rhodopsin and Gt, investigators determined a 2:1 molar ratio of photoactivated rhodopsin to Gt and calculated a 22 Å structure from projections of the negatively stained complexes. The molecular envelope accommodated two rhodopsin molecules together with one Gt heterotrimer, consistent with a heteropentameric structure for this complex (Jastrzebska et al. 2011b) (Figure 4a).
HETEROLOGOUS EXPRESSION OF GPCRs.
Most GPCRs are expressed at such low levels in native tissues that investigators must overexpress them to investigate their structure and function. Rhodopsin constitutes a notable exception because it is expressed at 1 mg per bovine retina and 5 mM in rod outer segments (Palczewski 2012). GPCRs in taste buds and olfactory receptor neurons are also expressed at high concentrations, but receptors in these tissues are scarce (DeMaria & Ngai 2010), making purification difficult. For these reasons, we developed an in vivo system to express heterologous GPCRs in rod photoreceptors of Xenopus (Zhang et al. 2005) and mice (Li et al. 2007, Salom et al. 2008). More recently, researchers achieved overexpression of heterologous GPCRs in C. elegans (Cao et al. 2012, Salom et al. 2012). However, simpler cultured insect (Schneider & Seifert 2010) and mammalian cells (Li et al. 2005) are most often employed for expressing heterologous GPCRs because they can perform the appropriate complex posttranslational modifications and their membranes mimic native membranes. GPCRs have been successfully expressed in yeast and cell-free systems, whereas expression in bacteria typically requires refolding (Lundstrom 2006, McCusker et al. 2007). To be useful, heterologously expressed GPCRs must also display the same pharmacology as native receptors.
Another breakthrough in the understanding of how an activated GPCR binds to G proteins came with the recent crystal structure of an active state complex composed of (a) agonist-occupied monomeric β2-adrenergic receptor-T4L-lyzosome fusion, (b) nucleotide-free Gs heterotrimer, and (c) a nanobody (Rasmussen et al. 2011) (Figure 4a). In this complex, the cytoplasmic end of TM-VI and an α-helical extension of the cytoplasmic end of TM-V were displaced by several Å compared with the inactive receptor. As predicted from biochemical studies (Oldham & Hamm 2008), the amino- and carboxy-terminal α-helices of the G protein exhibited conformational changes that propagated to its nucleotide-binding pocket. It was surprising that the GPCR was in a monomeric state because single-particle analyses of a similar preparation indicated that the receptor exists to some degree in a dimeric form (Westfield et al. 2011). But the most unexpected observation was a major displacement of the entire α-helical (AH) domain of Gα relative to the Ras-like GTPase domain (Rasmussen et al. 2011).
Recent models of photoactivated rhodopsin-Gt and β2-adrenergic receptor-Gs complexes provide important details about the interaction of a GPCR molecule with its corresponding G protein. Both photoactivated rhodopsin and the β2-adrenergic receptor interact with their corresponding G proteins through the C-terminal end of the α-domain (Figure 4a). Superposition of the Gt and Gs structures reveals a relatively small root-mean-square deviation (~0.5–1.0 Å ) for the β and γ subunits and the Ras subdomains of the α-domains. However, the positions of the AH subdomains of the α-domains differ markedly in Gt and Gs. This difference is due to the rigid body–type displacement of these subdomains (Figure 4a). (The averaged angle between these subdomains was ~130°, whereas the distance between the AH subdomains of Gt and Gs was found to be ~40 Å .) For the two domains to superpose, the Gsα-AH subdomain alone requires a full ~180° rotation (Figure 4a). This displacement could result from the nanobody that is stabilizing the β2-adrenergic receptor-Gs complex in the crystal.
As do rhodopsin and Gt, the M3 muscarinic receptor forms inactive-state complexes with Gq heterotrimers in intact cells (Qin et al. 2011). Using ~250 combinations of cysteine-substituted M3 muscarinic receptors and Gαq proteins, investigators tested which of these mutants undergo cross-link formation. This strategy identified M3 muscarinic receptor–Gαq contact sites that help build low- and high-resolution models of the complex (Hu et al. 2010).
In summary, allosteric communication between the ligand-binding site in photoactivated rhodopsin or β2-adrenergic receptors and the nucleotide-binding site of a G protein (Gs or Gt) ata distance of ~50–60 Å is achieved by precise conformational changes that include helical movements and a reorganization of internal water molecules. The complex between the GPCR and G protein suggests that binding of a G protein confers a stabilizing effect on the receptor whose rigidity resembles that of ground-state rhodopsin (Figure 4b). This observation sharply contrasts with the highly dynamic structure of photoactivated rhodopsin, which forms a broad constellation of conformers.
Other Proteins Interacting with GPCRs
GPCRs interact with receptor kinases, arrestins, and a variety of other proteins, in addition to their G proteins. Together they form large functional complexes that directly mediate receptor signaling or play key roles in trafficking and cellular localization. These multipart protein assemblages could influence pharmacological properties of these receptors (Ritter & Hall 2009).
Substrates favoring the activated ghrelin state initiated by agonist-induced arrestin recruitment cause ghrelin to adopt a different conformation from that observed in the absence of arrestin (Mary et al. 2012). Thus, G proteins affect the balance between active and inactive ghrelin receptors. Another study also showed that the ghrelin conformation stabilized by a G protein/agonist is distinct from that stabilized by arrestin-based agonists (Rahmeh et al. 2012). Molecular understanding of the interplay between GPCRs and their kinases has advanced significantly with more precise elucidation of receptor recognition and phospholipid binding that leads to kinase activation (Boguth et al. 2010). Huang & Tesmer (2011) have proposed that the interaction of receptor kinases and arrestins, despite differences in their folding, occurs via a common molecular mechanism. In the presence of ATP, the influence of photoactivated rhodopsin versus ground-state rhodopsin on rhodopsin kinase dynamics was negligible (Orban et al. 2012a). Few experimental techniques can assess the orientation of peripheral membrane proteins in their native environment. Sum-frequency generation vibrational spectroscopy was used to determine the membrane orientation of the GPCR kinase 2-G-βγ complex (Boughton et al. 2011). Rhodopsin kinase phosphorylates rhodopsin at different subsets of Ser and Thr residues at the C-terminal tail, producing phosphorylated receptor variants with divergent functional properties (Maeda et al. 2003). Moreover, researchers noted a similar phenomenon for the β2-adrenergic receptor in cell cultures (Liggett 2011, Nobles et al. 2011). Phosphorylation of GPCRs and the subsequent capping of these receptors by arrestins are essential because recent studies show that internalized GPCRs can continue either to stimulate or to inhibit sustained cAMP production by remaining associated with their cognate G protein subunits and adenylyl cyclase in endosomal compartments. Once internalized, these GPCRs can produce cellular responses distinct from those elicited at the cell surface (Jalink & Moolenaar 2010).
Large complexes with other proteins are also necessary to target GPCRs to subcellular compartments and their trafficking to and from the plasma membrane during internalization (Bockaert et al. 2010). For example, activation of Arf6 leads to β2-adrenergic receptor degradation and accumulation and also negatively controls Rab4-dependent fast recycling to prevent the resensitization of β2-adrenergic receptors (Macia et al. 2012). Another important physiological example is the interaction of GPCRs with voltage-gated calcium channels that can synergize neurotransmitter release (Altier & Zamponi 2011). In addition to Ca2+ influx and direct interaction with Ca2+-binding proteins (Navarro et al. 2012), rapid depolarization-induced charge movement in GPCRs is needed for neurotransmitter control (Kupchik et al. 2011). Opioid receptors form complexes with transient receptor potential (TRP) Ca2+ channels as well (Yekkirala 2013).
CELLULAR CONTEXT OF GPCR SIGNALING
Structural studies of GPCRs and other integral membrane proteins, such as those involving X-ray crystallography or nuclear magnetic resonance, require high concentrations of well-purified membrane proteins in detergent solution. These artificial conditions obviously limit the relevance of these studies to GPCR signaling in vivo, where these transmembrane proteins interact mostly with other integral membrane proteins and membrane-associated proteins. Although high-resolution methods are required to obtain atomic details of biochemical molecules, the resulting structures must be pieced together in both structural and functional contexts (Figure 5).
Rod Photoreceptor Cells
The visual system and its signaling pathways currently provide a good molecular framework—a GPCR signaling system with physiological relevance. Over the years, many methods were developed to investigate the visual system. It is the only system in which we can directly access the intact tissue and monitor its signal in vivo (Figure 6). In addition to the neural retina, the retinal pigment epithelium (RPE) layer is critically important in the eye. The RPE is responsible for (a) movement of metabolites between the choroidocapillaris and avascular photoreceptor cell layers; (b) metabolic transformation of spent all-trans-retinal back to its 11-cis-retinal light-sensitive form by a process called the visual (or retinoid) cycle; (c) daily phagocytosis of ~10% of photoreceptor outer segments; and (d ) other homeostatic functions. Photoreceptor outer segments are critical for phototransduction, namely transformation of single photons of light into a biochemical cascade of events that culminate in neurotransmission. Mouse ROS lengths were estimated to be ~24 μm with a diameter of ~1.2 μm (Liang et al. 2004). Particularly fascinating are structural studies of internal structures called disc membranes. For example, a mouse ROS contains ~800 membranous discs stacked on top of each other, increasing the total membrane surface area of the cell ~1,500-fold (Mayhew & Astle 1997). The cytoplasmic space used for phototransduction represents only ~30% of the space inside a ROS [see a recent review for a summary of this structure (Palczewski 2012)].
Figure 6.
High-resolution images and pictorial representation of mammalian photoreceptors and the retinal pigment epithelium (RPE). (a–d ) Fluorescence images obtained by two-photon excitation of endogenous fluorophores. Scale bars represent 20 μm. (a) Double-nucleated RPE cells obtained from an ex vivo unfixed C57BL/6J-TyrC-2J mouse eye. Highly fluorescent retinosomes are visible as bright spots located along the plasma membranes. (b) A transverse (xz) image of cones and rods in cynomolgus monkey peripheral retina assembled from a series of en face images. Ellipsoids of two cones directly on the xz axis are indicated by yellow arrows. (c) An en face (xy) image of the rod and cone outer segment mosaic. Outer segments of two cones are indicated with yellow arrows. (d ) En face (xy) image of the rod and cone inner segment mosaic. Two-photon images courtesy of G. Palczewska. (e) Pictorial representation of the RPE-photoreceptor interface. Melanosomes ( purple) are located in the RPE cell and processes. Other elements: lipofuscin (smaller dark red circles), phagosomes (larger blue circles), cone photoreceptor cell ( green), and rod cell (blue). The dashed line represents the external limiting membrane separating the nuclei and the ellipsoid with its abundant mitochondria. ( f–i ) Electron microscopic images. ( f ) Image of the RPE and a photoreceptor cell in extrafoveal rhesus monkey retina (from Anderson et al. 1980). ( g) The tip of the outer segment of a foveal cone engulfed by RPE apical processes in rhesus monkey retina (from Anderson & Fisher 1979). Some distal discs appear separate from the cell membrane. (h) The cone outer segment base from a rhesus monkey retina (from Anderson et al. 1978). Black arrows indicate a new evagination of the membrane. (i ) The nucleus of a Müller cell and the external limiting membrane. Müller cell villous processes extend beyond the external limiting membrane between rods and cones. ( j ) The cytotoxic effect of all-trans-retinal in light-induced photoreceptor degeneration. The illustrated signaling cascade implicates GPCRs, PLC/IP3/Ca2+ signaling, and NADPH oxidase in this process. Elevated signaling of Gq-coupled GPCRs is involved in mediating all-trans-retinal toxicity during light-induced photoreceptor degeneration, but the precise mechanism has yet to be clarified (black arrow with dotted line). Activation of Gq-coupled GPCRs stimulates PLC/IP3/Ca2+ pathways, which then lead to NADPH oxidase-mediated ROS production and photoreceptor degeneration (black arrows). Pharmacological interventions targeting Gq-coupled GPCRs, PLC/IP3/Ca2+, and NADPH oxidase protect photoreceptors from light-induced, all-trans-retinal-mediated degeneration (red bars). Modified illustration based on Chen et al. (2012, figure 8). APO/DPI, Apocynin/diphenylleneiodonium; ER, endoplasmic reticulum; NADPH, nicotinamide adenine dinucleotide phosphate; PLC, Phospholipase-C; ROS, reactive oxygen species
The main protein of ROS internal discs is rhodopsin, which constitutes ~50% of the membrane volume (Filipek et al. 2003a). In addition to being a photon catcher, rhodopsin is also a critical structural protein that establishes and preserves ROS morphology. The size of ROS discs is dictated by the level of rhodopsin expression (Liang et al. 2004), and absence of rhodopsin prevents ROS formation (Humphries et al. 1997).
Network Pharmacology in the Retina
The retinal transcriptome has now been analyzed, and most of the ROS proteome components have been identified by mass spectrometry (e.g., Wong et al. 2009). The retina contains a large number of GPCRs, including the visual pigments rhodopsin, short-wavelength cone opsin, melanopsin, and other receptors (Table 1). However, the intersections of multiple signaling cascades and the precise interplay between them are presently unknown. Localization of different GPCRs within the retina, their associated physiological responses, and genetic alterations in their expression will provide clues about their functions and the interdependence of these functions. These GPCRs also provide targets for therapeutics that can be used to prevent or ameliorate retinal degeneration.
Table 1.
Listed are genes detected in the C57BL/6J mouse eye with an average fragments per kilobase of exon per million fragments mapped (FPKM) value of 2 or greater that are categorized as GPCRs by gene ontology (see Mustafi et al. 2011 for details)
| Gene | B6 Eye Average FPKM |
Gene | B6 Eye Average FPKM |
Gene | B6 Eye Average FPKM |
|---|---|---|---|---|---|
| Rho | 6162.04 | Adra2c | 8.96 | Gpr158 | 3.72 |
| Rgr | 355.74 | Gpr146 | 8.91 | Tacr1 | 3.72 |
| Opn1sw | 125.13 | Vipr2 | 8.79 | Fzd8 | 3.56 |
| Drd4 | 93.84 | Fzd5 | 8.69 | Opn4 | 3.35 |
| Opn1mw | 62.97 | Gpr110 | 8.59 | Tshr | 3.24 |
| Nisch | 52.38 | Adrb1 | 8.43 | S1pr2 | 3.2 |
| Gprc5b | 29.82 | S1pr3 | 8.42 | Mrgprf | 3.18 |
| Gpr162 | 29.37 | Gabbr2 | 7.8 | Oprl1 | 3.15 |
| Darc | 28.91 | Lphn2 | 7.66 | F2rl1 | 3.13 |
| Gpr37 | 28.47 | Lpar1 | 7.47 | S1pr5 | 3.12 |
| Ednrb | 22.27 | P2ry2 | 7.2 | Gpr135 | 3.07 |
| Crcp | 21.88 | Adrb2 | 7.13 | Crhr1 | 3.02 |
| Gpr153 | 20.42 | Hrh3 | 7.11 | Eltd1 | 3.0 |
| Gabbr1 | 19.78 | Gpr143 | 6.8 | Mrgpre | 2.96 |
| Rrh | 19.29 | Celsr2 | 6.53 | Gpr27 | 2.92 |
| Gpr152 | 18.55 | Fzd7 | 6.34 | Ednra | 2.87 |
| Adora1 | 16.2 | Drd1a | 6.15 | Opn3 | 2.55 |
| Lphn1 | 15.98 | Adora2b | 6.09 | Gpr165 | 2.45 |
| Cxcr7 | 14.3 | Celsr3 | 5.82 | Cnr1 | 2.41 |
| Cd97 | 12.93 | Fzd4 | 5.39 | Gpr37l1 | 2.39 |
| Gpr19 | 12.21 | Gprc5c | 5.26 | Crhr2 | 2.24 |
| Fzd1 | 11.99 | Gpr179 | 5.12 | P2ry14 | 2.23 |
| Fzd6 | 11.34 | Gpr56 | 5.12 | Gpr176 | 2.21 |
| Gpr87 | 11.34 | Tacr3 | 4.95 | Celsr1 | 2.18 |
| Lgr4 | 11.09 | Ramp1 | 4.68 | Gpr22 | 2.17 |
| Drd2 | 10.82 | Adra2a | 4.6 | Lgr5 | 2.1 |
| Smo | 10.75 | Gpr85 | 4.56 | Gpr26 | 2.06 |
| S1pr1 | 10.66 | Lphn3 | 4.22 | Agtr2 | 2.02 |
| Glp2r | 9.94 | Htr3a | 4.14 | Calcrl | 2.02 |
| Ptger1 | 9.59 | Fzd2 | 3.89 | Gpr68 | 2.02 |
| Gpr124 | 9.56 | Fzd10 | 3.86 | Cckbr | 2.0 |
| F2r | 9.31 | Gpr98 | 3.79 |
Interrelated oxidative stress and Ca2+ imbalance both contribute to the pathology of retinal degeneration (Chen et al. 2012). NADPH oxidase inhibitors and PLC/IP3/Ca2+ signaling antagonists similarly protect retinas from light-mediated degeneration, suggesting that these activities are involved in the same signaling pathway (Figure 6j). Indeed, one could imagine a single signaling cascade/pathway for light-induced retinal degeneration involving GPCRs, PLC/IP3/Ca2+ signaling, and NADPH oxidase (Figure 6j). Pharmacological interventions targeting specific reactions in this pathway could provide novel therapeutic strategies for treating blinding retinal disorders such as Stargardt’s disease and age-related macular degeneration (Chen et al. 2012, Maeda et al. 2012). The serotonin 2A receptor is an excellent candidate for activating PLC, although data regarding its involvement in light-induced retinal degeneration are limited. Serotonin 2A receptor expression is readily detectible in the retina, and its activation leads mainly to elevations in cytosolic Ca2+ through PLC activation (Hoyer et al. 1994). This intersection of signaling pathways could be targeted to correct or prevent aberrant signaling leading to neurodegeneration.
CONCLUDING REMARKS
GPCRs have evolved to play an essential role in both sensory and endocrine control systems. The complexity of GPCR signaling continues to be revealed as investigators use new-generation high-throughput sequencing to identify the full repertoire of GPCRs in many organisms. Research has made steady progress to elucidate further the role of GPCRs in normal and pathological states. The past few years have provided much more detailed information about the assembly of GPCRs at the atomic level and their mode of binding to many antagonists and some agonists. This improved level of detail has led to testable hypotheses about GPCR activation. More than 50% of all publications in the past three years that discuss GPCRs explore the exciting possibility of expanding the GPCR pool by homo- and hetero-oligomerization (Park & Palczewski 2005). The next frontier in our understanding of GPCR signaling should uncover additional GPCR structures and their partner proteins. In addition, new research must further define the cellular context of these various interacting complexes, such as neurons, that pertain to the indistinct function and physiology and contribute to the physiology of the whole organism. Pharmacological manipulation of G protein signaling continues to be extremely important in defining many interacting signaling pathways. Most GPCRs are not essential to establish life, but they are critical for sustaining it. The next decade should witness exciting research breakthroughs in the GPCR field both in generation of new and improved structures and in our interpretation of these structures and their implications for signaling. Even now, we can elucidate the genetics of complex G protein–signaling pathways between different individuals of the same species owing to next-generation DNA sequencing. A rich array of experimental approaches and techniques are currently available; thus, various combinations of biochemical and structural methods should greatly enhance our understanding of GPCR structure/function and its applications to modern medicine.
SUMMARY POINTS.
GPCRs exist in many different structural forms to accommodate a broad range of external stimuli. Interest in their function and structure has been at the center of biological science for decades.
The past 15 years have witnessed tremendous advances in understanding of GPCR structures and those of a subset of complexes as determined by high-resolution methods such as electron microscopy, X-ray crystallography, and nuclear magnetic resonance. These studies show that highly divergent sequences yield GPCRs with similar overall topology but with high specificity in ligand binding. The mechanism of GPCR activation and GPCR interactions with partner proteins are likely to be highly evolutionally preserved with ligand and cognate G protein specificities among subgroups of receptors.
GPCRs are highly dynamic molecules, and water has emerged as an important element in their assembly and activation. Internal water molecules provide the elasticity essential for the ligand-binding signal to be transmitted 40 Å through the cell membrane to the cytoplasmic surface to enable interactions with G proteins and other partner proteins. This explanation is compatible with the multiple conformational changes elicited by different ligands and the thermodynamics governing the activation process.
Structural information derived from high-resolution methods must be aligned with physiological information about assemblies of GPCRs within cells such as neurons to avoid misinterpretation of potential artifacts arising from the study of highly purified, detergent-solubilized systems.
GPCRs were shown to exist in oligomeric forms that are critically important for their stability, signaling, and other cell-biological processes. Specific allosteric regulators (ligands of the allosteric-binding sites) could also target a specific subset of these structures. Because different signaling pathways intersect within a given cell, cultures of specific cell types can be used to identify pathology and devise preventive therapies that target GPCRs.
FUTURE ISSUES.
Determine high-resolution structures of native GPCRs and receptor–G protein and receptor-arrestin complexes with all relevant posttranslational modifications.
Elucidate the precise rearrangements of water molecules during GPCR activation.
Understand the structural complementarities of GPCR homo- and hetero-oligomerization.
Determine the intersections of different relevant GPCR signaling pathways in tissues of interest.
Obtain high-resolution structures of signaling complexes in native tissues. Determine how GPCR signaling complexes are compartmentalized within cells.
Understand the energy landscape of GPCR activation and folding. Identify key residues for folding and membrane insertion.
Use next-generation DNA and RNA sequencing to identify the impact of mutations and polymorphisms of GPCR expression levels.
ACKNOWLEDGMENTS
We thank Drs. Leslie T. Webster, Jr., Phoebe L. Stewart, David Salom, David Lodowski, Paul Park, Philip Kiser, and members of Dr. Palczewski’s laboratory for helpful comments on the manuscript. We also express our gratitude to Grazyna Palczewska for generating Figure 6a–i, Dr. Ray Stevens for generating Figure 1, and Debarshi Mustafi for data presented in Table 1. This work was supported by National Institutes of Health grants EY008061 and EY019478.
Glossary
- GPCR
G protein–coupled receptor
- 7-TM
7-transmembrane
- Rhodopsin
the red-colored GPCR composed of an opsin covalently linked to 11-cis-retinal. Responsible for dim-light vision
- Heterotrimeric G proteins
consist of three polypeptides: a GPCR-binding α-subunit with GTP to GDP hydrolytic activity and tightly bound βγ-subunits
- Transmembrane domain
any segment of a protein that passes completely through a cell membrane
- ECL
extracellular loop
- ICL
intracellular loop
- Orthosteric ligand-binding region
the primary ligand-binding site in a GPCR that is not subtype-selective for endogenous agonists
- GPCR ligands
agonists, antagonists/inverse agonists, or neutral ligands that bind to orthosteric GPCR sites, often causing GPCR activation or inhibition
- Gt (transducin)
the heterotrimeric G protein localized to the outer segment of rod photoreceptors that couples specifically to rhodopsin
- Bound water molecules
integral components of a biological macromolecule that do not readily exchange with external water and can act as cofactors
- Membrane proteins
water-insoluble proteins that reside at least partially in lipid membrane bilayers
- All-trans-retinal
vitamin A (retinol) aldehyde is metabolically produced from β, β-carotene but also results from photoisomerization of the visual pigment chromophore, 11-cis-retinal
- Hydrogen-deuterium exchange
a chemical labeling technique used to track dynamic changes in protein structural conformation
- Allosteric ligand-binding site
a separate ligand-binding site on a receptor that either positively or negatively modulates the receptor’s affinity for its primary ligand(s)
- Atomic force microscopy (AFM)
a high-resolution technique for imaging flat materials by probing them with a contacting cantilever
- Rod disc membranes
Internal membranes shaped like flattened sacks that occupy the outer segments of rod photoreceptor cells
- AH
α-helical domain
- Retinal pigment epithelium (RPE)
a cell layer just outside the neurosensory retina critical for metabolite transport and photoreceptor cell maintenance
- Oxidative stress
a condition that results when the production rate exceeds the destruction rate of toxic reactive oxygen intermediates
- NADPH oxidases
enzymes catalyzing formation of reactive oxygen species implicated in redox signaling. These constitute important therapeutic targets
- Age-related macular degeneration
includes both geographic atrophic (dry) and neovascular (wet) types of age-related degeneration of retinal photoreceptor cells with progressive visual loss
- Phospholipase C (PLC)
an enzyme that hydrolyzes phospholipids before the phosphate group
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Albizu L, Cottet M, Kralikova M, Stoev S, Seyer R, et al. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat. Chem. Biol. 2010;6:587–94. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altier C, Zamponi GW. Analysis of GPCR/ion channel interactions. Methods Mol. Biol. 2011;756:215–25. doi: 10.1007/978-1-61779-160-4_11. [DOI] [PubMed] [Google Scholar]
- Anderson DH, Fisher SK. The relationship of primate foveal cones to the pigment epithelium. J. Ultrastruct. Res. 1979;67:23–32. doi: 10.1016/s0022-5320(79)80014-3. [DOI] [PubMed] [Google Scholar]
- Anderson DH, Fisher SK, Erickson PA, Tabor GA. Rod and cone disc shedding in the rhesus monkey retina: a quantitative study. Exp. Eye Res. 1980;30:559–74. doi: 10.1016/0014-4835(80)90040-8. [DOI] [PubMed] [Google Scholar]
- Anderson DH, Fisher SK, Steinberg RH. Mammalian cones: disc shedding, phagocytosis, and renewal. Investig. Ophthalmol. Vis. Sci. 1978;17:117–33. [PubMed] [Google Scholar]
- Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc. Natl. Acad. Sci. USA. 2009a;106:8555–60. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel TE, Gupta S, Jastrzebska B, Palczewski K, Chance MR. Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc. Natl. Acad. Sci. USA. 2009b;106:14367–72. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert J, Perroy J, Bécamel C, Marin P, Fagni L. GPCR interacting proteins (GIPs) in the nervous system: roles in physiology and pathologies. Annu. Rev. Pharmacol. Toxicol. 2010;50:89–109. doi: 10.1146/annurev.pharmtox.010909.105705. [DOI] [PubMed] [Google Scholar]
- Boguth CA, Singh P, Huang CC, Tesmer JJG. Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 2010;29:3249–59. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornancin F, Pfister C, Chabre M. The transitory complex between photoexcited rhodopsin and transducin. Reciprocal interaction between the retinal site in rhodopsin and the nucleotide site in transducin. Eur. J. Biochem. 1989;184:687–98. doi: 10.1111/j.1432-1033.1989.tb15068.x. [DOI] [PubMed] [Google Scholar]
- Boughton AP, Yang P, Tesmer VM, Ding B, Tesmer JJG, Chen Z. Heterotrimeric G protein β1γ2 subunits change orientation upon complex formation with G protein-coupled receptor kinase 2 (GRK2) on a model membrane. Proc. Natl. Acad. Sci. USA. 2011;108:E667, 73. doi: 10.1073/pnas.1108236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczylko J, Saari JC, Crouch RK, Palczewski K. Mechanisms of opsin activation. J. Biol. Chem. 1996;271:20621–30. doi: 10.1074/jbc.271.34.20621. [DOI] [PubMed] [Google Scholar]
- Cao P, Sun W, Kramp K, Zheng M, Salom D, et al. Light-sensitive coupling of rhodopsin and melanopsin to G(i/o) and G(q) signal transduction in Caenorhabditis elegans. FASEB J. 2012;26:480–91. doi: 10.1096/fj.11-197798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–28. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- Chen Y, Okano K, Maeda T, Chauhan V, Golczak M, et al. Mechanism of all-trans-retinal toxicity with implications for Stargardt disease and age-related macular degeneration. J. Biol. Chem. 2012;287:5059–69. doi: 10.1074/jbc.M111.315432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–55. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–76. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, et al. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–15. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruela F, Vilardaga JP, Fernández-Dueñas V. Lighting up multiprotein complexes: lessons from GPCR oligomerization. Trends Biotechnol. 2010;28:407–15. doi: 10.1016/j.tibtech.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PW, Rasmussen SG, Parnot C, Fung JJ, Masood A, et al. A monoclonal antibody for G protein–coupled receptor crystallography. Nat. Methods. 2007;4:927–29. doi: 10.1038/nmeth1112. [DOI] [PubMed] [Google Scholar]
- Delille HK, Becker JM, Burkhardt S, Bleher B, Terstappen GC, et al. Heterocomplex formation of 5-HT2A-mGlu2 and its relevance for cellular signaling cascades. Neuropharmacology. 2012;62:2184–91. doi: 10.1016/j.neuropharm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- DeMaria S, Ngai J. The cell biology of smell. J. Cell Biol. 2010;191:443–52. doi: 10.1083/jcb.201008163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl. Acad. Sci. USA. 2012;109:119–24. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré AS, Robertson N, Errey JC, Ng I, Hollenstein K, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–93. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doze VA, Perez DM. G-protein-coupled receptors in adult neurogenesis. Pharmacol. Rev. 2012;64:645–75. doi: 10.1124/pr.111.004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, et al. Activation mechanism of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA. 2011;108:18684–89. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu. Rev. Biochem. 2008;77:127–48. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- Fichter KM, Flajolet M, Greengard P, Vu TQ. Kinetics of G-protein-coupled receptor endosomal trafficking pathways revealed by single quantum dots. Proc. Natl. Acad. Sci. USA. 2010;107:18658–63. doi: 10.1073/pnas.1013763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek S, Stenkamp RE, Teller DC, Palczewski K. G protein-coupled receptor rhodopsin: a prospectus. Annu. Rev. Physiol. 2003a;65:851–79. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek S, Teller DC, Palczewski K, Stenkamp R. The crystallographic model of rhodopsin and its use in studies of other G protein-coupled receptors. Annu. Rev. Biophys. Biomol. Struct. 2003b;32:375–97. doi: 10.1146/annurev.biophys.32.110601.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 2003;421:127–28. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 2004;564:281–88. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg M, Moreno JL, Holloway T, Provasi D, Baki L, et al. Decoding the signaling of a GPCR heteromeric complex reveals a unifying mechanism of action of antipsychotic drugs. Cell. 2011;147:1011–23. doi: 10.1016/j.cell.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxe K, Borroto-Escuela DO, Marcellino D, Romero-Fernandez W, Frankowska M, et al. GPCR heteromers and their allosteric receptor-receptor interactions. Curr. Med. Chem. 2012;19:356–63. doi: 10.2174/092986712803414259. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G protein-coupled receptors and neuronal functions. Annu. Rev. Neurosci. 2004;27:107–44. doi: 10.1146/annurev.neuro.27.070203.144206. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 1987;56:615–49. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Gilman AG, Rall TW. Factors influencing adenosine 3t,5t-phosphate accumulation in bovine thyroid slices. J. Biol. Chem. 1968a;243:5867–71. [PubMed] [Google Scholar]
- Gilman AG, Rall TW. The role of adenosine 3t,5t-phosphate in mediating effects of thyroid-stimulating hormone on carbohydrate metabolism of bovine thyroid slices. J. Biol. Chem. 1968b;243:5872–81. [PubMed] [Google Scholar]
- Granier S, Kobilka B. A new era of GPCR structural and chemical biology. Nat. Chem. Biol. 2012;8:670–73. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, et al. Structure of the δ-opioid receptor bound to naltrindole. Nature. 2012;485:400–4. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm HE, Deretic D, Hofmann KP, Schleicher A, Kohl B. Mechanism of action of monoclonal antibodies that block the light activation of the guanyl nucleotide-binding protein, transducin. J. Biol. Chem. 1987;262:10831–38. [PubMed] [Google Scholar]
- Harrison C, van der Graaf PH. Current methods used to investigate G protein coupled receptor oligomerisation. J. Pharmacol. Toxicol. Methods. 2006;54:26–35. doi: 10.1016/j.vascn.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Herrick-Davis K, Grinde E, Lindsley T, Cowan A, Mazurkiewicz JE. Oligomer size of the serotonin 5-hydroxytryptamine 2C (5-HT2C) receptor revealed by fluorescence correlation spectroscopy with photon counting histogram analysis: evidence for homodimers without monomers or tetramers. J. Biol. Chem. 2012;287:23604–14. doi: 10.1074/jbc.M112.350249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick-Davis K, Weaver BA, Grinde E, Mazurkiewicz JE. Serotonin 5-HT2C receptor homodimer biogenesis in the endoplasmic reticulum: real-time visualization with confocal fluorescence resonance energy transfer. J. Biol. Chem. 2006;281:27109–16. doi: 10.1074/jbc.M604390200. [DOI] [PubMed] [Google Scholar]
- Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci. 2009;34:540–52. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Hoyer D, Clarke DE, Fozard JR, Hartig PR, Martin GR, et al. International Union of Pharmacology classification of receptors for 5-hydroxytryptamine (Serotonin) Pharmacol. Rev. 1994;46:157–203. [PubMed] [Google Scholar]
- Hu J, Wang Y, Zhang X, Lloyd JR, Li JH, et al. Structural basis of G protein-coupled receptor/G protein interactions. Nat. Chem. Biol. 2010;6:541–48. doi: 10.1038/nchembio.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tesmer JJG. Recognition in the face of diversity: interactions of heterotrimeric G proteins and G protein-coupled receptor (GPCR) kinases with activated GPCRs. J. Biol. Chem. 2011;286:7715–21. doi: 10.1074/jbc.R109.051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 1997;15:216–19. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- Ianoul A, Grant DD, Rouleau Y, Bani-Yaghoub M, Johnston LJ, Pezacki JP. Imaging nanometer domains of beta-adrenergic receptor complexes on the surface of cardiac myocytes. Nat. Chem. Biol. 2005;1:196–202. doi: 10.1038/nchembio726. [DOI] [PubMed] [Google Scholar]
- Inagaki S, Ghirlando R, White JF, Gvozdenovic-Jeremic J, Northup JK, Grisshammer R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 2012;417:95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Palczewski K, Hofmann KP. Opsin/all-trans-retinal complex activates transducin by different mechanisms than photolyzed rhodopsin. Biochemistry. 1996;35:2901–8. doi: 10.1021/bi9524068. [DOI] [PubMed] [Google Scholar]
- Jalink K, Moolenaar WH. G protein-coupled receptors: the inside story. Bioessays. 2010;32:13–16. doi: 10.1002/bies.200900153. [DOI] [PubMed] [Google Scholar]
- Jastrzebska B, Debinski A, Filipek S, Palczewski K. Role of membrane integrity on G protein-coupled receptors: rhodopsin stability and function. Prog. Lipid. Res. 2011a;50:267–77. doi: 10.1016/j.plipres.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Fotiadis D, Jang GF, Stenkamp RE, Engel A, Palczewski K. Functional and structural characterization of rhodopsin oligomers. J. Biol. Chem. 2006;281:11917–22. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Golczak M, Fotiadis D, Engel A, Palczewski K. Isolation and functional characterization of a stable complex between photoactivated rhodopsin and the G protein, transducin. FASEB J. 2009;23:371–81. doi: 10.1096/fj.07-114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2013;27:1–13. doi: 10.1096/fj.12-225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Ringler P, Lodowski DT, Moiseenkova-Bell V, Golczak M, et al. Rhodopsin-transducin heteropentamer: three-dimensional structure and biochemical characterization. J. Struct. Biol. 2011b;176:387–94. doi: 10.1016/j.jsb.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai RS, Suzuki KG, Prossnitz ER, Koyama-Honda I, Nakada C, et al. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 2011;192:463–80. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol. Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya AI, Thaker TM, Preininger AM, Iverson TM, Hamm HE. Coupling efficiency of rhodopsin and transducin in bicelles. Biochemistry. 2011;50:3193–203. doi: 10.1021/bi200037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupchik YM, Barchad-Avitzur O, Wess J, Ben-Chaim Y, Parnas I, Parnas H. A novel fast mechanism for GPCR-mediated signal transduction–control of neurotransmitter release. J. Cell Biol. 2011;192:137–51. doi: 10.1083/jcb.201007053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert NA. GPCR dimers fall apart. Sci. Signal. 2010:3, pe12. doi: 10.1126/scisignal.3115pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004;343:1409–38. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- Li N, Salom D, Zhang L, Harris T, Ballesteros JA, et al. Heterologous expression of the adenosine A1 receptor in transgenic mouse retina. Biochemistry. 2007;46:8350–59. doi: 10.1021/bi700154h. [DOI] [PubMed] [Google Scholar]
- Li S, Huang S, Peng SB. Overexpression of G protein-coupled receptors in cancer cells: involvement in tumor progression. Int. J. Oncol. 2005;27:1329–39. [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J. Biol. Chem. 2003;278:21655–62. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Maeda T, Maeda A, Modzelewska A, et al. Rhodopsin signaling and organization in heterozygote rhodopsin knockout mice. J. Biol. Chem. 2004;279:48189–96. doi: 10.1074/jbc.M408362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett SB. Phosphorylation barcoding as a mechanism of directing GPCR signaling. Sci. Signal. 2011;4:pe36. doi: 10.1126/scisignal.2002331. [DOI] [PubMed] [Google Scholar]
- Liu W, Chun E, Thompson AA, Chubukov P, Xu F, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–36. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodowski DT, Palczewski K, Miyagi M. Conformational changes in the g protein-coupled receptor rhodopsin revealed by histidine hydrogen-deuterium exchange. Biochemistry. 2010;49:9425–27. doi: 10.1021/bi101502v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom KH. Structural Genomics on Membrane Proteins. CRC/Taylor & Francis; Boca Raton, FL: 2006. p. 383. [Google Scholar]
- Macia E, Partisani M, Paleotti O, Luton F, Franco M. Arf6 negatively controls the rapid recycling of the β2 adrenergic receptor. J. Cell Sci. 2012;125:4026–35. doi: 10.1242/jcs.102343. [DOI] [PubMed] [Google Scholar]
- Maeda A, Golczak M, Chen Y, Okano K, Kohno H, et al. Primary amines protect against retinal degeneration in mouse models of retinopathies. Nat. Chem. Biol. 2012;8:170–78. doi: 10.1038/nchembio.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Okano K, Park PS, Lem J, Crouch RK, et al. Palmitoylation stabilizes unliganded rod opsin. Proc. Natl. Acad. Sci. USA. 2010;107:8428–33. doi: 10.1073/pnas.1000640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Imanishi Y, Palczewski K. Rhodopsin phosphorylation: 30 years later. Prog. Retin. Eye Res. 2003;22:417–34. doi: 10.1016/s1350-9462(03)00017-x. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, et al. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–26. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary S, Damian M, Louet M, Floquet N, Fehrentz JA, et al. Ligands and signaling proteins govern the conformational landscape explored by a G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2012;109:8304–309. doi: 10.1073/pnas.1119881109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice P, Kamal M, Jockers R. Asymmetry of GPCR oligomers supports their functional relevance. Trends Pharmacol. Sci. 2011;32:514–20. doi: 10.1016/j.tips.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Mayhew TM, Astle D. Photoreceptor number and outer segment disk membrane surface area in the retina of the rat: stereological data for whole organ and average photoreceptor cell. J. Neurocytol. 1997;26:53–61. doi: 10.1023/a:1018563409196. [DOI] [PubMed] [Google Scholar]
- McCusker EC, Bane SE, O’Malley MA, Robinson AS. Heterologous GPCR expression: a bottleneck to obtaining crystal structures. Biotechnol. Prog. 2007;23:540–47. doi: 10.1021/bp060349b. [DOI] [PubMed] [Google Scholar]
- McMillin SM, Heusel M, Liu T, Costanzi S, Wess J. Structural basis of M3 muscarinic receptor dimer/oligomer formation. J. Biol. Chem. 2011;286:28584–98. doi: 10.1074/jbc.M111.259788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadegan T, Benko G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry. 2003;42:2759–67. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DJ, Wu N, Palczewski K. Vertebrate membrane proteins: structure, function, and insights from biophysical approaches. Pharmacol. Rev. 2008;60:43–78. doi: 10.1124/pr.107.07111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–67. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- Mustafi D, Kevany BM, Genoud C, Okano K, Cideciyan AV, et al. Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. FASEB J. 2011;25:3157–76. doi: 10.1096/fj.11-186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi D, Palczewski K. Topology of class A G protein-coupled receptors: insights gained from crystal structures of rhodopsins, adrenergic and adenosine receptors. Mol. Pharmacol. 2009;75:1–12. doi: 10.1124/mol.108.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro G, Hradsky J, Lluis C, Casadó V, McCormick PJ, et al. NCS-1 associates with adenosine A(2A) receptors and modulates receptor function. Front. Mol. Neurosci. 2012;5:53. doi: 10.3389/fnmol.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri M, Vanni S, Tavernelli I, Rothlisberger U. Role of aggregation in rhodopsin signal transduction. Biochemistry. 2010;49:4827–32. doi: 10.1021/bi100478j. [DOI] [PubMed] [Google Scholar]
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, et al. Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci. Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates J, Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr. Opin. Struct. Biol. 2011;21:802–7. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. USA. 2002;99:5982–87. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- Orban T, Huang CC, Homan KT, Jastrzebska B, Tesmer JJ, Palczewski K. Substrate-induced changes in the dynamics of rhodopsin kinase (G protein-coupled receptor kinase 1) Biochemistry. 2012a;51:3404–11. doi: 10.1021/bi300295y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban T, Jastrzebska B, Gupta S, Wang B, Miyagi M, et al. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012b;20:826–40. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. Oligomeric forms of G protein-coupled receptors (GPCRs) Trends Biochem. Sci. 2010;35:595–600. doi: 10.1016/j.tibs.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. Chemistry and biology of vision. J. Biol. Chem. 2012;287:1612–19. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Pardo L, Deupi X, Dolker N, López-Rodríguez ML, Campillo M. The role of internal water molecules in the structure and function of the rhodopsin family of G protein-coupled receptors. Chembiochem. 2007;8:19–24. doi: 10.1002/cbic.200600429. [DOI] [PubMed] [Google Scholar]
- Park D, O’Doherty I, Somvanshi RK, Bethke A, Schroeder FC, et al. Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2012;109:9917–22. doi: 10.1073/pnas.1202216109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Scheerer P, Hofmann KP, Choe H-W, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008a;454:183–87. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- Park PS, Filipek S, Wells JW, Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;43:15643–56. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PS, Lodowski DT, Palczewski K. Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annu. Rev. Pharmacol. Toxicol. 2008b;48:107–41. doi: 10.1146/annurev.pharmtox.48.113006.094630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PS, Palczewski K. Diversifying the repertoire of G protein-coupled receptors through oligomerization. Proc. Natl. Acad. Sci. USA. 2005;102:8793–94. doi: 10.1073/pnas.0504016102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellissier LP, Barthet G, Gaven F, Cassier E, Trinquet E, et al. G protein activation by serotonin type 4 receptor dimers: evidence that turning on two protomers is more efficient. J. Biol. Chem. 2011;286:9985–97. doi: 10.1074/jbc.M110.201939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin JP, Galvez T, Prézeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 2003;98:325–54. doi: 10.1016/s0163-7258(03)00038-x. [DOI] [PubMed] [Google Scholar]
- Prezeau L, Rives ML, Comps-Agrar L, Maurel D, Kniazeff J, Pin JP. Functional crosstalk between GPCRs: with or without oligomerization. Curr. Opin. Pharmacol. 2010;10:6–13. doi: 10.1016/j.coph.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Qin K, Dong C, Wu G, Lambert NA. Inactive-state preassembly of G(q)-coupled receptors and G(q) heterotrimers. Nat. Chem. Biol. 2011;7:740–47. doi: 10.1038/nchembio.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh R, Damian M, Cottet M, Orcel H, Mendre C, et al. Structural insights into biased G protein-coupled receptor signaling revealed by fluorescence spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:6733–38. doi: 10.1073/pnas.1201093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall TW, Sutherland EW. Formation of a cyclic adenine ribonucleotide by tissue particles. J. Biol. Chem. 1958;232:1065–76. [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–87. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–55. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat. Rev. Mol. Cell Biol. 2009;10:819–30. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison GA, Butcher RW, Sutherland EW. Cyclic AMP. Annu. Rev. Biochem. 1968;37:149–74. doi: 10.1146/annurev.bi.37.070168.001053. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–73. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Rovira X, Pin JP, Giraldo J. The asymmetric/symmetric activation of GPCR dimers as a possible mechanistic rationale for multiple signalling pathways. Trends Pharmacol. Sci. 2010;31:15–21. doi: 10.1016/j.tips.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Salom D, Cao P, Sun W, Kramp K, Jastrzebska B, et al. Heterologous expression of functional G-protein-coupled receptors in Caenorhabditis elegans. FASEB J. 2012;26:492–502. doi: 10.1096/fj.11-197780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. USA. 2006;103:16123–28. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D, Wu N, Sun W, Dong Z, Palczewski K, et al. Heterologous expression and purification of the serotonin type 4 receptor from transgenic mouse retina. Biochemistry. 2008;47:13296–307. doi: 10.1021/bi8018527. [DOI] [PubMed] [Google Scholar]
- Salon JA, Lodowski DT, Palczewski K. The significance of G protein-coupled receptor crystallography for drug discovery. Pharmacol. Rev. 2011;63:901–37. doi: 10.1124/pr.110.003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Schlegel W, Kempner ES, Rodbell M. Activation of adenylate cyclase in hepatic membranes involves interactions of the catalytic unit with multimeric complexes of regulatory proteins. J. Biol. Chem. 1979;254:5168–76. [PubMed] [Google Scholar]
- Schneider EH, Seifert R. Sf 9 cells: a versatile model system to investigate the pharmacological properties of G protein-coupled receptors. Pharmacol. Ther. 2010;128:387–418. doi: 10.1016/j.pharmthera.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Shimamura T, Hiraki K, Takahashi N, Hori T, Ago H, et al. Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region. J. Biol. Chem. 2008;283:17753–56. doi: 10.1074/jbc.C800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SO. Insights into the activation mechanism of the visual receptor rhodopsin. Biochem. Soc. Trans. 2012;40:389–93. doi: 10.1042/BST20110751. [DOI] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–60. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann R, Schröck K, Böselt I, Stäubert C, Russ A, Schöneberg T. Evolution of GPCR: change and continuity. Mol. Cell Endocrinol. 2011;331:170–78. doi: 10.1016/j.mce.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Struts AV, Salgado GFJ, Martinez-Mayorga K, Brown MF. Retinal dynamics underlie its switch from inverse agonist to agonist during rhodopsin activation. Nat. Struct. Mol. Biol. 2011;18:392–94. doi: 10.1038/nsmb.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating non-canonical unfolded protein response genes. Science. 2011;332:729–32. doi: 10.1126/science.1203411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J. Biol. Chem. 1958;232:1077–91. [PubMed] [Google Scholar]
- Tadagaki K, Jockers R, Kamal M. History and biological significance of GPCR heteromerization in the neuroendocrine system. Neuroendocrinology. 2012;95:223–31. doi: 10.1159/000330000. [DOI] [PubMed] [Google Scholar]
- Tate CG, Schertler GF. Engineering G protein-coupled receptors to facilitate their structure determination. Curr. Opin. Struct. Biol. 2009;19:386–95. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AA, Liu JJ, Chun E, Wacker D, Wu H, et al. GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods. 2011;55:310–17. doi: 10.1016/j.ymeth.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AA, Liu W, Chun E, Katritch V, Wu H, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–99. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidi PA, Przybyla JA, Hu CD, Watts VJ. Visualization of G protein-coupled receptor (GPCR) interactions in living cells using bimolecular fluorescence complementation (BiFC) Curr. Protoc. Neurosci. 2010 doi: 10.1002/0471142301.ns0529s51. Chapter 5: Unit 5.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West GM, Chien EY, Katritch V, Gatchalian J, Chalmers MJ, et al. Ligand-dependent perturbation of the conformational ensemble for the GPCR β2 adrenergic receptor revealed by HDX. Structure. 2011;19:1424–32. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc. Natl. Acad. Sci. USA. 2011;108:16086–91. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JP, Reboul E, Molday RS, Kast J. A carboxy-terminal affinity tag for the purification and mass spectrometric characterization of integral membrane proteins. J. Proteome Res. 2009;8:2388–96. doi: 10.1021/pr801008c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485:327–32. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau KW, Hardie RC. Phototransduction motifs and variations. Cell. 2009;139:246–64. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl. 2):ii246, 55. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- Yekkirala AS. Two to tango: GPCR oligomers and GPCR-TRP channel interactions in nociception. Life Sci. 2013;92:438–45. doi: 10.1016/j.lfs.2012.06.021. [DOI] [PubMed] [Google Scholar]
- Zhang L, Salom D, He J, Okun A, Ballesteros J, et al. Expression of functional G protein-coupled receptors in photoreceptors of transgenic Xenopus laevis. Biochemistry. 2005;44:14509–18. doi: 10.1021/bi051386z. [DOI] [PubMed] [Google Scholar]
- Zheng H, Pearsall EA, Hurst DP, Zhang Y, Chu J, et al. Palmitoylation and membrane cholesterol stabilize μ-opioid receptor homodimerization and G protein coupling. BMC Cell Biol. 2012;13:6. doi: 10.1186/1471-2121-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Nakatani E, Gronenberg LS, Tokimoto T, Wirth MJ, et al. Peptide-labeled quantum dots for imaging GPCRs in whole cells and as single molecules. Bioconjug. Chem. 2007;18:323–32. doi: 10.1021/bc0601929. [DOI] [PubMed] [Google Scholar]
- Zylbergold P, Hébert TE. A division of labor: asymmetric roles for GPCR subunits in receptor dimers. Nat. Chem. Biol. 2009;5:608–9. doi: 10.1038/nchembio0909-608. [DOI] [PubMed] [Google Scholar]
- Munger SD, Leinders-Zufall T, Zufall F. Subsystem organization of the mammalian sense of smell. Annu. Rev. Physiol. 2009;71:115–40. doi: 10.1146/annurev.physiol.70.113006.100608. [DOI] [PubMed] [Google Scholar]
- Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacol. Rev. 2011;63:291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G protein-coupled receptors. Nature. 2009;459:356–63. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SO. Structure and activation of the visual pigment rhodopsin. Annu. Rev. Biophys. 2010;39:309–28. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- Spiegel AM, Weinstein LS. Inherited diseases involving G proteins and G protein–coupled receptors. Annu. Rev. Med. 2004;55:27–39. doi: 10.1146/annurev.med.55.091902.103843. [DOI] [PubMed] [Google Scholar]
- Stevens RC. GPCR structure and activation. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–56. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]