Abstract
Fibrinogen like protein-1 (Fgl1) is a predominantly liver expressed protein that has been implicated as both a hepatoprotectant and a hepatocyte mitogen. Fgl1 expression is decreased in hepatocellular carcinoma (HCC) and its loss correlates with a poorly differentiated phenotype. To better elucidate the role of Fgl1 in hepatocarcinogenesis, we treated mice wild type or null for Fgl1 with diethyl nitrosamine and monitored for incidence of hepatocellular cancer. We find that mice lacking Fgl1 develop HCC at more than twice the rate of wild type mice. We show that hepatocellular cancers from Fgl1 null mice are molecularly distinct from those of the wild type mice. In tumors from Fgl1 null mice there is enhanced activation of Akt and downstream targets of the mammalian target of rapamycin (mTOR). In addition, there is paradoxical up regulation of putative hepatocellular cancer tumor suppressors; tripartite motif-containing protein 35 (Trim35) and tumor necrosis factor super family 10b (Tnfrsf10b). Taken together, these findings suggest that Fgl1 acts as a tumor suppressor in hepatocellular cancer through an Akt dependent mechanism and supports its role as a potential therapeutic target in HCC.
Keywords: Fibrinogen like protein-1, Hepatocellular carcinoma, Diethyl nitrosamine, Hepatoprotecatant, Mitogen
1. Introduction
Fgl1 is a 68kd protein that comprises two disulfide linked 34kd homodimers [1]. Although predominantly expressed in the liver, we recently demonstrated low-level transcription of Fgl1 in brown and white adipose in the setting of liver injury [2], and others have documented low-level expression in the pancreas [3]. Existing evidence supports a role for Fgl1 in liver regeneration and hepatoprotection: Fgl1 enhances tritiated thymidine uptake by primary hepatocytes and human hepatocellular carcinoma (HCC) cell lines [4], is upregulated following 70% partial hepatectomy in rodents [5], and the administration of recombinant Fgl1 protects hepatocytes from acute liver injury [6].
The pathogenesis of HCC involves a multistep process with inactivation of tumor suppressor genes (TSGs) and upregulation of proto-oncogenes [7]. A role for Fgl1 in HCC development has been suggested i) Fgl1 is frequently reduced or absent in HCC tissues [8,9], ii) restoration of Fgl1 expression in cultured HCC cell lines inhibits cell growth [8], iii) human FGL1 is located on the short arm of chromosome 8, a region rich in putative TSGs that are frequently deleted in HCC, and iv) additive loss of putative TSGs on human chromosome 8p, including Fgl1, is associated with increased growth of HCC, suggesting a synergistic effect of allelic loss of individual TSGs in promoting tumor growth [10]. These paradoxical effects of Fgl1 as both a mitogen in non-tumor hepatocytes and a growth inhibitor in HCC has been suggested to result from differing modes of Fgl1 dependent signaling: autocrine in hepatocyte proliferation versus intracrine in hepatocarcinogenesis [11].
Because previous studies on the role of Fgl1 in HCC has been limited to analysis of archived patient tumor samples [8,9] or cultured cells [8] or through shRNA driven reduction in cellular Fgl1 content in a murine model of HCC [10], we set out to determine whether Fgl1 directly influenced the development of HCC. Using our previously generated Fgl1 knockout mouse (Fgl1KO), we show that Fgl1KO mice develop HCC at a faster rate than Fgl1 wild type mates (Fgl1WT), following tumor induction with DEN and tumor promotion by low dose phenobarbital (Pb). We find that livers of Fgl1KO have more proliferative foci at 8 months and more than a two fold increase in prevalence of HCC at 12 months of DEN treatment. Although tumors from Fgl1WT and Fgl1KO were grossly and histologically similar, tumors from Fgl1KO were molecularly distinct as they exhibited 1) enhanced activation of Akt dependent signaling and 2) increased expression of putative HCC tumor suppressors Trim35 and Tnfrsf10b.
2. Materials and methods
2.1. Animals
Fgl1KO mice were as previously described [2] and available from the Jackson labs (B6; 129-Fgl1tm1Cuko/J, JAX Stock number: 0024004). Ethics Statement: All animal experiments were performed according to institutional guidelines of Harvard Medical School. All protocols with animal experimentation conformed to criteria outlined in the National Institutes of Health “Guide for the Care and Use of Laboratory Animals.” Protocols were reviewed and approved by the animal use committee of Harvard Medical School (protocol number 04193).
2.2. Hepatocarcinogenesis
Mice were injected with DEN (Sigma, St Louis, MO) 5 mg/kg at 15 days of age. Mice were fed a standard chow diet (LabDiet, Brentwood, MO) and were given free access to water supplemented with 0.07% phenobarbital as previously described [12]. Mice were euthanized at 6, 8, 10–12 months of age. Mouse liver and tumor tissue were collected and placed in 10% formalin or snap frozen at −80 °C for later analysis.
2.3. Western immunoblot assays
Liver and tumor tissue was homogenized in RIPA buffer (25 mM Hepes pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% deoxycholic acid, 0.1% SDS, and 2 mM EDTA). The resulting homogenate was centrifuged at 13,000 RPM and the supernatant was transferred to clean tubes. Protein concentrations were determined and lysates denatured with Laemmli sample buffer as previously described [13]. The following antibodies were used: eukaryotic transcription factor 4E binding protein 1(4EBP1), p4EBP1 (Thr37/46), protein kinase B (Akt), pAkt (Ser473), p38, p-p38 (Thr180/Tyr182), p-p70S6K (S371), p70S6K (Cell Signaling, Beverly, MA), UHRF1 (Santa Cruz Biotechnology, Dallas, TX), and β-actin (Sigma, St Louis, MO). Western immunoblot assays were performed as previously described [13]. Band intensity was quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
3. Nucleic acid assays
Quantitative PCR assays were performed as previously described [14]. mRNA levels were determined relative to cyclophilin A. Table S1 shows the primer sequences that were used for these assays.
3.1. Immunohistochemistry
Liver tissue fixed in 10% formalin was paraffin embedded and sectioned onto glass slides. Sections were deparaffinized in xylene and pretreated with epitope retrieval solution (IHC World, Elliot City, MD). Slides were blocked with 10% normal goat serum (Invitrogen, Carlsbad, CA). The following primary antibodies were used proliferating cell nuclear antigen (PCNA), ki-67, and β-catenin (Cell Signaling, Beverly, MA). The SuperPicture™ 3rd Generation IHC Detection Kit (Life Technologies, Carlsbad, CA) was used per manufacturer protocol.
4. Results
4.1. Disruption of Fgl1 enhances DEN induced hepatocarcinogenesis
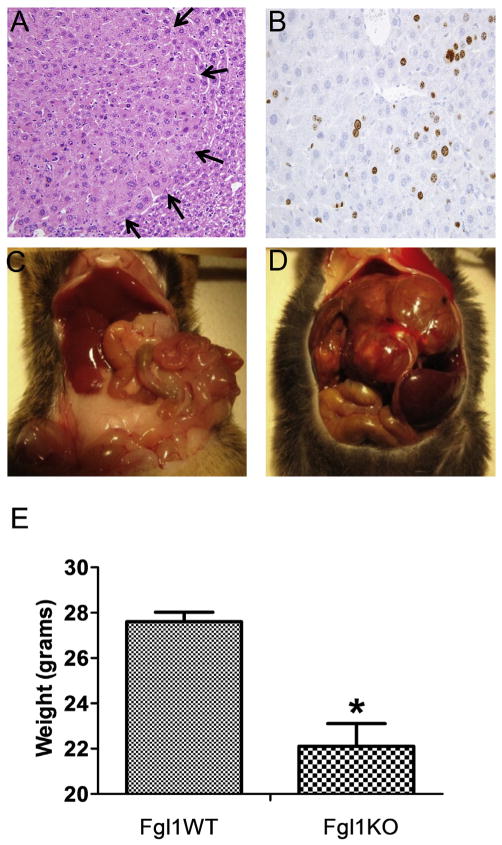
Fgl1KO mice did not develop spontaneous HCC even at 15 months of age (not shown). To determine whether there were differences in hepatocarcinogenesis between Fgl1WT and Fgl1KO we treated mice with DEN + Pb and evaluated for incidence of HCC at 6, 8 and 12 months. At 6 months gross and histologic analysis showed no evidence of any liver lesions (not shown). At 8 months we noted the appearance of numerous discrete foci in histologic sections of Fgl1KO livers (n = 4) with only one such lesion in Fgl1WT (n = 4). These foci contained proliferating cells as shown by the presence of ki-67 positive cells (Fig. 1A and B). At 10 months of the expected 12 months for the last cohort, we noted that a subset of Fgl1KO (3 of 12) displayed marked evidence of generalized deterioration characterized by hair loss, inability to groom, and weight loss. The 3 Fgl1KO mice and 3 random age matched littermates from the Fgl1WTcohort were euthanized. All 3 of the Fgl1KO mice harbored large and multiple liver lesions compared to 0 of the Fgl1WT mice (Fig. 1C and D). Analysis of the remaining cohort at 12 months post DEN, showed that 6 of the 9 Fgl1KO mice exhibited gross evidence of liver cancer compared to 1 of 6 of the Fgl1WT. In total, 10 months–12 months after tumor induction with DEN, 9 of 12 Fgl1 WT and 1 of 9 Fgl1WT developed HCC. At baseline, Fgl1KO mice were larger than their WT mates[2], however at 12 months, we noted that Fgl1KO mice were smaller despite the larger tumor burden when compared to Fgl1WT mice (22.1 gm ± 2.1 vs. 27.6 ± 0.9) (Fig. 1E).
Fig. 1.
Fgl1 null mice have a higher incidence of carcinoma compared to wild type. A). Representative images at 8 months after administration of diethyl nitrosamine. H&E staining of a lesion (left). Arrows denote boundary with normal hepatic parenchyma. B) Ki-67 staining shows evidence of proliferating cells. C) Representative non-tumor liver from Fgl1WT mouse. D) Large tumor burden in an Fgl1KO mouse E) Marked decrease in weight of Fgl1KO mouse.
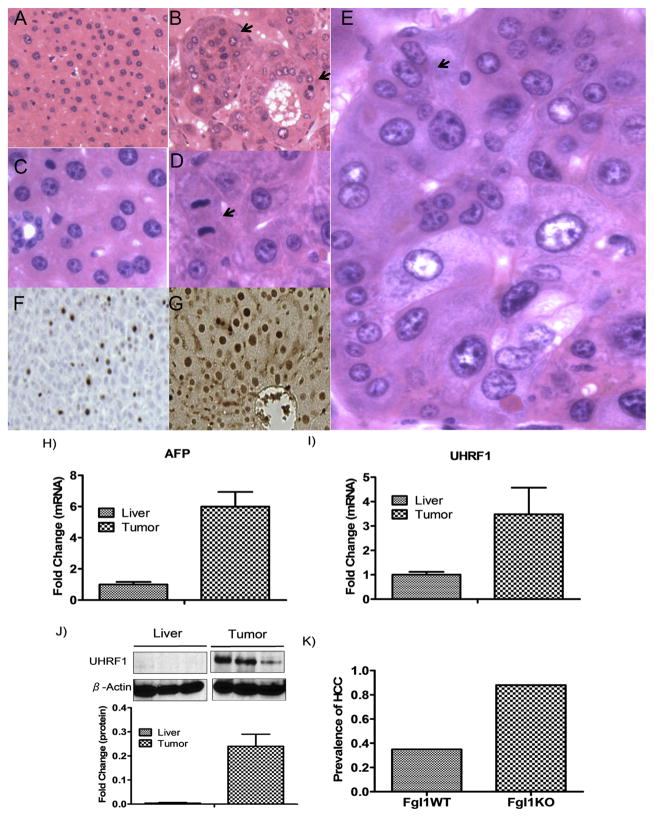
Histologic analysis confirmed that the liver lesions (including the solitary tumor from Fgl1WT) were HCCs, with numerous histopathologic features typical of human HCCs including nuclear atypia, mitotic figures, thickened cords and multinucleated cells (Fig. 2B, D and E). Not surprisingly these tumors showed positive staining for ki-67 and PCNA (Fig. 2G and F). Consistent with the diagnosis of HCC, the tumors had elevated expression of transcripts for alpha feto-protein (AFP) and the hepatocellular oncogene, ubiquitin-like with PHD and Ring fingers domain 1 (UHRF1) [15] when compared to non-tumor liver (Fig. 2H and I). UHRF1 protein usually undetectable in normal livers [16] was markedly elevated in the tumors (Fig. 2J) consistent with its known enhancement in rapidly dividing cells. To ensure that the findings were not spurious, we repeated the experiment to confirm increased incidence of HCC at 12 months after induction. Again we found that 13 out of 13 Fgl1KO and 6 of 11 Fgl1WT developed HCC. Over the course of these experiments 22 of 25 Fgl1KO developed tumors while only 7 of 20 Fgl1WT (Fig. 2K) had HCC. These data reveal that Fgl1KO are more prone to DEN induced carcinogenesis.
Fig. 2.
Liver Tumors are HCCs. A and C) H&E stains (original magnification x100 and x600, respectively) of non-tumor liver from Fgl1WT. H&E stains B (x100) D and E (x600) of Fgl1KO tumors. Note the abnormal architecture in the tumors with nested and pseudoglandular patterns (B; arrows) as well as mitotic figures (D, arrow), variation in nuclear size and multinucleate cells (E, arrow). F and G) Immunohistochemistry showing positive nuclear staining of Ki-67 and PCNA respectively in tumors of Fgl1KO. H and I) Tumors from Fgl1KO have elevated transcripts for HCC biomarker alfa-feto protein and oncogene UHRF1 compared to non-tumor liver (n = 4 livers, n = 6 tumors). J) Expression of UHRF1 protein in normal liver compared to marked increase in liver tumors (graph is from n = 3 livers, n = 6 tumors) (*p < 0.05). K) Prevalence of HCC is approximately 2.5 times in Fgl1KO compared to Fgl1WT.
4.2. Akt signaling is enhanced in Fgl1KO liver tumors
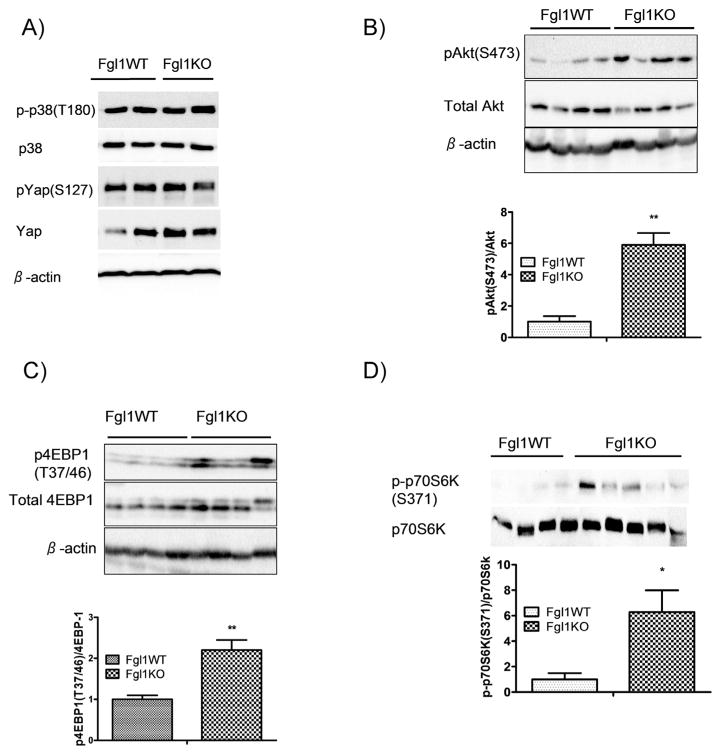
HCC is characterized by aberrant activation of a number of signaling pathways. Principal among these pathways are those regulated by protein kinase b (Akt), Yap/Hippo, p38 MAP kinase and β-catenin [17]. To examine differences in signaling pathways between the Fgl1KO and Fgl1WT tumors, we used lysates from Fgl1WT and Fgl1KO tumors from the second experimental cohort where we have more than 1 tumor from the Fgl1WT cohort for western immunoblot analysis. We first examined whether p38 MAPK was more active in the Fgl1KO compared to the WT and found no differences (Fig. 3A, top two panels). In addition, we observed no differences in activation of Yap, a transcriptional co-activator of the Hippo pathway that is also implicated in HCC (Fig. 3A, panels 3 and 4). We also found that β-catenin was entirely localized to the membrane in both Fgl1WT and Fgl1KO tumors, suggesting that this pathway was not aberrant in the HCCs (not shown). However, western immunoblot analysis for phosphorylated Akt (S473), showed a six fold increase in the ratio of phosphorylated to total Akt in Fgl1KO tumors compared to tumors from Fgl1WT (Fig. 3B). To confirm that Akt was more active in the Fgl1KO, we looked downstream of mTOR, a target of Akt and itself a serine/threonine kinase that is a master controller of growth and catabolic processes. We chose two mTOR targets 1) 4EPB1, a kinase with a regulatory role in cell proliferation and whose blockade of the 4EBP1/eIF-4E cascade delays while activation enhances hepatocarcinogensis [18] and 2) p70S6K, a serine/threonine kinase whose activity that regulates protein synthesis and cellular proliferation. Consistent with the enhanced phosphorylated Akt we found that tumors from Fgl1KO had a 2-fold increase in phosphorylated 4EBP1 compared to Fgl1WT tumors (Fig. 3C). We examined p70S6K phosphorylation on serine 371, a rapamycin sensitive, mTOR phosphorylation site [19] and found a 6 fold increased phosphorylation in Fgl1KO tumors when compared to Fgl1WT tumors. These observations suggest that, while Fgl1WT and Fgl1KO liver tumors were grossly and histologically similar, a distinct molecular characteristic of the Fgl1KO derived HCC is the enhanced activation of Akt dependent mTOR signaling.
Fig. 3.
Fgl1KO Tumors have increased activation of Akt dependent mTOR Signaling. A) Total and phospho p38 (top two panels) are unchanged in Fgl1WT compared to Fgl1KO tumors. Likewise total and phospho Yap levels (panels 3 and 4) pathways are not differentially activated in Fgl1KO and Fgl1WT tumors. B) Increased phosphorylation of Akt in tumors from Fgl1KO, compared to Fgl1WT tumors. Top). Representative blot (n = 4 for both Fgl1WT and Fgl1KO). Bottom). Quantitation shows nearly 6-fold increase in ratio of pAkt/Total Akt. C) Enhanced phosphorylation of 4EBP1 in tumors from Fgl1KO. Top). Representative blot. Bottom) quantitation shows a 2-fold difference in phosphorylated 4EPB1 to total 4EBP1 between Fgl1KO and Fgl1WT tumors. Data for graphs is from n = 6 Fgl1WT and n = 7 Fgl1KO tumors) (**p < 0.001). D) Increased phosphorylation on serine 371 of p70S6K in tumors from Fgl1KO. Graphs represent densitometric quantitation of p-P70S6K/total p70S6K. n = 4 for Fgl1WT and n = 5 for Fgl1KO. (*p < 0.05).
4.3. Deletion of Fgl1 results in increased expression of putative tumor suppressor genes
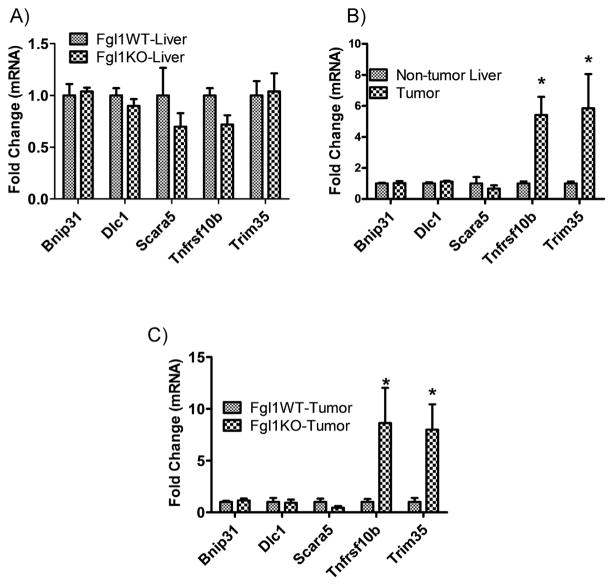
Human Fgl1 resides on the short arm of chromosome 8p within a region, which includes multiple candidate TSGs that are frequently deleted in HCC [10]. Tumors with these chromosomal deletions are often poorly differentiated and experimental data suggest that cumulative loss of individual tumor suppressors portends poor outcome. As such it has been posited that selection pressure with loss of candidate TSGs lead to the attenuation of multiple genes. Experimental proof has been gleaned from shRNA-mediated knockdown of these TSGs in a murine model of HCC in a cellular background of p53 loss and MYC overexpression [10]. In these experiments, knockdown of Fgl1 enhances hepatocarcinogenesis and supports the notion that Fgl1 is a TSG for HCC. Thus we asked whether complete loss, as opposed to reduction of Fgl1 alone, is required for HCC development and whether additional suppression of the human chromosome 8 TSGs accompanies Fgl1 loss in rodent HCC pathogenesis. We performed qPCR to determine the expression of selected human chromosome 8p TSG candidates in tumor and non-tumor liver tissue of Fgl1WT and Fgl1KO. We compared the expression of five candidate TSGs corresponding to the human chromosome 8 cluster: deleted in liver cancer 1 (Dlc1), scavenger receptor class A member 5 (Scara5), tripartite motif-containing protein 35(Trim 35), tumor necrosis factor super family 10b (Tnfrsf10b) and Bcl2/adenovirus protein-interacting protein 3-like (Bnip31) in livers of Fgl1WT and Fgl1KO animals at i) 6 months after treatment with DEN, a time point when we observe no proliferative foci or tumors and ii) 12 months after DEN a time point when tumors occur. At 6 months, we found no differences in the expression of TSGs (Fig. 4A) between the livers of the Fgl1WT and Fgl1KO mice. This suggested that any change in expression was not due to DEN treatment. At 12 months, when HCC was present in Fgl1KO mice, we compared the expression of TSGs from tumors with non-tumor liver. We found no significant down regulation of the TSGs but rather a paradoxical 4–5-fold increase in Trim35 and Tnfrsf10b in the Fgl1KO tumors (Fig. 4B). This suggested that these molecular changes occur only in the tumors not in the adjacent non-tumor liver. We next asked if similar up regulation of Trim35 and Tnfrsf10b was present in the Fgl1WT tumors or if these were molecular aberrations unique to HCC in livers of Fgl1KO mice. We once again found increased expression of Trim35 and Tnfrsf10b only in the tumors from Fgl1KO (Fig. 4C). Together these findings demonstrate that complete loss of Fgl1 alone without suppression of additional co-operating TSGs from human chromosome 8 cluster is sufficient for DEN induced HCC. It also illustrates a second point of molecular distinction between Fgl1KO and Fgl1WT tumors with the increased expression of Trim35 and Tnfrsf10b unique to the tumors in Fgl1KO.
Fig. 4.
Candidate genes from the human chromosome 8 tumor suppressor cluster are upregulated in Fgl1KO liver tumors. A) There are no differences in gene expression at 6 months after DEN administration when tumors and proliferative foci are absent (n = 6 in each group) B) Increased expression of Trim35 and Tnfrsf10b is exclusive to tumor tissue of Fgl1KO, with no enhancement in non-tumor liver tissue at 12 month of DEN + Pb treatment (left, n = 9 tumors, n = 12 livers). C) Increased expression of Trim35 and Tnfrsf10b expression is seen in Fgl1KO tumors only not those from Fgl1WT. (n = 5 in each group) (*p < 0.05).
5. Discussion
We show that in vivo disruption of Fgl1 in mice is associated with increased incidence of HCC in response to treatment with the tumor inducer DEN and the tumor promoter, Pb. Following administration of DEN there is a time dependent progression; first with development of proliferative foci and then eventual liver cancer. We find a reproducible increase in the incidence of HCC in the Fgl1KO after 12 months of carcinogen treatment. These results provide further support of the role of Fgl1 as a putative tumor suppressor.
Liver tumors in Fgl1KO, like those from the WT had histologic characteristics of human HCCs. They demonstrated nuclear clumping, thickened hepatocyte cords, and frequent mitotic figures. This model yielded no evidence of cholangiopathy or of a mixed cholangiocarcinoma/HCC phenotype. HCCs did express AFP, a biomarker of hepatocellular cancer and UHRF1 an oncogene for HCC [15]. Overall, tumors from the Fgl1KO were grossly and histologically consistent with what was expected following treatment with DEN + Pb.
Despite the histologic similarity between tumors from Fgl1KO and Fgl1WT, the tumors were molecularly distinct. Whereas we observed no differences in oncogenic pathways driven by beta catenin, Yap/Hippo or p38, we did find enhanced activation of Akt and its downstream targets in Fgl1KO tumors. However because there has been no cell surface nor intracellular receptor identified for Fgl1, we remain unsure as to whether the upstream signals that activated Akt originated from the cell surface or within the cell.
A second difference between HCCs from Fgl1KO and Fgl1WT was the paradoxical enhancement of transcripts for two members of the human chromosome 8 cluster of putative TSGs, Trim35 and Tnfrsf10b. Interestingly, the expressions of these genes were not enhanced in non-tumor liver or in tumors from the Fgl1WT mouse. Both genes encode for proteins involved in the regulation of apoptosis [20,21] and we speculate that up-regulation of these neighboring genes may have reflected compensation for the loss of Fgl1. The co-operative nature of the TSGs in the chromosome 8 tumor cluster in HCC progression has been elucidated and demonstrates that additional loss of putative TSGs enhances tumor growth and aggression [10]. In this vein the loss of Fgl1 alone within the tumor environment likely led to expression of Trim35 and Tnfrsf10b and would suggest that the combined loss of Fgl1 with either or both TSGs would lead to a more pronounced HCC phenotype than observed with Fgl1 alone.
A protective role of Fgl1 in human HCC pathogenesis is supported by its down regulation in a large number of HCCs [8,9]. This newly uncovered link between Fgl1 loss and Akt signaling supports a potential role of an Fgl1 regulated pathway as a therapeutic node. Akt and mTOR play roles in human HCCs, with 40–50% of human HCCs elaborating enhanced activity of this pathway [22–24]. Akt and mTOR inhibitors are under study as potential therapies for HCC, with Rapalogs that inhibit mTOR showing promise both in vitro and in vivo in pre-clinical studies [25], and are currently under investigation as therapy in humans for HCC [26–29]. Thus the finding of enhanced Akt/mTOR activity in tumors from the Fgl1KO supports a role for Fgl1 as therapeutic target in this pathway in HCC.
Taken together, our data suggests that Fgl1 functions as a tumor suppressor and that disruption of Fgl1 leads to increased rates of HCC in mice treated with DEN + Pb. The development of tumors is associated with increases in the HCC marker AFP as well as in the newly described oncogene UHRF1. The tumors from Fgl1KO uniquely exhibited increased activation of Akt-dependent mTOR signaling and paradoxical upregulation of Trim35 and Tnfrsf10b. Understanding of how Fgl1 inhibits Akt mediated signaling may provide further insights into the pathobiology of HCC.
Supplementary Material
Acknowledgments
The work was made possible through the generous support of the Sidney A. Swensrud Foundation. HN was supported by a T32 grant DK007533. DC was supported by NIH DK48873 and DK56626. Harvard Digestive Disease Center supported by NIH 5P30 DK034854 provided histologic services.
Abbreviations
- HCC
hepatocellular cancer
- DEN
diethyl nitrosamine
- Pb
phenobarbital
- Fgl1
fibrinogen like protein-1
- UHRF1
ubiquitin-like with PHD and ring finger domains-1
- Trim35
tripartite motif containing 35
- Tnfrsf10b
tumor necrosis factor receptor superfamily, member 10b
- AFP
alfa fetoprotein
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2015.07.078.
Footnotes
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2015.07.078.
References
- 1.Yamamoto T, Gotoh M, Sasaki H, Terada M, Kitajima M, Hirohashi S. Molecular cloning and initial characterization of a novel fibrinogen-related gene, HFREP-1. Biochem Biophys Res Commun. 1993;193:681–687. doi: 10.1006/bbrc.1993.1678. [DOI] [PubMed] [Google Scholar]
- 2.Demchev V, et al. Targeted deletion of fibrinogen like protein 1 reveals a novel role in energy substrate utilization. PLoS One. 2013;8:e58084. doi: 10.1371/journal.pone.0058084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hara H, Yoshimura H, Uchida S, Toyoda Y, Aoki M, Sakai Y, Morimoto S, Shiokawa K. Molecular cloning and functional expression analysis of a cDNA for human hepassocin, a liver-specific protein with hepatocyte mitogenic activity. Biochim Biophys Acta. 2001;1520:45–53. doi: 10.1016/s0167-4781(01)00249-4. [DOI] [PubMed] [Google Scholar]
- 4.Hara H, et al. Isolation and characterization of a novel liver-specific gene, hepassocin, upregulated during liver regeneration. Biochim Biophys Acta. 2000;1492:31–44. doi: 10.1016/s0167-4781(00)00056-7. [DOI] [PubMed] [Google Scholar]
- 5.Yan J, Ying H, Gu F, He J, Li YL, Liu HM, Xu YH. Cloning and characterization of a mouse liver-specific gene mfrep-1, up-regulated in liver regeneration. Cell Res. 2002;12:353–361. doi: 10.1038/sj.cr.7290137. [DOI] [PubMed] [Google Scholar]
- 6.Li CY, et al. Recombinant human hepassocin stimulates proliferation of hepatocytes in vivo and improves survival in rats with fulminant hepatic failure. Gut. 2010;59:817–826. doi: 10.1136/gut.2008.171124. [DOI] [PubMed] [Google Scholar]
- 7.Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- 8.Yan J, et al. LFIRE-1/HFREP-1, a liver-specific gene, is frequently down-regulated and has growth suppressor activity in hepatocellular carcinoma. Oncogene. 2004;23:1939–1949. doi: 10.1038/sj.onc.1207306. [DOI] [PubMed] [Google Scholar]
- 9.Yu HT, et al. Specific expression and regulation of hepassocin in the liver and down-regulation of the correlation of HNF1alpha with decreased levels of hepassocin in human hepatocellular carcinoma. J Biol Chem. 2009;284:13335–13347. doi: 10.1074/jbc.M806393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci U S A. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao MM, et al. Hepassocin regulates cell proliferation of the human hepatic cells L02 and hepatocarcinoma cells through different mechanisms. J Cell Biochem. 112:2882–2890. doi: 10.1002/jcb.23202. [DOI] [PubMed] [Google Scholar]
- 12.Awuah PK, Rhieu BH, Singh S, Misse A, Monga SP. Beta-Catenin loss in hepatocytes promotes hepatocellular cancer after diethylnitrosamine and phenobarbital administration to mice. PLoS One. 2012;7:e39771. doi: 10.1371/journal.pone.0039771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ukomadu C, Dutta A. Inhibition of cdk2 activating phosphorylation by mevastatin. J Biol Chem. 2003;278:4840–4846. doi: 10.1074/jbc.M208658200. [DOI] [PubMed] [Google Scholar]
- 14.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, UHRF1. Proc Natl Acad Sci U S A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mudbhary R, et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu J, et al. UHRF1 phosphorylation by cyclin A2/CDK2 is required for zebrafish embryogenesis. Mol Biol Cell. 2012;23:59–70. doi: 10.1091/mbc.E11-06-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, et al. 4EBP1/eIF4E and p70S6K/RPS6 axes play critical and distinct roles in hepatocarcinogenesis driven by AKT and N-Ras proto-oncogenes in mice. Hepatology. 2015;61:200–213. doi: 10.1002/hep.27396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurita S, et al. DNMT1 and DNMT3b silencing sensitizes human hepatoma cells to TRAIL-mediated apoptosis via up-regulation of TRAIL-R2/DR5 and caspase-8. Cancer Sci. 2010;101:1431–1439. doi: 10.1111/j.1349-7006.2010.01565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura F, et al. Cloning and characterization of a novel RING-B-box-coiled-coil protein with apoptotic function. J Biol Chem. 2003;278:25046–25054. doi: 10.1074/jbc.M303438200. [DOI] [PubMed] [Google Scholar]
- 22.Sieghart W, et al. Mammalian target of rapamycin pathway activity in hepatocellular carcinomas of patients undergoing liver transplantation. Transplantation. 2007;83:425–432. doi: 10.1097/01.tp.0000252780.42104.95. [DOI] [PubMed] [Google Scholar]
- 23.Villanueva A, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–83. 1983 e1–11. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahin F, Kannangai R, Adegbola O, Wang J, Su G, Torbenson M. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–8425. doi: 10.1158/1078-0432.CCR-04-0941. [DOI] [PubMed] [Google Scholar]
- 25.Buitrago-Molina LE, Vogel A. mTor as a potential target for the prevention and treatment of hepatocellular carcinoma. Curr Cancer Drug Targets. 2012;12:1045–1061. doi: 10.2174/156800912803988011. [DOI] [PubMed] [Google Scholar]
- 26.Zhu AX, et al. Phase 1/2 study of everolimus in advanced hepatocellular carcinoma. Cancer. 2011;117:5094–5102. doi: 10.1002/cncr.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiah HS, et al. Randomised clinical trial: comparison of two everolimus dosing schedules in patients with advanced hepatocellular carcinoma. Aliment Pharmacol Ther. 2013;37:62–73. doi: 10.1111/apt.12132. [DOI] [PubMed] [Google Scholar]
- 28.Rizell M, Andersson M, Cahlin C, Hafstrom L, Olausson M, Lindner P. Effects of the mTOR inhibitor sirolimus in patients with hepatocellular and cholangiocellular cancer. Int J Clin Oncol. 2008;13:66–70. doi: 10.1007/s10147-007-0733-3. [DOI] [PubMed] [Google Scholar]
- 29.Schoniger-Hekele M, Muller C. Pilot study: rapamycin in advanced hepatocellular carcinoma. Aliment Pharmacol Ther. 2010;32:763–768. doi: 10.1111/j.1365-2036.2010.04404.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.