Abstract
Because human epidermal melanocytes (HEMs) provide critical protection against skin cancer, sunburn, and photoaging, a genome-wide perspective of gene expression in these cells is vital to understanding human skin physiology. In this study we performed high throughput sequencing of HEMs to obtain a complete data set of transcript sizes, abundances, and splicing. As expected, we found that melanocyte specific genes that function in pigmentation were among the highest expressed genes. We analyzed receptor, ion channel and transcription factor gene families to get a better understanding of the cell signalling pathways used by melanocytes. We also performed a comparative transcriptomic analysis of lightly versus darkly pigmented HEMs and found 16 genes differentially expressed in the two pigmentation phenotypes; of those, only one putative melanosomal transporter (SLC45A2) has known function in pigmentation. In addition, we found 166 genes with splice isoforms expressed exclusively in one pigmentation phenotype, 17 of which are genes involved in signal transduction. Our melanocyte transcriptome study provides a comprehensive view and may help identify novel pigmentation genes and potential pharmacological targets.
Keywords: RNA-Seq, melanocytes, transcriptome, melanosomes, pigmentation, signal transduction
Introduction
Human epidermal melanocytes (HEMs) play a critical role in protecting our skin from sunburn, photoaging, and skin cancer [1]. HEMs are located on the basal layer of the epidermis and are responsible for the synthesis of the photoprotective pigment melanin [2]. Impaired melanocyte function can have severe consequences such as increased skin cancer risk, premature skin aging, or pigmentation disorders (i.e. vitiligo and albinism). Skin cancer is the most common form of cancer in the US and melanoma, resulting from melanocyte transformation, accounts for ~9000 deaths annually in the United States alone [3]. Thus, obtaining a better understanding of melanocyte function and human skin pigmentation is key to identifying novel targets for the treatment of skin cancer and pigmentation disorders.
Current insight into melanocyte function and human pigmentation has been based in part on comparative genomics studies using mouse coat color genes. Of the 378 loci that affect mouse coat color, 171 genes are cloned and 207 remain unidentified [4], suggesting that the molecular mechanisms that regulate pigmentation are far from being elucidated. With the recent advances in high-throughput sequencing technologies the identification of such pigmentation genes might become feasible.
The constitutive pigmentation of the skin is primarily determined by the amount of melanin produced in epidermal melanocytes and is closely correlated with the incidence of skin cancer, indicating that melanin has an important photoprotective function [5, 6]. Indeed, in the United States, the incidence of basal and squamous cell carcinomas is 50 times lower in African Americans compared to Caucasians, while melanoma rates are 13 times lower in dark compared to light skin [7-9]. Melanin is produced and stored in organelles named melanosomes using a specific set of melanocyte-specific proteins. Some of these pigmentation proteins are enzymes involved in melanin synthesis [e.g. tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1), and tyrosinase related protein 2 (DCT)], while others are structural proteins or have unknown functions. Microphthalmia transcription factor (MITF), considered the master regulator of melanocyte function, modulates the transcription of many of the pigmentation genes [10, 11]. Mutations in seven of the pigmentation genes result in ocular or oculocutaneous albinism, characterized by reduced pigment levels in the skin and eye [12].
The protective function of melanin and the identification of pigmentation proteins raised the question of what accounts for the difference between dark and light skin. It was hypothesized that the main melanogenic enzyme TYR is expressed at higher levels in melanocytes from dark skin, thus producing more melanin. However, early studies showed that the mRNA and protein levels of TYR were similar in light and dark skin, but the activity of the enzyme appeared to be different between the two skin types, being correlated with the amount of cellular melanin [13-16]. What regulates the activity of TYR is not well understood; one possibility is that it depends of the levels of TYRP1 and/or DCT, which might be higher in dark skin [17, 18]. Recent studies suggested that different pigmentation phenotypes could be the result of various combinations of single nucleotide polymorphisms (SNPs) in some pigmentation genes [19]. One particular SNP resulting in a point mutation in the melanosomal protein mutated in oculocutaneous albinism IV (OCA4 or SLC45A2) shows strong correlation with skin color [20, 21]. Moreover, the mRNA for the allele present in light skin was found to be higher than the allele present in dark skin [22]. Thus, the molecular mechanisms that determine and regulate skin color are yet to be revealed.
The advent of high-throughput RNA sequencing (RNA-Seq) has provided a more sensitive and dynamic way to study mRNA expression. RNA-Seq results contain less noise and have higher specificity compared with microarray experiments. Unlike microarray experiments, RNA-Seq provides quantitative data at single-base resolution, information on transcript size, and is not limited to the number of known genes and transcript isoforms at the time of the study [23, 24]. In addition, RNA-Seq data can be reanalyzed as sequence databases are updated. Expression profiling of human epidermal melanocytes using Affymetrix microarrays was used as a reference point for changes in melanoma lines in a study that identified only 14,500 transcripts in normal melanocytes [25]. Another microarray study sequenced melanocytes from diverse geographical origin and found highly homogenous gene expression profiles among melanocytes from skin with different pigmentation levels [18]. The recent expansion of the transcriptome to over 30,000 known protein coding transcripts [26] and the critical role of melanocytes in normal and abnormal skin physiology, make the reassessment of the melanocyte transcriptome an important task.
In this study we performed RNA-Seq on lightly and darkly pigmented human epidermal melanocytes (HEMs) using the Illumina HiSeq 2000 platform. The vast data set obtained was used to perform a comparative transcriptomic analysis of mRNA expression levels in lightly versus darkly pigmented HEMs. This analysis provides an unbiased approach to detect novel regulators of melanin synthesis, providing insight into genes critical to melanocyte function. By compiling the gene expression data into specific families of cellular signalling molecules, we bring to light potential pharmacological targets and genes important for melanocyte signal transduction. This genome-wide trancriptome analysis may serve as a valuable resource for investigating human melanocyte function and provide insight into therapeutic targets for skin cancer and pigmentation disorders.
Material and Methods
Sample collection, library preparation and sequencing
Lightly and darkly pigmented primary human epidermal melanocytes (HEMs) from neonatal foreskin (Life Technologies/Gibco) were cultured in Medium 254 and Human Melanocyte Growth Supplement (HMGS2, Life Technologies/Gibco). The four HEM lines used for this study (HEM-D1, -D2, -L1, -L2) were each derived from a different donor; all lines were propagated in culture under identical conditions for ≤15 population doublings. Total HEM RNA was isolated using the mirVana miRNA Isolation Kit (Ambion) and its quality was assessed using an Agilent 2100 Bioanalyzer: RNA Integrity Number (RIN) ≥ 8.7 for all samples, in agreement with Illumina recommended RIN ≥ 8. cDNA libraries were prepared with 4μg total RNA using standard Illumina protocols (TruSeq RNA Sample Preparation Kit) and resulted in cDNA fragments with 229 bp average size, or 355bp including adapter sequences. Each cDNA library was sequenced with 50 bp single-read chemistry using the IIlumina HiSeq 2000 system. All of the sequence files have been submitted to the GenBank Sequence Read Archive for public access [Accession No. SRP039354 http://dx.doi.org/10.7301/Z0MW2F2N].
Data analysis and annotation
The computational pipeline used for data analysis is shown in Supplemental Figure S1. The quality of the raw sequencing data was checked using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and the quality scores of the reads for each library and for each position in the read were above 30, which is defined as high quality (Supplemental Figure S2). The sequencing data in FASTQ format was then mapped against NCBI build 37.2 of the human genome using Bowtie 2 (version 2.1.0.0) [27]. RNA sequencing metrics were obtained using Picard tools (http://picard.sourceforge.net/) (Table 1). Spliced junctions were identified using Tophat (version 2.0.8) [28] and transcript abundance estimates were performed using Cufflinks (version 2.1.1) [29]. EdgeR was used to perform differential gene expression analysis between samples with different pigmentation levels [30, 31]. Genes differentially expressed were considered significant if they had a FDR adjusted p-value < 0.05 [32]. For the isoform analysis we imported the Cufflinks isoform expression data of lightly and darkly pigmented HEMs into Microsoft Excel. Using Excel functions we identified the isoforms only present in HEM-L or HEM-D and excluded those with high degree of variability (standard error mean > 35% the average FPKM).
Table 1. RNA sequencing metrics and summary analysis of gene expression in HEMs.
RNA sequencing metrics of human epidermal melanocytes (HEMs) for two darkly pigmented (HEM-D1 & HEM-D2) and two lightly pigmented (HEM-L1 & HEM-L2) samples.
| Library | Produced/Aligned Reads (million) | Sequenced bases (Mb) | mRNA bases (%) | Number of genes detected | Gene expression (FPKM) | |
|---|---|---|---|---|---|---|
| Range | Mean | |||||
| HEM-D1 | 78.7 / 76.4 | 3939.2 | 83.3 | 14,782 | 0.1-14645.4 | 27.02 |
| HEM-D2 | 181.0 / 177.1 | 9053.1 | 85.3 | 15,252 | 0.1- 8627.7 | 31.51 |
| HEM-L1 | 174.0 / 168.4 | 8700.0 | 80.0 | 15,060 | 0.1- 9180.9 | 28.63 |
| HEM-L2 | 188.1 / 183.8 | 9405.2 | 83.9 | 15,071 | 0.1- 5347.3 | 24.07 |
The mean and median FPKM, and the FPKM range was calculated for each HEM cDNA library, as well as the number of genes detected. A threshold of FPKM ≥ 0.1 was used for all calculations.
qPCR analysis
3μg of total RNA was extracted from the same HEM lines used for RNAseq using the RNeasy Plus Kit (Qiagen) and reverse transcribed (RT) using SuperScript III kit (Life Technologies). The resulting cDNA was used for qPCR validation of each biological replicate analysed by RNA-Seq. Reactions were prepared according to the manufacturer's protocol using SYBR Select Master Mix (Invitrogen) and cycled on a VIIA-7 Real-Time PCR System (Applied Biosystems). β-actin was used as an internal control and all reactions were run in triplicate. mRNA levels were quantified by calculating average 2-ΔCt values, where Ct is the cycle number for the control and target transcript at the chosen threshold. ΔCt = Cttarget - Ctβ-actin was calculated by subtracting the average Ct of β-actin from the average Ct of the target transcript. Primers were designed to span an exon-exon junction to avoid amplification of any contaminate DNA. Primers used for SLC45A2, OCA2, and SLC24A5 qPCR were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and were as follows: SLC45A2 (NM_016180.3) - F: CCCTGTACACTGTGCCCTTT and R: CTTCCCTCTCACGCTGTTGT, OCA2 (NM_000275.2) - F: GTGTGCAGGGATTGCAGAAC and R: ACATCCCAACAGTGCAGGAC, SLC24A5 (NM_205850.2) - F: GCAGCAGGAACAAGCATACC and R: ATGGAATACCAAGGCACAACA.
The relative difference in expression between the two pigmentation phenotypes (HEM-D and HEM-L) was calculated as fold change in HEM-L vs. HEM-D (HEM-L/HEM-D); for RNA-Seq it was calculated by dividing average FPKM of HEM-L by that of HEM-D and for qPCR by dividing the average 2-ΔCt of HEM-L by that of HEM-D.
Melanin quantification
HEMs were grown to 70 – 90% confluence in 35 mm Petri dishes and melanin collected and quantified as previously described [33]. Briefly, after cell lysis soluble and insoluble fractions were separated by centrifugation. The soluble fraction was used to determine the total amount of protein in each sample using a Bradford assays (Bio-Rad Laboratories). The insoluble fraction containing melanin was resuspended in 100 μl of 1 M NaOH and incubated at 85°C until melanin was fully dissolved (15 – 30 min). Melanin was quantified by comparing the optical density of samples at 405 nm with a standard curve generated with serial dilutions of synthetic melanin (Sigma) dissolved in 1 M NaOH. Average cellular melanin values were calculated as the ratio between total melanin and total protein from the same dish of cells. All melanin quantifications were performed in triplicate to account for dish-to-dish variability.
Statistical Analysis
P-values were calculated using an unpaired two-tailed Student's t-test with P < 0.05 considered significant. For the differential gene expression analysis a FDR adjusted p-value < 0.05 was considered significant [32]. qPCR and melanin quantification were performed using the same biological replicates used for RNA-Seq analysis.
Results and Discussion
RNA sequencing of human epidermal melanocytes
Because melanocytes in the human epidermis are important for the constitutive pigmentation of our skin, which correlates with the risk of skin cancer and is also affected by many pigmentation disorders, we sought to obtain a global gene expression profile of melanocytes from skin with different pigmentation levels. We used primary human epidermal melanocytes (HEMs) in culture to perform a genome-wide transcriptome analysis. We used samples derived from lightly pigmented (HEM-L) and from darkly pigmented melanocytes (HEM-D), which have ~ 7-fold higher cellular melanin content than the lightly pigmented ones (Fig. 1A). To ensure biological replication, we used RNA sequencing (RNA-Seq) on four different libraries: two libraries derived from individual donors of darkly pigmented melanocytes (HEM-D1 & HEM-D2) and two from individual donors of lightly pigmented melanocytes (HEM-L1 & HEM-L2). Each HEM cDNA library was derived by isolation of polyA(+) mRNA and reverse transcription with random hexamer primers, and sequenced using 50bp single-read chemistry. The total number of reads for each cDNA library varied between 78 million and 188 million for the four samples (Table 1). The sequenced fragments were aligned to the NCBI build 37.2 of the human genome using Bowtie 2 [27], splice junctions were detected using Tophat [28], and transcript abundances were calculated in Fragments Per Kilobase of exon per Million fragments mapped (FPKM) using Cufflinks [29] (Supplemental Figure S1). Because FPKM reflects the expression level of the genes to which the fragments correspond, we chose a significance threshold of FPKM ≥ 0.1 to prevent analysis of transcripts with expression levels close to zero (bottom 25th percentile of transcripts had FPKM values < 0.05). Using this criterion we detected between 14,782 and 15,252 genes for all four libraries (Table 1). We also identified over 21,000 transcript variants across all four libraries. The gene expression statistics for each HEM cDNA library are shown in Table 1, while a complete data set of transcript abundances, sizes, and alternative splicing for all four libraries is found in the on-line Supplemental Table S1.
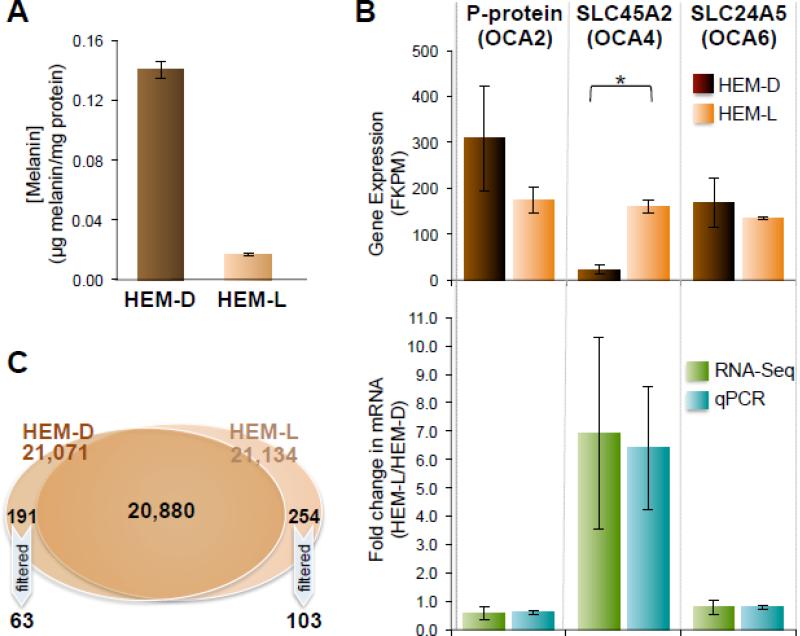
Figure 1. Differential gene and isoform expression in HEM-D vs. HEM-L.
A. Average melanin concentration of HEM-D vs. HEM-L. The average melanin concentration in the two darkly pigmented cell samples (HEM-D1 & HEM-D2) was ~ 7 fold higher than the average of the two lightly pigmented cell samples (HEM-L1 & HEM-L2) (n = 4, p < 0.002). Bars represent average ± SEM.
B. Expression of predicted melanosomal transporters in HEM-L vs. HEM-D. (top) Average transcript abundance of P-protein (OCA2), SLC45A2 (OCA4) and SLC24A5 (OCA6) in HEM-D and HEM-L determined by RNA-Seq (n = 2). SLC45A2 is expressed at significantly higher levels in HEM-L, compared to HEM-D (p < 0.015). Bars represent average FPKM ± SEM. (bottom) Comparison of fold change in transcript abundance of predicted melanosomal transporters in HEM-L vs. HEM-D, as determined by RNA-Seq or qPCR. SLC45A2 is expressed at higher levels in HEM-L by both RNA-Seq and qPCR, while P-protein and SLC24A5, with values less than one, have higher expression levels in HEM-D as measured by qPCR (n = 3, p < 0.01) but are not significantly different by RNA-Seq (n = 2, p > 0.36). The error bars represent the error propagation of the standard error mean from the average 2-ΔCt with respect to β-actin for qPCR, and ± the standard error mean from the average FPKM for RNA-Seq.
C. Distribution of the total number of transcripts identified from HEM-D and HEM-L. Venn diagram representing the distribution of the total number of transcripts between libraries obtained from darkly (HEM-D) and lightly (HEM-L) pigmented cells. The transcripts with FPKM < 0.1 in any of the libraries and variability >35% between the two libraries derived from the same pigmentation phenotype were filtered out.
The expression of different genes, as measured by FPKM, was in some cases variable between the four libraries. Genes that were expressed at different levels in the two dark HEM libraries compared to the two light ones were further analysed and discussed in Tables 5-7. For the rest of the genes that show variable expression levels, however, variability was not linked to the pigmentation phenotype of the cells and might represent biological variation between different individuals. Because we cannot exclude technical variability during library preparation, we used the average FPKM and the standard error mean for each gene to calculate the % variability among the four different samples. We generated a set of “high confidence” genes that had less than 35% variability among all biological replicates (included in Supplemental Table S1).
Table 5.
Differentially expressed genes in lightly (HEM-L) vs. darkly (HEM-D) pigmented melanocytes.
| Gene Name (Gene ID) | Relative expression in HEM-L vs. HEM-D | Fold Change | FDR p-value |
|---|---|---|---|
| apolipoprotein C-I (APOC1) | Up-regulated | 13 | 3.81 × 10−3 |
| apolipoprotein C-II (APOC2) | Up-regulated | 117 | 2.74 × 10−7 |
| family with sequence similarity 171, member A1 (FAM171A1) | Up-regulated | 12 | 1.81 × 10−2 |
| glyceraldehyde-3-phosphate dehydrogenase (GAPDHS) | Up-regulated | 8 | 1.63 × 10−2 |
| leucine rich repeat containing 61 (LRRC61) | Up-regulated | 20 | 2.23 × 10−2 |
| leucine rich repeat neuronal 1 (LRRN1) | Up-regulated | 47 | 1.12 × 10−2 |
| protein tyrosine phosphatase N, polypeptide 2 (PTPRN2) | Up-regulated | 24 | 3.23 × 10−2 |
| sarcoglycan, delta (SGCD) | Up-regulated | 15 | 2.74 × 10−2 |
| solute carrier family 45, member 2 (SLC45A2) | Up-regulated | 8 | 4.22 × 10−2 |
| tudor domain containing 12 (TDRD12) | Up-regulated | 27 | 2.69 × 10−3 |
| zinc finger protein 423 (ZNF423) | Up-regulated | 17 | 2.71 × 10−2 |
| claudin 1 (CLDN1) | Down-regulated | 10 | 1.43 × 10−2 |
| CXXC finger protein 4 (CXXC4) | Down-regulated | 19 | 1.59 × 10−2 |
| kinase insert domain receptor (KDR) | Down-regulated | 8 | 2.21 × 10−2 |
| myosin VIIB (MYO7B) | Down-regulated | 25 | 3.81 × 10−2 |
| protein tyrosine phosphatase, receptor type, O (PTPRO) | Down-regulated | 18 | 4.88 × 10−2 |
The number of reads that aligned to the identified genes in the HEM-L and HEM-D libraries were analyzed with the edgeR software to detect significant differences (FDR p-value < 0.05). The only pigmentation-associated gene differentially regulated (SLC45A2) is shown in bold.
Table 7.
Expression of cell signaling isoforms in HEM-L and HEM-D.
| A. Isoforms only expressed in HEM-D | |||
|---|---|---|---|
| Gene Name (Gene ID) | Accession Number | Gene Family | Transcript Variant |
| 5-hydroxytryptamine (serotonin) receptor 7 (HTR7) | NM_019859.3 | GPCR | Variant D Canonical sequence |
| frizzled class receptor 6 (FZD6) | NM_001164616.1 | GPCR | Variant 3 32aa missing |
| activin A receptor, type IC (ACVR1C) | NM_145259.2 | RK | Variant 1 Canonical sequence |
| histone linker H1 domain, spermatid-specific 1 (HILS1) | NR_024192.1 | TF | Variant 2 Non-coding |
| zinc finger protein 83 (ZNF83) | NM_001105550.1 | TF | Variant 3 Different 5’UTR |
| nuclear receptor subfamily 6, group A, member 1 (NR6A1) | NM_001489.4 | NR | Variant 2 5aa missing |
| B. Isoforms only expressed in HEM-L | |||
|---|---|---|---|
| Gene Name (Gene ID) | Accession Number | Gene Family | Transcript Variant |
| histamine receptor H4 (HRH4) | NM_001143828.1 | GPCR | Variant 2 88aa missing |
| regulator of G-protein signaling 3 (RGS3) | NM_001282922.1 | GAP | Variant 2 398aa missing |
| postmeiotic segregation increased 1(PMS1) | NM_001128143.1 | TF | Variant 2 39aa missing |
| ets variant 7 (ETV7) | NM_001207038.1 | TF | Variant 5 77aa missing |
| T-box 3 (TBX3) | NM_016569.3 | TF | Variant 2 Canonical sequence |
| hes family bHLH transcription factor 6 (HES6) | NM_018645.4 | TF | Variant 1 Canonical sequence |
| cut-like homeobox 1 (CUX1) | NM_001202546.1 | TF | Variant 7 39aa missing |
| myoneurin (MYNN) | NM_001185119.1 | TF | Variant 3 29aa missing |
| WD repeat & HMG-box DNA binding protein 1 (WDHD1) | NM_001008396.2 | TF | Variant 2 123aa missing |
| zinc finger protein (ZNF200) | NM_198087.2 | TF | Variant 3 1aa missing |
| zinc finger protein (ZNF662) | NM_207404.3 | TF | Variant 1 Canonical sequence |
| zinc finger protein (ZNF674) | NM_001039891.2 | TF | Variant 1 Canonical sequence |
| zinc finger protein 75D (ZNF75D) | NM_001185063.2 | TF | Variant 2 95aa missing |
| zinc finger protein (ZNF805) | NM_001145078.1 | TF | Variant 2 133aa missing |
Transcript isoforms from cell signaling gene families expressed exclusively in lightly pigmented (A) or darkly pigmented (B) HEMs Isoforms below expression threshold (FPKM < 0.1) and with high variability (SEM > 35% of its average FPKM) were excluded. Abbreviations: GPCR, G protein-coupled receptor; RK, receptor kinase; GAP, GTPase-accelerating protein; TF, transcription factor; NR, Nuclear receptor.
Genes highly expressed in human epidermal melanocytes
HEMs, due to their unique ability to produce melanin, require a constant turnover of melanosomes, specialized intracellular organelles in which melanin is synthesized, stored, and transferred to neighbouring keratinocytes. The abundant presence of these unique organelles in melanocytes suggests that in HEMs highly expressed genes also encode melanosomal components such as melanogenic enzymes, structural proteins, or regulators of the melanosomal environment and function. We determined the highest expressing genes in HEMs by arranging our results in the decreasing order of their average FPKM (Table 2). Not unexpectedly, we found that many melanocyte specific pigmentation genes (bolded in Table 2) are expressed at or above the level of the common housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin (Table 2; highlighted in dark grey background). In particular, six melanocyte specific pigmentation genes: tyrosinase (TYR), tyrosinase related protein 1 (TYRP1), tyrosinase related protein 2 (DCT), premelanosomal protein (PMEL17), glycoprotein NMB (GPNMB), and melanoma antigen 1 (MLANA) are among the 30 highest expressing genes in HEMs (highlighted in Table 2). PMEL17, a structural melanosomal protein important in early stage melanosome biogenesis, had the highest average expression in HEMs (FPKM = 8302.8 ± 2306.6) compared with all genes, almost an order of magnitude higher than β-actin (FPKM = 852.5 ± 140.8). TYR, TYRP1 and DCT are key enzymes involved in the melanin synthesis pathway; mutations in TYR and TYRP1 cause severe hypopigmentation and oculocutaneous albinism type 1 (OCA1) and type 3 (OCA3), respectively [34-36]. Widely expressed genes such as vimentin (VIM), ferritin heavy polypeptide (FTH1), ferritin light polypeptide (FTL), and prosaposin (PSAP) are also among the highest expressed genes.
Table 2.
Highly expressed genes in HEMs
| Gene Name (Gene ID) | Average FPKM ± SEM |
|---|---|
| premelanosome protein (PMEL17) | 8302.8 ± 2306.6 |
| tyrosinase-related protein 1 (TYRP1) | 7481.6 ± 882.4 |
| eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) | 3172.8 ± 734.0 |
| vimentin (VIM) | 2912.1 ± 526.4 |
| ferritin, heavy polypeptide (FTH1) | 2536.3 ± 416.5 |
| CD63 molecule (CD63) | 2355.4 ± 229.4 |
| ferritin, light polypeptide (FTL) | 1848.3 ± 268.5 |
| glycoprotein (transmembrane) nmb (GPNMB) | 1795.5 ± 447.3 |
| tumor protein, translationally-controlled 1(TPT1) | 1722.7 ± 362.5 |
| Prosaposin (PSAP) | 1682.8 ± 327.1 |
| ATPase, Na+/K+ transporting, alpha 1 polypeptide (ATP1A1) | 1628.1 ± 255.3 |
| tyrosinase-related protein 2 (TYRP2) | 1566.2 ± 538.9 |
| actin, gamma 1 (ACTG1) | 1562.1 ± 86.3 |
| melanoma antigen 1 (MLANA) | 1550.1 ± 244.1 |
| glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 1524.2 ± 141.1 |
| tyrosinase (TYR) | 1517.2 ± 65.0 |
| thymosin beta 10 (TMSB10) | 1367.6 ± 158.3 |
| eukaryotic translation elongation factor 1 gamma (EEF1G) | 1155.6 ± 157.7 |
| annexin A5 (ANXA5) | 1139.9 ± 93.8 |
| lectin, galactoside-binding, soluble, 3 (LGALS3) | 1087.2 ± 76.5 |
| spermidine/spermine N1-acetyltransferase 1 (SAT1) | 1033.1 ± 387.8 |
| lectin, galactoside-binding, soluble, 1 (LGALS1) | 1030.8 ± 63.7 |
| ubiquitin B (UBB) | 961.3 ± 161.7 |
| heat shock 70kDa protein 8 (HSPA8) | 944.6 ± 81.0 |
| cofilin 1 (non-muscle) (CFL1) | 922.6 ± 89.3 |
| ATP1A1 opposite strand (ATP1A1OS) | 853.4 ± 134.1 |
| actin, beta (ACTB) | 852.5 ± 140.8 |
| stearoyl-CoA desaturase (delta-9-desaturase) (SCD) | 848.4 ± 188.4 |
| TIMP metallopeptidase inhibitor 2 (TIMP2) | 835.1 ± 143.4 |
| G protein, beta polypeptide 2-like 1 (GNB2L1) | 813.6 ± 110.9 |
Highly expressed genes in melanocytes were obtained by averaging their FPKM from each library. For concision, ribosomal and non-coding genes we excluded from this table (their expression values can be found in Supplementary Table S1). Melanocyte specific pigmentation genes are highlighted in bold and, for comparison of expression levels, commonly used housekeeping genes (GAPDH and β-actin) are shown in dark grey background.
Receptors and ion channels highly expressed in human epidermal melanocytes
Receptors and ion channels play a crucial role in cellular signalling and are often used as drug targets. To identify receptors and ion channels highly expressed in HEMs and presumably important for their function, we compiled gene family lists of all known G protein-coupled receptors (GPCRs), receptor kinases, and ion channels using the IUPHAR database and the HUGO genes repository [37], then sorted these categories in the order of their average FPKM (Tables 3, 4, and Supplemental Table S5). As expected, we found that several genes known to be important for melanocyte function (shown in bold) among the highly expressed receptors and ion channels.
Table 3.
Top 25 highly expressed G protein-coupled receptors in HEMs
| Gene Name (Gene ID) | Average FPKM ± SEM |
|---|---|
| G protein-coupled receptor 143 (OA1) | 254.7 ± 44.0 |
| endothelin receptor type B (EDNRB) | 211.6 ± 43.0 |
| G protein-coupled receptor 56 (GPR56) | 148.8 ± 29.3 |
| G protein-coupled receptor 137B (GPR137B) | 117.4 ± 7.1 |
| 5-hydroxytryptamine (serotonin) receptor 2B (HTR2B) | 66.4 ± 37.5 |
| cysteinyl leukotriene receptor 2 (CYSLTR2) | 29.5 ± 14.4 |
| G protein-coupled receptor 107 (GPR107) | 25.4 ± 2.6 |
| coagulation factor II (thrombin) receptor (F2R) | 24.6 ± 9.2 |
| G protein-coupled receptor 108 (GPR108) | 21.5 ± 3.6 |
| G protein-coupled receptor 125 (GPR125) | 21.2 ± 1.5 |
| G protein-coupled receptor 175 (GPR175) | 14.6 ± 3.3 |
| G protein-coupled receptor, fam. C, group 5, member A (GPRC5A) | 13.3 ± 1.9 |
| melanocortin 1 receptor (MC1R) | 12.8 ± 4.9 |
| cadherin, EGF LAG seven-pass G-type receptor 2 (CELSR2) | 11.9 ± 3.0 |
| G protein-coupled receptor 124 (GPR124) | 11.3 ± 1.9 |
| CD97 molecule (CD97) | 10.9 ± 1.8 |
| encephalopsin (OPN3) | 10.1 ± 1.5 |
| G protein-coupled receptor, fam. C, group 5, member B (GPRC5B) | 9.5 ± 3.8 |
| smoothened, frizzled family receptor (SMO) | 8.8 ± 1.3 |
| lysophosphatidic acid receptor 6 (LPAR6) | 8.5 ± 3.9 |
| frizzled family receptor 6 (FZD6) | 8.5 ± 3.1 |
| G protein-coupled receptor 137 (GPR137) | 8.2 ± 1.3 |
| gonadotropin-releasing hormone (type 2) receptor 2 (GNRHR2) | 7.9 ± 0.7 |
| gamma-aminobutyric acid (GABA) B receptor, 1 (GABBR1) | 7.9 ± 1.4 |
| G protein-coupled receptor 155 (GPR155) | 7.8 ± 1.4 |
The highest expressed GPCRs in melanocytes were obtained by averaging their FPKM from each library. The mean expression levels of orphan GPCRs shown in dark grey background and receptors known to be important for melanocyte function (GPR143, EDNRB, and MC1R) are in bold.
Table 4.
Top 25 highly expressed ion channel genes in HEMs.
| Gene Name (Gene ID) | Average FPKM ± SEM |
|---|---|
| chloride intracellular channel 1 (CLIC1) | 242.8 ± 28.6 |
| purinergic receptor P2X, ligand-gated ion channel, 7 (P2RX7) | 78.4 ± 16.7 |
| TRP cation channel, subfamily V, member 2 (TRPV2) | 75.6 ± 16.2 |
| chloride channel, voltage-sensitive 3 (CLCN3) | 72.4 ± 13.5 |
| chloride intracellular channel 4 (CLIC4) | 71.4 ± 16.7 |
| TRP cation channel, subfamily M, member 1 (TRPM1) | 57.8 ± 9.4 |
| two pore segment channel 2 (TPCN2) | 52.1 ± 7.2 |
| chloride channel, voltage-sensitive 7 (CLCN7) | 50.6 ± 8.0 |
| polycystic kidney disease 2 (PKD2) | 31.6 ± 7.6 |
| bestrophin 1 (BEST1) | 31.3 ± 7.4 |
| pannexin 1 (PANX1) | 25.2 ± 2.7 |
| anoctamin 10 (ANO10) | 23.5 ± 3.1 |
| anoctamin 6 (ANO6) | 21.7 ± 4.0 |
| potassium channel, subfamily J, member 13 (KCNJ13) | 17.6 ± 9.4 |
| purinergic receptor P2X, ligand-gated ion channel, 4 (P2RX4) | 17.1 ± 1.7 |
| mucolipin 1 (MCOLN1, TRPML1) | 16.8 ± 3.3 |
| mucolipin 3 (MCOLN3, TRPML3) | 15.5 ± 5.6 |
| inositol 1,4,5-trisphosphate receptor, type 3 (ITPR3) | 12.6 ± 3.0 |
| chloride channel, voltage-sensitive 6 (CLCN6) | 11.8 ± 2.3 |
| potassium channel, delayed-rectifier, subfamily S, member 3 (KCNS3) | 10.7 ± 2.3 |
| TRP cation channel, subfamily M, member 7 (TRPM7) | 10.2 ± 3.1 |
| gap junction protein, gamma 1, 45kDa (GJC1) | 9.5 ± 2.7 |
| two pore segment channel 1 (TPCN1) | 8.8 ± 2.3 |
| chloride channel, voltage-sensitive 5 (CLCN5) | 5.1 ± 0.6 |
| gap junction protein, alpha 3, 46kDa (GJA3) | 4.8 ± 0.3 |
The highest expressed ion channels in melanocytes were obtained by averaging their FPKM from each library. Nine out of the 25 genes are chloride channels (shown in dark grey background). Ion channels reported to be important for melanocyte function are in bold.
We first sorted GPCRs and receptor kinases to find the highest expressed signalling receptor genes in HEMs. Both the endothelin receptor type B (EDNRB) and the melanocortin 1 receptor (MC1R) are among the top 25 highest expressing GPCRs in HEMs (Table 3). EDNRB plays an important role in melanocyte development and migration [38], while MC1R is critical for hair and skin pigmentation and plays a critical role in the UVB-induced pigmentation response [39-41]. The melanosomal receptor GPR143 (mutated in ocular albinism type 1, OA1), a receptor important for melanosome structure and biogenesis, was the highest expressing GPCR gene [42-44]. Interestingly, eleven of the top 25 GPCRs highly expressed in HEMs are orphan receptors (Table 3; shown in dark grey background). This finding is particularly exciting because identifying endogenous ligands for these receptors and associated signalling mechanisms might uncover novel pathways important for melanocyte function. Interestingly, the serotonin receptor, 5-hydroxytryptamine receptor 2B (HTR2B), and the predicted extraocular photoreceptor encephalopsin (OPN3) are also highly expressed, yet their physiological functions in melanocytes are not known. Critical melanocyte receptor kinases such as stem cell growth factor receptor c-Kit (KIT), tyrosine-protein kinase receptor TYRO3 (TYRO3), and the hepatocyte growth factor receptor (MET) are among the 10 highest expressed receptor kinase genes Supplemental Table S5 [45-47].
We next sorted the ion channels according to expression values in HEMs and found that among the top 25 highly expressed ion channel genes in HEMs are TRPM1, TRPM7, and TRPML3 (shown in bold in Table 4), known to function in regulating melanin content and pigmentation of human, mouse, and zebra fish [33, 48-50]. Interestingly, nine out of the top 25 highly expressed ion channel genes are chloride channels (Table 4; shown in dark grey background), suggesting that chloride regulation is important for melanocyte function. Among the highly expressed channels are the calcium activated chloride channel BEST1 [51], the intracellular CLIC1 channel [52], and the non-selective cation channel TRPV2 [53, 54]. Interestingly, TRPV2 ion channels were also found by a proteomics study in purified melanosomes [55]. In addition, the highly expressed chloride channel, CLCN7, and the two-pore channel, TPCN2, are associated with variations in pigmentation [56, 57], suggesting a role in melanosomal physiology and melanin content of melanocytes. The Transient Receptor Potential A1 (TRPA1) channel that was shown to be expressed in melanocytes and important for melanocyte responses to ultraviolet A radiation [58] is not among the highly expressed ion channels (TRPA1, FKPM = 0.3 ± 0.1), in agreement with the small size of the TRPA1 currents measured in HEMs by patch clamp [58]. This suggests that while high expression of ion channels could correlate with important physiological function, low expressing signalling proteins can also be critical for cell function. A complete list of GPCR, receptor kinase and ion channel gene expression in HEMs is found in Supplemental Table S2.
Transcription factor gene expression in human epidermal melanocytes
Transcription factors regulate a multitude of cellular processes by controlling the rates at which genes are transcribed. Using a previously compiled list of 1391 human transcription factor genes [59], we generated a table of the top 25 highest expressing transcription factor genes in HEMs (Supplemental Table S6). We found transcription factor genes important for melanocyte function such as microphthalmia-associated transcription factor (MITF), sex determining region Y-box 10 (SOX10), and Y box binding protein 1 (YBX1) to be among the top 25 highly expressed transcription factor genes. MITF is responsible for regulating a multitude of genes necessary for melanocyte development and differentiation [60]; SOX10 is a regulator of MITF and has a major role in melanocyte development [61, 62], while YBX1 is an activator of MIA (melanoma inhibitory activity) and plays an important part in the progression of malignant melanoma [63]. A complete list of transcription factor gene expression in HEMs is found in Supplemental Table S3.
Differentially expressed genes and isoforms in lightly versus darkly pigmented melanocytes
HEMs contain different amounts of melanin correlated with the pigmentation phenotype of the donor. Because melanin is photoprotective, we sought to identify differentially expressed genes and isoforms in lightly vs. darkly pigmented HEMs (HEM-L vs. HEM-D). We performed a global differential gene expression analysis using edgeR software [30, 31] and found 94 genes expressed at significantly different levels (false discover rate (FDR) adjusted p-value < 0.05), with 16 genes remaining after filtering genes expressed below threshold (FPKM ≥ 0.1) [32] (Table 5). Apolipoprotein C-II (APOC2) had the highest fold change in expression between HEM-L and HEM-D. Interestingly, solute carrier family 45, member 2 (SLC45A2) was the only known pigmentation gene that was significantly different between the two pigmentation phenotypes. Moreover, SLC45A2 was increased 8-fold in HEM-L compared to HEM-D (shown in bold in Table 5), as previous studies have suggested [22].
To confirm the EdgeR results and to further compare pigmentation gene expression in melanocytes, we analysed the average FPKM values of 11 melanocyte specific pigmentation genes (as defined in [64]), as a function of pigmentation levels and represented them in groups according to their predicted function (Table 6). As expected, all of the pigmentation genes were expressed at high levels, some of these transcripts had high variability between samples, but, with the exception of SLC45A2, we did not detect any pigmentation gene that was significantly different in darkly vs. lightly pigmented cells (Table 6). The similar mRNA expression levels of the rate limiting enzyme, tyrosinase (TYR) in HEM-L and HEM-D was confirmed with average FPKM ± SEM values of 1429.8 ± 18.0 (HEM-L) and 1604.6 ± 98.9 (HEM-D), respectively [14].
Table 6.
Expression of melanocyte specific pigmentation genes in lightly (HEM-L) vs. darkly (HEM-D) pigmented melanocytes.
| Gene Name (Gene ID) | Function (predicted) | Average HEM-L FPKM ± SEM | Average HEM-D FPKM ± SEM |
|---|---|---|---|
| P protein (OCA2) | 174.5 ± 28.7 | 308.3 ± 114.7 | |
| solute carrier family 45 member 2 (SLC45A2) | melanosomal transporter | 160.6 ± 14.0 | 23.2 ± 8.8 |
| solute carrier family 24 member 5 (SLC24A5) | 134.3 ± 3.4 | 167.6 ± 53.2 | |
| premelanosome protein (PMEL) | 7047.6 ± 1700.3 | 9558.0 ± 5087.4 | |
| melan-A (MLANA) | melanosome structure | 1677.3 ± 415.4 | 1423.0 ± 390.7 |
| G protein-coupled receptor 143 (OA1) | 238.9 ± 71.2 | 270.5 ± 77.9 | |
| glycoprotein (transmembrane) nmb (GPNMB) | melanosomal component | 1582.8 ± 302.6 | 2008.2 ± 1009.1 |
| tyrosinase (TYR) | 1429.8 ± 18.0 | 1604.6 ± 98.9 | |
| tyrosinase-related protein 1 (TYRP1) | melanogenic enzyme | 7250.1 ± 1930.9 | 7713.1 ± 914.7 |
| dopachrome tautomerase (DCT) | 1922.6 ± 1187.3 | 1209.8 ± 280.5 | |
| osteopetrosis associated transmembrane protein 1 (OSTM1) | pheomelanin synthesis | 105.4 ± 8.0 | 128.3 ± 62.2 |
The average FPKM for HEM-L and HEM-D libraries was calculated for melanocyte specific genes involved in pigmentation, classified according to their predicted function. The predicted melanosomal transporter SLC45A2, shown in bold, is the only gene expressed at significantly different levels in HEM-L vs. HEM-D.
SLC45A2 is predicted to function as a melanosomal transporter [65, 66], similar to P-protein and SLC24A5, but the role of these proteins in melanosomal function is not known. Moreover, mutations in all of these predicted transporters result in reduced melanin levels and oculocutaneous albinism: mutations in SLC45A2 result in oculocutaneous albinism type 4 (OCA4) [65], mutations in P-protein results in OCA2 [67], and mutations in SLC24A5 result in OCA6 [12]. To further explore differences in the expression levels of the melanosomal transporters between lightly and darkly pigmented melanocytes we represented their average FKPM as a function of pigmentation phenotype (Fig. 1B).
To validate the RNA-Seq data and to verify the identified difference between SLC45A2 mRNA expression in HEM-L vs. HEM-D, we measured by qPCR the mRNA levels of the three predicted melanosomal transporters: P-protein, SLC45A2, and SLC24A5 in cells derived from the same biological samples as used for RNA-Seq and corresponding to the two pigmentation phenotypes. We then calculated for each gene the average mRNA fold increase in HEM-L compared to HEM-D for RNA-Seq and qPCR (Fig. 1B). In our analysis genes enriched in HEM-L would have values greater than one, while genes expressed at higher levels in HEM-D would have values less than one. Both RNA-Seq and qPCR results showed similar fold changes and confirmed our previous observations that SLC45A2 (OCA4) is expressed at significantly higher levels in HEM-L. P-protein (OCA2) was found to be higher in HEM-D by qPCR (consistent with previous studies [22, 68]) as well as SLC24A5 (OCA6), but the difference was not statistically different by RNA-Seq. Interestingly, the SLC45A2 mRNA found in the HEM-L libraries was exclusively accounted for by the 374F allele, while HEM-D libraries contained the 374L allele, in agreement with previous findings [20-22].
In addition to our EdgeR analysis of differential gene expression, we investigated whether different isoforms of the same genes were differentially expressed for each pigmentation phenotype. Using Cufflinks isoform expression data, we isolated transcripts only expressed in HEM-L or HEM-D. After filtering out transcripts with a standard error mean greater than 35% of the average FPKM, we identified a total of 103 isoforms expressed only in HEM-L and 63 isoforms only HEM-D (Fig. 1C, Supplemental Table S4). From these isoforms, we isolated the genes likely to have a signalling role and to function as potential melanogensis regulators, shown in Table 7. Only four signalling isoforms were identified in HEM-D (Table 7A) and 13 isoforms in HEM-L (Table 7B). Three of the differentially expressed transcripts were G protein coupled receptors (GPCR), one was a receptor kinase (RK), one was a regulator of G protein signalling (RGS3) and 12 isoforms represented transcription factors. Among those, some isoforms encoded the canonical sequence of the gene, while others were splice variants resulting in truncated proteins.
Thus, our differential analysis results, despite the limited number of biological replicates, offered valuable insight into the expression of signalling proteins in melanocytes and revealed a single melanocyte specific gene and several genes and isoforms with unknown function in pigmentation being differentially expressed as a function of pigmentation phenotype.
Conclusions
In this study, we performed global gene expression analysis of HEMs with a focus on gene families related to signal transduction. To our knowledge this study is the first report of a comparative transcriptomic analysis of lightly versus darkly pigmented HEMs using high throughput sequencing technology. This work revealed a novel set of 15 genes and 166 transcript isoforms that are differentially expressed in lightly versus darkly pigmented HEMs that are potentially related to differences in skin pigmentation. SLC45A2 was the only melanocyte specific pigmentation gene differentially expressed, suggesting that SLC45A2 may be a key regulator of melanogenesis. These results provide a comprehensive view of the HEM transcriptome and a foundation for discovering novel pharmacological targets for treatment of pigmentation disorders and melanoma.
Supplementary Material
Research Highlights.
The transcriptome of human melanocytes was determined using RNA sequencing.
We compared melanocytes with different pigmentation levels from different skin types.
Our analysis focused on cell signaling genes like receptors and ion channels.
Pigmentation genes specific to melanocytes were expressed at very high levels.
Our study found 15 novel genes differentially expressed in light vs. dark human melanocytes.
Acknowledgments
We thank Dr. Jason G. Wood and Dr. Ece D. Gamsiz for helpful discussions. This work was funded by grants from Brown University (to E.O.), COBRE Pilot Project P30GM103410 (to E.O.), National Institute of General Medical Sciences of the National Institutes of Health Award No. R25GM083270 (to K.D.H.), and NSF Graduate Research Fellowship Award No. DGE-1058262 (to K.D.H.).
Abbreviations
- >HEMs
human epidermal melanocytes
- FPKM
Fragments Per Kilobase of exon per Million fragments mapped
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare no competing interests.
Authors' contributions: K.D.H. and E.O. designed the experiments, K.D.H. performed all experiments and analysed the data, K.D.H. and E.O. wrote the manuscript.
References
- 1.Heck DE, Gerecke DR, Vetrano AM, Laskin JD. Solar ultraviolet radiation as a trigger of cell signal transduction. Toxicol Appl Pharmacol. 2004;195:288–297. doi: 10.1016/j.taap.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 2.Costin GE, Hearing VJ. Human skin pigmentation: melanocytes modulate skin color in response to stress. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2007;21:976–994. doi: 10.1096/fj.06-6649rev. [DOI] [PubMed] [Google Scholar]
- 3.Skin cancer statistics. Centers for Disease Control and Prevention; [Google Scholar]
- 4.Montoliu L, Oetting WS, Bennett DC. Color Genes. European Society for Pigment Cell Research. 2013 [Google Scholar]
- 5.Tadokoro T, Kobayashi N, Zmudzka BZ, Ito S, Wakamatsu K, Yamaguchi Y, Korossy KS, Miller SA, Beer JZ, Hearing VJ. UV-induced DNA damage and melanin content in human skin differing in racial/ethnic origin. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2003;17:1177–1179. doi: 10.1096/fj.02-0865fje. [DOI] [PubMed] [Google Scholar]
- 6.Zeise L, Chedekel MR, Fitzpatrick TB. Melanin--its Role in Human Photoprotection: A Melanin Symposium Held on March 11 and 12, 1994. Valdenmar Pub.; Washington: 1995. [Google Scholar]
- 7.Halder RM, Bridgeman-Shah S. Skin cancer in African Americans. Cancer. 1995;75:667–673. doi: 10.1002/1097-0142(19950115)75:2+<667::aid-cncr2820751409>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 8.Preston DS, Stern RS. Nonmelanoma cancers of the skin. The New England journal of medicine. 1992;327:1649–1662. doi: 10.1056/NEJM199212033272307. [DOI] [PubMed] [Google Scholar]
- 9.Kricker A, Armstrong BK, English DR. Sun exposure and non-melanocytic skin cancer. Cancer causes & control : CCC. 1994;5:367–392. doi: 10.1007/BF01804988. [DOI] [PubMed] [Google Scholar]
- 10.Shibahara S, Yasumoto K, Amae S, Udono T, Watanabe K, Saito H, Takeda K. Regulation of pigment cell-specific gene expression by MITF. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2000;13(Suppl 8):98–102. doi: 10.1034/j.1600-0749.13.s8.18.x. [DOI] [PubMed] [Google Scholar]
- 11.Tachibana M. MITF: a stream flowing for pigment cells. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2000;13:230–240. doi: 10.1034/j.1600-0749.2000.130404.x. [DOI] [PubMed] [Google Scholar]
- 12.Montoliu L, Gronskov K, Wei AH, Martinez-Garcia M, Fernandez A, Arveiler B, Morice-Picard F, Riazuddin S, Suzuki T, Ahmed ZM, Rosenberg T, Li W. Increasing the complexity: new genes and new types of albinism. Pigment cell & melanoma research. 2014;27:11–18. doi: 10.1111/pcmr.12167. [DOI] [PubMed] [Google Scholar]
- 13.Naeyaert JM, Eller M, Gordon PR, Park HY, Gilchrest BA. Pigment content of cultured human melanocytes does not correlate with tyrosinase message level. The British journal of dermatology. 1991;125:297–303. doi: 10.1111/j.1365-2133.1991.tb14161.x. [DOI] [PubMed] [Google Scholar]
- 14.Iozumi K, Hoganson GE, Pennella R, Everett MA, Fuller BB. Role of tyrosinase as the determinant of pigmentation in cultured human melanocytes. The Journal of investigative dermatology. 1993;100:806–811. doi: 10.1111/1523-1747.ep12476630. [DOI] [PubMed] [Google Scholar]
- 15.Talwar HS, Griffiths CE, Fisher GJ, Russman A, Krach K, Benrazavi S, Voorhees JJ. Differential regulation of tyrosinase activity in skin of white and black individuals in vivo by topical retinoic acid. The Journal of investigative dermatology. 1993;100:800–805. doi: 10.1111/1523-1747.ep12476615. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Malek Z, Swope V, Collins C, Boissy R, Zhao H, Nordlund J. Contribution of melanogenic proteins to the heterogeneous pigmentation of human melanocytes. Journal of cell science. 1993;106(Pt 4):1323–1331. doi: 10.1242/jcs.106.4.1323. [DOI] [PubMed] [Google Scholar]
- 17.Alaluf S, Barrett K, Blount M, Carter N. Ethnic variation in tyrosinase and TYRP1 expression in photoexposed and photoprotected human skin. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2003;16:35–42. doi: 10.1034/j.1600-0749.2003.00005.x. [DOI] [PubMed] [Google Scholar]
- 18.Alonso S, Izagirre N, Smith-Zubiaga I, Gardeazabal J, Diaz-Ramon JL, Diaz-Perez JL, Zelenika D, Boyano MD, Smit N, de la Rua C. Complex signatures of selection for the melanogenic loci TYR, TYRP1 and DCT in humans. BMC evolutionary biology. 2008;8:74. doi: 10.1186/1471-2148-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturm RA. A golden age of human pigmentation genetics. Trends in genetics : TIG. 2006;22:464–468. doi: 10.1016/j.tig.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama K, Fukamachi S, Kimura H, Koda Y, Soemantri A, Ishida T. Distinctive distribution of AIM1 polymorphism among major human populations with different skin color. Journal of human genetics. 2002;47:92–94. doi: 10.1007/s100380200007. [DOI] [PubMed] [Google Scholar]
- 21.Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Human mutation. 2005;25:278–284. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- 22.Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, Brown DL, Duffy DL, Pastorino L, Bianchi-Scarra G, Leonard JH, Stow JL, Sturm RA. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. The Journal of investigative dermatology. 2009;129:392–405. doi: 10.1038/jid.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome research. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, Wu T, Niinobe M, Yoshikawa K, Hannigan GE, Halaban R. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer research. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 26.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic acids research. 2012;40:D130–135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy D.J. CYSGK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic acids research. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson M.D. MDJSGK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. The Annals of Applied Statistics. 2001;29:1165–1188. [Google Scholar]
- 33.Oancea E, Vriens J, Brauchi S, Jun J, Splawski I, Clapham DE. TRPM1 forms ion channels associated with melanin content in melanocytes. Sci Signal. 2009;2:ra21. doi: 10.1126/scisignal.2000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomita Y, Takeda A, Okinaga S, Tagami H, Shibahara S. Human oculocutaneous albinism caused by single base insertion in the tyrosinase gene. Biochemical and biophysical research communications. 1989;164:990–996. doi: 10.1016/0006-291x(89)91767-1. [DOI] [PubMed] [Google Scholar]
- 35.Boissy RE, Zhao H, Oetting WS, Austin LM, Wildenberg SC, Boissy YL, Zhao Y, Sturm RA, Hearing VJ, King RA, Nordlund JJ. Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as “OCA3”. American journal of human genetics. 1996;58:1145–1156. [PMC free article] [PubMed] [Google Scholar]
- 36.Manga P, Kromberg JG, Box NF, Sturm RA, Jenkins T, Ramsay M. Rufous oculocutaneous albinism in southern African Blacks is caused by mutations in the TYRP1 gene. American journal of human genetics. 1997;61:1095–1101. doi: 10.1086/301603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013:D545–552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosoda K, Hammer RE, Richardson JA, Baynash AG, Cheung JC, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 39.Rees JL. The melanocortin 1 receptor (MC1R): more than just red hair. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2000;13:135–140. doi: 10.1034/j.1600-0749.2000.130303.x. [DOI] [PubMed] [Google Scholar]
- 40.D'Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, Ito S, Fisher DE. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 41.Rouzaud F, Kadekaro AL, Abdel-Malek ZA, Hearing VJ. MC1R and the response of melanocytes to ultraviolet radiation. Mutation research. 2005;571:133–152. doi: 10.1016/j.mrfmmm.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Innamorati G, Piccirillo R, Bagnato P, Palmisano I, Schiaffino MV. The melanosomal/lysosomal protein OA1 has properties of a G protein-coupled receptor. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2006;19:125–135. doi: 10.1111/j.1600-0749.2006.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortese K, Giordano F, Surace EM, Venturi C, Ballabio A, Tacchetti C, Marigo V. The ocular albinism type 1 (OA1) gene controls melanosome maturation and size. Investigative ophthalmology & visual science. 2005;46:4358–4364. doi: 10.1167/iovs.05-0834. [DOI] [PubMed] [Google Scholar]
- 44.Bassi MT, Schiaffino MV, Renieri A, De Nigris F, Galli L, Bruttini M, Gebbia M, Bergen AA, Lewis RA, Ballabio A. Cloning of the gene for ocular albinism type 1 from the distal short arm of the X chromosome. Nature genetics. 1995;10:13–19. doi: 10.1038/ng0595-13. [DOI] [PubMed] [Google Scholar]
- 45.Alexeev V, Yoon K. Distinctive role of the cKit receptor tyrosine kinase signaling in mammalian melanocytes. The Journal of investigative dermatology. 2006;126:1102–1110. doi: 10.1038/sj.jid.5700125. [DOI] [PubMed] [Google Scholar]
- 46.Zhu S, Wurdak H, Wang Y, Galkin A, Tao H, Li J, Lyssiotis CA, Yan F, Tu BP, Miraglia L, Walker J, Sun F, Orth A, Schultz PG, Wu X. A genomic screen identifies TYRO3 as a MITF regulator in melanoma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17025–17030. doi: 10.1073/pnas.0909292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlson JA, Linette GP, Aplin A, Ng B, Slominski A. Melanocyte receptors: clinical implications and therapeutic relevance. Dermatologic clinics. 2007;25:541–557, viii-ix. doi: 10.1016/j.det.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu H, Delling M, Li L, Dong X, Clapham DE. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18321–18326. doi: 10.1073/pnas.0709096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNeill MS, Paulsen J, Bonde G, Burnight E, Hsu MY, Cornell RA. Cell death of melanophores in zebrafish trpm7 mutant embryos depends on melanin synthesis. The Journal of investigative dermatology. 2007;127:2020–2030. doi: 10.1038/sj.jid.5700710. [DOI] [PubMed] [Google Scholar]
- 50.Jin J, Wu LJ, Jun J, Cheng X, Xu H, Andrews NC, Clapham DE. The channel kinase, TRPM7, is required for early embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E225–233. doi: 10.1073/pnas.1120033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartzell HC, Qu Z, Yu K, Xiao Q, Chien LT. Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiological reviews. 2008;88:639–672. doi: 10.1152/physrev.00022.2007. [DOI] [PubMed] [Google Scholar]
- 52.Singh H. Two decades with dimorphic Chloride Intracellular Channels (CLICs) FEBS letters. 2010;584:2112–2121. doi: 10.1016/j.febslet.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Caterina MJ, Rosen TA, Tominaga M, Brake AJ, Julius D. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature. 1999;398:436–441. doi: 10.1038/18906. [DOI] [PubMed] [Google Scholar]
- 54.Benham CD, Gunthorpe MJ, Davis JB. TRPV channels as temperature sensors. Cell calcium. 2003;33:479–487. doi: 10.1016/s0143-4160(03)00063-0. [DOI] [PubMed] [Google Scholar]
- 55.Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, Mangini NJ, Huang H, Canfield VA, Cheng KC, Yang F, Abe R, Yamagishi S, Shabanowitz J, Hearing VJ, Wu C, Appella E, Hunt DF. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. Journal of proteome research. 2006;5:3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- 56.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA, Palsson A, Thorleifsson G, Palsson S, Sigurgeirsson B, Thorisdottir K, Ragnarsson R, Benediktsdottir KR, Aben KK, Vermeulen SH, Goldstein AM, Tucker MA, Kiemeney LA, Olafsson JH, Gulcher J, Kong A, Thorsteinsdottir U, Stefansson K. Two newly identified genetic determinants of pigmentation in Europeans. Nature genetics. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 57.Weinert S, Jabs S, Supanchart C, Schweizer M, Gimber N, Richter M, Rademann J, Stauber T, Kornak U, Jentsch TJ. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl-accumulation. Science. 2010;328:1401–1403. doi: 10.1126/science.1188072. [DOI] [PubMed] [Google Scholar]
- 58.Bellono NW, Kammel LG, Zimmerman AL, Oancea E. UV light phototransduction activates transient receptor potential A1 ion channels in human melanocytes. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2383–2388. doi: 10.1073/pnas.1215555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 60.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends in molecular medicine. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 61.Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR. Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. The Journal of biological chemistry. 2000;275:37978–37983. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- 62.Harris ML, Baxter LL, Loftus SK, Pavan WJ. Sox proteins in melanocyte development and melanoma. Pigment cell & melanoma research. 2010;23:496–513. doi: 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmid R, Meyer K, Spang R, Schittek B, Bosserhoff AK. Melanoma inhibitory activity promotes melanoma development through activation of YBX1. Pigment cell & melanoma research. 2013;26:685–696. doi: 10.1111/pcmr.12119. [DOI] [PubMed] [Google Scholar]
- 64.Borovansky JR, P. A. Melanins and melanosomes: biosynthesis, structure, physiological and pathological functions. Wiley-Blackwell; Weinheim, Germany: 2011. [Google Scholar]
- 65.Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. American journal of human genetics. 2001;69:981–988. doi: 10.1086/324340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer H, Vitavska O, Wieczorek H. Identification of an animal sucrose transporter. Journal of cell science. 2011;124:1984–1991. doi: 10.1242/jcs.082024. [DOI] [PubMed] [Google Scholar]
- 67.Gardner JM, Nakatsu Y, Gondo Y, Lee S, Lyon MF, King RA, Brilliant MH. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science. 1992;257:1121–1124. doi: 10.1126/science.257.5073.1121. [DOI] [PubMed] [Google Scholar]
- 68.Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome research. 2012;22:446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.