Abstract
Fish gills represent a complex organ composed of several cell types that perform multiple physiological functions. Among these cells, ionocytes are implicated in the maintenance of ion homeostasis. However, because the ionocyte represents only a small percent of whole gill tissue, its specific transcriptome can be overlooked among the numerous cell types included in the gill. The objective of this study is to better understand ionocyte functions by comparing the RNA expression of this cell type in freshwater and seawater acclimated rainbow trout. To realize this objective, ionocytes were captured from gill cryosections using laser capture microdissection after immunohistochemistry. Then, transcriptome analyses were performed on an Agilent trout oligonucleotide microarray. Gene expression analysis identified 108 unique annotated genes differentially expressed between freshwater and seawater ionocytes, with a fold change higher than 3. Most of these genes were up-regulated in freshwater cells. Interestingly, several genes implicated in ion transport, extracellular matrix and structural cellular proteins appeared up-regulated in freshwater ionocytes. Among them, several ion transporters, such as CIC2, SLC26A6, and NBC, were validated by qPCR and/or in situ hybridization. The latter technique allowed us to localize the transcripts of these ion transporters in only ionocytes and more particularly in the freshwater cells. Genes involved in metabolism and also several genes implicated in transcriptional regulation, cell signaling and the cell cycle were also enhanced in freshwater ionocytes. In conclusion, laser capture microdissection combined with microarray analysis allowed for the determination of the transcriptional signature of scarce cells in fish gills, such as ionocytes, and aided characterization of the transcriptome of these cells in freshwater and seawater acclimated trout.
Introduction
Fish gills have several functions implicated in the maintenance of ion and gas homeostasis (respiration, osmoregulation, acid-base regulation, and nitrogen secretion) [1]. Due to direct contact with the external medium and the possibility of contact with pollutants or pathogens, the fish gill also has a barrier role and presents certain mechanisms of xenobiotic biotransformation [2] and immune defense [3]. Thus, to accomplish all these functions, gill morphology is very complex. This organ is composed of filaments and lamellae covered by epithelia and supported by cartilage and pillar cells. Epithelia are subdivided into two regions: a primary epithelium covering the filament and a secondary epithelium covering the lamellae [4].
Epithelia are composed of two epithelial cell types directly in contact with the external medium, pavement cells and mitochondria-rich cells (MRCs), representing more than 90% and less than 10% of the epithelial surface area, respectively [1]. Scarce mucous cells were also observed in contact with the external medium. Under these epithelial cells, both undifferentiated and basal cells have been characterized [5].
MRCs, more recently named ionocytes, are a very important cell type implicated in ion transport to maintain fish blood homeostasis. Ionocytes absorb and secrete NaCl in freshwater (FW) and saltwater (SW) environments, respectively. These cells are also implicated in Ca2+ absorption, H+/HCO3 - flux for acid-base regulation and ammonia excretion. In fresh water, several ionocyte sub-types have been identified in different fish species [6]. These sub-types were characterized using ultrastructural morphology and/or several cell biology and physiological approaches [7–9]. In contrast, only one sub-type has been identified in seawater fish species [7–9]. This SW subtype is closely associated with the accessory cells that send out digitations within the apical part of the ionocytes. In salmonids, two FW ionocyte sub-types were identified using (i) ultrastructural studies, where α-ionocytes and β-ionocytes were observed at the base of the lamellae and in the interlamellar region, respectively [10], (ii) density gradient separation techniques combined with differential peanut lectin agglutinin (PNA) staining to identify PNA+ and PNA- cells [11] and (iii) triple-color immunofluorescence staining for NKA, NKCC1 and NHE3b [12].
For euryhaline fish, an important gill remodeling process occurs after transfer from fresh water to sea water or brackish water. The gill epithelium is transformed from a salt absorbing to a salt secreting epithelium. To understand salinity adaptation, several studies have performed large scale gene expression experiments on gill tissue in Dicentrarchus labrax [13], Anguilla anguilla [14], Anguilla japonica [15], Gillichthys mirabilis [16], Sarotherodon melanotheron [17], Fundulus heteroclitus [18]. These studies led to the characterization of important gill osmotic effectors and signaling pathways that develop during adaptation to osmotic stress. However, because ionocytes are scarce in gill tissue, the expression of genes specific to this cell type can be hidden by the global gene expression of fish gill cells. Several studies performed gill dissociation and ionocyte separation for candidate gene expression and proteomic approaches of eel MRCs [19–21]. However, no large-scale gene expression study has been conducted on these dissociated cells. To develop such a goal, we elected to develop a new approach that aimed to isolate gill ionocytes directly from tissue using laser capture microdissection (LCM) [3,22]. In contrast to cell dissociation, the LCM approach is not stressful for cells. This technique has become a valuable method for isolating individual cell types from heterogeneous tissues [23]. Preliminary studies have used laser capture microdissection to isolate intraepithelial lymphoid tissue from gills and revealed the expression of T-cell receptor transcripts in this area [3]. Combining LCM with microarray technology has the potential to identify the transcriptomic signature of specific cells. In this study, our objective was to compare the transcriptome signature of freshwater and seawater ionocytes using laser capture microdissection and microarray approach.
Materials and Methods
Ethics statement
Experimental research performed in this study was in accordance to the guiding principles for the use and care of laboratory animals and in compliance with the French and European regulations on animal welfare. Experimenters obtained authorization from the French Government to carry out animal experiments (Agreement No. 35–57 for IL). Experiments were conducted within INRA facilities that had authorization for animal experimentation (B29-777-02 and B35-238-6) and were approved by the Local Animal Care and Ethic Committee of INRA (approval n° B9029).
Fish conditioning and sampling
Immature rainbow trout (Oncorhynchus mykiss) bred in a local hatchery (Drennec, Sizun, France) were transferred to the laboratory facilities. After acclimation to the tanks for at least 1 week, the fish were directly transferred from fresh water to either fresh water (control group) or 35 ppt sea water. The protocol to transfer fish to SW was previously described [24]. The trout were maintained in a recirculated water system at 10–13°C under a natural photoperiod and fed commercially available pellets (BioMar, Nessac, France). Fish (90–140 g), kept either in fresh water or transferred into sea water, were collected at various time points following the salinity change. After euthanized with a lethal dose of phenoxyethanol, blood and gill tissue were sampled. Plasma was stored at -20°C until further use. The second gill arches were collected for laser capture microdissection (LCM) and in situ hybridization (ISH). For LCM, the gill arches were frozen in isopentane by submersion in liquid nitrogen and storage at -80°C until further use. For ISH, the gill arches were fixed for 8 hours at 4°C with 4% paraformaldehyde in phosphate buffered saline, then gradually dehydrated in methanol and stored in 100% methanol at −20°C.
Plasma parameters
Plasma sodium was analyzed using a model 410 flame photometer (Sherwood Scientific). Chloride and calcium were assessed using colorimetric kits (chloride with a mercuric-thiocyanate method and calcium with the Arsenazo III method (Biolabo, Maizy, France)). The values reported in the figures are mean ± s.e.m. The Mann-Whitney U-test was used to make comparison between freshwater and seawater at each time point after non-parametric analysis of variance (Kruskal-Wallis test). All these statistical analyses were done with Statistica software (Statsoft, Maisons-Alfort, France). Differences were considered as significant when P<0.05.
Fluorescent immunohistochemistry with Zenon labeling technique
Gill cryosections (10 μm thick with a Leica cryostat) were cut, mounted onto super frost glass slides and processed for immunostaining with Na/K-ATPase antibody (α5 antibody) to label the ionocytes. To reduce the quantity of incubations and washes, the antibody was conjugated with Zenon fragment. A mouse IgG labeling kit (Zenon Alexa Fluor 488, Molecular Probes, Life Technologies, Saint Aubin, France) was used, and the IgG labeling was performed according to the Zenon Complex Formation protocol and diluted, in PBS with RNase inhibitor, to the desired working concentration. After incubation with the labeling complex for 5 min at 4°C, the sections were washed with PBS-RNAse inhibitor, dehydrated (70% ethanol, 95% ethanol (2 x 30 sec), 100% ethanol (3 x 30 sec) and xylene (3 x 1 min)) and then air dried (2 min) before LCM. The mouse monoclonal anti- chicken Na/K-ATPase (α5 antibody) developed by Douglas M. Fambrough was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological sciences, Iowa City, IA 52242.
Laser capture microdissection
Arcturus® Veritas (Applied Biosystems, Life technologies, Saint Aubin, France) was used to capture fluorescently labeled ionocytes. The infrared laser was adjusted to produce spot sizes between 10 and 15 μm in diameter, which allowed consistent capture of cells onto CapSure LCM caps (Excilone, Elancourt, France). The cells on the caps were lysed (lysis buffer from the PicoPure® RNA isolation kit (Excilone, Elancourt, France)) for RNA extraction. To limit RNA degradation, the time between cell labeling and lysis was routinely no longer than 1 hour. During this time, a mean of 400 labeled ionocytes can be selected and captured from each animal. Ionocytes from four freshwater and four seawater trout were captured for this study.
RNA extraction and quality control
RNA was extracted using the PicoPure RNA isolation kit (Excilone) following the manufacturer’s recommendations and with 1 U of DNase (Invitrogen, Life Technologies, Saint Aubin, France) treatment for 15 min at room temperature. RNA was extracted from the gill sections and the ionocytes trapped in the CapSure LCM caps. The quality and purity of the gill section RNA were evaluated after cutting the frozen gill section. RNA quality was assessed by an Agilent 2100 Bioanalyzer using the RNA 6000 pico LabChip kit (Agilent Technologies, Massy, France).
RNA amplification
Ionocyte RNA from four freshwater and four seawater trout was amplified and purified using the Ovation picoSL WTA system V2 RNA Amplification kit (NuGEN Technologies, Leek, Netherlands) according to the manufacturer’s instructions. Linear amplification was obtained by reverse transcription of the RNA by a cDNA tag with a SPIA sequence. This amplification used a DNA/RNA chimeric primer, DNA polymerase and RNase H. RNase H removes the RNA portion of the heteroduplex SPIA tag sequence, revealing a binding site for the SPIA primer. The process of SPIA DNA/RNA primer binding, DNA replication, strand displacement and RNA cleavage is repeated in a processive manner. Amplified cDNA was quantified using a NanoDrop Spectrophotometer (Labtech, Palaiseau, France).
Microarray analysis
Ionocyte RNA from four individual freshwater and four individual seawater trout (biological replicates) was used for microarray analysis. Rainbow trout RNA expression profiling was conducted using an Agilent 8x60K high-density oligonucleotide microarray (GEO platform #GPL15840).
The one color labeling and hybridization steps of the amplified cDNA were performed according to the “Gene Expression FFPE Workflow” Agilent protocol (starting from step 5 with minor modifications). Briefly, 1.65 μg of amplified cDNA was labeled using the Genomic DNA ULS Labeling Kit (1 μl of ULS-Cy5 per μg of DNA) at a final volume of 20 μl. The labeled cDNA was then purified using an Agilent KREApure column according to the standard protocol. The yield (~100%) and the degree of labeling (between 1.5% and 3%) of Cy5-cDNA were checked in all samples using a Nanodrop ND–1000 spectrophotometer (Labtech, Palaiseau, France). Cy5-cDNA (600 ng) was denatured at 95°C in a final volume of 34 μl with Agilent 100X Blocking Agent and 2X Hi-RPM GE Hyb buffer. After adding 11 μl of Agilent-CGHblock, the labeled cDNA was finally hybridized on a sub-array. Hybridization was performed for 17 hours at 65°C in a rotating hybridization oven (20 RPM) prior to washing and scanning with an Agilent Scanner (Agilent DNA Microarray Scanner, Agilent Technologies, Massy, France) using the standard parameters for a gene expression 8x60K oligoarray (3 μm and 20 bits). Data were then obtained with the Agilent Feature Extraction software (10.7.1.1) according to the appropriate GE protocol (GE1_107_Sep09). Normalization and statistical analysis were performed using GeneSpring software (Agilent). Data were scale-normalized using the median value of each array, then log2 transformed before statistical analysis. Spots not significantly different from background were excluded from analysis. Probes were considered valid when corresponding spots remained present in at least 75% of the replicates of each experimental condition after the flagging procedure. To identify differentially expressed genes between gill ionocytes from trout maintained in fresh water (4 individual biological replicates) or acclimated to sea water (4 individual biological replicates), a T-test with a Benjamini-Hochberg correction (p-value<0.05) was performed on a set of genes previously selected with a Fold Change>3. Clustering analysis was performed and visualized using CLUSTER and TREEVIEW software [25]. For clustering, the data were log transformed, median-centered and an average linkage clustering was carried out. Microarray data were submitted to the GEO database with the reference GSE69409.
In situ hybridization and immunohistochemistry
The following bacterial clones were obtained from the USDA (Washington, DC, USA): 1RT64K03_A_F02 (SLC26A6), 1RT103M12 (Clcn2), and 1RT129G21_A_D11 (NBC). The bacteria containing plasmids were grown in LB-ampicillin medium. Plasmids were extracted (NucleoSpin Plasmid kit, Macherey-Nagel, Hoerdt, France) and cDNA inserts were amplified by PCR using vector-specific primers (Sigma), Go Taq DNA polymerase, and dNTPs (Promega). The PCR products were used as templates for digoxigenin-labeled probe synthesis using digoxigenin-conjugated UTP (Roche Diagnostics Corp,Meylan, France) and T7 or SP6 RNA polymerase (Promega, Harbonnières, France) for the antisense (specific signal) or sense (negative control) RNA probes.
In situ hybridization experiments were performed on the gill filaments. After fixation of the gill arches in 4% paraformaldehyde, the gill filaments, stored at -20°C in methanol, were cut above the gill septum, placed in baskets and deposited inside "In situ Pro VS automate" (Intavis Bioanalytic Instruments) to perform in situ hybridization using the following conditions. Briefly, the filaments were rehydrated in graded methanol-PBS, pre-hybridized for 1 hour at 65°C in hybridization solution (50% formamide, 5x SSC, 0.1% Tween20, 0.005% heparin, and 0.1 mg/ml tRNA) and hybridized for 16 hours at 65°C with 0.5–1 ng/μl of DIG-labeled cRNA probes. After several post-hybridization washes with hybridization solution at 65°C, then SSC/Tween20 solutions at 55°C, the gill filaments were incubated in blocking buffer (PBS, 0.2% TritonX100, 0.2% Tween20, and 2% serum) for 1 hour at room temperature. Then, the filaments were incubated with anti-DIG antibody coupled to alkaline phosphatase (1:1000 Roche Diagnostics Corp, Meylan, France) in blocking buffer for 6 hours. After several washes in PBS/Tween20 and detection buffer (100 mM Tris 1 M pH 9.5, 100 mM NaCl M, 50 mM MgCl2 1 M pH 9), the color reaction was performed in the presence of NBT/BCIP in detection buffer (Roche Diagnostics Corp, Meylan, France). After several PBS washes, post fixation in 4% paraformaldehyde in PBS and several rinses, the gill filaments were dehydrated and embedded in paraffin. The tissue samples were sectioned at 5 μm. All hybridizations were performed on several filaments of at least 3 fish per condition.
To identify the ionocytes, immunohistochemistry was performed on the gill filament sections using the UltraVision Kit and Labvision components (Microm France, Francheville, France). Briefly, deparaffined and rehydrated sections were treated for at least 5 min in 0.01 M PBS + 0.05% Tween20 (PBS-Tween20). The sections were incubated for 10–15 min in a hydrogen peroxide block to reduce endogenous peroxidase activity. After two washes in PBS-Tween20, the slides were then incubated for 5–10 min in Ultra V Block to block non-specific background staining. After one rinse with PBS, anti-Na/K-ATPase monoclonal antibody was applied overnight in a humidity chamber at 4°C. After 4 washes with PBS-Tween20, the sections were incubated for 10 min at room temperature with biotinylated secondary antibody (goat anti-polyvalent: anti-mouse and anti-rabbit) followed by 4 rinses and 10 min of incubation with a streptavidin peroxidase complex at room temperature. The chromogene selected for labeling was DAB (3,3’-diaminobenzidine), chosen according to manufacturer recommendations. The anti-Na/KATPase (α5 antibody) developed by Douglas M. Fambrough was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological sciences, Iowa City, IA 52242.
Quantitative real-time PCR
qPCR was performed on a StepOnePlus real-time PCR system (Applied Biosystems, Life Technologies). Reactions were performed in duplicate and in a total volume of 10 μl with 4 μl of diluted cDNA, 5 μl of Fast SYBR Green Master Mix (Applied Biosystems, Life Technologies) and 200–400 nM of each primer (see Table 1 for the list of primers). The RNA expression level was normalized by Fyn (clone 1rt99c24_c_b12, GenBank CA357459, Proto-oncogene tyrosine-protein kinase FYN (FYN)). Fyn was chosen because of its invariant expression in previous microarray experiments comparing freshwater and seawater gill transcriptomes. Primers targeting fyn (GenBank CA357459) and SLC26A6 (CA357980.p.om.8, www.sigenae.org) were designed using Primer3 software [26]. For Na/K-ATPase α1a, α1b and NaHCO3, we used primers already designed in [27,28]. A melting curve was generated to confirm product specificity. Expression data were calculated by the 2−ΔΔCt method. The values reported in the figures are mean ± s.e.m. Statistical differences between FW and SW ionocytes were examined by Mann-Whitney U-test using Statistica software (Statsoft, Maisons-Alfort, France)
Table 1. Primer pairs for real time quantitative RT-PCR.
| Gene name | Forward primer sequences | Reverse primer sequences |
|---|---|---|
| Na/K-ATPase α1a | GCAGACGCCTCTCGGAATT | CAATGAGAAAGATGATGGATGG |
| Na/K-ATPase α1b | GGAAGACGCCTATAGCCAAA | CGATGAGGAAGATGACAGCTTC |
| NBC | TGGACCTGTTCTGGGTAGCAA | AGCACTGGGTCTCCATCTTCAG |
| SLC26A6 | CTAAAGCCTCCCAGTTCACC | AGACCAACAGCCACCATCTC |
| FYN | CCGAGCACAGATAGGAGGAG | CACGCACACAGACACAAGTG |
Results and Discussion
The transfer of rainbow trout from fresh water to sea water induced a transient increase of plasma sodium, chloride and calcium, followed by the return of chloride and calcium to basal levels and a plateau to near freshwater plasma concentration for sodium (Fig 1). In accordance with a previous study [29], our results indicated that after 14 days in sea water, trout were able to regulate their hydromineral balance. Consequently, transcriptome analysis of ionocytes was compared between fish transferred for 14 days in seawater or freshwater tanks.
Fig 1. Plasma ion concentrations after sea water transfer.
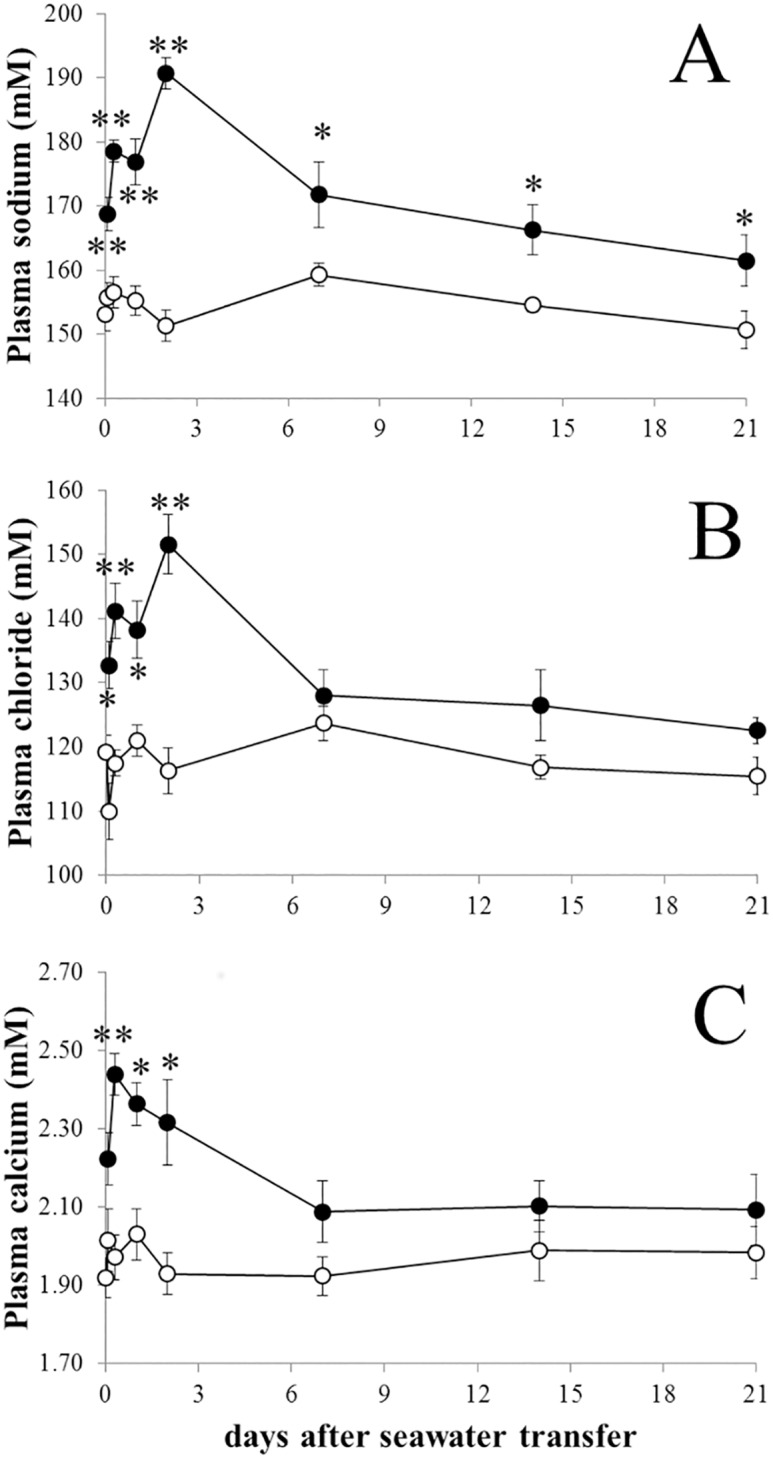
Mean plasma sodium (A), chloride (B), and calcium (C) from time 0 to 21 days post sea water transfer. Open and filled circles represent respectively freshwater and seawater transferred trout. Values represent means ± s.e.m. of six fishes. Differences between freshwater and seawater at each time point were assessed with non-parametric Mann-Whitney U-test after non-parametric analysis of variance (Kruskal-Wallis test). * and **, significantly different from the corresponding values in freshwater at P<0.05 and P<0.01.
To compare ionocyte transcriptomes in both FW and SW, an immuno-laser capture microdissection was performed on the trout gill sections. Because the Na/K-ATPase pump (NKA) is highly expressed in ionocytes, whatever the salinity, an NKA antibody against the α-subunits was used as a marker for identifying ionocytes on the tissue sections.
Using the selected genes with a fold change >3 between freshwater and seawater ionocytes as well as a t-test with a Benjamini-Hochberg correction (p-value<0.05), 138 genes appeared differentially expressed. Among these genes, 127 were up and 11 down-regulated in freshwater compared to seawater ionocytes (Fig 2 and S1 Table). The genes up-regulated in FW ionocytes corresponded to 98 annotated genes (certain genes were represented by several oligos, such as SLC4A4 (5 probes); S1 Table) and 14 non-annotated genes. In seawater ionocytes, among the 11 oligonucleotides with higher expression, 10 corresponded to annotated genes.
Fig 2. Hierarchical clustering analysis of gene differentially expressed between FW and SW ionocytes.
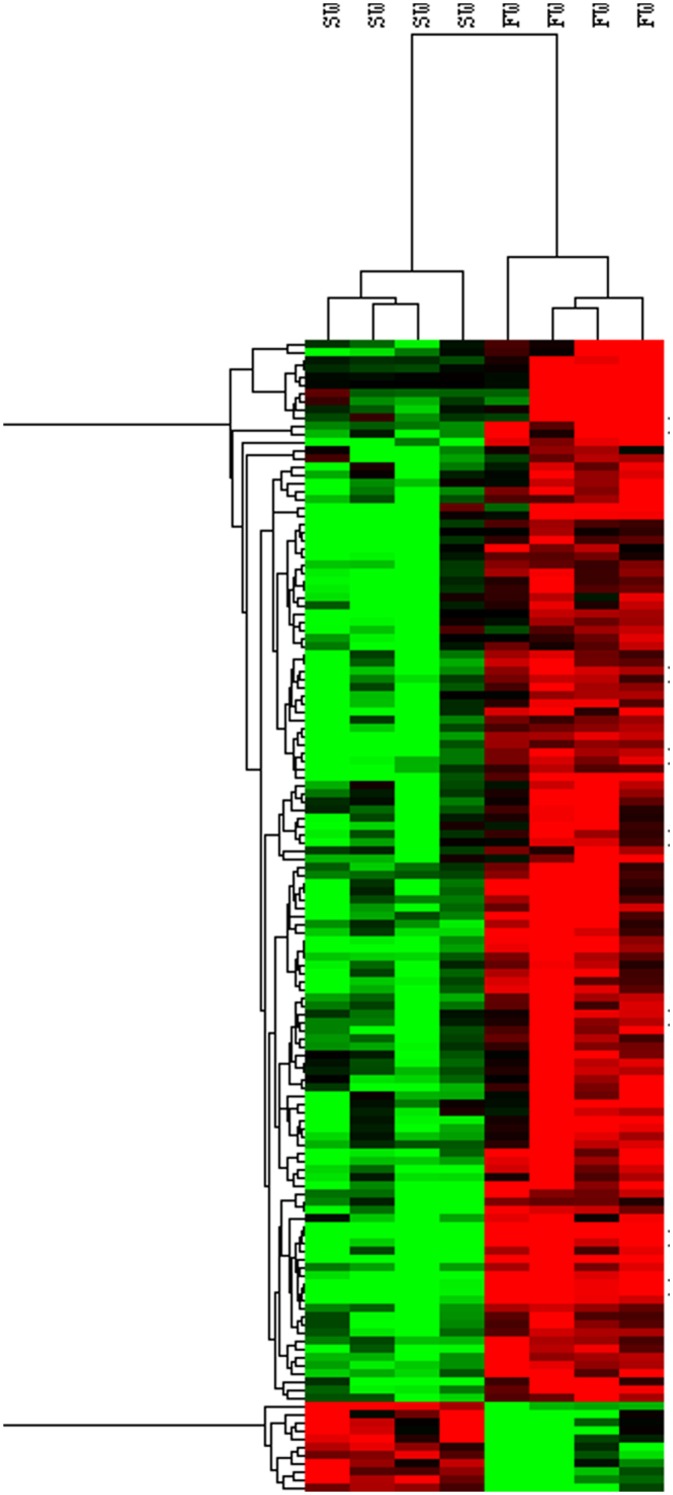
Analysis was performed based on a log2-transformed ratio value of 138 genes differentially expressed. Row and columns represent genes and samples respectively. Expression levels of log2-transformed ratio are represented by a color tag: red and green for high and low levels of expression respectively.
Interestingly, when comparing RNA expression profiles between FW and SW ionocytes, most genes appeared up-regulated in FW ionocytes. Gene expression in other species did not present such difference in gill expression between FW and SW acclimated fish [13,14]. However, in these studies, RNA expression of whole gill tissue was used to perform transcriptome analysis. In our study, we attained transcript expression of one cell type present in less than 10% of whole gill tissue.
Interestingly, the most differentially expressed genes (fold change >9) were ionic transporters (S1 Table). This is consistent with the role of ionocytes in ion transport (FW and SW ionocytes absorb and secrete salt, respectively). Other genes; grouped according to their functions using uniprot (http://uniprot.org), gene ontology, literature; revealed more abundant genes involved in structural organization and regulation in FW ionocytes (Table 2). Some of these genes were implicated not only in extracellular matrix composition and cytoskeleton organization but also in cell adhesion and motility. Another set of genes was grouped into biological functions related to metabolism. The first group was related to energetic metabolism, implicating several mitochondrial enzymes in ATP synthesis, and the second group was principally involved in amino acid and protein metabolism. Finally, several genes implicated in transcriptional regulation, cell signaling and the cell cycle were also expressed more in FW ionocytes.
Table 2. Annotated genes exhibiting differential expression between FW and SW ionocytes.
| Gene identification | function |
|---|---|
| Sodium/potassium-transporting ATPase subunit alpha–1 | ion transport |
| Chloride channel protein 2 | ion transport |
| Solute carrier family 26 member 6 | ion transport |
| Electrogenic sodium bicarbonate cotransporter 1 | ion transport |
| Sodium/potassium-transporting ATPase subunit beta–233 | ion transport |
| Zinc transporter | ion transport |
| P3 protein | ion transport |
| Major facilitator superfamily domain-containing protein 1 | transport |
| Collagen alpha–1(X) | extracellular matrix |
| Collagen alpha–1(I) chain | extracellular matrix |
| Collagen alpha–2(I) chain | extracellular matrix |
| Fibronectin | extracellular matrix |
| Sparc | extracellular matrix |
| sparc/osteonectin | extracellular matrix |
| Bridging integrator 3 | cytoskeleton |
| Coronin–6 | cytoskeleton |
| Cytoplasmic dynein 1 intermediate chain 2 | cytoskeleton |
| Sorting nexin–3 | cytoskeleton |
| Spastin | cytoskeleton |
| Transgelin | cytoskeleton |
| Tubulin alpha-1D chain | cytoskeleton |
| Vesicle-associated membrane protein 5 | cytoskeleton |
| Uncharacterized protein ENSP00000361571 | cytoskeleton |
| CD9 protein | cell adhesion |
| Carcinoembryonic antigen-related cell adhesion molecule 5 | cell adhesion |
| Liprin-beta–1 | cell adhesion |
| Vascular cell adhesion protein 1 | cell adhesion |
| Claudin–4 | junction |
| Peripheral myelin protein 22 | junction |
| Isocitrate dehydrogenase [NADP], mitochondrial | energetic metabolism |
| NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | energetic metabolism |
| NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | energetic metabolism |
| Cytochrome b-c1 complex subunit 6, mitochondrial | energetic metabolism |
| Up-regulated during skeletal muscle growth protein 5 | energetic metabolism |
| Aconitate hydratase, mitochondrial | energetic metabolism |
| Aldehyde dehydrogenase, mitochondrial | response to oxygen level |
| Hemoglobin subunit beta–1 | response to oxygen level |
| NAD(P) transhydrogenase, mitochondrial | response to oxygen level |
| Selenoprotein M | response to oxygen level |
| 5-methyltetrahydropteroyltriglutamate–-homocysteine methyltransferase | amino acid synthesis |
| Ornithine decarboxylase | amino acid synthesis |
| Proline synthetase co-transcribed bacterial homolog protein | amino acid synthesis |
| Fetuin-B | protein metabolism |
| Serine protease inhibitor Kazal-type 2 | protein metabolism |
| ATP-binding cassette sub-family F member 1 | protein metabolism |
| COP9 signalosome complex subunit 5 | protein metabolism |
| DCN1-like protein 4 | protein metabolism |
| Eukaryotic translation initiation factor 4E-1A | protein metabolism |
| Proteasome subunit beta type–7 | protein metabolism |
| 26S proteasome non-ATPase regulatory subunit 5 | protein metabolism |
| Uridine 5-monophosphate synthase | protein metabolism |
| RING finger protein 152 | protein metabolism |
| Transmembrane protease serine 9 | protein metabolism |
| NEDD8-conjugating enzyme UBE2F | protein metabolism |
| Ubiquilin–4 | protein metabolism |
| Eukaryotic translation initiation factor 3 subunit B | protein metabolism |
| 40S ribosomal protein S7 | protein metabolism |
| Glycerol-3-phosphate dehydrogenase 1-like protein | Carbohydrate-lipid metabolism |
| Arylsulfatase B | glucopolysaccharide catabolism |
| Glycogen phosphorylase, muscle form | glycogen metabolism |
| Elongation of very long chain fatty acids protein 1 | lipid metabolism |
| Putative phospholipase B-like 1 | lipid metabolism |
| Splicing factor, arginine/serine-rich 6 | mRNA splicing |
| U1 small nuclear ribonucleoprotein A | mRNA splicing |
| Exosome complex exonuclease RRP44 | RNA catabolic process |
| Eukaryotic peptide chain release factor subunit 1 | RNA catabolic process |
| Early growth response protein 1 | regulation of transcription |
| far upstream element (FUSE) binding protein 1 | regulation of transcription |
| GON-4-like protein | regulation of transcription |
| Histone deacetylase 1 | regulation of transcription |
| Hepatic leukemia factor | regulation of transcription |
| LIM/homeobox protein Lhx6 | regulation of transcription |
| Protein LLP homolog | regulation of transcription |
| Protein max | regulation of transcription |
| Polycomb protein SCMH1 | regulation of transcription |
| TSC22 domain family protein 3 | regulation of transcription |
| Forkhead box protein N3 | regulation of transcription |
| Histone acetyltransferase MYST2 | regulation of transcription |
| Thioredoxin-related transmembrane protein 1 | regulation of transcription |
| C4b-binding protein alpha chain | immune system |
| Putative HLA class I histocompatibility antigen, alpha chain H | immune system |
| T-cell immunomodulatory protein (Fragment) | immune system |
| T-cell receptor beta chain C region | immune system |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit | cell cycle_anti-apoptosis |
| Probable Bax inhibitor 1 | cell cycle_anti-apoptosis |
| RNA-binding protein 24 | cell cycle_differentiation |
| cAMP-regulated phosphoprotein 19 | cell cycle_division |
| Salmo salar Gametogenetin-binding protein 2 | cell cycle_division |
| Guanine nucleotide-binding protein G(i) subunit alpha–2 | cell cycle_division |
| Protein NDRG3 | cell cycle-division |
| Heme-binding protein 2 | cell cycle_pro-apoptosis |
| Frizzled–1 | cell signaling |
| Protein Wnt–11 | cell signaling |
| Growth factor receptor-bound protein 2 | cell signaling |
| Homer protein homolog 2 | cell signaling |
| Hepatocyte growth factor receptor | cell signaling |
| 3-phosphoinositide-dependent protein kinase 1 | cell signaling |
| Gamma-aminobutyric acid receptor-associated protein-like 2 | cell signaling |
| Arylamine N-acetyltransferase, pineal gland isozyme NAT–10 | detoxification |
| Post-GPI attachment to proteins factor 3 | GPI anchor metabolic process |
| Metalloreductase STEAP3 | iron homeostasis |
| Transposable element Tcb2 transposase | DNA integration |
Genes in italic were upregulated in SW and others genes were upregulated in FW.
Transporters
Because ionocytes are the major cell type implicated in transepithelial ion transport in the gill, the first genes examined in the list of differentially expressed genes between freshwater and seawater ionocytes were the ion transporters. Indeed, certain transporters appeared up-regulated in freshwater ionocytes.
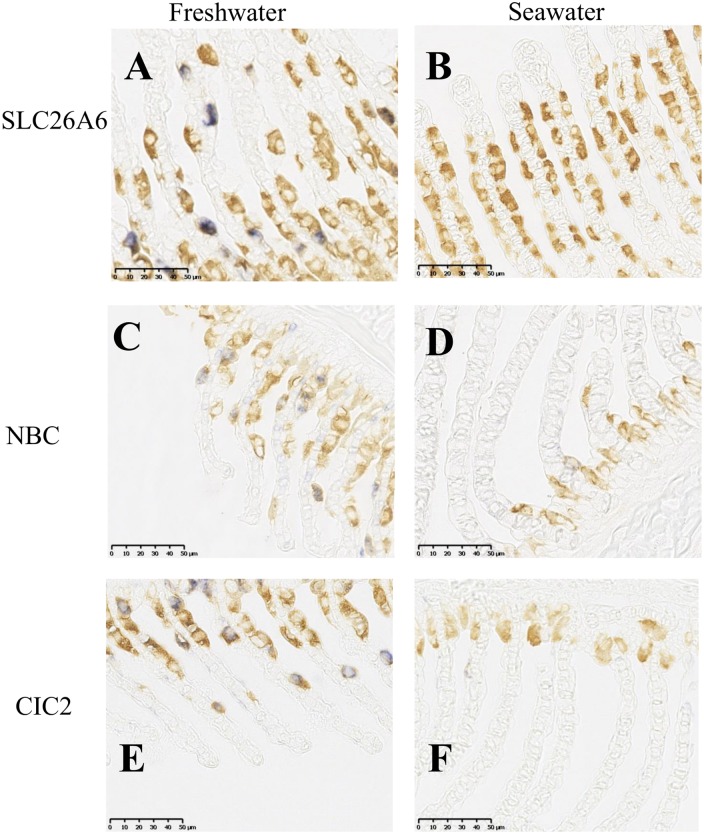
One of the most interesting pieces of information provided by this transcriptomic analysis is related to chloride channel 2 (CIC2), which was expressed more in freshwater ionocytes in our microarray analysis. Interestingly, in situ hybridization (i) confirms this result with no detectable CIC2 mRNA in the SW ionocytes and (ii) shows expression only in FW ionocytes with no detectable labeling in pavement cells or other gill cells (Fig 3). CIC2 is an anion channel located in the surface membranes of excitable and epithelial mammal cells [30,31]. One important function of CIC2 is chloride efflux at the basolateral membrane in the gastro-intestinal tract of guinea pigs [32,33]. Our results suggest that CIC2 acts as a channel participating in chloride absorption in FW rainbow trout gills and more particularly in basolateral flux from cells to the blood. In accordance with our results, two studies on zebrafish revealed the presence of CLC-2c in ionocytes [34,35]. CIC3 was previously reported to be a good candidate for involvement in basolateral chloride transport at the level of the gill epithelium in other teleost fish species, Oreochromis mossambicus [36,37], Tetraodon nigroviridis [38] and Dicentrarchus labrax [39]. However, in our study, the RNA expression of CIC3 in the trout microarray was not significantly different between FW and SW ionocytes. Overall, our data suggest that basolateral chloride transport in the FW ionocytes of rainbow trout is most likely associated with CIC2 as in zebrafish, and not CIC3, as in other fish species.
Fig 3. In situ hybridization of 3 genes (purple labelling) in fish gill from freshwater (A, C, E) or seawater (B, D, F) acclimated rainbow trout associated with immunocytochemistry of ionocyte with Na/K-ATPase antibody (brown labelling).
AB: SLC26A6, CD: NBC, EF: CIC2.
The present transcriptomic analysis also indicated that another chloride transporter, SLC26A6 (a Cl-/HCO3 exchanger from the SLC26 family), had higher mRNA expression in freshwater ionocytes. This result was obtained using a microarray experiment and validated with qPCR (Fig 4C). Furthermore, in situ hybridization showed expression of this exchanger only in freshwater ionocytes (Fig 3). Our results corroborated another recent study showing higher expression of this transporter in the gills of rainbow trout in fresh water compared to salt water [40]. Overall, using complementary approaches, the present study allowed clear identification of SLC26A6 in ionocytes with no detectable labeling in pavement cells or other freshwater gill cells.
Fig 4. Quantitative real-time PCR of 4 genes in freshwater and seawater ionocytes.
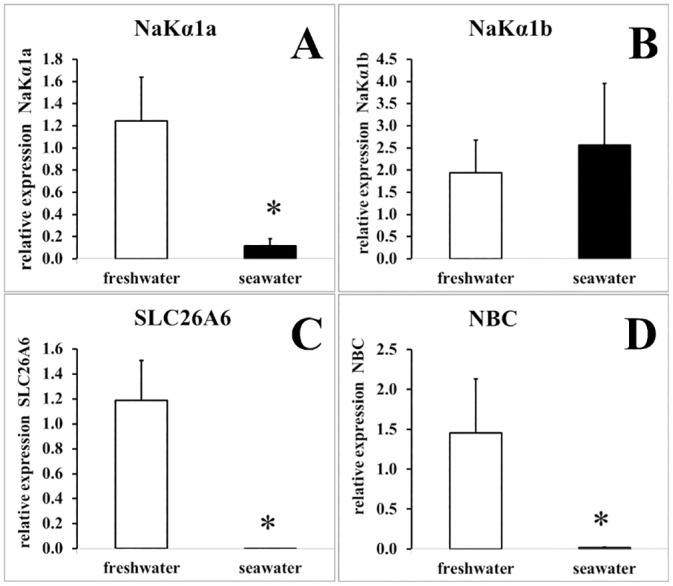
Values represent means ± s.e.m. of four fishes. *, significantly different from the corresponding values in freshwater at P<0.05, Mann-Whitney U-test.
Expression analysis of the different subunits of the Na/K-ATPase pump, which provides the driving force to absorb (in FW) and secrete (in SW) NaCl, also provided interesting information. Among the subunits of the Na/K-ATPase pump, the expression levels of the α1a isoform and one β-subtype (NKAβ233, the duplicate copy of NKA-β1 in Anguilla anguilla) were higher in FW than in SW ionocytes (Table 2 and Fig 4A). The co-expression of NKA α1a and this β1-subtype suggests their link to Na/K-ATPase activity in freshwater ionocytes. The pump consists of three subunits, α, β, and γ [41]. Among the α-subtypes, α1a and α1b were significantly decreased and increased, respectively, in salmonid gills after transfer from FW to SW [42–44]. In accordance with previous studies using whole gill tissue, the transcript expression of the Na/K-ATPase α1a isoform was significantly reduced in seawater ionocytes in the present study (Fig 4A). In contrast with these previous studies, our data showed that mRNA expression of isoform α1b in isolated ionocytes was not significantly different between FW and SW, neither in microarray nor in qPCR analysis (Fig 4B). This difference in transcript expression between whole gill tissue and ionocytes suggests either a higher number of ionocytes in SW versus FW or higher RNA expression of this α1b isoform in other gill cell types.
For gill sodium flux in rainbow trout, several ion transporters were proposed in FW fish [6]. One model implicated a Na+ channel and a Na+/HCO3 - cotransporter (NBC) in the apical and basolateral side of ionocytes, respectively [45]. Our results confirm the involvement of a NBC (SLC4A4) in trout gill ionocytes (microarray in Table 2, ISH in Fig 3, and qPCR in Fig 4D). Furthermore, this co-transporter appears to be predominantly involved in freshwater environments, as gill mRNA expression is significantly reduced after at least 2 weeks in sea water. Our results are in agreement with others studies showing NBC in FW ionocytes of Osorezan dace [46], zebrafish [47] and Mozambique tilapia [48]. No sodium channel isoforms were differentially expressed in our microarray experiment. Recently, members to the amiloride-sensitive ENaC/degenerin family of ion channel were identified as putative Na+ channels involved in apical sodium transport in rainbow trout gill cells: acid-sensitive channels (ASICs) [49]. However the two trout subunits of ASICs identified in gill are not present on our microarray, further experiments will be necessary to compare their gene expression between FW and SW ionocyte. Interestingly, several Epithelial Na+ Channel (ENaC) regulators, identified in other animals, appeared up-regulated in FW ionocytes in this study. Thus, we observed an increase in "sorting nexin3" RNA expression, which encodes a protein known to increase the cell surface expression of ENaCs [50]. Another gene, TSC22 domain family protein 3 (or Glucocorticoid-Induced Leucine Zipper protein) has been shown to stimulate ENaC cell surface expression and activity [51]; in the present study, we observed an increase in its expression in FW ionocytes. Overall, these data do not demonstrate the presence of an active epithelial Na+ channel but instead show possible involvement of regulatory pathways related to that channel in FW.
Other transporters that were enhanced in FW ionocytes included two solute carrier proteins (P3 protein (SLC10A3) and a zinc transporter (SLC39A11)). Solute carrier family 10 comprises influx transporters for various molecules, such as bile acids, steroidal hormones and drugs [52]. SLC10A3 exhibits amino acid identity with other SLC10 proteins, however its function and substrate specificity are not yet known [52]. SLC39A11 is a zinc transporter from the ZIP family. This family increases the intracellular zinc concentration via uptake across the plasma membrane or efflux from intracellular compartments [53]. Higher SLC39A11 expression in FW ionocytes suggests an important role of zinc in certain cellular processes. Another transporter, MFSD1 (major facilitator superfamily domain-containing protein 1), was also up-regulated in FW. However, like SLC10A3, the function and solute transported are unknown. Further investigation is necessary to characterize the role and importance of each of these transporters in ionocyte functions.
Extracellular matrix (ECM) and structural cellular proteins
Interestingly, several genes related to cell structure and morphogenesis (extracellular matrix and cytoskeleton) presented a higher expression level in FW ionocytes than in SW ionocytes (Table 2). This is not unexpected, as up-regulation of these expression transcripts can not only be related to differences in the morphological ultrastructure of ionocytes in fresh water and sea water [8,10] but also to their localization. In FW, ionocytes are present in the lamellae and filament, whereas in SW, most ionocytes are located in the filament. Thus, the morphology and localization of the ionocytes are different between FW and SW, which may be associated with the contrasting transcriptomic signatures of structural proteins in FW and SW ionocytes.
Among the structural genes, several extracellular matrix component proteins were up-regulated in FW ionocytes, including collagens (COL1A1, COL1A2, and COL10A1) and fibronectin (FN1). The RNA expression of two proteins associated with the extracellular matrix was also higher in FW; these genes were from the Secreted Protein Acidic and Rich in Cysteine (SPARC) family, SPARC and testican–2 (or SPOCK2). These proteins modulate cellular interaction with the extracellular matrix by regulating ECM turnover, growth factor signaling and receptor activity, and extracellular protease activity [54]. Most of these genes were up-regulated in Salmo salar smolts compared to parr [55]. The smolt is one of last stages of smoltification (when salmon are ready to live in sea water), in contrast to the parr that lives only in fresh water, and our results appear opposed to this study. However, in Seear et al. (2010), transcriptome analysis was performed on whole gill tissue, whereas we worked with isolated ionocytes. Furthermore, in this previous study, smolts were only pre-acclimated to sea water because they were maintained in fresh water. The measurement of collagen and SPARC mRNA in smolts transferred to SW will be necessary to determine the importance of these genes in FW and SW. In our microarray results, the higher expression of two protease inhibitors, serine protease inhibitor Kazal-type 2 (SPINK2) and Fetuin-B (FETUB), secreted in the extracellular portion of FW ionocytes, strengthens the importance of this extracellular matrix for these salt-absorbing cells.
In the cytoskeleton, the transcriptional expression of a protein implicated in cytoskeleton F-actin organization, bridging integrator 3 (BIN3) [56], was significant in FW vs SW ionocytes. Two actin-associated proteins were also up-regulated in FW ionocytes, transgelin (TAGLN) and coronin–6 (CORO6). Down regulation of transgelin has been shown to have an oncogenic effect and to modify extracellular remodeling and invasion [57]. Furthermore, coronin–6 modulates anchoring between the receptor (the acetylcholine receptor, in particular) and the actin cytoskeleton [58]. Several genes related to microtubules, such as tubulin alpha-1D chain (TUBA1D), and implicated in intracellular trafficking were also up-regulated in FW ionocytes: cytoplasmic dynein 1 intermediate chain 2 (DYNC1L2) [59], sortin nexin (SNX3), [50], vesicle-associated membrane protein 5 (VAMP5) [60] and spastin (SPAST) [61]. All genes suggest an important role of the cytoskeleton in FW ionocyte functions.
Cell adhesion and motility
In relation to cell structure and morphogenesis, other genes implicated in cell adhesion and motility were up-regulated in freshwater ionocytes, including vascular cell adhesion protein 1 VCAM1), liprin-beta–1 (PPFIBP1), CD9 antigen (CD9) [62–64], as well as carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) [65]. Unlike in SW, where most of the gill ionocytes were localized on filaments, the ionocytes in FW rainbow trout were also localized in all parts of the lamellae [66]. Gill epithelial proliferation occurred mainly at the base of lamellae with minor proliferation at the base of the filament [67]. To be operational, ionocytes have to migrate from the progenitor compartment. This distance is more important to access the lamellae, which may explain the higher transcript expression for adhesion and motility in FW ionocytes.
Cell-cell adhesion also includes proteins that form tight junctions in the epithelium. Among these tight junction proteins, several claudins have been characterized [68]. In our study, higher RNA expression of claudin–4 (CLDN4) in FW ionocytes was in accordance with other studies showing a significant elevation of claudin 4 protein in the gills of tilapia, killifish, and southern flounder acclimated to FW versus SW [18,69,70]. Another protein from the apical intercellular junction in epithelial cells [71] was up-regulated in FW ionocytes: peripheral myelin protein 22 (PMP22). As another junction protein, expression of human PMP22 in cultured kidney cells (MDCK) elevated the transepithelial resistance but at the same time increased permeability of nonionic molecules [72]. Further experiments will be necessary to understand the role of PMP22 in gill epithelium.
Energetic metabolism
Several enzymes implicated in mitochondrial ATP synthesis presented higher transcriptional expression in FW ionocytes, suggesting higher energy demand than the SW ionocytes. Among the genes implicated were (i) two enzymes in tricarboxylic acid cycle, isocitrate dehydrogenase (IDH2) and aconitate hydratase, and (ii) several proteins of the electron transfer chain, enzymes of complex I (NADH dehydrogenase [ubiquinone] flavoprotein 1 and 2 (NDUFV1 and NDUFV 2)) and complex III (Cytochrome b-c1 complex subunit (UQCRH)). Higher expression of glycogen phosphorylase (the cytoplasmic enzyme implicated in glycogen catabolism and consequent glucose increase) could link this to ATP production. A component and regulator of mitochondrial ATP synthase was also up-regulated, diabetes-associated protein in insulin-sensitive tissues (DAPIT also known as USMG5) [73]. This demand in ATP synthesis can be explained by higher ion-ATPase activity in fresh water. In FW and SW, Na/K-ATPase pumps are present in the basolateral plasma membrane of ionocytes. However, in FW rainbow trout, another ATPase pump has been shown to play a role in Na+ uptake, the H-ATPase pump [74]. The changes in carbohydrate metabolism of gills during the gradual adaptation of rainbow trout to sea water was measured by a previous study [75] that suggested a higher ATP demand to adapt to salt water. However, this study was performed on whole gill tissue during adaptation. The ionocytes represent less than 10% of the whole gill, thus their metabolic activity is most likely hidden by the global activity of whole gill tissue.
An increase of electron transport chain (ETC), in the mitochondria, for higher energy production can result in the formation of more reactive oxygen species (ROS) that contribute to oxidative stress [76]. Two mitochondrial enzymes identified as protective proteins during oxidative stress were expressed more in FW ionocytes, aldehyde dehydrogenase–2 (ALDH2) and NAD(P) transhydrogenase (NTT). ALDH2 detoxifies aldehydes produces by ROS [77], and NNT generates NADPH, a cofactor required for antioxidant-related enzyme activity, such as glutathione peroxidase and glutathione reductase [78]. Among the genes with higher expression in FW ionocytes, one other antioxidant protein was identified: hemoglobin beta (HBB) [79].
Amino acid and protein metabolism
Several genes related to amino acid synthesis were also up-regulated in FW compared to SW ionocytes. These genes included 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (metE) and proline synthetase co-transcribed bacterial homolog protein (PROSC). Similar to data reported by Whitehead et al., ornithine decarboxylase 1 (ODC–1), implicated in polyamine synthesis, was up-regulated under hypoosmotic conditions.
Several genes implicated in protein synthesis were up-regulated in FW and SW ionocytes. In FW ionocytes, these genes included 2 genes involved in translation initiation, eukaryotic translation initiation factor 4E-1A (EIF4E1A) and ATP-binding cassette sub-family F member 1 (ABCF1), and for SW, another translation initiation factor and a ribosome protein, eukaryotic translation initiation factor 3 subunit B (EIF3B) and 40S ribosomal protein S7 (RPS7), respectively.
Other genes implicated in protein regulation were only enhanced in FW ionocytes; thus, several genes of the ubiquitin proteasome system (UPS) were up-regulated in FW versus SW ionocytes. Protein modified by ubiquitination, which is performed by activating (E1), conjugating (E2) and ligating (E3) enzymes, can be degraded or implicated in other functions, such as DNA repair, trafficking and endosomal sorting. The fate of ubiquitylated protein depends on the number of conjugated ubiquitin chains (mono or polyubiquitin chains) and on the amino acid residues of ubiquitin involved [80,81]. For example, a protein conjugated to a polyubiquitin chain by ubiquitin lysine 48 (but possibly by all non-lysine 63 linkage) is targeted to the 26S proteasome to be degraded [80]. In our microarray results, one E3 ubiquitin-protein ligase mediating 'Lys–48'-linked polyubiquitination of target proteins (RING finger protein 152 (RNF152)) [82] was up-regulated in FW ionocytes. Furthermore, two non-ATPase regulatory subunits of the 26S proteasome were concomitantly more expressed (PSMD5 and PSMD 7). Ubiquilins are ubiquitin-like proteins that play a role in the regulation of proteasomal protein degradation [83], and ubiquilin 4 (UBQLN4) appears up-regulated in FW ionocytes. All of these genes suggest protein degradation after ubiquitin conjugation. A higher rate of proteolysis in FW ionocytes is also suggested by the up regulation of one serine-protease, transmembrane protease serine 9 (TMPRSS9) [84].
Another UPS protein was more expressed in FW ionocytes, it was a E2-conjugating enzyme, NEDD8-conjugating enzyme (UBE2F). This enzyme interacts with E3 ubiquitin ligase, RBX2 [85]. Several factors can modify this enzyme interaction, defective in cullin neddylation protein 1-like protein 1 (DCUN1D) [86] and COP9 signalosome [87]. Among the genes up-regulated in FW ionocytes, we observed DCN1-like protein 4 (DCUN1D4) and the COP9 signalosome complex subunit 5 (COPS5).
Conclusion
This study on gill ionocyte from acclimated trout (freshwater and seawater) has allowed (i) to compare transcriptome of cells absorbing versus cell secreting salts and (ii) to develop a LCM technique on trout gill epithelium. This study is the first to provide information specific to the functions of ionocytes (and not whole gill tissue) related to FW versus SW acclimation. Higher expression of genes in FW cells suggests higher activity of ionocyte in fresh water compared to salt water. Interestingly, new ion transporters, such as CIC2 and SLC10A3, have been identified. Further characterization of these ion transporters is necessary. In perspective, the development of this LCM technique in fish gills can be adapted to (1) compare transcriptomic cell signatures of others cells (i.e., pavement cells and ionocyte subtypes with specific antibodies) but also to (2) monitor certain cell types during external challenges, such as the modification of salinity.
Supporting Information
Ratios>3 identified transcripts that were upregulated in freshwater ionocyte and ratios<3 identified transcripts that were upregulated in seawater ionocytes.
(DOCX)
Acknowledgments
P. Prunet and P.Y. Rescan are gratefully acknowledged for their critical reading of the manuscript. The authors would like to thank the experimental facilities staff of INRA-LPGP (Rennes, France) and INRA-PEIMA (Sizun, France) for fish rearing.
Data Availability
All microarray data are available from the GEO database (accession number GSE69409).
Funding Statement
This work was funded by the European Community's Seventh Framework (FP7/2007-2013) under grant agreement no. 222719-LIFECYCLE (to IL) and by INRA-PHASE (to IL) and IFR140 grants (to IL, AF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Evans DH, Piermarini PM, Choe KP (2005) The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85: 97–177. [DOI] [PubMed] [Google Scholar]
- 2. Leguen II, Carlsson C, Perdu-Durand E, Prunet P, Part P, et al. (2000) Xenobiotic and steroid biotransformation activities in rainbow trout gill epithelial cells in culture. Aquat Toxicol 48: 165–176. [DOI] [PubMed] [Google Scholar]
- 3. Haugarvoll E, Bjerkas I, Nowak BF, Hordvik I, Koppang EO (2008) Identification and characterization of a novel intraepithelial lymphoid tissue in the gills of Atlantic salmon. J Anat 213: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laurent P, Dunel S (1978) Anatomical relationships of the ionocytes (chloride cells) with the branchial venous compartment: definition of two types of epithelium in fish gills. C R Acad Sci Hebd Seances Acad Sci D 286: 1447–1450. [PubMed] [Google Scholar]
- 5. Wilson JM, Laurent P (2002) Fish gill morphology: inside out. J Exp Zool 293: 192–213. [DOI] [PubMed] [Google Scholar]
- 6. Dymowska AK, Hwang PP, Goss GG (2012) Structure and function of ionocytes in the freshwater fish gill. Respir Physiol Neurobiol 184: 282–292. 10.1016/j.resp.2012.08.025 [DOI] [PubMed] [Google Scholar]
- 7. Hwang PP, Lee TH, Lin LY (2011) Ion regulation in fish gills: recent progress in the cellular and molecular mechanisms. Am J Physiol Regul Integr Comp Physiol 301: R28–47. 10.1152/ajpregu.00047.2011 [DOI] [PubMed] [Google Scholar]
- 8. Pisam M, Rambourg A (1991) Mitochondria-Rich Cells in the Gill Epithelium of Teleost Fishes: An Ultrastructural Approach In: Kwang WJ, Martin F, editors. International Review of Cytology: Academic Press; pp. 191–232. [Google Scholar]
- 9. Takei Y, Hiroi J, Takahashi H, Sakamoto T (2014) Diverse mechanisms for body fluid regulation in teleost fishes. Am J Physiol Regul Integr Comp Physiol 307: R778–792. 10.1152/ajpregu.00104.2014 [DOI] [PubMed] [Google Scholar]
- 10. Pisam M, Le Moal C, Auperin B, Prunet P, Rambourg A (1995) Apical structures of "mitochondria-rich" alpha and beta cells in euryhaline fish gill: their behaviour in various living conditions. Anat Rec 241: 13–24. [DOI] [PubMed] [Google Scholar]
- 11. Goss GG, Adamia S, Galvez F (2001) Peanut lectin binds to a subpopulation of mitochondria-rich cells in the rainbow trout gill epithelium. Am J Physiol Regul Integr Comp Physiol 281: R1718–1725. [DOI] [PubMed] [Google Scholar]
- 12. Hiroi J, McCormick SD (2012) New insights into gill ionocyte and ion transporter function in euryhaline and diadromous fish. Respir Physiol Neurobiol 184: 257–268. 10.1016/j.resp.2012.07.019 [DOI] [PubMed] [Google Scholar]
- 13. Boutet I, Long Ky CL, Bonhomme F (2006) A transcriptomic approach of salinity response in the euryhaline teleost, Dicentrarchus labrax. Gene 379: 40–50. [DOI] [PubMed] [Google Scholar]
- 14. Kalujnaia S, McWilliam IS, Zaguinaiko VA, Feilen AL, Nicholson J, et al. (2007) Transcriptomic approach to the study of osmoregulation in the European eel Anguilla anguilla. Physiological Genomics 31: 385–401. [DOI] [PubMed] [Google Scholar]
- 15. Tse WKF, Sun J, Zhang HM, Law AYS, Yeung BHY, et al. (2013) Transcriptomic and iTRAQ proteomic approaches reveal novel short-term hyperosmotic stress responsive proteins in the gill of the Japanese eel (Anguilla japonica). Journal of Proteomics 89: 81–94. 10.1016/j.jprot.2013.05.026 [DOI] [PubMed] [Google Scholar]
- 16. Evans TG, Somero GN (2008) A microarray-based transcriptomic time-course of hyper- and hypo-osmotic stress signaling events in the euryhaline fish Gillichthys mirabilis: osmosensors to effectors. J Exp Biol 211: 3636–3649. 10.1242/jeb.022160 [DOI] [PubMed] [Google Scholar]
- 17. Tine M, de Lorgeril J, D'Cotta H, Pepey E, Bonhomme F, et al. (2008) Transcriptional responses of the black-chinned tilapia Sarotherodon melanotheron to salinity extremes. Mar Genomics 1: 37–46. 10.1016/j.margen.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 18. Whitehead A, Roach JL, Zhang S, Galvez F (2012) Salinity- and population-dependent genome regulatory response during osmotic acclimation in the killifish (Fundulus heteroclitus) gill. J Exp Biol 215: 1293–1305. 10.1242/jeb.062075 [DOI] [PubMed] [Google Scholar]
- 19. Tse WK, Au DW, Wong CK (2006) Characterization of ion channel and transporter mRNA expressions in isolated gill chloride and pavement cells of seawater acclimating eels. Biochem Biophys Res Commun 346: 1181–1190. [DOI] [PubMed] [Google Scholar]
- 20. Tse WK, Chow SC, Lai KP, Au DW, Wong CK (2011) Modulation of ion transporter expression in gill mitochondrion-rich cells of eels acclimated to low-Na(+) or-Cl(-) freshwater. J Exp Zool A Ecol Genet Physiol 315: 385–393. 10.1002/jez.681 [DOI] [PubMed] [Google Scholar]
- 21. Tse WK, Chow SC, Wong CK (2012) Eel osmotic stress transcriptional factor 1 (Ostf1) is highly expressed in gill mitochondria-rich cells, where ERK phosphorylated. Front Zool 9: 3 10.1186/1742-9994-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nowak B, Cadoret K, Feist SW, Bean TP (2013) Laser-capture dissection and immunohistochemistry reveals chloride and mucous-cell specific gene expression in gills of seawater acclimated Atlantic salmon Salmo salar. Journal of Fish Biology 83: 1459–1467. 10.1111/jfb.12235 [DOI] [PubMed] [Google Scholar]
- 23. Ladanyi A, Sipos F, Szoke D, Galamb O, Molnar B, et al. (2006) Laser microdissection in translational and clinical research. Cytometry A 69: 947–960. [DOI] [PubMed] [Google Scholar]
- 24. Leguen I, Odjo N, Le Bras Y, Luthringer B, Baron D, et al. (2010) Effect of seawater transfer on CYP1A gene expression in rainbow trout gills. Comp Biochem Physiol A Mol Integr Physiol 156: 211–217. 10.1016/j.cbpa.2010.02.002 [DOI] [PubMed] [Google Scholar]
- 25. Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 27. Leguen I, Cauty C, Odjo N, Corlu A, Prunet P (2007) Trout gill cells in primary culture on solid and permeable supports. Comp Biochem Physiol A Mol Integr Physiol 148: 903–912. [DOI] [PubMed] [Google Scholar]
- 28. Perry SF, Furimsky M, Bayaa M, Georgalis T, Shahsavarani A, et al. (2003) Integrated responses of Na+/HCO3- cotransporters and V-type H+-ATPases in the fish gill and kidney during respiratory acidosis. Biochim Biophys Acta 1618: 175–184. [DOI] [PubMed] [Google Scholar]
- 29. Prunet P, Boeuf G, Houdebine LM (1985) Plasma and pituitary prolactin levels in rainbow trout during adaptation to different salinities. J Exp Zool 235: 187–196. [DOI] [PubMed] [Google Scholar]
- 30. Stolting G, Fischer M, Fahlke C (2014) CLC channel function and dysfunction in health and disease. Front Physiol 5: 378 10.3389/fphys.2014.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thiemann A, Grunder S, Pusch M, Jentsch TJ (1992) A chloride channel widely expressed in epithelial and non-epithelial cells. Nature 356: 57–60. [DOI] [PubMed] [Google Scholar]
- 32. Catalan M, Cornejo I, Figueroa CD, Niemeyer MI, Sepulveda FV, et al. (2002) ClC–2 in guinea pig colon: mRNA, immunolabeling, and functional evidence for surface epithelium localization. American journal of physiology Gastrointestinal and liver physiology 283: G1004–1013. [DOI] [PubMed] [Google Scholar]
- 33. Catalan M, Niemeyer MI, Cid LP, Sepulveda FV (2004) Basolateral ClC–2 chloride channels in surface colon epithelium: regulation by a direct effect of intracellular chloride. Gastroenterology 126: 1104–1114. [DOI] [PubMed] [Google Scholar]
- 34. Guh YJ, Lin C-H, Hwang PP (2015) Osmoregulation in zebrafish: ion transport mechanisms and functional regulation. EXCLI Journal 14: 627–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perez-Rius C, Gaitan-Penas H, Estevez R, Barrallo-Gimeno A (2015) Identification and characterization of the zebrafish ClC–2 chloride channel orthologs. Pflugers Arch 467: 1769–1781. 10.1007/s00424-014-1614-z [DOI] [PubMed] [Google Scholar]
- 36. Tang C-H, Lee T-H (2011) Ion-Deficient Environment Induces the Expression of Basolateral Chloride Channel, ClC-3-Like Protein, in Gill Mitochondrion-Rich Cells for Chloride Uptake of the Tilapia Oreochromis mossambicus. Physiological and Biochemical Zoology 84: 54–67. 10.1086/657161 [DOI] [PubMed] [Google Scholar]
- 37. Miyazaki H, Uchida S, Takei Y, Hirano T, Marumo F, et al. (1999) Molecular cloning of CLC chloride channels in Oreochromis mossambicus and their functional complementation of yeast CLC gene mutant. Biochem Biophys Res Commun 255: 175–181. [DOI] [PubMed] [Google Scholar]
- 38. Tang CH, Hwang LY, Lee TH (2010) Chloride channel ClC–3 in gills of the euryhaline teleost, Tetraodon nigroviridis: expression, localization and the possible role of chloride absorption. J Exp Biol 213: 683–693. 10.1242/jeb.040212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bossus M, Charmantier G, Blondeau-Bidet E, Valletta B, Boulo V, et al. (2013) The ClC–3 chloride channel and osmoregulation in the European sea bass, Dicentrarchus labrax. J Comp Physiol B 183: 641–662. 10.1007/s00360-012-0737-9 [DOI] [PubMed] [Google Scholar]
- 40. Boyle D, Clifford AM, Orr E, Chamot D, Goss GG (2015) Mechanisms of Cl(-) uptake in rainbow trout: cloning and expression of slc26a6, a prospective Cl(-)/HCO3(-) exchanger. Comp Biochem Physiol A Mol Integr Physiol 180: 43–50. 10.1016/j.cbpa.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 41. Toyoshima C, Kanai R, Cornelius F (2011) First Crystal Structures of Na+,K+-ATPase: New Light on the Oldest Ion Pump. Structure 19: 1732–1738. 10.1016/j.str.2011.10.016 [DOI] [PubMed] [Google Scholar]
- 42. Madsen SS, Kiilerich P, Tipsmark CK (2009) Multiplicity of expression of Na+,K+-ATPase {alpha}-subunit isoforms in the gill of Atlantic salmon (Salmo salar): cellular localisation and absolute quantification in response to salinity change. J Exp Biol 212: 78–88. 10.1242/jeb.024612 [DOI] [PubMed] [Google Scholar]
- 43. McCormick SD, Regish AM, Christensen AK (2009) Distinct freshwater and seawater isoforms of Na+/K+-ATPase in gill chloride cells of Atlantic salmon. J Exp Biol 212: 3994–4001. 10.1242/jeb.037275 [DOI] [PubMed] [Google Scholar]
- 44. Richards JG, Semple JW, Bystriansky JS, Schulte PM (2003) Na+/K+-ATPase alpha-isoform switching in gills of rainbow trout (Oncorhynchus mykiss) during salinity transfer. J Exp Biol 206: 4475–4486. [DOI] [PubMed] [Google Scholar]
- 45. Parks SK, Tresguerres M, Goss GG (2007) Interactions between Na+ channels and Na+-HCO3- cotransporters in the freshwater fish gill MR cell: a model for transepithelial Na+ uptake. Am J Physiol Cell Physiol 292: C935–944. [DOI] [PubMed] [Google Scholar]
- 46. Hirata T, Kaneko T, Ono T, Nakazato T, Furukawa N, et al. (2003) Mechanism of acid adaptation of a fish living in a pH 3.5 lake. Am J Physiol Regul Integr Comp Physiol 284: R1199–1212. [DOI] [PubMed] [Google Scholar]
- 47. Lee YC, Yan JJ, Cruz SA, Horng JL, Hwang PP (2011) Anion exchanger 1b, but not sodium-bicarbonate cotransporter 1b, plays a role in transport functions of zebrafish H+-ATPase-rich cells. Am J Physiol Cell Physiol 300: C295–307. 10.1152/ajpcell.00263.2010 [DOI] [PubMed] [Google Scholar]
- 48. Furukawa F, Watanabe S, Inokuchi M, Kaneko T (2011) Responses of gill mitochondria-rich cells in Mozambique tilapia exposed to acidic environments (pH 4.0) in combination with different salinities. Comp Biochem Physiol A Mol Integr Physiol 158: 468–476. 10.1016/j.cbpa.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 49. Dymowska AK, Schultz AG, Blair SD, Chamot D, Goss GG (2014) Acid-sensing ion channels are involved in epithelial Na+ uptake in the rainbow trout Oncorhynchus mykiss. American Journal of Physiology-Cell Physiology 307: C255–C265. 10.1152/ajpcell.00398.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boulkroun S, Ruffieux-Daidie D, Vitagliano JJ, Poirot O, Charles RP, et al. (2008) Vasopressin-inducible ubiquitin-specific protease 10 increases ENaC cell surface expression by deubiquitylating and stabilizing sorting nexin 3. Am J Physiol Renal Physiol 295: F889–900. 10.1152/ajprenal.00001.2008 [DOI] [PubMed] [Google Scholar]
- 51. Soundararajan R, Pearce D, Ziera T (2012) The role of the ENaC-regulatory complex in aldosterone-mediated sodium transport. Mol Cell Endocrinol 350: 242–247. 10.1016/j.mce.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Claro da Silva T, Polli JE, Swaan PW (2013) The solute carrier family 10 (SLC10): beyond bile acid transport. Mol Aspects Med 34: 252–269. 10.1016/j.mam.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeong J, Eide DJ (2013) The SLC39 family of zinc transporters. Mol Aspects Med 34: 612–619. 10.1016/j.mam.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bradshaw AD (2012) Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol 44: 480–488. 10.1016/j.biocel.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seear PJ, Carmichael SN, Talbot R, Taggart JB, Bron JE, et al. (2010) Differential gene expression during smoltification of Atlantic salmon (Salmo salar L.): a first large-scale microarray study. Mar Biotechnol (NY) 12: 126–140. [DOI] [PubMed] [Google Scholar]
- 56. Ramalingam A, Duhadaway JB, Sutanto-Ward E, Wang Y, Dinchuk J, et al. (2008) Bin3 deletion causes cataracts and increased susceptibility to lymphoma during aging. Cancer Res 68: 1683–1690. 10.1158/0008-5472.CAN-07-6072 [DOI] [PubMed] [Google Scholar]
- 57. Thompson O, Moghraby JS, Ayscough KR, Winder SJ (2012) Depletion of the actin bundling protein SM22/transgelin increases actin dynamics and enhances the tumourigenic phenotypes of cells. BMC Cell Biol 13: 1 10.1186/1471-2121-13-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen Y, Ip FC, Shi L, Zhang Z, Tang H, et al. (2014) Coronin 6 regulates acetylcholine receptor clustering through modulating receptor anchorage to actin cytoskeleton. J Neurosci 34: 2413–2421. 10.1523/JNEUROSCI.3226-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pfister KK, Fisher EM, Gibbons IR, Hays TS, Holzbaur EL, et al. (2005) Cytoplasmic dynein nomenclature. J Cell Biol 171: 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takahashi M, Tajika Y, Khairani AF, Ueno H, Murakami T, et al. (2013) The localization of VAMP5 in skeletal and cardiac muscle. Histochem Cell Biol 139: 573–582. 10.1007/s00418-012-1050-0 [DOI] [PubMed] [Google Scholar]
- 61. Errico A, Ballabio A, Rugarli EI (2002) Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet 11: 153–163. [DOI] [PubMed] [Google Scholar]
- 62. Yanez-Mo M, Tejedor R, Rousselle P, Sanchez -Madrid F (2001) Tetraspanins in intercellular adhesion of polarized epithelial cells: spatial and functional relationship to integrins and cadherins. J Cell Sci 114: 577–587. [DOI] [PubMed] [Google Scholar]
- 63. Serra-Pages C, Medley QG, Tang M, Hart A, Streuli M (1998) Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem 273: 15611–15620. [DOI] [PubMed] [Google Scholar]
- 64. Bai R, Bai H, Kuse M, Ideta A, Aoyagi Y, et al. (2014) Involvement of VCAM1 in the bovine conceptus adhesion to the uterine endometrium. Reproduction 148: 119–127. 10.1530/REP-13-0655 [DOI] [PubMed] [Google Scholar]
- 65. Kuespert K, Pils S, Hauck CR (2006) CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol 18: 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Avella M, Masoni A, Bornancin M, Mayergostan N (1987) Gill morphology and sodiuminflux in the rainbow-trout (Salmo-gairdneri) acclimated to artificial fresh-water environments. Journal of Experimental Zoology 241: 159–169. [Google Scholar]
- 67. Zenker WGE, Ferguson HW, Barker IK, Woodward B (1987) Epithelial and pillar cell replacement in gills of juvenile trout, Salmo-gairdneri richardson. Comparative Biochemistry and Physiology a-Physiology 86: 423–428. [DOI] [PubMed] [Google Scholar]
- 68. Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, et al. (2008) Structure and function of claudins. Biochimica et Biophysica Acta (BBA)—Biomembranes 1778: 631–645. [DOI] [PubMed] [Google Scholar]
- 69. Tipsmark CK, Baltzegar DA, Ozden O, Grubb BJ, Borski RJ (2008) Salinity regulates claudin mRNA and protein expression in the teleost gill. Am J Physiol Regul Integr Comp Physiol 294: R1004–1014. 10.1152/ajpregu.00112.2007 [DOI] [PubMed] [Google Scholar]
- 70. Tipsmark CK, Luckenbach JA, Madsen SS, Kiilerich P, Borski RJ (2008) Osmoregulation and expression of ion transport proteins and putative claudins in the gill of Southern Flounder (Paralichthys lethostigma). Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 150: 265–273. [DOI] [PubMed] [Google Scholar]
- 71. Notterpek L, Roux KJ, Amici SA, Yazdanpour A, Rahner C, et al. (2001) Peripheral myelin protein 22 is a constituent of intercellular junctions in epithelia. Proc Natl Acad Sci U S A 98: 14404–14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Roux KJ, Amici SA, Fletcher BS, Notterpek L (2005) Modulation of epithelial morphology, monolayer permeability, and cell migration by growth arrest specific 3/peripheral myelin protein 22. Mol Biol Cell 16: 1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ohsakaya S, Fujikawa M, Hisabori T, Yoshida M (2011) Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria. J Biol Chem 286: 20292–20296. 10.1074/jbc.M110.198523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lin H, Randall D (1991) Evidence for the presence of an electrogenic proton pump on the trout gill epithelium. Journal of Experimental Biology 161: 119–134. [Google Scholar]
- 75. Soengas JL, Barciela P, Aldegunde M, Andres D (1995) Gill carbohydrate-metabolism of rainbow-trout is modified during gradual adaptation to sea-water. Journal of Fish Biology 46: 845–856. [Google Scholar]
- 76. Kohen R, Nyska A (2002) Oxidation of biological systems: oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol Pathol 30: 620–650. [DOI] [PubMed] [Google Scholar]
- 77. Chen CH, Sun L, Mochly-Rosen D (2010) Mitochondrial aldehyde dehydrogenase and cardiac diseases. Cardiovasc Res 88: 51–57. 10.1093/cvr/cvq192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Freeman H, Shimomura K, Cox RD, Ashcroft FM (2006) Nicotinamide nucleotide transhydrogenase: a link between insulin secretion, glucose metabolism and oxidative stress. Biochem Soc Trans 34: 806–810. [DOI] [PubMed] [Google Scholar]
- 79. Nishi H, Inagi R, Kato H, Tanemoto M, Kojima I, et al. (2008) Hemoglobin is expressed by mesangial cells and reduces oxidant stress. J Am Soc Nephrol 19: 1500–1508. 10.1681/ASN.2007101085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grabbe C, Husnjak K, Dikic I (2011) The spatial and temporal organization of ubiquitin networks. Nature reviews Molecular cell biology 12: 295–307. 10.1038/nrm3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hicke L (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195–201. [DOI] [PubMed] [Google Scholar]
- 82. Zhang S, Wu W, Wu Y, Zheng J, Suo T, et al. (2010) RNF152, a novel lysosome localized E3 ligase with pro-apoptotic activities. Protein Cell 1: 656–663. 10.1007/s13238-010-0083-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Seok Ko H, Uehara T, Tsuruma K, Nomura Y (2004) Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. Febs Letters 566: 110–114. [DOI] [PubMed] [Google Scholar]
- 84. Cal S, Quesada V, Garabaya C, Lopez-Otin C (2003) Polyserase-I, a human polyprotease with the ability to generate independent serine protease domains from a single translation product. Proc Natl Acad Sci U S A 100: 9185–9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, et al. (2009) E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell 33: 483–495. 10.1016/j.molcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, et al. (2008) SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem 283: 33211–33220. 10.1074/jbc.M804440200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, et al. (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ratios>3 identified transcripts that were upregulated in freshwater ionocyte and ratios<3 identified transcripts that were upregulated in seawater ionocytes.
(DOCX)
Data Availability Statement
All microarray data are available from the GEO database (accession number GSE69409).