Abstract
Background
Stripe rust, a highly destructive foliar disease of wheat (Triticum aestivum), causes severe losses, which may be accompanied by reduced photosynthetic activity and accelerated leaf senescence.
Methods
We used suppression subtractive hybridization (SSH) to examine the mechanisms of resistance in the resistant wheat line L693 (Reg. No. GP-972, PI 672538), which was derived from a lineage that includes a wide cross between common and Thinopyrum intermedium. Sequencing of an SSH cDNA library identified 112 expressed sequence tags.
Results
In silico mapping placed one of these tags [GenBank: JK972238] on chromosome 1A. Primers based on [GenBank: JK972238] amplified a polymorphic band, which co-segregated with YrL693. We cloned a candidate gene encoding wheat copper-binding protein (WCBP1) by amplifying the polymorphic region, and we mapped WCBP1 to a 0.64 cM genetic interval. Brachypodium, rice, and sorghum have genes and genomic regions syntenic to this region.
Discussion
Sequence analysis suggested that the resistant WCBP1 allele might have resulted from a deletion of 36-bp sequence of the wheat genomic sequence, rather than direct transfer from Th. intermedium. qRT-PCR confirmed that WCBP1 expression changes in response to pathogen infection.
Conclusions
The unique chromosomal location and expression mode of WCBP1 suggested that WCBP1 is the putative candidate gene of YrL693, which was involved in leaf senescence and photosynthesis related to plant responses to stripe rust infection during the grain-filling stage.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0612-4) contains supplementary material, which is available to authorized users.
Keywords: Quantitative reverse transcription PCR, Puccinia striiformis, Triticum aestivum, Suppression subtractive hybridization, Photosynthesis
Background
Stripe rust, caused by the fungus Puccinia striiformis f. sp. tritici (Pst), is one of the most destructive diseases affecting wheat (Triticum aestivum L.) in many regions of the world. In China, stripe rust infects millions of hectares of wheat annually [1], causing large yield losses over the past two decades due to the emergence of new virulent races in the fungus population [2, 3].
As a foliar disease, stripe rust has a considerable effect on grain yield, depending on the disease response [4] and the host growth stage at which Pst infection is initiated [5]. Stripe rust epidemics can begin at any growth stage in wheat [6, 7], and therefore, they have a marked effect on plant vigor in addition to grain yield [8]. Severe stripe rust infection also alters the distribution of assimilates among the various organs of the plant [9, 10].
Stripe rust infection is closely linked to both leaf photosynthesis and senescence, and many of the expressed sequence tags (ESTs) induced by Puccinia striiformis are related to those two processes [11–14]. A recent study in wheat showed that the senescence-associated gene (SAG) TaSAG120 plays an important role in the defense against stripe rust [15]. In addition, the multi-gene pathogen resistance factor ‘Lr34/Yr18/Pm38’ involves leaf senescence [16]. These findings imply that senescence-and photosynthesis-related processes might be part of the resistance response to stripe rust infection.
Wheat line L693 (Reg. No. GP-972, PI 672538) is from F6:7 families of a cross between MY11 and YU25, the latter being a derivative of a wide cross involving Thinopyrum intermedium [17]. Genetic analysis demonstrated that stripe rust resistance observed in L693 is conferred by a single dominant gene, YrL693, which was mapped to chromosome 1B by linkage analysis with simple sequence repeat (SSR) markers and Chinese Spring nulli-tetrasomic lines [18, 19]. Although the pedigree of L693 includes Th. intermedium, the question of whether YrL693 is derived directly from Th. intermedium remains unanswered.
The objectives of the present study were to assess whether resistant plants maintained the normal timing of leaf senescence during grain filling and to examine the physiological effects of any leaf senescence traits that were affected by infection. We also explored the molecular mechanism of resistance to stripe rust in wheat and attempted to determine the origin of YrL693 by high-density mapping and comparative genomic analysis. We demonstrated that it is possible to map resistance genes using the identified ESTs between isogenic lines after pathogen infection, and we outline a putative novel genetic mechanism of resistance conferred by this gene.
Results
Identification of unique ESTs induced by Pst infection
L693 was highly resistant and L661 highly susceptible to Pst race CYR32 (Fig. 1a). The response tests performed at the adult (Fig. 1b) and seedling stages (Fig. 1a) showed similar results. Random sequencing of bacterial colonies from the suppression subtractive hybridization (SSH) library identified 112 ESTs, with an average length of 178 bp, which were submitted to NCBI under the accession numbers [GenBank: JK972179] though [GenBank: JK972287] and [GenBank: JK974072] through [GenBank: JK974074] (Additional file 2: Table S1). Of these, 87 unique ESTs (uniESTs) with an average length of 197 bp (Table 1), including 21 contigs with an average length of 226 bp and 66 singletons with an average length of 188 bp, were found. Most of the uniESTs were between 50 bp and 300 bp in length (Additional file 1: Figure S2) and were further identified through clustering and assembly of putative full-length cDNAs based on the BLAST analysis option in NCBI GenBank (Additional file 2: Table S2).
Fig. 1.
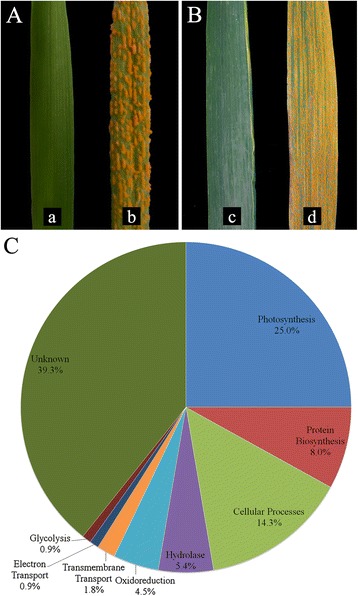
Seedling (a) and adult (b) responses to a) L693, b) L661, c) L693, d) L661. c Functional classes of differentially expressed sequences induced by Pst inoculation
Table 1.
ESTs in the SSH library prepared from combined leaf samples obtained from wheat seedlings
| Number obtained | Average length (bp) | |
|---|---|---|
| ESTs | 112 | 178 |
| Singletons | 66 | 188 |
| Contigs | 21 | 226 |
| UniESTs | 87 | 197 |
Both L693 and L661 were sampled 24, 48 and 96 h after inoculation with Pst CYR32. cDNA prepared from L693 leaves infected with Pst CYR32 was used as the tester, and cDNA prepared from L661 leaves infected with Pst CYR32 was used as the driver
Functional categorization of the ESTs, as well as gene network analysis, predicted that Pst infection in L693 induces differential regulation of leaf senescence-associated-genes (SAGs).
Of the 112 sequences, 68 showed high sequence similarity to known or annotated genes from cDNA libraries from wheat, rice, barley, pea and Arabidopsis thaliana (Fig. 1c, Additional file 2: Table S2). Of the genes that were upregulated in L693, 25 % were involved in photosynthesis-like and leaf senescence-like pathways, 14 % in cellular process pathways, 8 % in protein biosynthesis pathways, 5 % in the hydrolase pathway, 5 % in the oxidoreduction pathway, 2 % in the transmembrane transport pathway, 1 % in the electron transport pathway, and 1 % in glycolysis. The remaining 44 (39 %) of upregulated sequences had unspecified or unknown functions, 14 of which closely matched the sequences of putative senescence-associated genes (Fig. 1c and Additional file 2: Table S2).
Polymorphisms between ESTs and linkage analysis of stripe rust resistance
Out of 62 primers designed, one primer pair (LS36) (Additional file 2: Table S3) produced polymorphic amplicons between L693 and L661 as well as between the resistant parent YU25 and the susceptible parent MY11 (Fig. 2a). The design of the LS36 primers was from the sequence of [GenBank: JK972238], which we mapped to wheat chromosome 1A by in silico mapping. The L693, L661, YU25 and MY11 lines produced two amplicons, one 106 bp amplicon, which was the same among all genotypes, and a second amplicon that was polymorphic between L693 and L661 and between YU25 and MY11. In L661 and susceptible parents MY11 and CM107, this amplicon was 133 bp, and in the resistant lines L693, L658, L696 and L699, as well as YU25 and sister line YU24, the amplicon was 97 bp. Amplification of genomic DNA from the nullisomic-tetrasomic and ditelosomic lines (Fig. 2b) revealed that the amplicon common among all lines and CS was absent in the nullisomic 1A lines (N1AT1B and N1AT1D).
Fig. 2.
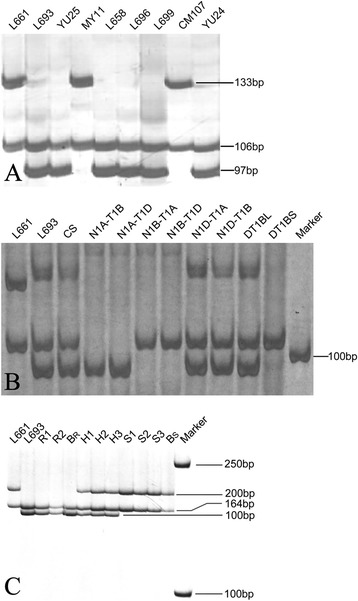
Identification of polymorphism between stripe rust-resistant and stripe rust-susceptible genotypes. The Ls36 primer designed from an EST sequence [GenBank: JK972238] produced polymorphic amplicons among the various genotypes. a a 133 bp amplicon was amplified in the susceptible L661 line and its susceptible parents MY11 and CM107. A 97 bp amplicon was amplified in resistant L693 line, resistant sister lines L658, L696, and L699, and their resistant parent, YU25. A 97 bp amplicon was also amplified from YU24, one of the resistant sister lines of YU25. A 106 bp fragment was amplified in all genotypes. b chromosomal localizations of the amplicons that were polymorphic between L693 and L661, with CS nulli-tetrasomic lines of homoeologous group 1 and ditelosomic lines of wheat chromosome 1B. PCR was performed to map the gene using genomic DNA from the CS and various aneuploids. PCR products were resolved on 6 % polyacrylamide gels. No PCR product was generated from nullisomic 1B (N1BT1A and N1BT1D) or the ditelosomic 1BS (DT1BS) lines. c Silver-stained polyacrylamide gels showing polymorphic markers generated using the LSc18 primer linked to the stripe rust resistance gene in L693. L661, susceptible parent; L693, resistant parent; R1 and R2, resistant F2 individuals; BR, the resistant F2 DNA pool; H1, H2 and H3, resistant F2 individuals; S1, S2 and S3, susceptible F2 individuals; BS, the susceptible F2 DNA pool. L661, S1, S2, S3 and Bs showed amplification of 173 bp and 200 bp fragments; L693, R1, R2 and BR showed amplification of 164 bp and 173 bp fragments; H1, H2 and H3 showed amplification of all three fragments, indicating heterozygosity
The polymorphic 97 bp amplicon was located on chromosome arm 1BL, as no PCR products were generated from the nullisomic 1B lines (N1BT1A and N1BT1D) or the ditelosomic 1BS line (DT1BS). Sequencing of the polymorphic amplicon found that there was an extra repeat of 36 bp sequence in the susceptible allele compared with the resistant allele. Sequence similarity searches with the polymorphic amplicon found a contig (3837062) with high sequence similarity located on wheat chromosome 1B.
We designed four pairs of primers for linkage analysis based on differences in the number of copies of the 36 bp sequence (Additional file 2: Table S4). Analysis using the LSc18 primer pair (Fig. 2c) detected a 100 bp product closely linked to the YrL693 resistance gene in L693. Analysis of 523 F2:3 lines with additional primer pairs (Additional file 2: Table S5) further demonstrated a co-dominant marker on chromosome 1B co-segregating with YrL693.
Fine mapping and comparative genomic analysis of WCBP1 indicated that it co-segregated with YrL693
Genomic in situ hybridization (GISH) failed to detect an alien chromosome segment from Th. intermedium in L693 (Additional file 1: Figure S6; Additional file 3: Methods S1). A high-density integrated genetic map was constructed (Fig. 3). The linkage map consisted of nine SSR markers and eight EST-STS markers (Additional file 2: Table S8). The WCBP1 locus fell within a genetic interval of 0.64 cM. WCBP1 co-segregated with the YrL693 locus. Two flanking EST-STS markers, BF474347 and BE443300, were linked to the WCPB1 locus at 0.096 and 0.544 cM, respectively, and mapped to the chromosome 1B bin C–1BL-6-0.32. Therefore, WCBP1 is located in the same chromosomal region as YrL693. The contigs of the two SSR markers Xwmc269-1B and Xcfd65-1B on one side of WCBP1 mapped to wheat chromosomal arm 1BL, whereas those of the seven other SSR markers on the other side of WCBP1 mapped to 1BS according to the published SSR linkage map [20].
Fig. 3.
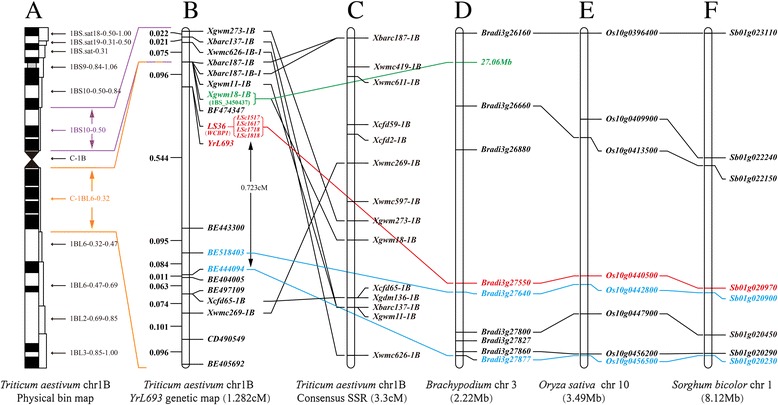
Genetic and comparative genomic linkage map of the candidate gene WCBP1 and YrL693. a Physical bin map of WCPB1; WCBP1 was mapped to the bin C-1BL6-0.32. b Genetic map of WCBP1 with a total length of 1.28 cM on wheat chromosome 1B. The genetic distance in cM is shown on the left and genetic markers are shown on the right. c The consensus SSR map for a total of 3.3 cM of wheat chromosome 1B. d, e, f, Orthologous regions of WCBP1 in Brachypodium distachyon chromosome 3, and Oryza sativa japonica chromosome 10, and Sorghum bicolor chromosome 1, respectively
Orthologs of wheat ESTs BE518403 and BE444094 were detected on B. distachyon chromosome 3, rice chromosome 10, and sorghum chromosome 1. Comparative genomic analysis established the collinearity of WCBP1 genomic regions with a 2.22 Mb (Bradi3g26160–Bradi3g27877) region on B. distachyon chromosome 3, a 3.49 Mb region (Os10g0396400–Os10g0456500) on rice chromosome 10, and a 8.12 Mb region (Sbo1g020230–Sb01g023110) on sorghum chromosome 1, regions that carry 112, 260, and 297 genes, respectively. The gene order was highly conserved among wheat, B. distachyon, and rice (Fig. 4), but showed an intra-chromosomal inversion in the 8.12 Mb chromosome region of sorghum. We also found an orthologous genomic region (from 27,059,150 to 27,060,251) of wheat contig 1BS-3450437 carrying the SSR marker Xgwm18-1B at 27.06 Mb on B. distachyon chromosome 3 (E-value = 5.34E−34), but there was no corresponding ortholog in either rice or sorghum (Fig. 3). An ortholog of WCBP1 was found on Bradi3g27550 (E-value ≈ 0) at 28 Mb on B. distachyon chromosome 3, on Os10g0440500 (E-value = 5.34E−168) at 16 Mb on rice chromosome 10, and on Sb01g020970 (E-value = 1.14E−130) at 24.05 Mb on sorghum chromosome 1 (Figs. 3 and 4).
Fig. 4.
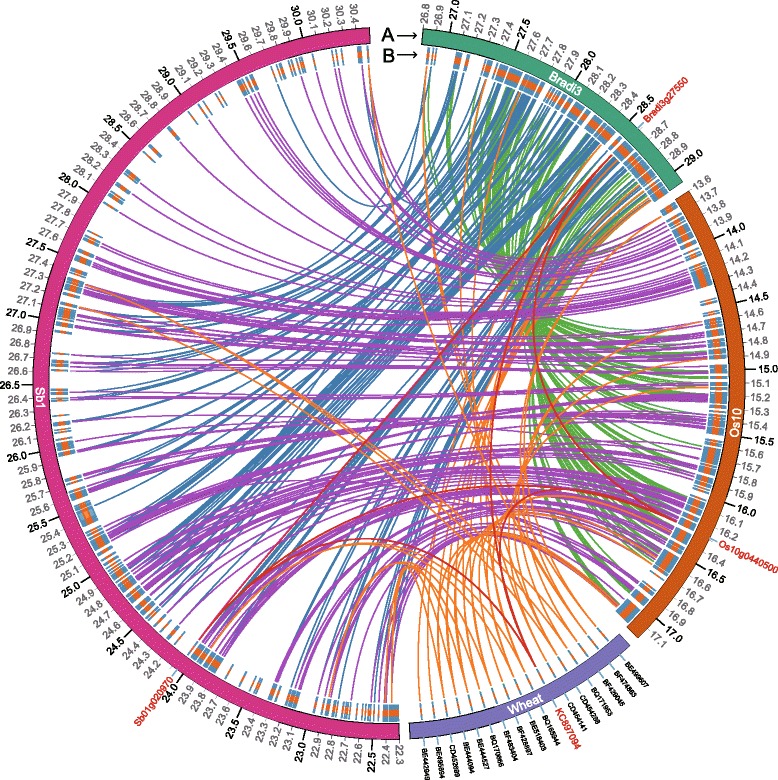
Comparative genomic mapping of ESTs in the WCBP1 (red) chromosomal region of wheat chromosome 1B (purple) with Brachypodium distachyon chromosome 3 (green), Oryza sativa japonica chromosome 10 (orange), and Sorghum bicolor chromosome 1 (red). The gene order in all species is clockwise
PCR-based cloning and functional analysis of the candidate stripe rust resistance gene
We obtained a candidate resistance gene by sequencing PCR products amplified by seven primers (Additional file 2: Table S6) from genomic DNA and then comparing the amplified sequence with that of the contig (3837062). This gene was 1,398 bp in the resistant lines L693 and YU25 but 1,434 bp in the susceptible lines L661 and MY11 (Additional file 1: Figure S4). The genomic sequence of the candidate resistance gene isolated from L693 and YU25 included four exons (154, 329, 79, and 479 bp) and three introns (112, 126, and 119 bp). The fourth exon contained a single repeat of the 36 bp sequence in the resistant L693 line and its parent YU25 as well as two repeats in the susceptible L661 line and its parent MY11. We also identified and verified a single nucleotide polymorphism (SNP) in the first exon, with an adenine (A) in the resistant lines and a cytosine (C) in susceptible lines L661 and MY11 (Additional file 1: Figure S5).
The open reading frame (ORF) (Fig. 5a) of the candidate resistance gene encodes a putative wheat copper binding protein 1 (WCBP1) predicted to contain 346 amino acids in resistant lines (AGS38338) and 358 amino acids in susceptible lines. The SNP in the first exon (Additional file 1: Figure S5) does not change the deduced amino acid sequence. The 36 bp repeated sequence in the fourth exon (Fig. 5b and Additional file 1: Figure S3) caused an addition of 12 amino acids (K K K D K G A G D G G E) in the susceptible line (Fig. 5b) at amino acids 242–253. The deduced amino acid sequence forms a protein containing two predicted copper-binding domains, at amino acids 34–39 (L H C A G C, exon 1), and at amino acids 170–175 (L H C D G C, exon 3) (Fig. 5b). The full-length, deduced WCBP1 protein shows sequence similarity with known heavy metal-binding proteins from diverse species. Phylogenetic analysis indicated that WCBP1 shared the highest similarity with the EMS53947 protein (Fig. 5d), but they did not cluster together.
Fig. 5.
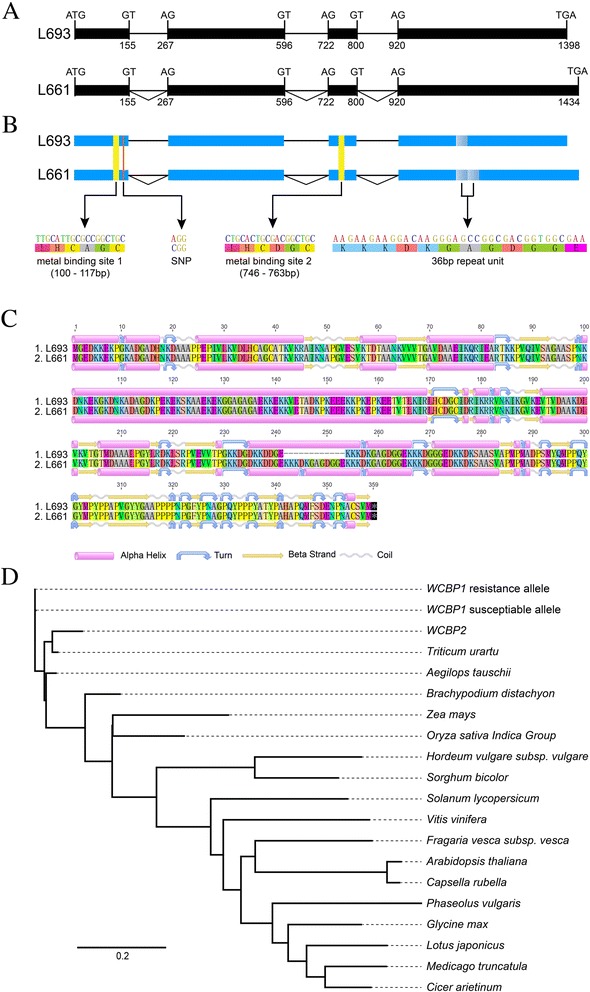
The WCBP1 gene, its putative protein structure, and its phylogenetic tree. a The predicted mRNA structure of WCBP1 is shown above the genomic sequence, including introns (thin lines) and exons (thick lines). ATG represents the methionine start codon, and TGA represents the stop codon. b Coding regions of two heavy metal copper-binding domains in exons 1 and 3. The major differences between the resistance-and susceptibility-associated WCBP1 alleles are a SNP in exon 1 and a 36 bp deletion in exon 4. c Alignment of the amino acid sequences and secondary structures of the proteins putatively encoded by the resistance-and susceptibility-associated alleles of WCBP1. d Phylogenetic tree constructed using the neighbor-joining algorithm in MEGA 5.05 following WCBP1 protein sequence alignment using the CLUSTALW program. Accession numbers for the other heavy metal copper-binding proteins were as follows: EMS53947 (Triticum urartu) (11.37 %), EMT15307 (Aegilops tauschii) (11.37 %), XP_003573974 (Brachypodium distachyon) (11.37 %), EAY78671 (Oryza sativa indica group) (3.72%), XP_002465840 (Sorghum bicolor) (6.65 %), AFK36536 (Medicago truncatula) (4.72 %), XP_004497534 (Cicer arietinum) (4.72%), XP_004955283 (Setaria italica) (4.58 %), BAK03814 (Hordeum vulgare subsp. vulgare) (4.15 %), XP_004304460 (Fragaria vesca subsp. vesca) (4.01%), DAA49855 (Zea mays) (3.93 %), NP_195958 (Arabidopsis thaliana) (3.79 %), EOA20845 (Capsella rubella) (3.79 %), XP_004246628 (Solanum lycopersicum) (3.72 %), XP_003542527 (Glycine max) (3.72 %), AFK35929 (Lotus japonicus) (3.72 %), EAY78671 (Oryza sativa indica group) (3.72%), XP_002276537 (Vitis vinifera) (3.72%) and AFW90521 (Phaseolus vulgaris) (3.29 %)
Sequence analysis identified a highly similar gene, WCBP2, on chromosome 1A. This gene had the same DNA sequence in L693 and L661. WCBP1 and WCBP2 show 91.6 % DNA sequence similarity. Moreover, no polymorphism in the length of fragments was detected in genomic DNA of resistant and susceptible varieties.
WCBP1 and WCBP2 are differentially regulated in association with resistance
To examine possible involvement of WCBP1 in defense against stripe rust, we measured its transcript levels at different time points following pathogen inoculation (Additional file 2: Table S7). WCBP1 [NCBI GenBank: KC897094] was upregulated in leaves following Pst inoculation of both the resistant and the susceptible parent. Transcript abundance of WCBP1 was significantly higher (P <0.05) in leaves from L661 and L693 at 24 h post inoculation than prior to inoculation (Fig. 6). However, at later time points, the WCBP1 transcript abundance decreased in L661 but remained higher in L693. At 96 h post-inoculation, the WCBP1 transcript abundance was significantly lower in L661 compared to L693 (P <0.01, Fig. 6a).
Fig. 6.
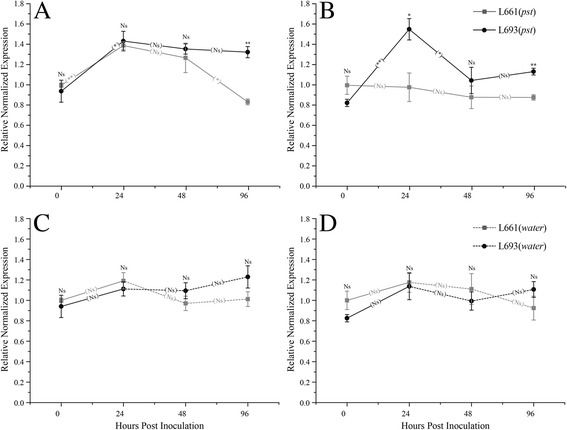
qRT-PCR expression analysis of WCBP1 and WCBP2. Amplification of WCBP1 (a) and WCBP2 (b) transcripts following Pst infection. C, Amplification of WCBP1 (c) and WCBP2 (d) transcripts following mock inoculation with water. The experiment was performed using three biological replicates with three technical replicates per biological sample. Relative expression was calculated using GAPDH as a reference gene to infer steady-state mRNA levels, and relative expression was normalized by setting the control sample from L661 at 0 h to 1. Bars represent the standard error, and the significance was determined using independent sample t-tests. Asterisks represent significant differences as follows: **P ≤0.01, *P ≤0.05; Ns, no significant difference. The raised asterisks or the Ns designation represent differences in gene expression between L693 and L661 at each time point. Asterisks or the Ns designation in the trend line represent the differences in gene expression between two adjacent time points in the same genotype
We also measured the transcript levels of WCBP2. Prior to inoculation, L693 and L661 showed similar WCBP2 transcript levels, but after inoculation, WCBP2 was upregulated only in the resistant L693 line. At 24 h post-inoculation, WCBP2 transcript abundance in L693 increased significantly compared with its level prior to inoculation (P <0.01), and WCBP2 transcript levels were significantly higher (P <0.01) in L693 than in L661 at both 24 h and 96 h post-inoculation (Fig. 6b). In addition, we found that mock inoculation with water had no significant effects on the transcript level of either WCBP1 or WCBP2 in L693 or L661 at any time point (Fig. 6c and d).
The effects of resistance on photosynthesis, chlorophyll fluorescence dynamics, and yield
Although the net photosynthetic rates (Pn) at all time points were significantly higher (P <0.01) in L693 than L661, these genotypes showed similar trends (Fig. 7). Notably, there were no significant differences (P >0.05) between any two consecutive measurements in L693, whereas changes in L661 were highly significant (P <0.01) (Fig. 7a). At the heading stage, the stomatal conductance (Gs) in L693 was significantly lower than that in L661 (Fig. 7b). The Gs of L693 increased sharply during the early stages and decreased toward the end of the experiment, while the values obtained for L661 declined sharply during the early stages of the experiment and then increased. The intercellular CO2 concentrations (Ci) observed in L693 were significantly lower (P <0.01) than those in L661 at all time points, except at 20 days after heading (Fig. 7c). In L693, there was a notable increase in Ci, whereas this value decreased sharply between heading and 10 days after heading in L661.
Fig. 7.
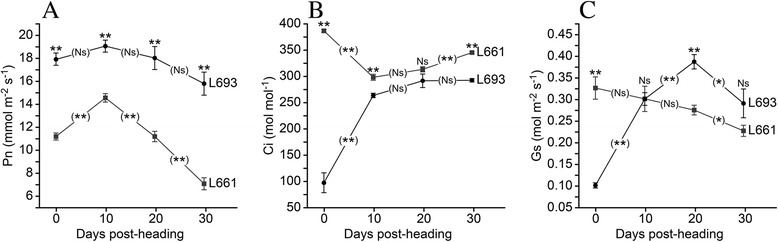
Photosynthetic parameters used to assess the different responses of L693 and L661 to Pst inoculation. a, Net photosynthetic rate (P n); b, intercellular CO2 concentration (C i); c, stomatal conductance (G s). The parameters were measured once every 10 days after heading in the field, using a Li-Cor 6400. The disease had fully developed at heading, and 10 plants per genotype were used for these measurements. Bars represent the standard error, and significance was determined using an independent sample t-test. Asterisks represent significant differences as follows: **P ≤0.01 and *P ≤0.05, and Ns indicates no significant difference. Raised asterisks or the Ns label represent the differences in gene expression between L693 and L661 at each time point. Asterisks or the Ns label on the trend line represent differences in gene expression between two adjacent time points in the same genotype
Although the changes in the maximal photochemical efficiency of PSІІ in the dark-adapted leaves (Fv/Fm) were similar between L693 and L661, the Fv/Fm value obtained for L693 was significantly higher than that obtained for L661 at 30 days after heading. A significant decrease in Fv/Fm occurred in L661 between 20 days and 30 days after heading, whereas only a slight decrease occurred in this timeframe in L693 (Fig. 8a). The efficiencies of excitation capture by the open PSII reaction centers (Fv‘/ Fm’) observed in L693 were significantly lower than, similar to, and higher than those recorded in L693 prior to 20 days, at 20 days and at 30 days after heading, respectively (Fig. 8b). The photochemical quenching coefficient (qP) was similar in L693 and L661. Although the qP values obtained in L693 were greater than those in L661, the differences were not significant at the P = 0.05 level (Fig. 8c). The changes in the quantum yield of photochemical energy conversion in PSII (ΦPSII) (Fig. 8d), the quantum yield of regulated non-photochemical energy loss in PSII (NPQ) (Fig. 8e), and the quantum yield of non-regulated, non-photochemical energy loss in PSII (NO) (Fig. 8f) were quite similar in L693 and L661. The value of ΦPSII recorded in L693 was significantly lower at the heading stage and significantly greater at 30 days after heading, compared with that in L661 (Fig. 8d). In L693, the value of NPQ obtained was significantly greater than that in L661 at the heading stage (Fig. 8e), and the value of NO was significantly lower than that in L661 at 30 days after heading (Fig. 8f).
Fig. 8.
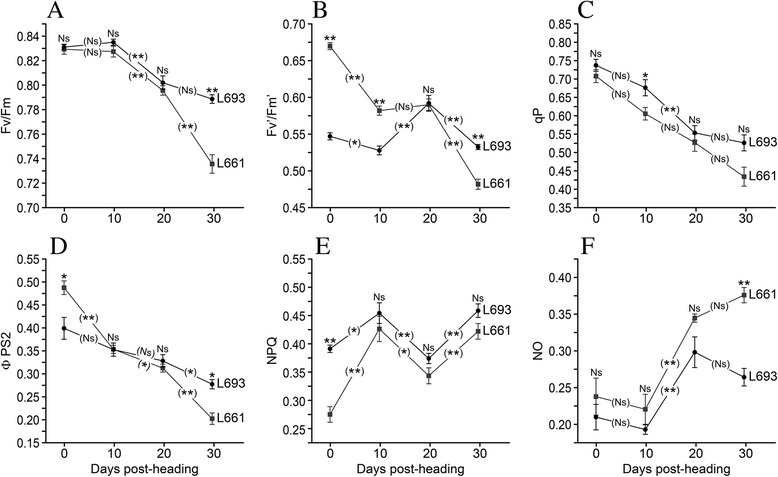
Chlorophyll fluorescence-related parameters used to determine differences in photosystem II (PSII) activity between the resistant line L693 and the susceptible line L661 following Pst inoculation and after heading. a, Values for the maximal photochemical efficiency of PSII (F v /F m); b, the efficiency of excitation capture by the open PSII reaction centers (F’ v /F’ m); c, the photochemical quenching coefficient (qP); d, quantum yield of photochemical energy conversion in PSII (ΦPSII); e, quantum yield of regulated non-photochemical energy loss in PSII (NPQ); and f, quantum yield of non-regulated non-photochemical energy loss in PSII (NO) in wheat flag leaves. The parameters ΦPSII, NPQ and NO were derived from averages of related parameters according to a previously described method [36]. The disease had fully developed at heading, and 10 plants per genotype were used for the measurements. Bars represent the standard error, and significance was determined using independent sample t-tests. Asterisks represent significant differences as follows: **P ≤0.01, *P ≤0.05, and Ns for no significant difference. Raised asterisks or NS represent the differences in gene expression between L693 and L661 at each time point. The asterisks or the Ns label on the trend line represent differences in gene expression between two adjacent time points in the same genotype
Yields were compared between the various WCBP1 genotypes and between the resistant plants and the susceptible plants in the F2 population (Table 2). L693 exhibited a significantly greater 1000-grain weight (Sw) and total grain weight per ear (Tw) than that of L661 (P <0.05). The number of grains per ear (Ns) was also greater (P = 0.054). A total of 471 plants from the genetic mapping population was analyzed to determine yields (Additional file 2: Table S5), and resistant plants displayed significantly higher Sw, Ns and Tw values compared to those of susceptible plants (Table 2). In addition, homozygous resistant and heterozygous WCBP1 genotypes showed significantly higher Sw, Ns and Tw values than those of homozygous susceptible WCBP1 genotypes.
Table 2.
Comparison of the main yield parameters, including the number of grains per ear (N s), total grain weight per ear (T w) and 1000-grain weight (S w), between the L693 and L661 parents and among the different genotypes in the segregating F2 population by independent sample t-tests
| Yield component | Parent | F2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Genotype | No. of individuals | Mean | Std. error | P-value | Phenotype | Genotype | No. of individuals | Mean | Std. error | P1-value | P2-value | |
| Ns | R | L693 (+WCBP1/+WCBP1) | 15 | 59.167 | 4.121 | 0.054 | R | +WCBP1/+WCBP1 | 237 | 33.810 | 1.344 | 0.000** | 0.149 |
| R | +WCBP1/-WCBP1 | 115 | 32.699 | 0.852 | 0.000** | ||||||||
| S | L661 (+WCBP1/+WCBP1) | 12 | 45.267 | 5.195 | S | -WCBP1/-WCBP1 | 119 | 22.684 | 0.983 | ||||
| Tw | R | L693 (+WCBP1/+WCBP1) | 15 | 2.020 | 0.222 | 0.003** | R | +WCBP1/+WCBP1 | 237 | 0.841 | 0.046 | 0.000** | 0.449 |
| R | +WCBP1/-WCBP1 | 115 | 0.766 | 0.032 | 0.000** | ||||||||
| S | L661 (+WCBP1/+WCBP1) | 12 | 0.966 | 0.219 | S | -WCBP1/-WCBP1 | 119 | 0.463 | 0.031 | ||||
| Sw | R | L693 (+WCBP1/+WCBP1) | 15 | 33.338 | 2.352 | 0.021* | R | +WCBP1/+WCBP1 | 237 | 25.537 | 0.944 | 0.001** | 0.055 |
| R | +WCBP1/-WCBP1 | 115 | 21.019 | 0.492 | 0.042* | ||||||||
| S | L661 (+WCBP1/+WCBP1) | 12 | 20.254 | 4.351 | S | -WCBP1/-WCBP1 | 119 | 18.379 | 1.618 | ||||
R indicates the resistant phenotype, and S indicates the susceptible phenotype. In the different genotypes, R indicates the dominant resistance gene, and r indicates the recessive, susceptible allele. P, value describing the differences between L693 and L661; P1, value describing the differences between the RR and Rr genotypes compared to rr; P2, value describing the differences between the RR and Rr genotypes. Asterisks represent significant differences: **P ≦0.01, *P ≦0.05
Discussion
A gene encoding a heavy metal copper-transport protein is involved in response to stripe rust infection
In this study, we identified WCBP1 and WCBP2, two genes that encode predicted copper-binding proteins. Genes encoding heavy metal copper-binding proteins have proven interesting in multiple studies. For example, similar genes exhibited significant changes in expression in all of eight lines near-isogenic for stripe rust resistance [21]. In rice, the Pi21 gene, encoding a putative heavy metal copper-binding domain protein, confers resistance to blast disease [22]. Furthermore, the rice gene Xa13, encoding a heavy metal copper-transport protein, confers resistance to the bacterial pathogen Xanthomonas oryzae [23]. In humans, Hah1 encodes a metallochaperone protein belonging to a family of proteins containing metal-binding domains and is related to the Menkes/Wilson disease protein [24]. These findings indicate that a great diversity of copper-binding protein genes can confer resistance to various fungal and bacterial diseases in different species.
qPCR analysis (Fig. 6a) indicated that the transcription of the WCBP1 susceptible allele was inhibited. In contrast, WCBP2 is constitutively expressed in both L693 and L661, and its sequence is identical in these lines (Fig. 6b). However, its expression was only activated by inoculation with Pst in L693. WCBP1 co-segregated with resistance in F2:3 populations (Additional file 2: Table S5). Expression of both WCBP1 and WCBP2 was not upregulated (P <0.05) by mock inoculation with water (Fig. 6c and d), indicating that these genes respond specifically during the resistance response to stripe rust.
A gene encoding a blue copper-binding protein was induced by various oxidative stresses in Arabidopsis and transgenic tobacco plants [25–27]. Indeed, there is an oxidative burst during the primary hypersensitive response [21], which implies that the oxidative burst potentially resulting from Pst attack may induce the expression of WCBP1.
WCBP1 enhances photosynthetic competence without altering the timing of leaf senescence and protects wheat yields against Pst infection.
Another interesting category of differentially expressed genes is the leaf senescence-associated genes (SAGs). Previous studies implied that leaf senescence might be involved in the defense against pathogenic fungi [16]. Transcriptome analysis also confirmed that TaSAG120 [GenBank: JN558557] is involved in the plant defense against Pst infection [15]. A total of 37.5 % (42/112) of sequences were identified as belonging to photosynthesis-like and leaf senescence-like pathways (Fig. 1c). Previous studies showed that many pathogenesis-related (PR) genes are induced during leaf senescence in Arabidopsis, and the types and numbers of SAGs present within this family vary from species to species and from genotype to genotype in the same species infected with the same fungal pathogen [24, 28, 29]. Fourteen of the SAG-related ESTs (Additional file 2: Table S2) have very high sequence similarity to TaSAG120 (99.4 %). This finding further suggests that unique transcripts of SAGs are specific to certain Yr genotypes.
YrL693 confers effective resistance at the seedling and adult stages (Fig. 1a and b). The physiological effects of stripe rust on wheat yields were estimated by measuring photosynthetic parameters at the adult stage because yield losses caused by Pst infection occur mostly from disease at the adult stage [5]. Previous studies suggested that a leaf senescence-associated pathway might be involved in the response to stripe rust infection [14–16]. Our data (Fig. 7) suggest that Pn was regulated by the photosynthetic apparatus rather than by stomatal factors in L693, indicating that L693 could actively regulate the photosynthetic pathway following infection with Pst. This hypothesis was also supported by observed differences in gene expression.
The increase in photosynthetic competence observed in L693 could be due either to an increase in the photochemical efficiency of PSII or an increase in the efficiency of excitation capture by open PSII reaction centers. The latter phenomenon would result from other aspects of photosynthesis, such as photosynthetic electron transport and light utilization, and could therefore be affected by pathogen infection. The onset of leaf senescence generally occurs at 30 days after heading [30]. At this time point, leaves from the resistant wheat line L693 exhibited significantly higher Fv/Fm, Fv‘/Fm’ and ΦPSII values as well as significantly lower NO compared with that of L661 leaves, although the changes in Fv/Fm, Fv’/Fm’, qP, ΦPSII, and NPQ were very similar (Fig. 8), especially at 20–30 days after heading. These findings indicated that the higher photosynthetic competence observed in L693 resulted from a higher photochemical efficiency of PSII, which is an important, actively regulated mechanism involved in adaptation to disease stress. This result showed that the putative candidate stripe rust resistance gene WCBP1 did not alter the onset or progression of leaf senescence (Fig. 7). The susceptible allele possibly accelerated the progress of leaf senescence within a normal onset period; therefore, the maintenance of a higher photochemical efficiency in PSII is a direct and important factor in protection of yield against Pst infection.
YrL693 may have originated from a DNA sequence change in the wheat genome
We originally thought that YrL693 originated from Th. intermedium because the pedigree of L693 includes Th. intermedium [18, 31]. However, the fine genetic map of YrL693 shows similar marker order and total length that is consistent with the chromosome map of 1B (Fig. 3). This result indicates that there is likely no alien chromosome segment in YrL693. Comparative genomic analysis of YrL693 (Fig. 4) also demonstrated that an alien chromosome segment is unlikely. Moreover, genes orthologous to WCBP1 were also identified in B. distachyon, rice, and sorghum (Figs. 3 and 4). Additionally, we did not detect Th. intermedium sequences in the genomic region of YrL693 using closely linked markers. Furthermore, the genetic linkage map showed that YrL693 is located in the centromeric region, whereas it is commonly thought that insertion or translocation of an alien chromosome segment into the centromeric region very rarely occurs in wheat [32]. All available evidence supports the hypothesis that YrL693 was not transferred from Th. intermedium, so we conclude that YrL693 is a wheat gene.
WCBP1 is a candidate for the YrL693 stripe rust resistance gene
The sequence of the WCBP1 allele associated with resistance contains a 36 bp deletion compared to the sequence in the susceptible line (Fig. 2 and Additional file 1: Figure S3). This finding suggests that resistance originated with the 36 bp deletion, similar to the event observed for the blast resistance Pi21 gene in rice [22]. These findings implied that DNA sequence changes, especially deletions, might be important mechanisms for evolution of disease resistance in plants. Previous studies confirmed that the Yr36 resistance gene encodes a kinase with a putative lipid-binding domain [33], whereas Yr18 encodes an adenosine triphosphate-binding cassette transporter belonging to a pleiotropic drug resistance subfamily [16]. These findings indicate that different Yr genes have diverse and specific functions.
Conclusions
In this study, we identified the gene WCBP1, which may act as a candidate gene for the wheat stripe rust resistance gene YrL693 based on the following evidences. First, the DNA sequence of WCBP1 was polymorphic between L693 and L661, as well as between the parents YU25 and MY11, and results from the polymorphic amplification demonstrated polymorphism between the WCBP1 alleles in all parental pairs. Second, genetic mapping of YrL693 showed that WCBP1 co-segregated with YrL693. Third, qPCR analysis confirmed that the expression of WCBP1 differed significantly between resistant and susceptible alleles at 96 h after Pst-inoculation. Moreover, the expression of WCBP1 in resistant/susceptible plants was upregulated only upon Pst infection, while it did not change significantly upon mock-inoculation with water. The fact that WCBP1 was not annotated previously can be explained by its putative divergence in being specific to certain Yr genotypes. It is therefore reasonable to hypothesize that WCBP1 is a candidate gene for YrL693, but positive confirmation will come with appropriate transformation experiments.
Materials and methods
Plant materials
The isogenic wheat lines L693 with YrL693 and L661 without YrL693 were used to examine differential gene expression after the plants were inoculated with Pst. Their parents, YU25 and MY11, were employed as controls for the analysis of the candidate resistance gene. Th. intermedium was employed for in situ hybridization to check for alien chromatin in YU25 and L693. A set of Chinese Spring (CS) nulli-tetrasomic (NT) and relevant ditelosomic lines were used to identify the chromosomal location of the candidate resistance gene. Genotypes of 523 F2:3 families derived from the cross L661/L693 were used to construct the fine genetic map of the candidate resistance gene.
Gene expression in plants challenged with Pst
Seedlings at the three-leaf stage, grown in a greenhouse, were challenged with urediniospores of Pst race CYR32. Pst inoculation was conducted according to a previously described method [34]. Leaves from L693 and L661 plants were harvested at 24, 48 and 96 h post-inoculation. A mixture of L693 RNA samples containing equal amounts of RNA extracted at each time point was used as the tester. A mixture of L661 RNA collected at the same time points was used as the driver for the construction of the cDNA library. Non-inoculated leaves from L693 and L661 plants at 0 h, as well as leaves mock-inoculated with water, were harvested for use as controls for gene expression analyses. A sample was collected from L661 at 18 days post-inoculation to represent full disease development (Fig. 1a).
Construction of the SSH library
Total RNA was extracted from L693 and L661 leaves using the RNA extraction reagent TRIzol (Invitrogen, Carlsbad, CA, USA). The Dynabeads Oligo dT25 system (Dynal A.S., Oslo, Norway) was then employed to purify mRNA and to construct cDNA libraries produced by RT-PCR. An SSH library was constructed using the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA, USA) according to a previously described protocol [30, 35]. Positive colonies were sequenced using an ABI Prism 3100 automated sequencer (Perkin Elmer ABD, Santa Clara, CA, USA). The resulting ESTs were analyzed against the GenBank database using BLASTX and BLASTN. The threshold probability for a sequence match was set at 10−5.
Plant growth conditions
All wheat materials were planted in the field at the Yaan Agricultural Research Station of Sichuan Agricultural University (27°17′ N, 120°16′ E) during the 2010–2011 wheat-growing season (total rainfall, approximately 445 mm). Seeds were sown in a clay soil on October 29, 2010. Each experiment was executed as a randomized complete block design with three replicates. Approximately 25 seeds from each line were planted in 2.5 m rows with 25 cm spacing. The susceptible wheat line SY95-71, serving as a vector of the pathogen, was planted in every third row of each population to ensure that all plants had an equal chance of infection. SY95-71 was artificially inoculated with Pst race CYR32 at the seedling stage according to a previously described method [36]. The average air temperatures from sowing to grain maturity ranged from 8.8 to 22.5 °C. The heading date of both L693 and L661 was approximately March 15, 2011. The plants received 3, 4, 4 and 8 g N m−2 ammonium nitrate at the following growth stages, respectively: one-node, meiosis, heading and anthesis. After planting, 10 % Imidacloprid wettable powder (1-[6-chloro-3-pyridylmethyl]-nitroimidazolidin-2-ylideneamine; Yangnong Chemical Group Company, Jiangsu) was applied to control pests. Thirty plants from each genotype showing identical growth and developmental characteristics at heading were marked for all subsequent measurements and observations.
Measurement of photosynthetic indices and PSII activity
Photosynthetic parameters and chlorophyll fluorescence were measured in L693 and L661 using a portable photosynthesis system (Li-6400-02B, Li-Cor, USA) with a red-blue light source and a modulated photosynthesis system (Li-Cor 6400XT, Lincoln, NE, USA), respectively. Data collection was carried out once every 10 days, starting from heading on March 15, 2011, through April 14, 2011. At each sampling date, the mean of three independent measurements from each plant represented the plant phenotypic value, and the average values from 10 plants were employed for comparisons between genotypes. Measurements, parameter recording, and parameter calculations followed previously described methods [30, 37, 38].
In silico mapping of candidate genes
The genomic locations of the wheat ESTs were determined in silico via BLASTN searches against the mapped expressed wheat sequences using GrainGenes 2.0 [39] at a 10−5 threshold probability and via a BLASTX search against the wheat draft genome [40]. Sequences without a BLASTX hit or those showing an e-value of less than 10−5 were excluded from the in silico mapping to the wheat genome and the Brachypodium genome.
PCR-based mapping of polymorphic ESTs and linkage analysis for resistance
Primers were designed using Primer3 [41] to amplify ESTs from wheat genomic DNA. Wheat genomic DNA was extracted using a previously described method [42]. Primer sequences were listed in Additional file 2: Table S3. PCR amplification and product analysis were performed according to previously described methods [19, 43].
Polymorphic primers were used to genotype the segregating population of 523 F2:3 families (Additional file 2: Table S5). Linkage analysis of markers and resistance genes was performed with JoinMap 4 [44]. The linkage group was declared at a LOD threshold of 3.0. Subsequently, the chromosomal locations of the linked markers were confirmed using Chinese Spring nullisomic-tetrasomic and the ditelosomic lines of chromosome 1B [45].
PCR-based cloning of putative stripe rust resistance genes
The primers designed with Primer3 (Additional file 2: Table S4 and S6) according to the contig sequence containing the co-segregated markers and the polymorphic amplicon sequence [46] were employed to amplify the entire length of the candidate gene. These primers were listed in Additional file 2: Table S6. Each amplicon was purified using the GeneJET PCR Purification Kit (Thermo Scientific, Beijing) and cloned into the pGEM T Easy Vector (Promega). JM109-competent cells were transformed and cultured according to the manufacturer’s protocol. Plasmids retrieved using the GeneJET Plasmid Miniprep Kit (Thermo Scientific) were sequenced by Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
qRT-PCR of the candidate gene and the key unigene conferring resistance
We used M-MLV (Invitrogen) to synthesize complementary DNA (cDNA) for real-time qRT-PCR (Invitrogen). Each 20 μl reaction mixture contained 4 μg of RNA. For qRT-PCR assays, 2 μl of the diluted cDNA (1:20) and 9 μl of RealMasterMix (SYBR Green) (TIAGEN) were included in 20 μl reaction volumes. qRT-PCR was conducted using the MiniOpticon Time PCR Detection System with the following program: pre-incubation at 95 °C for 5 min, followed by amplification for 40 cycles at 95 °C for 30 s and 62 °C for 30 s. The melting curve was set at 65 to 95 °C, with a 0.5 °C increase per step, and cooling was set at 40 °C for 30 s. Each qRT-PCR run was replicated with three independent biological samples, and three technical repeats were included for each biological sample. The primer sequences and descriptions of the genes are provided in Additional file 2: Table S7. The crossing point value and results of the melting curve analyses were obtained using Roche CFX Manager Software for the MiniOpticon System 3.0 (Hercules, CA, USA). Melting curve data showing only a single peak, which denotes primer specificity (Additional file 1: Figure S1a), were collected for all samples. The repeatability and reliability of amplifications are demonstrated by the raw data shown in Additional file 1: Figure S1b.
Statistical analysis
Significant differences in the mean physiological parameters, gene expression, and yield between L693 and L661 and between plants carrying and lacking WCBP1 were observed. Differences between two adjacent time points for the same genotype were determined using independent sample t-tests in IBM SPSS Statistics 19 (SPSS Inc., Chicago, IL, USA).
Acknowledgments
We are grateful for the financial support of the National Natural Science Foundation of China (30971787, 31271721, 31171557 and 31571661), the State Key Laboratory for Biology of Plant Disease and Insect Pests, China (SKLOF201410), the Provincial Science and Technology Foundation for Young Scientists of Sichuan, China (2010JQ0042) and the Ministry of Science and Technology, China (2011CB100403 and 2013CB127701).
Abbreviations
- bp
Base pair
- ESTs
Expressed sequence tags
- ORF
Open reading frame
- PR
Pathogenesis-related
- Pst
Puccinia striiformis f. sp. tritici
- qRT-PCR
Quantitative reverse transcription PCR
- SAG
Senescence-associated gene
- SNP
Single nucleotide polymorphism
- SSH
Suppression subtractive hybridization
- SSR
Simple sequence repeat
- WCBP
Wheat copper-binding protein
Additional files
Length distributions of the wheat leaf ESTs (A) and the assembled uniESTs (B), including contigs (C) and singletons (D). Figure S2. Primers and the amplified region of genomic DNA. To clone the WCBP1 gene, primers were designed according to the Chinese Spring draft genome sequence. Full-length amplification was performed using the 1B-1 forward primer and the 1B-6 reverse primer. Figure S3. Comparison of WCBP1 gene sequences. WCBP1 encodes a heavy metal copper-binding protein with two copper sensing domains. The four sequences were cloned using PCR with primers designed according to the polymorphic fragment. L693 is the stripe rust-resistant line; L661 is the stripe rust-susceptible line; YU25 is the resistant parent of L693 and L661; MY11 is the susceptible parent of L693 and L661. Figure S4. Quality map of the sequences flanking the SNP site. A represents L693; B represents YU25; C represents L661; D represents MY11, which showed that the SNP existed among the different genotypes. Figure S5. GISH analysis of mitotic metaphase chromosomes of TAI 7047, L661, L693 and YU25 using genomic DNA of Th. Intermedium as a probe (green). A TAI 7047, B L661, C L693, D YU25. Chromosomes were counterstained with DAPI (blue). Figure S6. Results demonstrating the high quality and good replication of q-PCR. (A), the melt curve of the q-PCR product during the amplification of WCBP2. (B), The amplification curve shows the consistent crossing points of the three technological replicates of the WCBP2 gene and the reference gene GAPDH. This chart shows L661 at 0 h. (ZIP 837 kb)
. Detailed information on 112 ESTs. A BLAST search was performed using BLASTX in NCBI. In silico mapping was performed using the IWGSC draft wheat sequence database. Table S2. BLAST searches performed for the forward subtraction clones found in the stripe rust-resistant genotype L693. Table S3. The primers used to amplify ESTs. Primer pairs were only designed for 62 out of the 112 ESTs because the remaining ESTs were too short to be used for primer design. Table S4. Primers were designed according to the sequence of the contig (3837062) carrying the polymorphic amplicon, which was located on wheat chromosome 1B (http://www.wheatgenome.org/). These amplicons were used to determine the linkage relationship between the markers and the stripe rust resistance gene in L693. Table S5. Results of the amplification of 5 WCBP1-linked genetic markers in an F2 population derived from crosses between L661 and L693 and between L693 and L661. The genotypes (RR, Rr, rr) of the F2 individuals were determined according to the phenotypes of the corresponding F2:3 families; "A" and "B" for the individual F2 populations were derived from crosses between L661 and L693 and between L693 and L661, respectively. R represents the same amplicon as the resistant parent L693; S represents the same amplicon as the susceptible parent L661; H represents the heterozygous condition. The yield components of the F2 plants, including the number of grains per ear (N s), the total grain weight per ear (T w) and the 1000-grain weight (S w), are provided. ‘\’ represents a missing yield component in the F2 plants. Table S6. The sequences of seven primer pairs designed according to the sequence of a cloned candidate gene in silico; these primers were used for PCR-based gene cloning from genomic DNA. Table S7. Primers for q-PCR analysis. (ZIP 94 kb)
Methods S1. Genomic in situ hybridization (GISH). (PDF 13 kb)
Footnotes
Xin Li, Taiguo Liu and Wanquan Chen contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XL, TL, WC, and PL designed the study; XL and SZ performed SSH under the instruction of LL; XL and TL carried out the comparative genomic analysis; and XL and WC performed the genetic analysis and genetic map construction. The qRT-PCR was performed by XL, MZ and HR. XL, SZ, HZ, and LW performed the majority of experiments with contributions from ZT, ZC, RZ and PL. PL wrote the manuscript with input from XL, TL and WC. All authors read and approved the final manuscript.
Authors’ information
Not applicable.
Availability of data and materials
Not applicable.
Contributor Information
Xin Li, Email: sadoneli@gmail.com.
Taiguo Liu, Email: tgliu@ippcaas.cn.
Wanquan Chen, Email: wqchen112@ippcaas.cn.
Shengfu Zhong, Email: 18481012@qq.com.
Huaiyu Zhang, Email: zhyu@sicau.edu.cn.
Zongxiang Tang, Email: zxtang@sicau.edu.cn.
Zhijian Chang, Email: wrczj@126.com.
Ling Wang, Email: 731022275@qq.com.
Min Zhang, Email: 691020460@qq.com.
Liqin Li, Email: lilq88@qq.com.
Hefei Rao, Email: 654786574@qq.com.
Zhenglong Ren, Email: lpg052000@aliyun.com.
Peigao Luo, Phone: 86-28-86290978, Email: lpglab@sicau.edu.cn.
References
- 1.Liu J, Chang Z, Zhang X, Yang Z, Li X, Jia J, et al. Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor Appl Genet. 2013;126:265–74. doi: 10.1007/s00122-012-1979-3. [DOI] [PubMed] [Google Scholar]
- 2.Wan AM, Chen XM, He ZH. Wheat stripe rust in China. Aust J Agric Res. 2007;58:605–19. doi: 10.1071/AR06142. [DOI] [Google Scholar]
- 3.Kang Z, Zhao J, Han D, Zhang H, Wang X, Wang C et al., editors. Status of wheat rust research and control in China. St Petersburg, Russia: BGRI 2010 Technical Workshop Oral Presentations; 2010.
- 4.Yang F-p, Yang W-x, Shang X-w. A study on the effect of wheat stripe rust on wheat yield. Journal of Gansu Agricultural University. 2008;2:011. [Google Scholar]
- 5.Murray GM, Ellison PJ, Watson A, Cullis BR. The relationship between wheat yield and stripe rust as affected by length of epidemic and temperature at the grain development stage of crop growth. Plant Pathol. 1994;43:397–405. doi: 10.1111/j.1365-3059.1994.tb02701.x. [DOI] [Google Scholar]
- 6.Hendrix J, Fuchs E. Influence of fall stripe rust infection on tillering and yield of wheat. Plant Dis Rep. 1970;54:347–9. [Google Scholar]
- 7.Finckh MR, Mundt CC. Plant competition and disease in genetically diverse wheat populations. Oecologia. 1992;91:82–92. doi: 10.1007/BF00317245. [DOI] [PubMed] [Google Scholar]
- 8.Doodson J, Manners J, Myers A. Some effects of yellow rust (Puccinia striiformis) on the growth and yield of a spring wheat. Ann Bot. 1964;28:459–72. [Google Scholar]
- 9.Doodson JK, Manners JG, Myers A. Some effects of yellow rust (Puccinia striiformis) on 14carbon assimilation and translocation in wheat. J Exp Bot. 1965;16:304–17. doi: 10.1093/jxb/16.2.304. [DOI] [Google Scholar]
- 10.Siddique MQ, Manners JG. Some effects of general yellow rust (Puccinia striiformis) infection on 14carbon assimilation, translocation and growth in a spring wheat. J Exp Bot. 1971;22:792–9. doi: 10.1093/jxb/22.4.792. [DOI] [Google Scholar]
- 11.Mallard S, Nègre S, Pouya S, Gaudet D, LU ZX, Dedryver F. Adult plant resistance-related gene expression in ‘Camp Remy. Mol Plant Pathol. 2008;9:213–25. doi: 10.1111/j.1364-3703.2007.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Tang C, Zhang G, Li Y, Wang C, Liu B, et al. cDNA-AFLP analysis reveals differential gene expression in compatible interaction of wheat challenged with Puccinia striiformis f. Sp. tritici. BMC Genomics. 2009;10:289. doi: 10.1186/1471-2164-10-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Liu W, Chen X, Tang C, Dong Y, Ma J, et al. Differential gene expression in incompatible interaction between wheat and stripe rust fungus revealed by cDNA-AFLP and comparison to compatible interaction. BMC Plant Biol. 2010;10:9. doi: 10.1186/1471-2229-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Hu Y, Wang C, Ji W. Gene expression in wheat induced by inoculation with Puccinia striiformis west. Plant Mol Biol Rep. 2011;29:458–65. doi: 10.1007/s11105-010-0245-6. [DOI] [Google Scholar]
- 15.Zhang H, Yang B, Wang Y, Wang C, Liu X, Ji W. Molecular characterisation and expression of a pathogen-induced senescence-associated gene in wheat (Triticum aestivum) Australasian Plant Pathol. 2013;42:53–61. doi: 10.1007/s13313-012-0184-9. [DOI] [Google Scholar]
- 16.Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–3. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Xu M, Xiang Z, Li X, Chen W, Luo P. Registration of the novel wheat lines L658, L693, L696, and L699, with resistance to Fusarium Head blight, stripe rust, and powdery mildew. J Plant Registrations. 2015;9:121–4. doi: 10.3198/jpr2014.01.0003crg. [DOI] [Google Scholar]
- 18.Luo P, Hu X, Chang Z, Zhang M, Zhang H, Ren Z. A new stripe rust resistance gene transferred from Thinopyrum intermedium to hexaploid wheat (Triticum aestivum) Phytoprotection. 2009;90:57–63. doi: 10.7202/044023ar. [DOI] [Google Scholar]
- 19.Huang Q, Li X, Chen WQ, Xiang ZP, Zhong SF, Chang ZJ, et al. Genetic mapping of a putative Thinopyrum intermedium-derived stripe rust resistance gene on wheat chromosome 1B. Theor Appl Genet. 2014;127:843–53. doi: 10.1007/s00122-014-2261-7. [DOI] [PubMed] [Google Scholar]
- 20.Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, et al. Development and mapping of Microsatellite (SSR) markers in wheat. Theor Appl Genet. 2005;110:550–60. doi: 10.1007/s00122-004-1871-x. [DOI] [PubMed] [Google Scholar]
- 21.Coram TE, Huang X, Zhan G, Settles ML, Chen X. Meta-analysis of transcripts associated with race-specific resistance to stripe rust in wheat demonstrates common induction of blue copper-binding protein, heat-stress transcription factor, pathogen-induced WIR1A protein, and ent-kaurene synthase transcripts. Funct Integr Genomics. 2010;10:383–92. doi: 10.1007/s10142-009-0148-5. [DOI] [PubMed] [Google Scholar]
- 22.Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- 23.Yuan M, Chu Z, Li X, Xu C, Wang S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell. 2010 Online:2010 [doi:10.1105/tpc.110.078022]]. [DOI] [PMC free article] [PubMed]
- 24.Quirino BF, Noh Y-S, Himelblau E, Amasino RM. Molecular aspects of leaf senescence. Trends Plant Sci. 2000;5:278–82. doi: 10.1016/S1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- 25.Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998;116:409–18. doi: 10.1104/pp.116.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JD, Arteca RN, Pell EJ. Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 1999;120:1015–24. doi: 10.1104/pp.120.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang K, Im Y, Chung G, Cho B. Activity of the Arabidopsis blue copper-binding protein gene promoter in transgenic tobacco plants upon wounding. Plant Cell Rep. 2002;20:987–91. doi: 10.1007/s00299-002-0436-1. [DOI] [Google Scholar]
- 28.Quirino BF, Normanly J, Amasino RM. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol. 1999;40:267–78. doi: 10.1023/A:1006199932265. [DOI] [PubMed] [Google Scholar]
- 29.Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MFO, Yariv I, et al. Large‐scale identification of leaf senescence‐associated genes. Plant J. 2003;36:629–42. doi: 10.1046/j.1365-313X.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- 30.Luo PG, Deng KJ, Hu XY, Li LQ, Li X, Chen JB, et al. Chloroplast ultrastructure regeneration with protection of photosystem II is responsible for the functional ‘stay-green’ trait in wheat. Plant Cell Environ. 2013;36:683–96. doi: 10.1111/pce.12006. [DOI] [PubMed] [Google Scholar]
- 31.Luo PG, Luo HY, Chang ZJ, Zhang HY, Zhang M, Ren ZL. Characterization and chromosomal location of Pm40 in common wheat: a new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor Appl Genet. 2009;118:1059–64. doi: 10.1007/s00122-009-0962-0. [DOI] [PubMed] [Google Scholar]
- 32.Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS. Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica. 1996;91:59–87. doi: 10.1007/BF00035277. [DOI] [Google Scholar]
- 33.Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, et al. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science. 2009;323:1357–60. doi: 10.1126/science.1166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo PG, Hu XY, Ren ZL, Zhang HY, Shu K, Yang ZJ. Allelic analysis of stripe rust resistance genes on wheat chromosome 2BS. Genome. 2008;51:922–7. doi: 10.1139/G08-079. [DOI] [PubMed] [Google Scholar]
- 35.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Nalt Acad Sci. 1996;93:6025–30. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo PG, Ren ZL, Zhang HQ, Zhang HY. Identification, chromosome location, and diagnostic markers for a new gene (YrCN19) for resistance to wheat stripe rust. Phytopathol. 2005;95:1266–70. doi: 10.1094/PHYTO-95-1266. [DOI] [PubMed] [Google Scholar]
- 37.Kramer DM, Johnson G, Kiirats O, Edwards GE. New fluorescence parameters for the determination of q(a) redox state and excitation energy fluxes. Photosynth Res. 2004;79:209–18. doi: 10.1023/B:PRES.0000015391.99477.0d. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Liang Y, Hu X, Wang X, Tan F, Zhang H, et al. Physiological characterization of ‘stay green’ wheat cultivars during the grain filling stage under field growing conditions. Acta Physiol Plant. 2010;32:875–82. doi: 10.1007/s11738-010-0475-0. [DOI] [Google Scholar]
- 39.Carollo V, Matthews DE, Lazo GR, Blake TK, Hummel DD, Lui N, et al. GrainGenes 2.0. An improved resource for the small-grains community. Plant Physiol. 2005;139:643–51. doi: 10.1104/pp.105.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Wheat Genome Sequencing Consortium (IWGSC) Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 41.Wernimont AK, Huffman DL, Lamb AL, O’Halloran TV, Rosenzweig AC. Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat Struct Biol. 2000;7:766–71. doi: 10.1038/78999. [DOI] [PubMed] [Google Scholar]
- 42.Tai TH, Tanksley SD. A rapid and inexpensive method for isolation of total DNA from dehydrated plant tissue. Plant Mol Biol Rep. 1990;8:297–303. doi: 10.1007/BF02668766. [DOI] [Google Scholar]
- 43.Bassam BJ, Caetano-Anollés G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–3. doi: 10.1016/0003-2697(91)90120-I. [DOI] [PubMed] [Google Scholar]
- 44.Van Ooijen J. JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations of diploid species. Wageningen, Netherlands: Kyazma BV; 2006. [Google Scholar]
- 45.Devos K, Sorrels J, Anderson T, Miller T, Reader A, Lukaszewski A, et al. Chromosome aberrations in wheat nullisomic-tetrasomic and ditelosomic lines. Cereal Res Commun. 1999;27:231–9. [Google Scholar]
- 46.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Bioinformatics methods and protocols. Springer Verlag; 1999. p. 365-86. [DOI] [PubMed]


