Abstract
Background
Previous study showed that downregulated BCL11B expression in T cell acute lymphoblastic leukemia (T-ALL) cell line Molt-4 inhibited cell proliferation and induce apoptosis, which may be related to PHTF1 gene overexpression. The objective of this study was to investigate the expression of PHTF1 and related genes in ALL and further explore its function in T-ALL cell lines.
Methods
Real-time PCR was used to determine the gene expression level of PHTF1 in hematologic malignancies. The PHTF1, BCL11B, FEM1B and Apaf-1 gene expression levels and correlations were analyzed in patients with primary ALL (including T-ALL and B-ALL) and healthy individuals (HIs). Inhibition and overexpression of PHTF1 by lentiviral transduction were performed using the Molt-4 and Jurkat cell lines. Cell growth and apoptosis were measured by the Cell Counting Kit-8 assay and flow cytometry, respectively. Upon PHTF1 overexpression, the BCL11B, FEM1B and Apaf-1 gene expression levels were determined by real-time PCR.
Results
PHTF1 overexpression was found in both T-ALL (p = 0.004) and B-ALL (p < 0.001) groups compared with HIs group. A trend toward a negative correlation between the PHTF1 and BCL11B genes was detected for the T-ALL group, while positively correlated expression was found for the PHTF1 and BCL11B genes in HIs (P = 0.001). FEM1b and Apaf-1 overexpression was found in recently diagnosed ALL patients compared with HIs (p < 0.05). Positively correlated expression was found for the PHTF1, FEM1b and Apaf-1 genes in patients with ALL (p < 0.05) and HIs (p < 0.05). Direct up-regulation of PHTF1 expression inhibited the proliferation of Jurkat and Molt-4 cells and effectively induced apoptosis in Molt-4 cells. Direct inhibition of PHTF1 expression had no significant effect on the proliferation or apoptosis of Jurkat and Molt-4 cells. FEM1b and Apaf-1 overexpression, which did not obviously alter the BCL11B expression level, was detected in PHTF1-transduced T-ALL cell lines.
Conclusions
PHTF1 overexpression is responsible for regulating cell proliferation and apoptosis in T-ALL cell lines. PHTF1 may be a tumor-suppressor like gene and a therapeutic target for triggering the PHTF1-FEM1b-Apaf-1 apoptosis pathway.
Keywords: PHTF1, BCL11B, FEM1b, Apaf-1, ALL, Overexpression, Proliferation, Apoptosis
Background
T cell acute lymphoblastic leukemia (T-ALL) results from clonal malignant T cell proliferation, is an aggressive malignancy that does not respond well to chemotherapy and has a poorer prognosis [1, 2]. The cellular biology and pathogenesis of T-ALL are relatively complex, and these might be related to the different original malignant T cell clone [3–5]. Complex acquired genetic aberrations include chromosomal translocations, gene rearrangements and mutations, resulting in the abnormal expression of oncogenes such as Notch1, TAL1 (T-cell acute lymphoblastic leukemia 1), and BCL11B (B-cell chronic lymphocytic leukemia/lymphoma 11B), which may be associated with advanced disease and resistance to treatment [6–10].
The B-cell leukemia/lymphoma 11B (BCL11B) gene is a member of the BCL family and plays a crucial role in the development, proliferation, differentiation, and subsequent survival of T cells [11]. BCL11B gene alterations are related to the malignant T cell transformation that occurs in hematological malignancies [6, 12–15]. Remarkably, the BCL11B gene is responsible for regulating apoptosis and cell proliferation [16–18]. Previous studies [16–18] have shown that inhibition of BCL11B expression by siRNA selectively inhibits proliferation and effectively induces apoptosis in T-ALL cell lines (Jurkat and Molt-4) but not in normal mature T and CD34+ cells [17, 19]. Additionally, global gene expression profiling has revealed that BCL11B siRNA-mediated apoptosis in Molt-4 cells might be related to the PHTF1 gene [10].
PHTF1 (putative homeodomain transcriptional factor) is a putative homeobox gene located at 1p11-p13 in the human genome [20]. This gene is evolutionarily conserved [21] and mainly expressed in the testis [20]. As a transcription factor, the PHTF1 gene is mainly involved in biological processes such as DNA- dependent transcription and the regulation of biological processes. However, studies on the PHTF1 gene in leukemia have not been reported. FEM1b (feminization-1 homolog b) has been identified as a binding partner for PHTF1 [22]. Previous in vitro experiments have suggested that human FEM1b is involved in apoptosis. FEM1b is a proapoptotic protein that interacts with the apoptosis-inducing proteins Fas, tumor necrosis factor receptor-1 (TNFR1), and apoptotic protease activating factor-1 (Apaf-1) [23]. Therefore, we hypothesized that the BCL11B gene and the PHTF1-FEM1b-Apaf-1 pathway may work together in tumor cell apoptosis. In this study, we analyzed the expression level of PHTF1 and its related genes for the first time in patients with ALL. To further explore its function in T-ALL cell lines, we performed experiments involving the down regulation or overexpression of PHTF1 in T-ALL cell lines using growth and apoptosis assays in vitro.
Results
Expression characteristics and correlation analysis of the PHTF1 and BCL11B genes in T-ALL and B-ALL patients and HIs
In order to characterize the expression of PHTF1 in primary T-ALL samples, we detected the expression level of PHTF1 in peripheral blood mononuclear cells (PBMCs) from 9 cases with T-ALL (median: 2.73 %, mean rank: 20.33, P = 0.004), as well as 13 cases with B-ALL (median: 3.34 %, mean rank: 21.54, P < 0.001). Overexpression of PHTF1 was found in both groups in comparison with HIs group, and there was no significant difference between the two ALL subtypes (Fig. 1a).
Fig. 1.
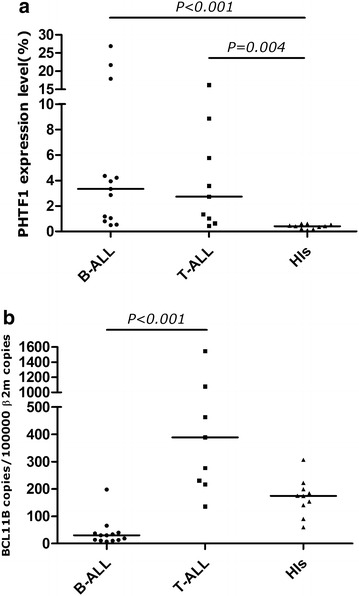
PHTF1 and BCL11B expression level in the different ALL subtypes and healthy individuals. a PHTF1 expression level in the different ALL subtypes and healthy individuals. PHTF1 overexpression was detected in recently diagnosed T-ALL and B-ALL patients. P = 0.004 for T-ALL versus HIs, P < 0.001 for B-ALL versus HIs. b BCL11B expression level in the different ALL subtypes and healthy individuals. BCL11B overexpression was detected in patients with T-ALL. P < 0.001 for T-ALL versus B-ALL
As previously reported [24], the BCL11B mRNA expression level in PBMCs from patients with T-ALL (median: 389.04 copies/105β2M copies, mean rank: 26.56, P < 0.001) was significantly higher than that in patients with B-ALL (median: 30.39 copies/105 β2M copies, mean rank: 7.69) (Fig. 1b).
Spearman’s rank correlation analysis of the expression levels of the PHTF1 and BCL11B genes was performed for patients with T-ALL and B-ALL. No significant correlation was found for the PHTF1 and BCL11B genes in the B-ALL patients (rs = 0.044, P = 0.887) (Fig. 2a). However, a negative correlation trend was detected for the PHTF1 and BCL11B genes in the T-ALL patient (rs = −0.083, P = 0.831) (Fig. 2b). A positively correlated expression level for the PHTF1 and BCL11B genes was found in the HIs (rs = 0.891, P = 0.001) (Fig. 2c).
Fig. 2.
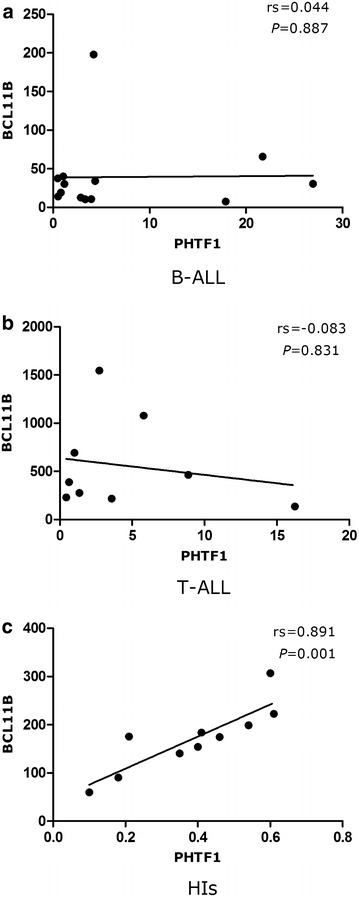
Correlation analyses of the PHTF1 and BCL11B expression levels. a No significant correlation was found for the PHTF1 and BCL11B genes in B-ALL patients. b A negative correlation trend was detected for the PHTF1 and BCL11B genes in the T-ALL patients. c A positively correlated expression level for the PHTF1 and BCL11B genes was found in the HIs
Overexpression characteristics and positive correlation of the PHTF1, FEM1b and Apaf-1 genes in ALL patients and HIs
FEM1b and Apaf-1 overexpression was found in recently diagnosed ALL (FEM1b median: 11.19 %, mean rank: 15.90, P = 0.014) (Apaf-1 median: 7.47 %, mean rank: 16.87, P = 0.001) patients compared with HIs (FEM1b median: 4.31 %, mean rank: 8.65) (Apaf-1 median: 2.52 %, mean rank: 7.20) (Fig. 3).
Fig. 3.
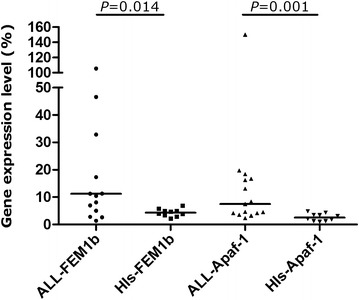
FEM1b and Apaf-1 expression levels in the ALL patients and healthy individuals. FEM1b and Apaf-1 overexpression was found in recently diagnosed ALL patients versus HIs. P = 0.014 for FEM1b, P = 0.001 for Apaf-1
Positively correlated expression for the PHTF1, FEM1b and Apaf-1 genes was found in ALL patients (PHTF1 vs. FEM1b: rs = 0.864, P < 0.001 and FEM1b vs. Apaf-1: rs = 0.682, P = 0.005) (Fig. 4a, b) and HIs (PHTF1 vs. FEM1b: rs = 0.939, P < 0.001 and FEM1b vs. Apaf-1 rs = 0.830 P = 0.003) (Fig. 4c, d).
Fig. 4.
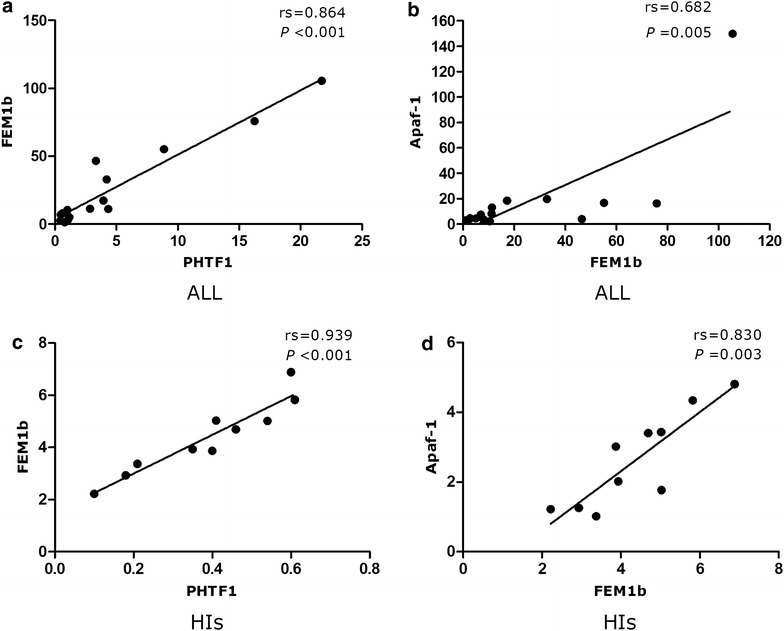
Correlation analysis of the PHTF1, FEM1b and Apaf-1 expression levels. Positively correlated expression for the PHTF1, FEM1b and Apaf-1 genes was found in ALL patients (a PHTF1 vs. FEM1b, b FEM1b vs. Apaf-1) and HIs (c PHTF1 vs. FEM1b, d FEM1b vs. Apaf-1)
Inhibition or overexpression of PHTF1 by lentiviral transduction in T-ALL cell lines
To investigate the potential role of PHTF1 in leukemogenesis, we first transfected Jurkat and Molt-4 cells with an active shRNA pair. We found that the PHTF1 expression level in Jurkat (Fig. 5a) and Molt-4 (Fig. 5b) cells was downregulated approximately 3-fold compared with a scrambled lentiviral transduction control group (CON 1).
Fig. 5.
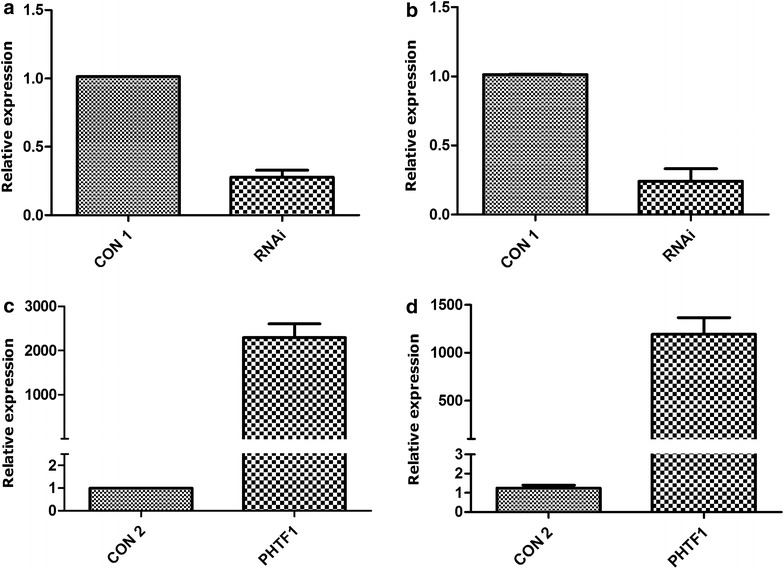
Knockdown and overexpression of PHTF1 in T-ALL cell lines. a, b PHTF1 knockdown with PHTF1 shRNA lentivirus in Jurkat (a) and Molt-4 (b) cells. PHTF1 was down-regulated approximately 3-fold in both Jurkat and Molt4 cells. P < 0.01 for bar 1 versus bar 2 and bar 3 versus bar 4. c, d PHTF1 overexpression by PHTF1 lentivirus in Jurkat (c) and Molt-4 (d) cells. PHTF1 was up-regulated approximately 2291-fold in Jurkat cells and approximately 1100-fold in Molt4 cells. P < 0.05 for bar 1 versus bar 2 and bar 3 versus bar 4. The results represent the mean ± SEM (n = 3)
Similarly, we transduced Jurkat and molt-4 cells using a lentivirus expressing the PHTF1 gene. The PHTF1 expression level was approximately 2291-fold (Jurkat) (Fig. 5c) and approximately 1100-fold (Molt-4) (Fig. 5d) higher compared with the lentiviral transduction control group (CON 2).
PHTF1 overexpression inhibits the proliferation of Jurkat and Molt-4 cells
The effect of PHTF1 on Jurkat and Molt-4 cell growth was next evaluated. Compared with the negative control group, PHTF1 had markedly lower proliferation in both Jurkat (n = 3, P = 0.004) (Fig. 6a) and Molt-4 cells (n = 3, P = 0.007) (Fig. 6b). However, we found no obvious proliferation inhibition after PHTF1 knockdown in Jurkat (Fig. 6c) and Molt-4 cells (Fig. 6d).
Fig. 6.
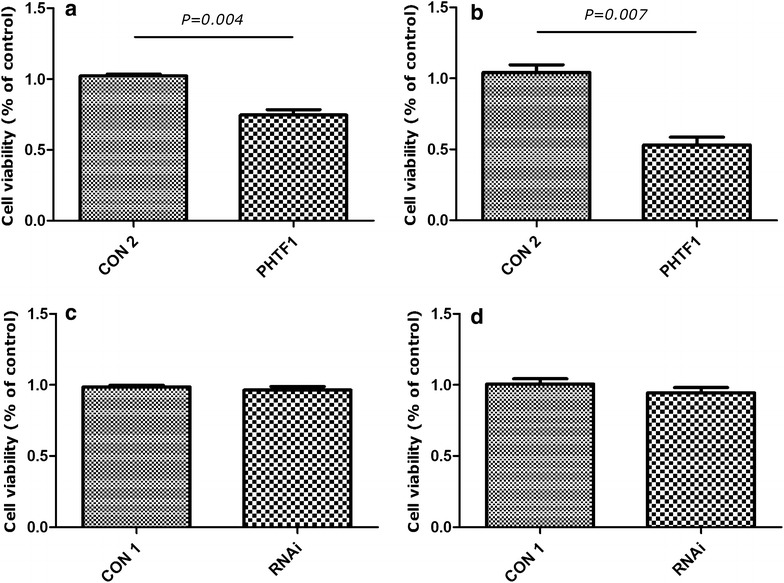
PHTF1 overexpression inhibits the proliferation of Jurkat and Molt-4 cells. a Jurkat cells infected with PHTF1 (PHTF1) or control (CON 2) lentivirus were seeded in a 96-well plate and incubated for 72 h. The CCK8 assay demonstrated that cells infected with PHTF1 have lower viability compared with control. The results shown are the mean ± SEM (n = 3). P = 0.004 for bar 1 versus bar 2. b Molt-4 cells infected with PHTF1 (PHTF1) or control (CON 2) lentivirus were seeded in a 96-well plate and incubated for 72 h. The CCK8 assay demonstrated that cells infected with PHTF1 have lower viability compared with controls. The results represent the mean ± SEM (n = 3). P = 0.007 for bar 1 versus bar 2. c Jurkat cells infected with PHTF1 shRNA (RNAi) or control (CON 1) lentivirus were seeded in a 96-well plate and incubated for 72 h. The CCK8 assay demonstrated that cells infected with PHTF1 shRNA have no significant difference compared with controls. The results represent the mean ± SEM (n = 3). NS no significance. d Molt4 cells infected with PHTF1 shRNA (RNAi) or control (CON 1) lentivirus were seeded in a 96-well plate and incubated for 72 h. The CCK8 assay demonstrated that cells infected with PHTF1-shRNA have no significant difference compared with controls. The results represent the mean ± SEM (n = 3). NS no significance
PHTF1 overexpression remarkably induces apoptosis in Molt-4 cells
We then explored the effects of PHTF1 overexpression on T-ALL apoptosis in in vitro assays. The ratio of apoptosis in Molt-4 cells was 78.67 ± 29.26 %, and it was 15.50 ± 5.97 % for the negative control group (n = 3, P = 0.021) (Fig. 7a). Interestingly, the apoptosis ratio for Jurkat cells was 15.87 ± 5.65 %, while it was 9.57 ± 2.35 % for the negative control group. No significant difference between these two groups was found (n = 3, P = 0.150) (Fig. 7b). Notably, we could observe an increase in the trend toward apoptosis for Jurkat cells after PHTF1 upregulation; however, we found no significant apoptosis after PHTF1 knockdown in the Jurkat (Fig. 7c) and Molt-4 cells (Fig. 7d).
Fig. 7.
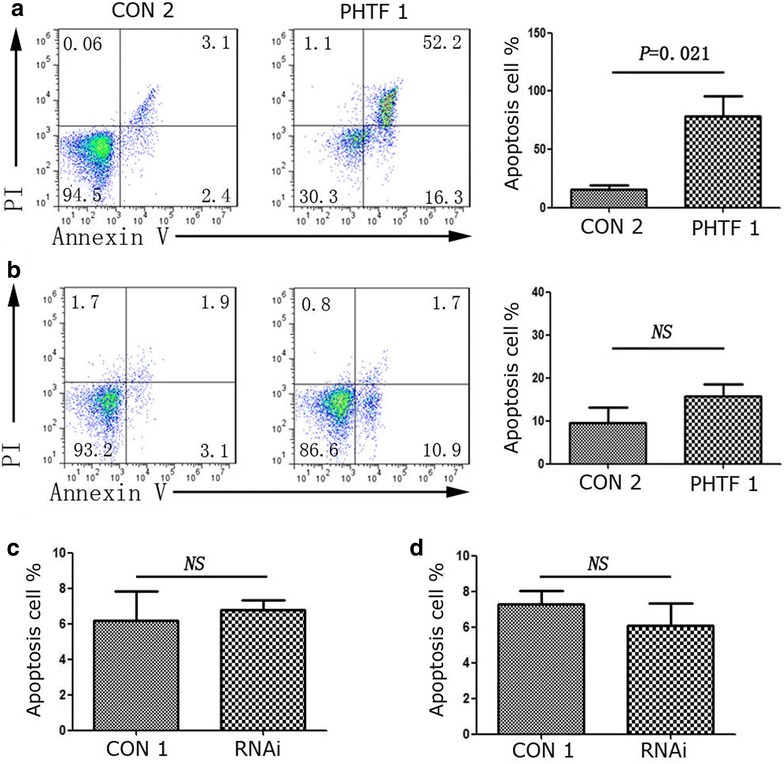
PHTF1 overexpression induces apoptosis in Molt-4 cells. a Representative FACS plots and summary (right) of apoptotic Molt 4 cells after transduction with PHTF1 (PHTF1, left) or control (CON 2, middle) lentivirus. Data represent the mean ± SEM (n = 3). P = 0.021 for bar 1 versus bar 2. b Representative FACS plots and summary (right) of apoptotic Jurkat cells after transduction with PHTF1 (PHTF1, left) or control (CON 2, middle) lentivirus. Data represent the mean ± SEM (n = 3). NS no significance. c, d Summary of apoptotic Jurkat (c) or Molt-4 (d) cells after transduction with PHTF1 shRNA (RNAi) or control (CON 1) lentivirus. Data represent the mean ± SEM (n = 3). NS no significance
PHTF1-related gene expression in Jurkat and Molt-4 overexpressing cells
To further examine the mechanism by which PHTF1 regulates apoptosis, we investigated the expression of PHTF1-related genes in PHTF1 (PHTF1) or control (CON 2) infected Jurkat and Molt-4 cells. In infected Jurkat cells, the BCL11B expression level was unchanged, while FEM1b and Apaf-1 were upregulated approximately 2.4- and 4.4-fold, respectively (Fig. 8a). In transfected Molt-4 cells, the BCL11B expression level was also unchanged, while FEM1b and Apaf-1 were upregulated approximately 6.3- and 29.4-fold, respectively (Fig. 8b).
Fig. 8.
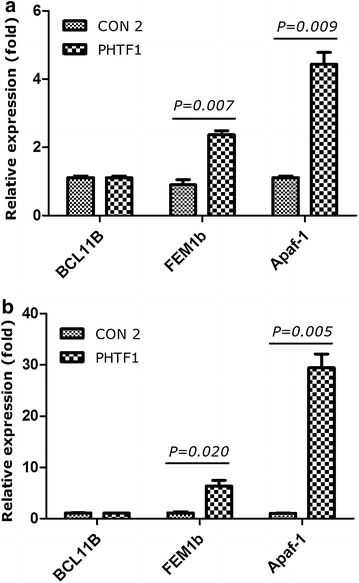
Genes activated with PHTF1 overexpression. a Expression level of BCL11B, FEM1b and Apaf-1 in Jurkat cells after PHTF1 overexpression by transduction with PHTF1 lentivirus (PHTF1) or in the presence of control lentivirus (CON 2). The BCL11B expression level was unchanged versus control; FEM1b and Apaf-1 were up-regulated 2.4- and 4.4-fold, respectively, versus control. Data shown are the mean ± SEM (n = 3), P = 0.007 for bar 3 versus bar 4; P = 0.009 for bar 5 versus bar 6. b Expression levels of BCL11B, FEM1b and Apaf-1 in Molt-4 cells after PHTF1 overexpression by PHTF1 lentivirus (PHTF1) or control lentivirus (CON 2). The BCL11B expression level was unchanged versus control; FEM1b and Apaf-1 were up-regulated 6.3- and 29.4-fold, respectively, versus control. Data shown are the mean ± SEM (n = 3), P = 0.020 for bar 3 versus bar 4; P = 0.005 for bar 5 versus bar 6
Discussion
Despite significant improvement in our understanding of T-ALL biology and pathogenesis [25], knowledge of the T cell activative signaling pathways involved in T-ALL remains limited. Thus, novel molecular insights and therapeutic approaches are urgently needed. Thus, based on previous finding that BCL11B siRNA-mediated apoptosis in the Molt-4 T-ALL cell line might be related to the PHTF1 gene [10], we focused our attention on the characteristics of BCL11B and PHTF1 gene expression in T-ALL patients. We found a positive correlation between the PHTF1 and BCL11B genes in healthy individuals. In contrast, a trend toward a negative correlation was found for the PHTF1 and BCL11B genes in the T-ALL patient although there was no statistical significance, which may be due to the limited number of samples. However, this result may indicate that there are different expression patterns for both of these genes in T-ALL, and for further confirmation of the relationship between PHTF1 and BCL11B is needed in a larger cohort of T-ALL samples. Low BCL11B expression is associated with poor prognosis, particularly in the standard risk group for thymic T-ALL [26]. PHTF1 and BCL11B genetic disorders may contribute to T-ALL pathogenesis.
To further explore its function in T-ALL cell lines, we downregulated and overexpressed PHTF1 in T-ALL cell lines and examined cell line growth and apoptosis using in vitro assays. PHTF1 up-regulation inhibits the proliferation of Jurkat and Molt-4 cells and effectively induces apoptosis in Molt-4 cells. The BCL11B expression level was unchanged, while FEM1b and Apaf-1 were upregulated. Interestingly, compared with the Jurkat cells, the remarkable apoptosis of Molt-4 cells may be related to the higher FEM1b and Apaf-1 expression level. Based on the reports regarding the expression characteristics of tumor suppressor genes in leukemia, such as wilms tumor 1 (WT1), which was consistently found to be highly expressed in peripheral blood (PB) or bone marrow (BM) in acute myeloid leukemia (AML) and is used for inhibiting tumor targeting [27], we hypothesized that PHTF1 is involved in negative regulation of tumor growth. Therefore, we considered that PHTF1 has tumor-suppressive activity and triggers the PHTF1-FEM1b-Apaf-1 apoptosis pathway using in vitro assays. However, direct inhibition of PHTF1 expression, and the BCL11B expression level was unchanged (data not shown), had no significant effect on the proliferation or apoptosis of Jurkat and Molt-4 cells. Therefore, we considered that PHTF1 might be the downstream gene of the BCL11B.
In order to further characterize the role of PHTF1 in T-ALL, it is of interest to analyze the downstream genes regulated by PHTF1. Because overexpression of PHTF1 was found in both groups (T-ALL and B-ALL) in comparison with HIs group, we characterized the expression of FEM1b and Apaf-1, which are mainly involved in apoptosis in ALL patients and HIs. In a previous study [22], western blotting and immunofluorescence assays revealed the presence of PHTF1 and FEM1b in the same cells, and association between these proteins was demonstrated by co-immunoprecipitation. Previous in vitro experiments have suggested that the human FEM1b gene is involved in apoptosis. Human FEM1b is 99 % identical to the mouse protein, and it is reportedly capable of associating with the intracellular tail of the death membrane receptors Tnfrsf6 (tumor necrosis factor receptor superfamily, member 6; also known as Fas) and Tnfrsf1a (also known as TNFR1) [28]. Proteasome inhibitor treatment of SW620, HCT-116, and DLD-1 cells led to upregulation of the FEM1b protein and was associated with apoptosis induction. Blockade of FEM1b upregulation with morpholino antisense oligonucleotides suppressed proteasome inhibitor-induced apoptosis in these cells. The authors of this study have demonstrated that FEM1b can induce apoptosis when overexpressed in some cell lines. The proapoptotic protein FEM1b could represent a novel molecular target for overcoming apoptosis resistance in colon cancer therapy [29]. As a binding protein for FEM1b, Apaf-1 is a central component of the intrinsic apoptosis pathway. High Apaf-1 expression elevates erythroid apoptosis in iron overload myelodysplastic syndrome [30]. Zermati et al. have suggested that Apaf-1 deficiency contributes to tumor progression not only by decreasing activation of the apoptotic caspase but also by reducing DNA damage-induced cell cycle arrest, thus weakening the cytostatic effects of chemotherapy and radiotherapy [31]. FEM1b and Apaf-1 overexpression was found in samples from patients recently diagnosed with ALL, and a positively correlated expression level for the PHTF1, FEM1b and Apaf-1 genes was found in ALL patients and HIs. Thus, PHTF1, FEM1b, and Apaf-1 might be involved in the same apoptosis pathway. The data provide the first molecular biological characterization of these genes at the gene expression level. To further explore its function in B-ALL,further studies will be performed to address this question.
Conclusions
In conclusion, PHTF1 overexpression is responsible for regulating the cell proliferation and apoptosis of T-ALL cell lines. Based on these preliminary findings, and reports regarding the expression characteristics of tumor suppressor genes in leukemia, such as wilms tumor 1 (WT1), which was consistently found to be highly expressed in peripheral blood (PB) or bone marrow (BM) in acute myeloid leukemia (AML) and is used for inhibiting tumor targeting, we hypothesized that PHTF1 is involved in negative regulation of tumor growth. PHTF1 may be a tumor-suppressor like gene and a therapeutic target for triggering the PHTF1-FEM1b-Apaf-1 apoptosis pathway.
Methods
Samples
Nine newly diagnosed and untreated patients with T-ALL, and thirteen newly diagnosed and untreated patients with B-ALL were recruited. The diagnoses for all patients were based on cytomorphology, immunohistochemistry, cytoimmunological and cytogenetic analysis. Peripheral blood mononuclear cells (PBMCs) from ten healthy individuals (HIs) served as controls. The details of the samples are listed in Table 1. Peripheral blood was collected by heparin anticoagulation, and PBMCs were separated using the Ficoll-Hypaque gradient centrifugation method. All procedures were conducted in accordance with the guidelines of the Medical Ethics committees of the Health Bureau of Guangdong Province, China.
Table 1.
The details of samples used in study
| Diagnosis | Subtype | Numbers | Age (year) | |||
|---|---|---|---|---|---|---|
| Total | Male | Female | Range | Median | ||
| ALL | 22 | 17 | 5 | 15–55 | 22.5 | |
| T-ALL | 9 | 7 | 2 | 17–27 | 20 | |
| B-ALL | 13 | 10 | 3 | 15–55 | 24 | |
| HIs | 10 | 6 | 4 | 22–35 | 25.5 | |
Cell culture
Cells from the well-characterized human T-cell acute lymphoblastic leukemia (T-ALL) cell lines Jurkat and Molt-4 were obtained from American Type Culture Collection (ATCC, Manassas, Virginia, USA) and maintained in RPMI-1640 media (Gibco, New York, USA) containing 10 % fetal bovine serum (FBS, NATOCOR, Argentina) at 37 °C and 5 % CO2.
Lentivirus production and transduction
Two recombinant lentivirus vectors based on the U6-shRNA-Ubi-EGFP and U6-NC-Ubi-EGFP vectors (purchased from Shanghai GeneChem Co., Ltd) were constructed to target the human PHTF1 gene (NM_006608) and as a scrambled negative control, respectively. The target sequence of the PHTF1 shRNA was ACCTAAACTCTCAGGTAAA, and that for the negative control was TTCTCCGAACGTGTCACGT.
Two recombinant lentivirus vectors based on the Ubi-PHTF1-3FLAG-SV40-EGFP and Ubi-3FLAG-SV40-EGFP vectors (purchased from Shanghai GeneChem Co., Ltd) were constructed to express the human PHTF1 gene (NM_006608) and green fluorescence protein gene (negative control), respectively.
The transduced cells were divided into four groups: shRNA transduction (RNAi), scrambled control (CON 1), PHTF1 gene transduction (PHTF1), and transduction control (CON 2). The lentiviral titers ranged from 1 to 2 × 108 TU/mL. Transduction was performed by ‘spin-infection’, and cells and lentiviruses (MOI = 5 for Jurkat, MOI = 10 for molt4) plus 10 μg/mL polybrene were mixed and spun at 900 × g for 40 min. The transfected cells were then cultured for 2–3 days prior to analysis by flow cytometry. For GFP cell analysis, GFP+ cells were cultured for 3 days and then FACS purified using the Aria II System (Becton–Dickinson Immunocytometry Systems, San Jose, CA, USA).
RNA extraction and cDNA synthesis
RNA was extracted using the TRIzol kit (Invitrogen, Carlsbad, CA, USA), and it was then reverse-transcribed into first-strand cDNA using random hexamer primers and the reverse transcriptase Superscript II Kit (TaKaRa, Dalian, PR China) according to the manufacturer’s instructions.
Real-time polymerase chain reaction
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) quantitative detection of the β2 microglobulin (β2M), BCL11B, PHTF1, FEM1b, and Apaf-1 genes in cDNA from PBMCs was performed using TaqMan real-time PCR. To precisely determine the BCL11B copy number, a duplex vector, including a fragment of the BCL11B and β2M genes was constructed and used as a reference (the duplex vector was a gift from Prof. C. A. Schmidt, Ernst-Moritz-Arndt University Greifswald, Germany). Another vector including the PHTF1, FEM1b, and Apaf-1 genes was constructed based on DNA concentration, and it was measured by spectrophotometry and confirmed by quantitative gel electrophoresis. Standard dilutions of the vector ranging from 107 to 101 copies were prepared.
Briefly, PCR was performed in a 25 μL total volume containing 2 μL cDNA, 25 pmol of each primer, 10 nmol of each dNTP, 1.5 U AmpliTaq Gold (Applied Biosystems, Branchburg, NJ, USA), 5 pmol 6FAM-TAMRA probe, and PCR buffer containing 4.5 mM MgCl2. After an initial denaturation at 95 °C for 5 min, 50 cycles of 95 °C for 15 s and 64 °C for 1 min were performed.
Primers and probes for β2M, BCL11B, PHTF1, FEM1b, and Apaf-1 gene amplification were synthesized by Invitrogen (Carlsbad, CA, USA) (Table 2). The absolute amounts of BCL11B and β2M were measured in two independent assays, and the BCL11B content per 100,000 β2M copies was calculated using the following formula: n = 100,000 × BCL11B/β2M. The amounts of the PHTF1, FEM1b, and Apaf-1 copies were calculated using the following formulas: n = PHTF1/β2M × 100 %, n = FEM1b/β2M × 100 %, and n = Apaf-1/β2M × 100 %.
Table 2.
Sequences of primers and probes for real-time PCR (TaqMan method)
| Primer/probe | Sequence |
|---|---|
| BCL11B-F | 5′-CACCCCCGACGAAGATGACCAC-3′ |
| BCL11B-R | 5′-CGGCCCGGGCTCCAGGTAGATG-3′ |
| BCL11B-P | 5′-FAM-TCACCCACGAAAGGCATCTGTCCCAAGCA-TAMRA-3′ |
| β2M-F | 5′-CTCGCGCTACTCTCTCTTTCT-3′ |
| β2M-R | 5′-TACATGTCTCGATCCCACTTAACTAT-3′ |
| β2M-P | 5′-FAM-CTCACGTCATCCAGCAGAGAATGGAAAGTCA-TAMRA-3′ |
| PHTF1-F | 5′-GGAAAGTGATGACTGCAGAAACC-3′ |
| PHTF1-R | 5′-AACACCATTCATTCGCTTTGG-3′ |
| PHTF1-P | 5′-FAM-CCTGCTTGTTCACATGCACAGGTGC-TAMRA-3′ |
| FEM1B-F | 5′-CACTCCATCATCATTAGCCTAGTTGA-3′ |
| FEM1B-R | 5′-TGTACTTTTGTCTAGCGGAGTCTTATTCT-3′ |
| FEM1B-P | 5-FAM-CCGGAGCTCACACTGACATGACGAATAA-TAMRA-3′ |
| APAF1-F | 5′-TGCGCTGCTCTGCCTTCT-3′ |
| APAF1-R | 5′-CATGGGTAGCAGCTCCTTCTTC-3′ |
| APAF1-P | 5′-FAM-TGAGCTTCTTCATTTGTGTGCTCCGCT-TAMRA-3′ |
The absolute amounts of BCL11B and β2M were measured in two independent assays, and the BCL11B content per 100,000 β2M copies was calculated using the following formula: n = 100,000 × BCL11B/β2M [12]. In this study, we used this method to measure BCL11B expression. The standard for other genes (PHTF1, FEM1B and Apaf-1) was constructed using another triplex plasmid (synthesized by Invitrogen), and we determined the β2M, PHTF1, FEM1B and Apaf-1 copy number, which were compared relative to the gene expression level of the β2M reference gene between different clinical samples.
Cell proliferation assay
Cell proliferation was detected using the Cell Counting Kit-8 (CCK-8) assay according to the manufacturer’s instructions. Briefly, Jurkat and Molt-4 cells transfected with or without lentiviruses at a density of 5 × 105 cells/mL were seeded into a 96-well plate (100 µL/well). CCK-8 reagent (10 µL) was then added to each well, and the cells were cultured for 3 h at 37 °C and 5 % CO2. After incubation, the absorption value (450 nm) of each well was measured with a 680-type microplate reader (Bio-Rad, Berkeley, CA, USA).
Apoptosis assays
Annexin-V binding assays were conducted 72 h after lentiviral transduction using the Apoptosis Detection Kit (eBioscience, San Diego, CA, USA) according to the manufacturer’s instructions. Briefly, cells were washed twice with PBS, resuspended in 100 μL binding buffer containing APC-conjugated Annexin V, and incubated in the dark for 15 min. Cells were then washed and suspended in 200 μL binding buffer containing PI. Annexin V positive cells were analyzed with an Accuri C6 flow cytometer (Becton–Dickinson Immunocytometry Systems, San Jose, CA, USA).
Statistical analyses
Differences in mRNA expression between two clinical groups were analyzed by the Mann–Whitney U test. Data are presented as medians. Spearman’s rank correlation analysis was used to analyze the PHTF1, BCL11B, FEM1b, and Apaf-1 mRNA level in different clinical samples. An independent-sample t test was used for all comparisons, and data are represented as mean ± SD. P values <0.05 were considered statistically significant.
Authors’ contributions
XD, YQL and XH conceived of the study, participated in its design and coordination and helped draft the manuscript. XH, SXG, LJZ, and XLW performed experiments and analyses. JYW, ZSL, MML and CXD were responsible for patient treatment and acquiring of clinical data. All authors read and approved the final manuscript.
Acknowledgements
The project was sponsored by Grants from the National Natural Science Foundation of China (No. 81100384, No. 81270648, No. 91129720, and No. 30771980). We thank Peng Li and Yiren Xiao who contributed essential materials for the in vitro studies. We thank Dr. Shannon T. Bailey who made significant language revisions.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Abbreviations
- ALL
acute lymphoblastic leukemia
- Apaf-1
apoptotic protease activating factor-1
- ATCC
American Type Culture Collection
- B-ALL
B-cell acute lymphoblastic leukemia
- β2M
β2 microglobulin
- BCL11B
B-cell leukemia/lymphoma 11B
- CCK8
Cell Counting Kit-8
- FEM1b
feminization-1 homolog b
- HIs
healthy individuals
- PBMCs
peripheral blood mononuclear cells
- PHTF1
putative homeodomain transcriptional factor
- qRT-PCR
real-time quantitative reverse transcription-polymerase chain reaction
- siRNA
small interference RNA
- shRNA
small hairpin RNA
- T-ALL
T-cell acute lymphoblastic leukemia
- TNFR1
tumor necrosis factor receptor-1
Contributor Information
Xin Huang, Email: huangmingxin2001@sina.com.
Suxia Geng, Email: gsx76@126.com.
Jianyu Weng, Email: wengjianyu1969@163.com.
Zesheng Lu, Email: zeshenglu@126.com.
Lingji Zeng, Email: bigdps@qq.com.
Minming Li, Email: 406155968@qq.com.
Chengxin Deng, Email: siulier@163.com.
Xiuli Wu, Email: 378246347@qq.com.
Yangqiu Li, Email: yangqiuli@hotmail.com.
Xin Du, Email: miyadu@hotmail.com.
References
- 1.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 3.Zheng HT, Wang X, Ma Y, Xu B, Chen S, Yang LJ, Wu XL, Przybylski GK, Huang SM, Ye TZ, Li YQ. The TCR gammadelta repertoire and relative gene expression characteristics of T-ALL cases with biclonal malignant Vdelta1 and Vdelta2 T cells. DNA Cell Biol. 2014;33:49–56. doi: 10.1089/dna.2013.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gimenes-Teixeira HL, Lucena-Araujo AR, Dos Santos GA, Zanette DL, Scheucher PS, Oliveira LC, Dalmazzo LF, Silva-Junior WA, Falcao RP, Rego EM. Increased expression of miR-221 is associated with shorter overall survival in T-cell acute lymphoid leukemia. Exp Hematol Oncol. 2013;2:10. doi: 10.1186/2162-3619-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S, Huang X, Zheng HT, Geng SX, Wu XL, Yang LJ, Weng JY, Du X, Li YQ. The evolution of malignant and reactive γδ+ T cell clones in a relapse T-ALL case after allogeneic stem cell transplantation. Mol Cancer. 2013;12:73. doi: 10.1186/1476-4598-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oshiro A, Tagawa H, Ohshima K, Karube K, Uike N, Tashiro Y, Utsunomiya A, Masuda M, Takasu N, Nakamura S, Morishima Y, Seto M. Identification of subtype-specific genomic alterations in aggressive adult T-cell leukemia/lymphoma. Blood. 2006;107:4500–4507. doi: 10.1182/blood-2005-09-3801. [DOI] [PubMed] [Google Scholar]
- 7.Vanura K, Vrsalovic MM, Le T, Marculescu R, Kusec R, Jager U, Nadel B. V(D)J targeting mistakes occur at low frequency in acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:725–736. doi: 10.1002/gcc.20677. [DOI] [PubMed] [Google Scholar]
- 8.Lin C, Zheng HT, Wang C, Yang LJ, Chen SH, Li B, Zhou YQ, Tan H, Li YQ. Mutations increased overexpression of Notch1 in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 2012;12:13. doi: 10.1186/1475-2867-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel B, Kang Y, Cui K, Litt M, Riberio MS, Deng C, Salz T, Casada S, Fu X, Qiu Y, Zhao K, Huang S. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:349–361. doi: 10.1038/leu.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Du X, Li YQ. The role of BCL11B in hematological malignancy. Exp Hematol Oncol. 2012;1:22. doi: 10.1186/2162-3619-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 12.Przybylski GK, Dik WA, Wanzeck J, Grabarczyk P, Majunke S, Martin-Subero JI, Siebert R, Dölken G, Ludwig WD, Verhaaf B, van Dongen JJ, Schmidt CA, Langerak AW. Disruption of the BCL11B gene through inv 14 q11.2q32.31 results in the expression of BCL11B-TRDC fusion transcripts and is associated with the absence of wild-type BCL11B transcripts in T- ALL. Leukemia. 2005;19:201–208. doi: 10.1038/sj.leu.2403619. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson A, Nordigården A, Jönsson JI, Söderkvist P. Bcl11b mutations identified in murine lymphomas increase the proliferation rate of hematopoietic progenitor cells. BMC Cancer. 2007;7:195. doi: 10.1186/1471-2407-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strehl S, Konig M, Spath K. Juxtaposition of the BCL11B gene to a novel region at 17q by a t (14; 17) (q32; Q21) in childhood T-Cell lymphoblastic lymphoma [abstract] Blood. 2007;110:101B. [Google Scholar]
- 15.Su XY, Della-Valle V, Andre-Schmutz I, Lemercier C, Radford-Weiss I, Ballerini P, Lessard M, Lafage-Pochitaloff M, Mugneret F, Berger R, Romana SP, Bernard OA, Penard-Lacronique V. HOX11L2/TLX3 is transcriptionally activated through T-cell regulatory elements downstream of BCL11B as a result of the t (5; 14) (q35; q32) Blood. 2006;108:4198–4201. doi: 10.1182/blood-2006-07-032953. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Chen S, Yang LJ, Chen SH, Zhou YB, Schmidt CA, Li YQ. Effects of down-regulating BCL11B expression on the proliferation, apoptosis and global gene expression profiling of Molt-4 cells [abstract] Blood. 2009;114:4505. doi: 10.1182/blood-2009-02-206219. [DOI] [Google Scholar]
- 17.Grabarczyk P, Przybylski GK, Depke M, Völker U, Bahr J, Assmus K, Bröker BM, Walther R, Schmidt CA. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26:3797–3810. doi: 10.1038/sj.onc.1210152. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Chen S, Shen Q, Chen SH, Yang LJ, Grabarczyk P, Przybylski GK, Schmidt CA, Li YQ. Down regulation of BCL11B expression inhibits proliferation and induces apoptosis in malignant T cells by BCL11B-935-siRNA. Hematology. 2011;16:236–242. doi: 10.1179/102453311X13025568941961. [DOI] [PubMed] [Google Scholar]
- 19.Shen Q, Huang X, Chen S, Yang L, Shen SH, Li B, Wu X, Grabarczyk P, Przybylski GK, Schmidt CA, Li Y. BCL11B suppression does not influence CD34+ cell differentiation and proliferation. Hematology. 2012;17(6):329–333. doi: 10.1179/1024533212Z.000000000145. [DOI] [PubMed] [Google Scholar]
- 20.Manuel A, Beaupain D, Romeo PH, Raich N. Molecular characterization of a novel gene family (PHTF) conserved from Drosophila to mammals. Genomics. 2000;64(2):216–220. doi: 10.1006/geno.1999.6079. [DOI] [PubMed] [Google Scholar]
- 21.Raich N, Mattei MG, Romeo PH, Beaupain D. PHTF, a novel atypical homeobox gene on chromosome 1p13, is evolutionarily conserved. Genomics. 1999;59:108–109. doi: 10.1006/geno.1999.5836. [DOI] [PubMed] [Google Scholar]
- 22.Oyhenart J, Benichou S, Raich N. Putative homeodomain transcription factor 1 interacts with the feminization factor homolog fem1b in male germ cells. Biol Reprod. 2005;72:780–787. doi: 10.1095/biolreprod.104.035964. [DOI] [PubMed] [Google Scholar]
- 23.Subauste MC, Sansom OJ, Porecha N, Raich N, Du L, Maher JF. Fem1b, a proapoptotic protein, mediates proteasome inhibitor-induced apoptosis of human colon cancer cells. Mol Carcinog. 2010;49:105–113. doi: 10.1002/mc.20594. [DOI] [PubMed] [Google Scholar]
- 24.Huang X, Chen SH, Shen Q, Yang LJ, Li B, Zhong LY, Geng SX, Du X, Li YQ. Analysis of the expression pattern of the BCL11B gene and its relatives in patients with T-cell acute lymphoblastic leukemia. J Hematol Oncol. 2010;3:44. doi: 10.1186/1756-8722-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo XD, Tan H, Zhou YQ, Xiao TT, Wang CY, Li YQ. Notch1 signaling is involved in regulating Foxp3 expression in T-ALL. Cancer Cell Int. 2013;13:34. doi: 10.1186/1475-2867-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartram I, Gökbuget N, Schlee C, Heesch S, Fransecky L, Schwartz S, Stuhlmann R, Schäfer-Eckhart K, Starck M, Reichle A, Hoelzer D, Baldus CD, Neumann M. Low expression of T-cell transcription factor BCL11b predicts inferior survival in adult standard risk T-cell acute lymphoblastic leukemia Patients. J Hematol Oncol. 2014;7:51. doi: 10.1186/s13045-014-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyu X, Xin Y, Mi R, Ding J, Wang X, et al. Overexpression of Wilms tumor 1 gene as a negative prognostic indicator in acute myeloid leukemia. PLoS One. 2014;9(3):e92470. doi: 10.1371/journal.pone.0092470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan SL, Tan KO, Zhang L, Yee KS, Ronca F, Chan MY, Yu VC. F1Aalpha, a death receptor-binding protein homologous to the Caenorhabditis elegans sex-determining protein, FEM-1, is a caspase substrate that mediates apoptosis. J Biol Chem. 1999;274:32461–32468. doi: 10.1074/jbc.274.45.32461. [DOI] [PubMed] [Google Scholar]
- 29.Subauste MC, Ventura-Holman T, Du L, Subauste JS, Chan SL, Yu VC, Maher JF. RACK1 downregulates levels of the pro-apoptotic protein Fem1b in apoptosis-resistant colon cancer cells. Cancer Biol Ther. 2009;8(23):2297–2305. doi: 10.4161/cbt.8.23.10262. [DOI] [PubMed] [Google Scholar]
- 30.Gu S, Zhao Y, Guo J, Xu F, Fei C, Zhang X, Xiao C, Chang C, Li X. High expression of APAF-1 elevates erythroid apoptosis in iron overload myelodysplastic syndrome. Tumour Biol. 2014;35(3):2211–2218. doi: 10.1007/s13277-013-1294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zermati Y, Mouhamad S, Stergiou L, Besse B, Galluzzi L, Boehrer S, Pauleau AL, Rosselli F, D’Amelio M, Amendola R, Castedo M, Hengartner M, Soria JC, Cecconi F, Kroemer G. Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol Cell. 2007;28(4):624–637. doi: 10.1016/j.molcel.2007.09.030. [DOI] [PubMed] [Google Scholar]


