Abstract
When environmental temperatures exceed a certain threshold, the upregulation of the ovine HSP90AA1 gene is produced to cope with cellular injuries caused by heat stress. It has been previously pointed out that several polymorphisms located at the promoter region of this gene seem to be the main responsible for the differences in the heat stress response observed among alternative genotypes in terms of gene expression rate. The present study, focused on the functional study of those candidate polymorphisms by electrophoretic mobility shift assay (EMSA) and in vitro luciferase expression assays, has revealed that the observed differences in the transcriptional activity of the HSP90AA1 gene as response to heat stress are caused by the presence of a cytosine insertion (rs397514115) and a C to G transversion (rs397514116) at the promoter region. Next, we discovered the presence of epigenetic marks at the promoter and along the gene body founding an allele-specific methylation of the rs397514116 mutation in DNA extrated from blood samples. This regulatory mechanism interacts synergistically to modulate gene expression depending on environmental circumstances. Taking into account the results obtained, it is suggested that the transcription of the HSP90AA1 ovine gene is regulated by a cooperative action of transcription factors (TFs) whose binding sites are polymorphic and where the influence of epigenetic events should be also taken into account.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-015-0629-5) contains supplementary material, which is available to authorized users.
Keywords: HSP90AA1, Sheep, Polymorphisms, Epigenetic marks, Luciferase, EMSA, Allele-specific methylation
Introduction
Promoters are sequence elements that interact with a significant number of transcription factors and epigenetic modifications to regulate gene expression (Landolin et al. 2010). Moreover, they are responsible for the integration of different favourable mutations including those which result beneficial for environmental condition changes (Gagniuc and Ionescu-Tirgoviste 2013).
When exposed to elevated temperatures and other sources of stress, cells and tissues from a wide variety of organisms synthesize proteins known as heat shock proteins (HSPs), to cope with cellular injuries caused by those stressors. In eukaryotes, members of the HSP90 gene family have undergone major duplication events. In all vertebrates studied so far, there are two known cytoplasmic isoforms, namely hsp90α or inducible form and hsp90β which is the constitutive one. HPS90AA1 gene encodes a protein (hsp90α) not only related with the maintenance of cellular homeostasis, with the capacity to shield, fold, or unfold substrates in a context-dependent manner (Verghese et al. 2012), but also directly involved in the heat shock response. This results in the immediate enhanced transcription of a battery of cytoprotective genes, in the presence of thermal and other environmental stresses.
Although the transcriptional enhancement of HSP90AA1 is mainly due to HSF1 (heat shock factor 1), many other heat stress-related transcription factors also modulate gene expression in response to environmental stress and could act simultaneously by cross-talk (Guisbert et al. 2013; LeBlanc et al. 2012; Pirkkala et al. 2001). Polymorphic changes (INDELs, SNPs, etc.) at the promoter region can cause alterations in gene expression levels if they affect binding motifs of transcription activators or repressors. Mutations can alter transcription factor binding sites not only blocking totally or partially their binding to the DNA sequence but also creating or destroying DNA motifs of epigenetic changes, i.e., CpG sites. Alterations at the transcription level can have consequences at the phenotypic level in characteristics related with the functional pathways in which genes are involved.
Marcos-Carcavilla and co-workers (2008) described the HSP90AA1 promoter as a rich CG region, which opens the possibility that this highly inducible gene could have an alternative regulatory region apart from the identified TATA-box. Although the HSP90AA1 gene is ubiquitously expressed, there are tissues as testes and brain where the highest expression rates of this gene have been found (Csermely et al. 1998). Therefore, it is possible that different gene regulatory pathways exist in different tissues.
In previous works (Salces-Ortiz et al. 2013, 2015b), differences in the expression of the HSP90AA1 ovine gene depending on some polymorphisms and environmental conditions have been assessed by q-PCR methods. However, due to the linkage disequilibrium (LD) existing among some of the candidate polymorphisms, it was not possible to distinguish the causal mutation producing such changes at the transcription level of the gene. Moreover, five SNPs located at the gene promoter are susceptible of allele-specific methylation which might be other alternative mechanism regulating gene transcription levels.
The aims of this work were (1) to develop a functional study of the HSP90AA1 promoter to detect which polymorphism(s) is (are) responsible for temperature-dependent changes in the transcription rate of the gene previously detected, (2) to determine if allele-specific methylation (ASM) pattern of susceptible polymorphisms at the promoter region exists and (3) to characterize the CpG island detected along the promoter and gene body.
Methods
Ethics statement
The current study was carried out under a Project License from the INIA Scientific Ethic Committee. Animal manipulations were performed according to the Spanish Policy for Animal Protection RD 53/2013, which meets the European Union Directive 86/609 about the protection of animals used in experimentation. We hereby confirm that the INIA Scientific Ethic Committee (IACUC) specifically approved this study.
Animal material and nucleic acid isolation
Genomic DNA samples were extracted from ovine lymphocytes according to the salting-out procedure described by Miller et al. (1988): Sample identification, age and their alternative genotypes are described in Table 1.
Table 1.
Sample identification, breed, age number of samples (N) and alternative genotypes studied
| ID | Tissue | Breed | Age | Number | Genotype (g.660G>C) |
|---|---|---|---|---|---|
| MCAB | Blood-control | Manchega | Adult | 120 | CC, GG, CG |
| MHSB | Blood-heat stress | Manchega | Adult | 16 | CC, GG, CG |
| MBT | Blood-trios | Manchega | Adult | 6 | CC, GG, CG |
PCR and genotyping
The polymerase chain reaction was performed, and the resulting PCR fragments were sequenced as in Salces-Ortiz et al. (2013). A promoter fragment of 499 bp was obtained, containing 11 SNPs [g.660G>C (rs397514116), g.601A>C (rs397514117), g.528A>G (rs397514269), g.524G>T (rs397514270), g.522A>G (rs397514271), g.468G>T (rs397514272), g.444A>G (rs397514273), g.304A>G (rs397514277), g.296A>G (rs397514274), g.295C>T (rs397514275), g.252C>G (rs397514276)] and four INDELs [g.703_704ins(2)A (ss1570034695), g.667_668insC (rs397514115.1), g.666_667insC (rs397514115.2), g.516_517insG (rs307514268)]. All polymorphisms received their names according to their relative positions based on their distance to the transcription start site (TSS, position +1). Primers used in the PCR and PCR conditions are previously described in Salces-Ortiz et al. (2013).
Cell culture
As model system, we chose human HepG2 hepatoma cell line, where HSP90α levels and regulatory mechanisms are well characterized and based on the fact that transcription factor binding sequences are conserved through mammals. They were used for in vitro experiments and electrophoretic mobility shift assay (EMSA) extracts. Cells were maintained in culture with Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA), supplemented with 10 % fetal bovine serum (FBS) and antibiotics, incubated at 37 °C and 5 % CO2.
Cells were plated at 5000 cells/cm2 until 80 % confluence. Then, media was removed and they were washed with PBS 1x to be used.
Cell culture plates were treated with two different temperature treatments. Basal temperature-treated cells were incubated at 37 °C and 5 % CO2. Heat stress-treated cells were incubated 2 h at 42 °C and 1 h at 37 °C before nuclear extract procedure was made, always providing 5 % CO2.
Electrophoretic mobility shift assay
A double-stranded probe with the sequences of each SNP was used to determine the differences in binding due to genotype and methylation. Oligonucleotide sequences are shown in Supplementary Table 1. The forward oligonucleotide was labelled in 5′ with IRDye700 (Li-Cor Biosciences, Lincoln, NE, USA) during the synthesis. We compared cell extracts binding, obtained under two different temperature treatments (see cell culture section), between genotypes. For the competition experiments, an excess of unlabelled un-methylated probe (40 μM) was added to the mix prior to the addition of the labelled oligonucleotide (5 μM). Band intensities were quantified in an Odyssey Infrared Imaging System (Li-Cor Biosciences). The inverse of band intensity versus the excess of unlabelled oligonucleotide was represented, and the slope of the resulting straight line indicated the affinity of each probe for the proteins in the nuclear extract.
Nuclear extracts were obtained from human HepG2 hepatoma cell line using the Nuclear Extract kit from Active Motif (California, USA) following manufacturer’s instructions. Protein quantification was obtained by Bradford method (Bradford 1976).
In vitro methylation of probes
EMSA oligonucleotide probes were used to compare methylation patterns (Supplementary Table 1) and then were incubated with M. SssI (New England Biolabs, Ipswich, MA, USA) by double methylation according to a protocol described previously (Hu et al. 2013). To determinate the efficiency of methylation, we used Sau3AI and MspJI restriction enzymes (New England Biolabs, Ipswich, MA, USA) following manufacturer’s instructions.
Promoter-reporter constructs
We amplified from −1449 to +61 (TSS defined as +1) of the ovine HSP90AA1 proximal promoter from genomic DNA. Primers used in PCR introduced a KpnI and XhoI restriction sites into the 5′ ends to enable directional ligation into the same sites in pGL3-Basic (Promega). The PCR products were previously cloned in the pGEM®-T basic vector (Promega) to remove polyA generated during PCR. The HSP90AA1 promoter fragment cloned and pGL3-Basic vector were digested with restriction enzymes, gel purified and bounded together with T4 ligase. Sequences of all plasmids were verified by sequencing. Relevant primer sequences are presented in Supplementary Table 2.1.
Site-directed mutagenesis of the genotypes not carried out by any of the animals previously sequenced (Salces-Ortiz et al. 2013, 2015a, b) were obtained by overlap extension PCR and confirmed by sequencing. Primer sequences used are shown in Supplementary Table 2.1.
Transient transfections and luciferase reporter assay
HepG2 cells were transiently transfected in six-well plates using jetPEI transfection reagent (PolyPlus-transfection SA, Illkirch, France) according to the manufacturer’s instructions. For transfections, 2 μg/well of each reporter vector were used. The Renilla gene (0.1 μg) served as an internal control for transfection efficiency. After 48 h, cells were lysed with passive lysis buffer (Applied Biosystems) and luciferase activity was measured with the Dual-Glo luciferase assay system (Promega) following manufacturer’s instructions. Three independent experiments were carried out for each construction and experimental condition.
Cell culture plates were also incubated with two different temperature treatments: basal temperature-treated cells (37 °C) and heat shock induced (2 h at 42 °C and 1 h at 37 °C before cell lysis).
DNA methylation analysis by sequencing
DNA methylation status was determined using sodium bisulphite treatment. Bisulphite treatment was performed with ≤2 μg of whole blood genomic DNA from samples recorded in Table 1 using EpiTect Plus Bisulphite Conversion (Qiagen, Valencia, CA, USA) and EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA) following manufacturer’s guidelines.
Genomic DNA and DNA bisulphite-treated concentrations were determined using a NanoDrop ND-1000 UV/Vis spectrophotometer (NanoDrop Technologies, Inc., DE, USA).
Due to the bisulphite treatment, DNA strands do not have to be complementary; therefore, the top and bottom strands from the two fragments of interest were amplified and analysed separately to confirm ASM pattern (Hajkova et al. 2002). Primers were designed manually with the assistance of “Methyl Primer Express” software (Applied Biosystems, CA, USA) to replace all Cs for Ts except at CpG sites and NetPrimer software (Biosoft International, CA, USA) to avoid possible hairpin structures and primer dimmers and cross-dimmers (Warnecke et al. 2002). Primers are listed in Supplementary Table 2.2 together with amplicon sizes, Tm and PCR kits used for each amplification fragment.
The polymerase chain reactions were optimized from 150 to 200 ng of bisulphite-treated DNA (Supplementary Table 2.2). The resulting PCR fragments were purified with ExoSAP-IT (USB Corporation) and High Pure PCR Product Purification kit (Roche Diagnostics, Indianapolis, IN, USA) and sequenced with specific primers (Supplementary Table 2.2).
Bisulphite sequencing has some associated technical difficulties and potential artefacts. This may involve the formation of stable secondary structures around the methylated CpG site, creating a localized region of dsDNA and preventing access by bisulphate (Warnecke et al. 2002). Accordingly, we resolved this constraint by, first, re-sequencing and, second, RFLP analysis with restriction enzymes (TaqI, BtsIMuI and BstBI).
Statistical analysis
Differences in EMSA band intensities were analysed with ImageJ software using the Gel Analysis method. Average intensity bands were scored as the inversed of the most intensity band, which was used as reference.
The results from luciferase assays were analysed using the GLM procedure of the SAS statistical package (SAS/STAT® statistical package) fitting a model in which the dependent variable was the relative luminescence unit (RLU) obtained for each gene and genotype in each transfection. Transfection, replicate nested to genotype and genotype were included in the model as fixed effects. Least square means, 95 % confidence intervals and t test for means comparisons were calculated.
Bioinformatics analysis of CpG islands and identification of short interspersed elements
The nucleotide sequence surrounding the transcription start site (TSS) of the HSP90AA1 gene was explored by three putative CpG island softwares: Methyl Primer Express (Applied Biosystems, Bedford, MA, USA), EMBL-EBI (http://www.ebi.ac.uk/tools/emboss) and USCnorris (http://www.uscnorris.com/cpgislands2/cpg.aspx). To differentiate between transposable elements and CpG islands, we used the software RepeatMasker 4.0.5 (2014) version of Repbase 19.03 (for all species except human).
Results
Functional study
Differences in transcription factor binding affinities
We investigated the formation of protein complex binding to the polymorphic sequence of the promoter, using a purified protein assay system to elucidate the differences in gene expression ratios. We designed labelled oligonucleotides for the alternative alleles of a total of ten polymorphisms selected from previous expression works (Salces-Ortiz et al. 2013, 2015b). Three of them, showing high levels of linkage disequilibrium (SNPs g.601A>C, g.524G>T and g.468G>T), did not show any expression differences in a previous expression assay (Salces-Ortiz et al. 2013). We could confirm this fact analysing each SNP independently by electrophoretic mobility shift assay (EMSA) under control and heat stress-treated cell extracts. There were no differences in protein complex binding among their alleles, or they were unspecific (data not shown).
We used the same procedure assay to evaluate g.444A>G and g.522A>G which showed in Salces-Ortiz and colleague’s work (2013) subtle differences in gene expression, the first one under heat stress and the second one under thermoneutral conditions. EMSAs from both mutations did not show any specific protein affinity to any of the alleles from those polymorphisms with any of the two different cell extracts (data not shown).
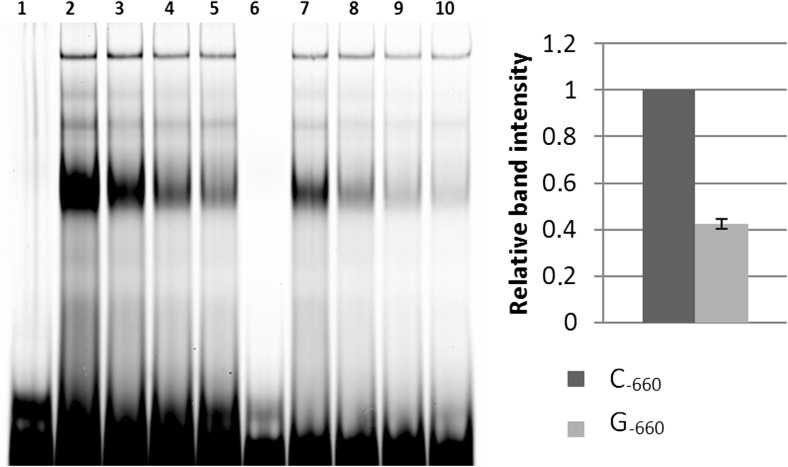
Other LD block was that constituted by g.703_704ins(2)A, g.660G>C and g.528A>G polymorphisms (Salces-Ortiz et al. 2013). In the case of the SNP g.660G>C, which has been largely suspicious as the causal mutation of the differences in gene expression observed under heat shock and thermoneutral conditions, EMSA suggested a difference in the ability of g.660G>C alternative alleles to bind nuclear proteins under thermoneutral conditions (Fig. 1). Competition experiments carried out with nonspecific oligonucleotides revealed the presence of a specific binding. The binding affinity to G−660 was 42 ± 3.7 % with respect to the affinity to the C−660 allele (100 %) and disappeared very easily when competing with increasing amounts of an unlabelled C−660 oligonucleotide. Similar binding patterns were obtained in EMSAs with heat shock-treated cell extracts (data not shown).
Fig. 1.
Electrophoretic mobility shift assay (EMSA) using nuclear extracts from HepG2 cells under thermoneutral conditions. Nuclear extracts from cultured cells were incubated with C−660-labelled oligonucleotide probe alone (lane 2), in the presence of increasing excess unlabelled C−660 probe (lanes 3-10×, 4-25×, 5-50×), G−660-labelled (lane 7), G−660-labelled with increasing excess unlabelled C-660 probe (lanes 8-10×, 9-25×, 10-50×). In lanes 1 and 6, nuclear extracts were not added
We performed the same EMSA experiment for g.703_704 ins(2)A, and also, differences in band intensities were observed (Fig. 2) when using cell extracts under thermoneutral conditions. In this case, the band with the double adenine insertion (I−704) has 79 ± 12 % of the intensity compared to the deletion one (D−704, 100 %). As a glucocorticoid receptor has been predicted to bind the sequence with the deletion allele (Oner et al. 2012), we made a competition with 50-fold excess of cold oligo with the specific binding sequence of the glucocorticoid receptor and the band almost disappeared with a final intensity of 33 ± 6 %. Similar binding patterns were obtained in EMSAs with heat shock-treated cell extracts (data not shown).
Fig. 2.
Electrophoretic mobility shift assay (EMSA) using nuclear extracts from HepG2 cells under thermoneutral conditions. Nuclear extracts from cultured cells were incubated with the D−704-labelled oligonucleotide probe alone (lane 2), in the presence of excess unlabelled D−704 probe (lane 3), in the presence of excess unlabelled I−704 probe (lane 4), the I−704-labelled oligonucleotide probe alone (lane 6), in the presence of excess unlabelled I−704 probe (lane 7), in the presence of excess unlabelled I−704 probe (lane 8) and D−704 in the presence of excess unlabelled canonical GR sequence binding oligonucleotide (lane 9). In lanes 1 and 5, nuclear extracts were not added
EMSA, performed based on g.528A>G alternative alleles, did not show any specific band affinity among its alternative alleles with any of the different cell extracts (data not shown).
In addition, we also studied the INDELs g.666_667insC and g.667_668insC in combination with C−660 as they are only 6/7 bp away and there was oligo’s minimum length limitation. We compared the double deletion with one and two cytosine insertions. When cell extracts were subjected to thermoneutral temperatures, we could observe that the sequence with an insertion had a specific band not present in the deletion and with low affinity in the double insertion lane (Fig. 3, purple arrow). EMSA suggested a difference in the ability of the alleles to bind nuclear proteins. As Fig. 3 shows, three specific binding complexes were formed. This suggests that proteins represented by those bands compete for the same binding sequence and their different affinity could be due to the number of cytosines present in the binding sequence. For example, the band indicated with the blue arrow showed that the protein complex formed with the single or double insertion has stronger binding properties than the deletion one. In the same way, the double insertion also showed stronger binding properties than the single insertion genotype. The same pattern can be observed in the complex indicated with the purple arrow. However, the sequence with a single insertion showed lower binding properties than the other two genotypes in the complex indicated with the grey arrow.
Fig. 3.
Electrophoretic mobility shift assay (EMSA) using nuclear extracts from HepG2 cells under thermoneutral conditions. Nuclear extracts from cultured cells were incubated with D−668D−667-labelled oligonucleotide probe alone (lane 2), in the presence of excess unlabelled I−668D−667 probe (lane 3), in the presence of excess unlabelled I−668I−667 probe (lane 4), the I−668D−667-labelled oligonucleotide probe alone (lane 6), in the presence of excess unlabelled D−668D−667 probe (lane 7), in the presence of excess unlabelled I−668I−667 probe (lane 8), the I−668I−667-labelled oligonucleotide probe alone (lane 10), in the presence of excess unlabelled D−668D−667 probe (lane 11) and in the presence of excess unlabelled I−668D−667 probe (lane 12). In lanes 1, 5 and 9, nuclear extracts were not added. The grey arrow indicates the binding of a complex of proteins with more efficiency to D−668D−667 and I−668I−667 whereas the protein indicated with the purple arrow seems to have more efficiency to I−668D−667. The blue arrow indicates a protein that binds in all three possible genotypes, even though it seems to have more preference to I−668D−667, as during competition, it does not disappear
EMSA experiment performed with cell extracts under heat stress showed similar band intensities. In this case, lower affinity was observed comparing with the control band, probably due to the unfolding of transcription factors caused by their denaturation under high temperatures. In addition, in the I−668D−667 lane, all complexes seem to disappear except the binding of a unique protein or complex (Supplementary Fig. 1).
In vitro expression assay
Luciferase assays were performed only with those polymorphisms whose alternative genotypes showed differences in EMSA transcription binding affinities, i.e. g.703_704ins(2)A, g.667_668insC and g.660G>C and an additional INDEL not tested in previous expression studies (Salces-Ortiz et al. 2013, 2015b), g.666_667insC, due to its low frequency in the samples used (Fig. 4).
Fig. 4.
Luciferase assays for several polymorphisms at the HSP90AA1 promoter. Each alternative allele was transiently expressed in HepG2 cells for luciferase assays. Firefly luciferase activity was normalized with Renilla luciferase activity. Data are represented compared to pGL3-Basic and the means SD are for three replicates. a g.660G>C alternative genotypes under thermoneutral conditions. b g.703_704ins(2)A alternative genotypes under thermoneutral conditions. Ns non-significant c g.667_668insC and g.666_667insC alternative genotypes under thermoneutral conditions. d g.667_668insC and g.666_667insC alternative genotypes under heat stress conditions
The results based on luciferase assays for the transversion g.660G>C showed that the promoter containing the G−660 allele had a 17.6 % decrease in its activity (P = 0.0048) comparing with that containing the C−660 one under thermoneutral temperatures (Fig. 4a).
In the case of g.703_704ins(2)A, luciferase activity did not show significant in vitro expression differences between its alleles when cells were cultured only under thermoneutral temperature (Fig. 4b).
Luciferase assays involving g.667_668insC and g.666_667insC alternative alleles were carried out only with C−660, as that is the allele associated with single or double C insertion. This one has been corroborated in all sheep already genotyped (836 animals from 31 different sheep breeds from different locations of Europe, Africa and Asia (Salces-Ortiz et al. 2015a).
Cytosine insertions did not show differences in promoter activity under thermoneutral conditions (Fig. 4c), which agree with the expression results showed in the work of Salces-Ortiz and co-workers (2015b). However, they showed significant differences after the heat stress treatment (Fig. 4d). Thus, insertion of one or two cytosines produce similar increase of promoter activity, 28.3 % (0.0114) and 30.3 % (P = 0.0078), respectively.
Epigenetic mark patterns in the HSP90AA1 gene
Detection of two fragments SINE in the sequence of gene
Two fragments from short interspersed elements (SINEs) were identified (Supplementary Table 3). One of them had 35 bp at the intergenic region studied (1090 bp upstream of Inr), which has 89 % of sequence identity to the 3′ Bov-tA2 or SINE2/tRNA of ruminants (212 bp). In addition, 50 bp on the third intron of the HSP90AA1 gene has 66 % of identity with MIRc SINE2/tRNA of mammals (268 bp) (Supplementary Table 4) (Fig. 5). Bov-tA2 or SINE2/tRNA of ruminants could be a good candidate as an alternative transcriptional regulator mechanism if the entire sequence is found in this region (not yet sequenced). Anyhow, the exact transcriptional control of this intergenic SINE found remains still unknown.
Fig. 5.
Diagram of the short interspersed elements (SINEs) predicted in the HSP90AA1 gene by RepeatMasker software. The two SINEs predicted are highlighted in pink. One SINE (35 bp long) is located in the intergenic region (1090 bp upstream of the Inr) and the second one (50 bp long) located in the third intron of the HSP90AA1. Exons 1, 2 and 3 are marked in grey
The HSP90AA1 CpG island
Next, we analysed the ovine HSP90AA1 gene promoter structure. Based on Ovis aries HSP90AA1 sequence (DQ983231.1), a CpG island (CGI) was predicted to be associated with the 5′ promoter region according to Gardiner-Garden and Frommer criteria (Gardiner-Garden and Frommer 1987) (i.e. GCm >50 % and y value >0.6, where the intervals between CpGs are 200 bp; Supplementary Fig. 2).
The HSP90AA1 promoter region is significantly CpG-rich, and according to the bisulphite sequencing results, it is non-methylated (Deaton and Bird 2011). This analysis allowed us to define the limits of the CpG island: −745 to +1445 in relation with putative TSS position. Therefore, the CpG island (CGI) present at the HSP90AA1 gene has a length of 2199 bp covering part of its promoter and gene body structure (Supplementary Fig. 2).
Allele-specific DNA methylation pattern in blood
Five SNPs (g.660G>C, g.601A>C, g.528A>G, g.522A>G and g.304A>G) at the promoter region of the HSP90AA1 ovine gene could lead to methyl-CpGs depending on their alternative alleles. However, only one of them, g.660G>C, in its G allele form, creates an allele-specific methylation (ASM), which is flanked downstream by other non-polymorphic methyl-CpG site located 632 bp before TSS(A+1) (g.632_methyl-CpG) (Fig. 6). Bisulphite sequencing results showed that the cytosine adjacent to g.660G>C results un-methylated with the genotype CC, hemi-methylated when carrying CG and methylated if GG was present.
Fig. 6.
Description of the HSP90AA1 promoter CpG island motifs. HSE (purple), TATA-box (pink) and TSS(A+1) (grey and underlined) at the Inr (yellow). Furthermore, HSP90AA1 promoter polymorphisms (g.703_704ins(2)A, g.667_668insC, g.666_667insC, g.660G>C, g.601A>C, g.528A>G, g.524G>T, g.522A>G, g.516_517insG, g.468G>T, g.444A>G, g.304A>G, g.296A>G, g.295C>T, g.252C>G) are positioned in the figure. Polymorphism’s names are based on their distances to TSS. Polymorphism susceptible of allele-specific methylation are underlined In addition, g.660G>C is highlighted in orange and g.632_methyl-CpG in green
To check whether there were differences in methylation patterns in blood comparing DNA control versus heat stress and if methylation segregates between sexes, 16 animals and two family trios were analysed, respectively. The trios genotypes for the g.660G>C were the following: (1) ♂CC (un-methylated) × ♀GG (methylated) = CG (hemi-methylated) and (2) ♂CG (hemi-methylated) × ♀CC(un-methylated) = CG (hemi-methylated) (Extracted from the trios used in Salces-Ortiz and co-workers (2013).
We observed that ASM in DNA from blood was independent of both temperature of the sample collection day and parental origin.
We found five hemi-methylated CpG sites (5′ CpG island boundary) immediately upstream g.660G>C and two hemi-methylated sites immediately 5′ to the first methylated CpG site (3′ CpG island boundary) (Supplementary Fig. 2).
Transcription factor binding affinities involving methylation in blood
As mentioned above, g.660G>C creates an ASM. When the G allele is present, it forms a CpG where the cytosine is methylated in DNA extracted from blood. To investigate if this methylation could interfere with the binding of transcription factors, methylated and non-methylated oligonucleotides at polymorphic CpG site were used in EMSA. We observed that there was no difference in transcription factor binding pattern even though it seemed to have more initial binding affinity to methylated CpG (Supplementary Fig. 3). However, when the oligo containing the CpG methylated was competed with 50-fold excess of cold un-methylated oligo of any of both alleles, there was the same degree of decrease in the intensity of the bands, which indicates that no differences in binding complexes with or without methylation exist. We also designed a long oligonucleotide (60 bp) where two methylations, the CpG site formed by the G allele of g.660G>C and the g.632_methyl-CpG, were included. Even though, due to the length of the oligonucleotide, undesirable secondary structures were produced and EMSAs assays were not conclusive (data not shown).
Discussion
The present study was based on previous results obtained in two expression assays carried out by Salces-Ortiz and colleagues (2013, 2015b) which were limited by the high co-segregation of the polymorphisms studied. In the present study, we could finally distinguish which of the polymorphisms of the linked block, already selected as a candidate, has a role in the transcription mechanisms causing differences in HSP90AA1 expression levels.
In previous works (Marcos-Carcavilla et al. 2010; Salces-Ortiz et al. 2013, 2015b), expression rate changes were associated with alternative genotypes of g.660G>C (in high linkage disequilibrium, with g.703_704ins(2)A and g.528A>G, LD = 0.54–0.98) and with the INDEL g.667_668insC (in moderate linkage disequilibrium with the SNP g.660G>C, LD = 0.23).
Even though q-PCR is a strong, simple and widely used technology, it has some limitations. In our case, it was impossible to distinguish which of these polymorphisms was the one responsible for the expression differences observed under basal and heat stress conditions. Besides, we could analyse the possible implications of an additional INDEL (g.666_667insC), close to both candidate polymorphisms, not tested in the expression analyses due to the absence of a representative number of samples carrying their alternative genotypes.
In vitro expression
In this study, and based on the knowledge that mammalian transcription factor binding sites are well conserved, we could probe the direct effect of the different polymorphisms along the HSP90AA1 ovine promoter. Functional studies performed in the present study have shed some light on the transcription factors responsible for the differences in the promoter activity, previously observed by quantitative expression assays.
We have observed in vitro that the SNP g.660G>C is involved in the gene expression modulation, under basal conditions. As it has been previously explained, even small differences in the activation of this gene can cause great differences in the overall equilibrium of the cell homeostasis as HSP90 is one of the most abundant proteins in the cell (Taipale et al. 2010). So, the small differences in the expression levels under thermoneutral temperatures here observed can also be essential to cope with other types of stresses not directly related with temperature (Han et al. 2009; Kim et al. 2013; Sud et al. 2007; Tian et al. 2014; Yang et al. 2011). We have assessed that only the SNP g.660G>C, highly linked with g.703_704ins(2)A and g.528A>G, is responsible for the transcription differences of the gene observed under thermoneutral conditions previously associated with this LD block.
In addition, we have found that two cytosine insertions (g.667_668insC and g.666_667insC) are the responsible for the highest upregulation of the gene as response to heat stress previously associated to g.667_668insC by q-PCR (Salces-Ortiz et al. 2015b). Hsp gene inducible expression is regulated by heat shock transcription factors (HSFs) that exist as inactive proteins mostly in the cytoplasm. In response to physiological and environmental stimuli, heat stress in particular, these proteins bind to the promoter targets sequences (HSEs) triggering the transcription of heat shock genes and the formation of heat shock proteins (HSPs) (Morimoto 1998; Pirkkala et al. 2001). Besides the HSE, there is a number of promoter sequence structures involved in regulating HSP90AA1 gene expression, as distal transcription enhancers. The polymorphisms g.667_668insC, g.666_667insC and g.660G>C are enough upstream to be part of distal enhancers sequence binding sites with affinity to rich cytosine sequences such as SP family transcription factors. Those distal enhancers could modulate the gene expression under heat stress in a synergistic effect with the HSE (Rieping and Schoffl 1992) by interacting with coactivators, stimulating chromatin remodelling (Korfanty et al. 2014) and histone acetylation. These modifications produce a hairpin that allows the approach of those distal enhancers with the core promoter. By this way, this complex promotes the triggering assembly of the RNA polymerase which allows the transcription regulation (Fig. 7) (Khrapunov et al. 2006). In addition, we do not discard that the presence of DNA-binding proteins or mediator proteins, which make possible the hairpin or any other intermediary mechanism which takes part of this complex, would be mainly expressed under heat shock conditions.
Fig. 7.
A schematic drawing of the effect that g.667_668insC, g.666_667insC and g.660G>C could have in the HSP90AA1 expression regulation. They are located in a putative distal enhancer binding sequence which allows activators to bind depending on their genotype. Those activators recruit different mediator transcription factors which form a hairpin that allows an approach with the core promoter and promotes transcription. Adapted from original drawing from Pearson Education, Inc. (publishing as Pearson Benjamin Cummings, Copyright © 2008)
Cis-regulated allele-specific methylation in blood
We have previously shown that g.660G>C alternative alleles are associated with HSP90AA1 expression differences in blood. Gene expression in animals carrying the GG genotype was reduced when compared to those carrying the CC genotype (Salces-Ortiz et al. 2013, 2015b). Therefore, there is an imbalance between the expression of the G−660 and C−660 alleles. Bisulphite sequencing of peripheral blood DNA samples confirmed the presence of a cis-regulated ASM caused by this transversion, which is flanked downstream by a non-polymorphic methyl-CpG site (g.632_methyl-CpG).
The effects of methylated CpG over gene activity are based on two general mechanisms that contribute gene silencing. Either methylation at promoter specific sites can prevent transcription factors binding, or methylation attracts methyl binding proteins (MBPs) (Nan et al. 1997). Since EMSAs did not show differences in the band intensities when methylated vs un-methylated oligos were compared, we can hypothesize that if a transcription factor binds, this DNA sequence could do it independently of the methylation existence. The repression effect of methylation can act without altering the binding of transcription factors (Salvatore et al. 1998). Thus, both mechanisms, methylation repression and TF binding can be independent processes that only in few cases are directly correlated (Medvedeva et al. 2014). Therefore, a MBP, primary factor in most cases, can act as indirect repressor without altering the binding of transcription factors (Nan et al. 1997; Salvatore et al. 1998).
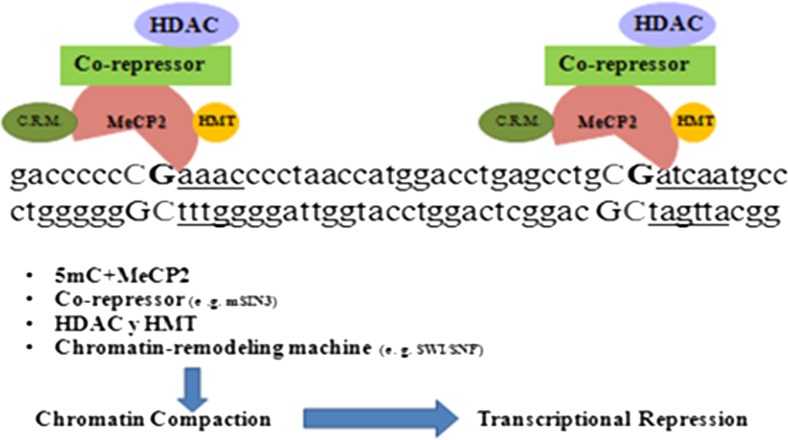
The identification of methylation-dependent transcriptional repressors is determined by the detection of proteins with affinity for methylation (Nan et al. 1997). MeCP2 (methyl-CpG binding protein 2) is an abundant MBP with high affinity for a specific DNA sequence. It depends on the density and location of methyl-CpGs in gene promoters and interferes with regulatory components of the transcription complex (Nan et al. 1997). The sequence fragment spanning from g.660G>C to g.632_methyl-CpG could constitute two putative MeCP2 binding sites (Fig. 8) (Klose et al. 2005; Nan et al. 1993). On the basis of the sequence characteristics (Klose et al. 2005; Nan et al. 1993) and the results obtained in the EMSAs, we cannot discard that MeCP2 would be indirectly repressing transcription at this point.
Fig. 8.
Possible mechanisms of transcriptional repression by MeCP2 (adapted from (Wade 2005). Interaction of MeCP2 with two methylated DNA sites results in local recruitment of chromatin-remodeling machine. This factor alters histone-DNA contacts. Furthermore, histone deacetylation by HDAC (histone deacetylases) and histone methylation by HMT (histone methyltransferases) facilitate formation of repressive chromatin conformation and therefore contribute to transcriptional repression
We also have addressed an ASM in peripheral blood, where the adjacent cytosine to g.660G>C is always methylated when the G allele is present. Moreover, we have observed that g.632 methyl-CpG is always methylated. It will be interesting to analyse if this pattern is conserved in other tissues or on the contrary if a tissue-specific methylation behaviour of these CpGs exists.
Another important question is whether the presence of the polymorphism g.660G>C removes a methylation site (wild allele would be G−660) or creates a de novo methylation position (wild allele would be C−660). We suggest the latter hypothesis is the correct as methylated cytosines are highly mutable to thymine in CpG dinucleotides (Coulondre et al. 1978), so it is unlikely to lead afterwards to a guanine mutation. Furthermore, the C−660 allele is the most common one in 31 sheep breeds, goats and other species of wild ruminants analysed in a previous work (Salces-Ortiz et al. 2015a, b).
We have described the epigenetic pattern shown in blood from adult animals. We have not discovered the cause that produces these epigenetic changes present in the ovine HSP90AA1 promoter. Moreover, it seems not probable that these modifications are due because of the heat. However, HSP90AA1 gene is ubiquitously expressed; there are tissues as testes and brain where the highest expression rates of this gene have been found (Csermely et al. 1998). Therefore, it is possible that different gene regulatory pathways exist in different tissues. We need to keep studying the specific methylation patterns in different tissues and at different ages and deepen in the knowledge of regulatory mechanisms that could affect the HSP90AA1 expression.
Electronic supplementary material
Additional supporting information associated with this article includes:
Electroforetic Motility Shift Assay (EMSA) using nuclear extracts from HepG2 cells under thermoneutral and heat stress conditions. Nuclear extracts from thermoneutral cultured cells were incubated with the D-668D-667-labelled oligonucleotide probe (lane 2) and nuclear extracts from heat shock cultured cells were incubated with the D-668D-667-labelled oligonucleotide probe (lane 3). The I-668D-667-labelled oligonucleotide probe was incubated with nuclear extracts from thermoneutral cultured cells (lane 5) and from heat shock cultured cells (lane 6). The I-668I-667-labelled oligonucleotide probe was incubated with nuclear extracts from thermoneutral cultured cells (lane 8) and from heat shock cultured cells (lane 9). In lanes 1, 4 and 7, nuclear extracts were not added. (DOCX 130 kb)
HSP90AA1 methylation analysis from bllos samples by bisulfite genomic sequencing (Primers are in light grey). Exons are dark grey shaded. CpGs of the CGI are highlighted in green and the CGI limits in blue: −745 to +1445 respect putative TSS(A+1). Uncertain genotyped methylated CpGs are in yellow. Some core promoter motifs are depicted: BRE is double underlined, TATA-box simple underlined and TSS (A+1) at the Inr highlighted in pink. g.660G > C and g.632_methyl-CpG are circled. (GIF 348 kb)
Electroforetic Motility Shift Assay (EMSA) comparing un-methylated CpG (CpG) vs methylated CpG (meCpG) associated with g.660G > C. Nuclear extracts from HepG2 cultured cells were incubated with the CpG-labelled oligonucleotide probe (lane 2), in the presence of increasing excess unlabelled CpG probe (lane 3-50x), in the presence of increasing excess unlabelled meCpG probe (lane 4-50x), meCpG-labelled (lane 6), meCpG-labelled with increasing excess unlabelled meCpG probe (lane 7-50x) and meCpG-labelled with increasing excess unlabelled CpG probe (lane 8-50x). In lanes 1 and 5 nuclear extracts were not added. (DOCX 142 kb)
EMSA oligonucleotides used in the present work. (XLSX 7 kb)
Primers designed for the different experiments performed in the work. (DOCX 19 kb)
Output of the RepeatMasker software. This software predicts the presence of two short interspersed elements in the HSP90AA1 ovine gene. (DOCX 13 kb)
Output obtained from using the RepeatMasker software. This software predicts: a fragment of Bov-tA2 SINE/tRNA-Core-RTE and a fragment of MIRc SINE/MIR. (DOCX 11 kb)
Acknowledgments
This work was supported by the RTA2009-00098 INIA project. Judit Salces-Ortiz was supported by an INIA doctoral grant. We thank AGRAMA breeders association and CITA (Centro de Investigación y Tecnología Agroalimentaria de Aragón) for providing the biological samples and Facultad de Medicina, Universidad de Cantabria, for providing the technical support.
References
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/S0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagniuc P, Ionescu-Tirgoviste C. Gene promoters show chromosome-specificity and reveal chromosome territories in humans. BMC Genomics. 2013;14:278. doi: 10.1186/1471-2164-14-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Guisbert E, Czyz DM, Richter K, McMullen PD, Morimoto RI. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 2013;9:e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, el-Maarri O, Engemann S, Oswald J, Olek A, Walter J. DNA-methylation analysis by the bisulfite-assisted genomic sequencing method. Methods Mol Biol. 2002;200:143–154. doi: 10.1385/1-59259-182-5:143. [DOI] [PubMed] [Google Scholar]
- Han G, Ma H, Chintala R, Fulton DJ, Barman SA, White RE. Essential role of the 90-kilodalton heat shock protein in mediating nongenomic estrogen signaling in coronary artery smooth muscle. J Pharmacol Exp Ther. 2009;329:850–855. doi: 10.1124/jpet.108.149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, et al. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapunov S, Brenowitz M, Rice PA, Catalano CE. Binding then bending: a mechanism for wrapping DNA. Proc Natl Acad Sci U S A. 2006;103:19217–19218. doi: 10.1073/pnas.0609223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Kim JY, Ko AR, Kang TC. Reduction in heat shock protein 90 correlates to neuronal vulnerability in the rat piriform cortex following status epilepticus. Neuroscience. 2013;255:265–277. doi: 10.1016/j.neuroscience.2013.09.050. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Korfanty J, et al. Crosstalk between HSF1 and HSF2 during the heat shock response in mouse testes. Int J Biochem Cell Biol. 2014;57C:76–83. doi: 10.1016/j.biocel.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Landolin JM, et al. Sequence features that drive human promoter function and tissue specificity. Genome Res. 2010;20:890–898. doi: 10.1101/gr.100370.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc S, Hoglund E, Gilmour KM, Currie S. Hormonal modulation of the heat shock response: insights from fish with divergent cortisol stress responses. Am J Physiol Regul Integr Comp Physiol. 2012;302:R184–R192. doi: 10.1152/ajpregu.00196.2011. [DOI] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, et al. Structural and functional analysis of the HSP90AA1 gene: distribution of polymorphisms among sheep with different responses to scrapie. Cell Stress Chaperones. 2008;13:19–29. doi: 10.1007/s12192-007-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, et al. A SNP in the HSP90AA1 gene 5′ flanking region is associated with the adaptation to differential thermal conditions in the ovine species. Cell Stress Chaperones. 2010;15:67–81. doi: 10.1007/s12192-009-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva YA, et al. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics. 2014;15:119. doi: 10.1186/1471-2164-15-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Oner Y, Calvo JH, Serrano M, Elmaci C. Polymorphisms at the 5 ′ flanking region of the HSP90AA1 gene in native Turkish sheep breeds. Livest Sci. 2012;150:381–385. doi: 10.1016/j.livsci.2012.07.028. [DOI] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Rieping M, Schoffl F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet. 1992;231:226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- Salces-Ortiz J, Gonzalez C, Moreno-Sanchez N, Calvo JH, Perez-Guzman MD, Serrano MM. Ovine HSP90AA1 expression rate is affected by several SNPs at the promoter under both basal and heat stress conditions. PLoS ONE. 2013;8:e66641. doi: 10.1371/journal.pone.0066641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Ortiz J, Gonzalez C, Martinez M, Mayoral T, Calvo JH, Serrano M. Looking for adaptive footprints in the HSP90AA1 ovine gene. BMC Evol Biol. 2015;15:7. doi: 10.1186/s12862-015-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Ortiz J, Ramón M, González C, Pérez-Guzmán MD, Garde JJ, García-Álvarez O, et al. Differences in the ovine HSP90AA1 gene expression rates caused by two linked polymorphisms at its promoter affect rams sperm DNA fragmentation under environmental heat stress conditions. PLoS ONE. 2015;10(21):e0116360. doi: 10.1371/journal.pone.0116360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore P, Benvenuto G, Caporaso M, Bruni CB, Chiariotti L. High resolution methylation analysis of the galectin-1 gene promoter region in expressing and nonexpressing tissues. FEBS Lett. 1998;421:152–158. doi: 10.1016/S0014-5793(97)01553-6. [DOI] [PubMed] [Google Scholar]
- Sud N, et al. Nitric oxide and superoxide generation from endothelial NOS: modulation by HSP90. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1444–L1453. doi: 10.1152/ajplung.00175.2007. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Tian WL, et al. High expression of heat shock protein 90 alpha and its significance in human acute leukemia cells. Gene. 2014;542:122–128. doi: 10.1016/j.gene.2014.03.046. [DOI] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano KA. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA. SWItching off methylated DNA. Nat Genet. 2005;37:212–213. doi: 10.1038/ng0305-212. [DOI] [PubMed] [Google Scholar]
- Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/S1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- Yang Z, et al. Novel insights into the role of HSP90 in cytoprotection of H2S against chemical hypoxia-induced injury in H9c2 cardiac myocytes. Int J Mol Med. 2011;28:397–403. doi: 10.3892/ijmm.2011.682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electroforetic Motility Shift Assay (EMSA) using nuclear extracts from HepG2 cells under thermoneutral and heat stress conditions. Nuclear extracts from thermoneutral cultured cells were incubated with the D-668D-667-labelled oligonucleotide probe (lane 2) and nuclear extracts from heat shock cultured cells were incubated with the D-668D-667-labelled oligonucleotide probe (lane 3). The I-668D-667-labelled oligonucleotide probe was incubated with nuclear extracts from thermoneutral cultured cells (lane 5) and from heat shock cultured cells (lane 6). The I-668I-667-labelled oligonucleotide probe was incubated with nuclear extracts from thermoneutral cultured cells (lane 8) and from heat shock cultured cells (lane 9). In lanes 1, 4 and 7, nuclear extracts were not added. (DOCX 130 kb)
HSP90AA1 methylation analysis from bllos samples by bisulfite genomic sequencing (Primers are in light grey). Exons are dark grey shaded. CpGs of the CGI are highlighted in green and the CGI limits in blue: −745 to +1445 respect putative TSS(A+1). Uncertain genotyped methylated CpGs are in yellow. Some core promoter motifs are depicted: BRE is double underlined, TATA-box simple underlined and TSS (A+1) at the Inr highlighted in pink. g.660G > C and g.632_methyl-CpG are circled. (GIF 348 kb)
Electroforetic Motility Shift Assay (EMSA) comparing un-methylated CpG (CpG) vs methylated CpG (meCpG) associated with g.660G > C. Nuclear extracts from HepG2 cultured cells were incubated with the CpG-labelled oligonucleotide probe (lane 2), in the presence of increasing excess unlabelled CpG probe (lane 3-50x), in the presence of increasing excess unlabelled meCpG probe (lane 4-50x), meCpG-labelled (lane 6), meCpG-labelled with increasing excess unlabelled meCpG probe (lane 7-50x) and meCpG-labelled with increasing excess unlabelled CpG probe (lane 8-50x). In lanes 1 and 5 nuclear extracts were not added. (DOCX 142 kb)
EMSA oligonucleotides used in the present work. (XLSX 7 kb)
Primers designed for the different experiments performed in the work. (DOCX 19 kb)
Output of the RepeatMasker software. This software predicts the presence of two short interspersed elements in the HSP90AA1 ovine gene. (DOCX 13 kb)
Output obtained from using the RepeatMasker software. This software predicts: a fragment of Bov-tA2 SINE/tRNA-Core-RTE and a fragment of MIRc SINE/MIR. (DOCX 11 kb)