Abstract
Human melanocortin receptors (hMCRs) have been challenging targets to develop ligands that are explicitly selective for each of their subtypes. To modulate the conformational preferences of the melanocortin ligands and improve the biofunctional agonist/antagonist activities and selectivities, we have applied a backbone N-methylation approach on Ac-Nle-c[Asp-His-d-Nal(2′)-Arg-Trp-Lys]-NH2 (Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2), a nonselective cyclic peptide antagonist at hMC3R and hMC4R and an agonist at hMC1R and hMC5R. Systematic N-methylated derivatives of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2, with all possible backbone N-methylation combinations, have been synthesized and examined for their binding and functional activities toward melanocortin receptor subtypes 1, 3, 4, and 5 (hMCRs). Several N-methylated analogues are selective and potent agonists or antagonists for hMC1R or hMC5R or have selective antagonist activity for hMC3R. The selective hMC1R ligands show strong binding for human melanoma cells. We have also discovered the first universal antagonist (compound 19) for all subtypes of hMCRs.
Graphical abstract
INTRODUCTION
The melanocortin system1–3 remains a challenging target for rational peptide and peptidomimetic design because the 3D topographical requirements for specific melanocortin receptor subtype recognition have not been fully elucidated.4–7 Nevertheless, the numerous multifaceted physiological functions of the five known subtypes of human melanocortin receptors (hMC1–5R), including skin pigmentation,8,9 control of the immune system,10,11 erectile function,12,13 blood pressure and heart rate,14,15 control of feeding behavior and energy homeostasis,3,6,16–21 modulation of aggressive/defensive behavior,22,23 and mediation of pain,24–26 continue to provide a strong stimulus for further development of potent and selective melanocortin agonists and antagonists. Until recently, much of this work was focused on hMC4R due to its direct involvement in the regulation of feeding behavior and energy homeostasis3,6,16–21 as well as sexual behavior.1,6,12,13,27 The hMC3 receptor has been suggested to play a complementary role in weight control.20,21,28 Although the full scope of physiological functions of this receptor is yet to be unraveled, the current understanding based on the observed stimulation of food intake by peripheral administration of an MC3R-selective agonist29 and MC3R agonist-induced inhibition of spontaneous action of pro-opiomelanocortin (POMC) neurons30 suggests that hMC3R is an inhibitory autoreceptor on POMC neurons.3 In addition, development of selective ligands for the hMC1 and hMC5 receptors is receiving increasing attention lately due to the roles of these receptors in regulating pain and skin pigmentation,6,7 controlling the immune system (hMC1R),8,11 regulating exocrine gland function,31 and coordinating central and peripheral signals for aggression (hMC5R).22,23
Another consideration in the development of selective ligands for the melanocortin system bears intrinsic challenges due to conserved amino acid sequences and their structural similarity contained in the seven transmembrane GPCR fold of the melanocortin receptors.1–3,32,33 Unlike many other G protein coupled receptor (GPCR) targets, the hMCRs, known to have the shortest N-terminus among GPCRs, have separate natural agonist and antagonist molecules for functional regulation.1–3,32,33 This aspect imposes a second dimension for the development of melanocortin ligands that achieve selectivity for receptor subtype as well as the desired agonist, antagonist, and bioavailability properties.
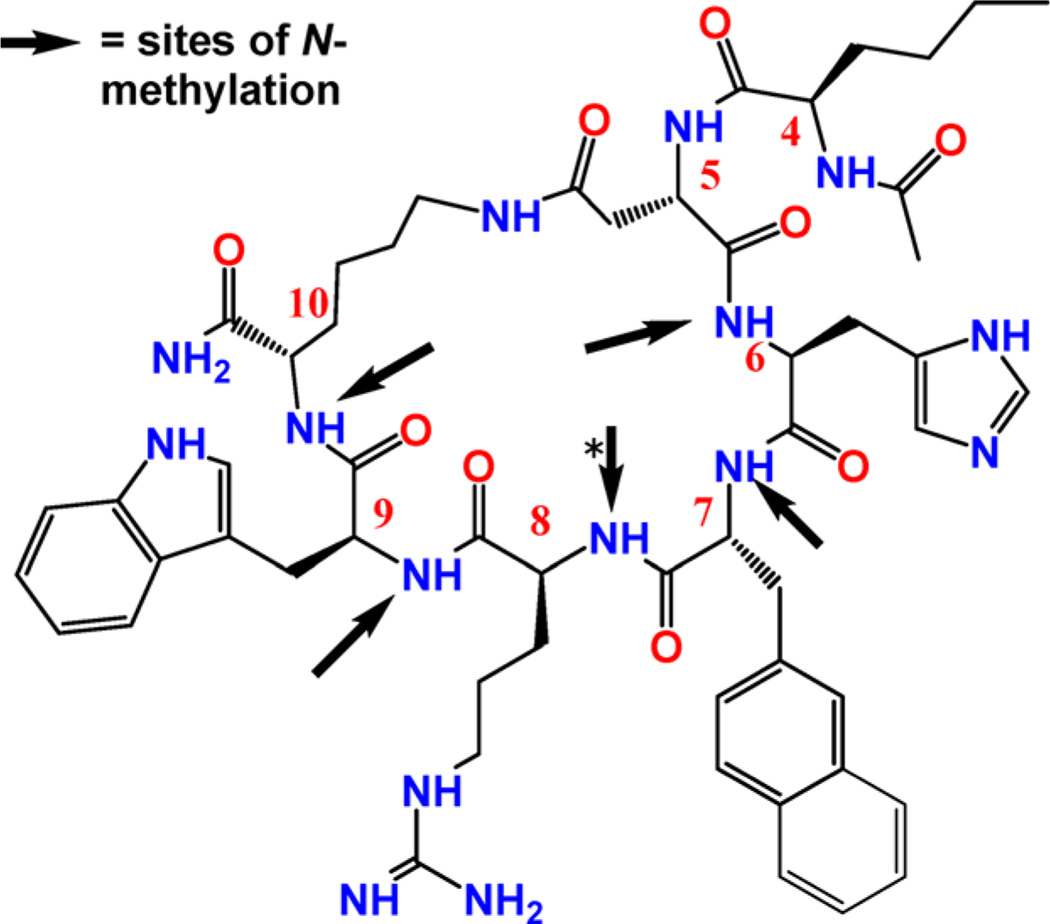
To accomplish this, we have envisioned the application of an N-methylation strategy34–36 to the melanocortin ligands. Recently, N-methylation of backbone amide NHs of peptides has been shown to substantially improve the physicochemical, structural, and biological properties of peptides.34–40 N-Methylation is one of the simplest ways to include conformational restraints into the peptide backbone, as it introduces steric restrictions, allows for cis-peptide bonds, and also prevents hydrogen-bond formation. In addition, N-methylated amino acids often act as turn-promoting moieties (akin to prolines)34,40 and thus this strategy helps to generate secondary structure features in peptides without changing the constituent peptide sequence. The specific impact of N-methylation on melanocortin peptide ligands had not been investigated systematically until we used this strategy on Ac-Nle-c[Asp-His-d-Phe-Arg-Trp-Lys]-NH2 (MT-II) to develop completely selective hMC1R agonists.41,42 Interestingly, we have also observed that some of the N-methylated Ac-Nle4-c[Asp5,dPhe7, Lys10]-NH2 (MT-II) peptides possessed antagonist activity, which is a switch in functional selectivity, although the amino acid sequence remained unchanged. Here, we have modulated the peptide conformation and the functional side chain disposition of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 (peptide 1, SHU9119),43 a peptide with antagonist activities for hMC3R and hMC4R and with agonist activities for hMC1R and hMC5R, by systematic N-methylation of the backbone amide NHs to gain further insight into the intricate correlations among amino acid sequence, conformation, and activity–selectivity. A combinatorial library consisting of N-methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 (peptide 1, SHU9119) and analogues with single and multiple N-methylation in the core cyclic sequence of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 has been designed, synthesized (Figure 1 and Table 1), and characterized chemically, and in vitro studies have been performed for binding affinities and efficacies at the hMCRs (Figure 2b–e and Tables 1–3). Syntheses were performed in analogy to the synthesis of the library of N-methylated Ac-Nle4-c[Asp5,d-Phe7,Lys10]-NH2 (MT-II) peptides.41
Figure 1.
Structure of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 peptide. The arrows indicate the sites of N-methylation in analogues synthesized, with all combinations examined systematically (see text).
Table 1.
HPLC and Mass Data for N-Methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 Peptidesa
| # | Sequence | Calculated Mass (M+H)+ |
ESI-MS (M+H)+ |
RP-HPLC 10–100% in 30 min [tR] min |
RP-HPLC 10–50% in 30 min [tR] min |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Nle | D | H | DNal(2′) | R | W | K | 1074.6 | 1074.7 | 15.58 | 26.70 |
| 2 | Nle | D | H | DNal(2′) | R | W | K | 1088.6 | 1088.8 | 16.16 | 28.13 |
| 3 | Nle | D | H | DNal(2′) | R | W | K | 1088.6 | 1088.8 | 14.80 | 24.52 |
| 4 | Nle | D | H | DNal(2′) | R | W | K | 1088.6 | 1088.8 | 13.71 | 22.73 |
| 5 | Nle | D | H | DNal(2′) | R | W | K | 1088.6 | 1088.8 | 14.07 | 23.80 |
| 6 | Nle | D | H | DNal(2′) | R | W | K | 1088.6 | 1089.1 | 15.82 | 27.57 |
| 7 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.7 | 15.86 | 27.28 |
| 8 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.7 | 13.89 | 23.08 |
| 9 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.7 | 14.43 | 24.21 |
| 10 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.6 | 15.71 | 27.28 |
| 11 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.6 | 14.02 | 22.90 |
| 12 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.7 | 14.09 | 23.52 |
| 13 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.6 | 15.52 | 26.33 |
| 14 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.7 | 13.93 | 23.28 |
| 15 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.7 | 13.93 | 23.12 |
| 16 | Nle | D | H | DNal(2′) | R | W | K | 1102.6 | 1102.7 | 15.25 | 25.94 |
| 17 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 13.80 | 22.75 |
| 18 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 14.55 | 24.51 |
| 19 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 15.98 | 27.65 |
| 20 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 13.98 | 23.27 |
| 21 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 14.17 | 23.77 |
| 22 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 14.49 | 24.48 |
| 23 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 13.91 | 23.05 |
| 24 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 13.69 | 22.57 |
| 25 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 14.87 | 25.08 |
| 26 | Nle | D | H | DNal(2′) | R | W | K | 1116.6 | 1116.7 | 14.00 | 23.24 |
| 27 | Nle | D | H | DNal(2′) | R | W | K | 1130.6 | 1130.7 | 14.35 | 24.57 |
| 28 | Nle | D | H | DNal(2′) | R | W | K | 1130.6 | 1130.7 | 13.97 | 22.95 |
| 29 | Nle | D | H | DNal(2′) | R | W | K | 1130.6 | 1130.7 | 15.05 | 25.65 |
| 30 | Nle | D | H | DNal(2′) | R | W | K | 1130.6 | 1130.8 | 14.45 | 24.09 |
| 31 | Nle | D | H | DNal(2′) | R | W | K | 1130.6 | 1130.7 | 13.40 | 22.05 |
| 32 | Nle | D | H | DNal(2′) | R | W | K | 1144.6 | 1144.7 | 14.17 | 23.73 |
Amino acids with N-methyl groups are highlighted in gray. The calculated mass (M + H)+ for each peptide provides the theoretically calculated mass based on light isotope composition; ESI-MS (M + H)+ represents the experimentally determined mass. For all peptides, purity was characterized based on RP-HPLC measurements using a C18 column and employing a linear gradient of acetonitrile (MeCN) and water. All peptides showed a purity ≥95%. The gradients 10−100% and 10–50% indicate solvent gradients of MeCN in water from 10 to 100% in 30 min and from 10 to 50% in 30 min, respectively. Last two columns give RP-HPLC retention times [tR] (in min) for each peptide.
Figure 2.
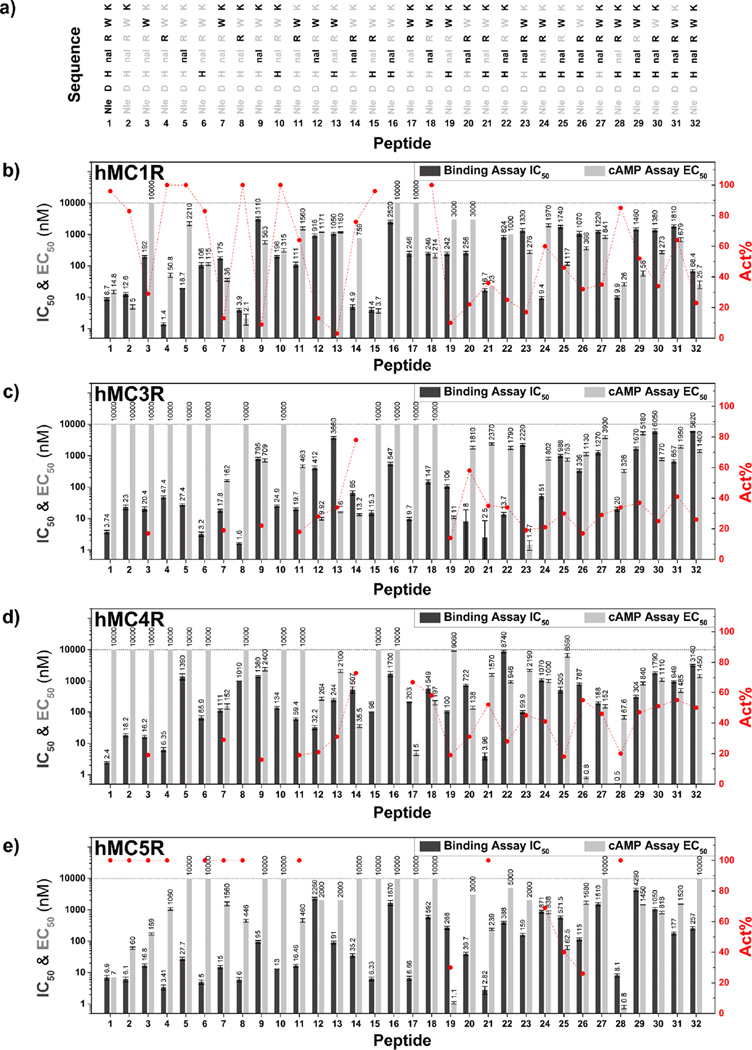
(a) Amino acid sequences for Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 (peptide 1) and N-methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 analogues (peptides 2–32; the site of N-methylation is highlighted in bold. (b–e) Histograms showing the results for IC50 from binding assays (black bars) and EC50 from cAMP assays (light gray bars) at hMC1R, hMC3R, hMC4R, and hMC5R, respectively. Although IC50 and EC50 values on the y axis are in real number units, for convenient representation and comparison, log10 scaling is used on the y axis. The specific IC50 and EC50 values for each peptide are indicated above its corresponding histogram bar. Act% values (>0%) of the ligands for the cAMP assay are plotted in red with a second y dimension on the right. The location of the red spots demonstrates increased and decreased agonist activity. For more details, including the binding efficiency, refer to Tables 2 and 3. IC50, concentration of peptide at 50% specific binding (N = 4); EC50, effective concentration of peptide that was able to generate 50% maximal intracellular cAMP accumulation (N = 4).
Table 3.
cAMP Assay Results of N-Methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 Analogues at hMCRsa
| hMC1R | hMC3R | hMC4R | hMC5R | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # sequence | EC50, nM | %max | EC50, nM | %max | EC50, nM | %max | EC50, nM | %max | |||||||
| 1 | Nle | D | H | DNal(2′) | R | W | K | 14.8±2.0 | 96 | >10000 | NA | >10000 | NA | 7 | 100 |
| 2 | Nle | D | H | DNal(2′) | R | W | K | 5.0±0.8 | 83 | >10000 | NA | >10000 | NA | 60±5 | 100 |
| 3 | Nle | D | H | DNal(2′) | R | W | K | >10000 | 29 | >10000 | 17 | >10000 | 19 | 169±14 | 100 |
| 4 | Nle | D | H | DNal(2′) | R | W | K | 50.8±8 | 100 | >10000 | NA | >10000 | NA | 1060±100 | 100 |
| 5 | Nle | D | H | DNal(2′) | R | W | K | 2210±310 | 100 | >10000 | NA | >10000 | NA | >10000 | NA |
| 6 | Nle | D | H | DNal(2′) | R | W | K | 115±12 | 83 | >10000 | NA | >10000 | NA | >10000 | 100 |
| 7 | Nle | D | H | DNal(2′) | R | W | K | 36±4 | 13 | 162±12 | 19 | 152±30 | 29 | 1560±210 | 100 |
| 8 | Nle | D | H | DNal(2′) | R | W | K | 2.1±0.8 | 100 | >10000 | NA* | >10000 | NA | 446±50 | 100 |
| 9 | Nle | D | H | DNal(2′) | R | W | K | 563±70 | 9 | 709±87 | 22 | 2400±290 | 16 | >1 0000 | NA |
| 10 | Nle | D | H | DNal(2′) | R | W | K | 315±40 | 100 | >10000 | NA | >10000 | NA | >10000 | NA |
| 11 | Nle | D | H | DNal(2′) | R | W | K | 1560±170 | 64 | 463±52 | 18 | >10000 | 19 | 460±61 | 100 |
| 12 | Nle | D | H | DNal(2′) | R | W | K | 1171±21 | 13 | 9.92±1 | 28 | 264±31 | 21 | >2000 | NA |
| 13 | Nle | D | H | DNal(2′) | R | W | K | 1160±52 | 3 | 16±0.8 | 34 | 2100±231 | 31 | >2000 | NA |
| 14 | Nle | D | H | DNal(2′) | R | W | K | 759 | 76 | 13.2±0.9 | 78 | 35.5±3.9 | 73 | >10000 | NA |
| 15 | Nle | D | H | DNal(2′) | R | W | K | 3.7±0.6 | 96 | >10000 | NA* | >10000 | NA | >10000 | NA |
| 16 | Nle | D | H | DNal(2′) | R | W | K | >10000 | NA | >10000 | NA | >10000 | NA | >10000 | NA |
| 17 | Nle | D | H | DNal(2′) | R | W | K | >10000 | NA | >10000 | NA* | 5±0.9 | 67 | >10000 | NA |
| 18 | Nle | D | H | DNal(2′) | R | W | K | 214±35 | 100 | >10000 | NA | 197±29 | 58 | >10000 | NA |
| 19 | Nle | D | H | DNal(2′) | R | W | K | >3000 | 10* | 11.±1 | 14* | 9060±110 | 19* | 1.1±0.1 | 30* |
| 20 | Nle | D | H | DNal(2′) | R | W | K | >3000 | 22 | 1810±190 | 58 | 138±15 | 31 | >3000 | NA |
| 21 | Nle | D | H | DNal(2′) | R | W | K | 23 | 36 | 2370±180 | 35 | 1570±170 | 52 | 239±20 | 100 |
| 22 | Nle | D | H | DNal(2′) | R | W | K | >1000 | 25 | 1790±213 | 34 | 946±78 | 28 | >5000 | NA |
| 23 | Nle | D | H | DNal(2′) | R | W | K | 276±32 | 17 | 1.47±0.5 | 19 | 2190±178 | 45 | >2000 | NA |
| 24 | Nle | D | H | DNal(2′) | R | W | K | 1970±213 | 60 | 802±91 | 21 | 1000±120 | 41 | 838±90 | 69 |
| 25 | Nle | D | H | DNal(2′) | R | W | K | 117±13 | 46 | 753±89 | 30 | 6590±912 | 18 | 62.5±9 | 40 |
| 26 | Nle | D | H | DNal(2′) | R | W | K | 366±41 | 32 | 1130±134 | 17 | 0.8±0.05 | 55 | 1680±192 | 26 |
| 27 | Nle | D | H | DNal(2′) | R | W | K | 841±91 | 35 | 3930±410 | 29 | 152±19 | 46 | >10000 | NA |
| 28 | Nle | D | H | DNal(2′) | R | W | K | 26±3 | 85 | 326±41 | 34 | 67.6±9 | 20 | 0.8±0.1 | 100 |
| 29 | Nle | D | H | DNal(2′) | R | W | K | 58±11 | 52 | 5180±589 | 37 | 840±93 | 47 | 1450±14 | NA |
| 30 | Nle | D | H | DNal(2′) | R | W | K | 273±31 | 34 | 770±77 | 25 | 1110±141 | 51 | 818±83 | NA |
| 31 | Nle | D | H | DNal(2′) | R | W | K | 679±98 | 64 | 1950±200 | 41 | 485±67 | 55 | 1520±19 | NA |
| 32 | Nle | D | H | DNal(2′) | R | W | K | 25.7±7 | 23* | 1400±150 | 26 | 1450±1 62 | 50 | >1 0000 | NA |
Amino acids with N-methyl groups are highlighted in gray. EC50, effective concentration of peptide that was able to generate 50% maximal intracellular cAMP accumulation (N = 4). Percent activity, percent of cAMP produced at 10 µM ligand concentration in relation to Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II). NA, 0% cAMP accumulation observed at 10 µM. The peptides were tested at a range of concentrations from 10−10 to 10−5 M.
Asterisks show the lignds and receptors for which we have determined pA2 values. The pA2 values for the peptide 8, 15, and 17 at the hMC3R are 9.1, 8.2, and 8.5, respectively; The pA2 values for the peptide 19 at the hMC1R, hMC3R, hMC4R, hMC5R are 7.5, 8, 8, and 7.4, respectively. The pA2 value for peptide 32 at the hMC1R is 8.8.
RESULTS AND DISCUSSION
SAR Studies of N-Methylated Ac-Nle4-c[Asp5,d-Nal-(2′)7,Lys10]-NH2 Analogues
Effects of N-methylation on Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 were evaluated on stably cloned hMCR (hMC1R, hMC3R, hMC4R, and hMC5R) cell lines (HEK 293 cells) via competition binding assays and functional activity assays (cAMP assay). Competitive binding assays with [125I]-[Nle4,d-Phe7]-α-MSH (NDP-α-MSH) on whole cells and [3H]-adenylate cyclase assays for cAMP were examined along with the application of standard Ac-Nlec[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II) and Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 (SHU9119). The results are shown in Figure 2 and Tables 2 and 3. For the pharmacological characterization of the ligands in this study, a binding efficiency >70% is defined as competitive binding and binding efficiency <70% is considered to be noncompetitive binding. For the characterization of the potency of the ligand, we used Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II) as the standard full agonist (Act% = 100%). Therefore, the cAMP functional activities (cAMP level) induced by ligands are expressed as a percent of the activity that is generated by the standard Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II) and are defined as Act%. Act% < 30% is defined as no activity, Act% between 50 and 80%, partial agonist, and Act % > 80, agonist.
Table 2.
Binding Assay Results of N-Methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 Analogues at hMCRsa
| hMC1R | hMC3R | hMC4R | hMC5R | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # sequence | IC50, nM | Bind-Effic | IC50, nM | Bind-Effic | IC50, nM | Bind-Effic | IC50, nM | Bind-Effic | |||||||
| 1 | Nle | D | H | DNal(2′) | R | W | K | 8.7±0.9 | 100 | 3.74±0.5 | 97 | 2.4±0.2 | 98 | 6.9±1.2 | 98 |
| 2 | Nle | D | H | DNal(2′) | R | W | K | 12.6±1.8 | 100 | 23±4 | 97 | 18.2±2.1 | 97 | 6.1±1.1 | 98 |
| 3 | Nle | D | H | DNal(2′) | R | W | K | 192±20 | 77 | 20.4±3 | 77 | 16.2±2 | 70 | 16.8±2.3 | 77 |
| 4 | Nle | D | H | DNal(2′) | R | W | K | 1.4±0.15 | 90 | 47.4±5.2 | 95 | 6.35±1.0 | 90 | 3.41±0.6 | 96 |
| 5 | Nle | D | H | DNal(2′) | R | W | K | 18.7±0.30 | 80 | 27.4±2.1 | 90 | 1390±310 | 90 | 27.7±3.4 | 90 |
| 6 | Nle | D | H | DNal(2′) | R | W | K | 106±21 | 90 | 3.2±0.5 | 90 | 65.9±10 | 90 | 5±0.8 | 90 |
| 7 | Nle | D | H | DNal(2′) | R | W | K | 175±20 | 86 | 17.8±2 | 95 | 111±13 | 89 | 15±1.8 | 94 |
| 8 | Nle | D | H | DNal(2′) | R | W | K | 3.9±0.5 | 80 | 1.6±0.1 | 90 | 1010±1.1 | 90 | 6±1.0 | 90 |
| 9 | Nle | D | H | DNal(2′) | R | W | K | 3110±456 | 82 | 795±90 | 90 | 1380±110 | 101 | 95±10 | 87 |
| 10 | Nle | D | H | DNal(2′) | R | W | K | 196±16 | 90 | 24.9±2.1 | 99 | 134±15 | 90 | 13±0.12 | 90 |
| 11 | Nle | D | H | DNal(2′) | R | W | K | 111±20 | 89 | 19.7±2 | 93 | 59.4±6 | 89 | 16.46±2.1 | 93 |
| 12 | Nle | D | H | DNal(2′) | R | W | K | 916±120 | 92 | 412±60 | 92 | 32.2±5 | 88 | 2260±230 | 72 |
| 13 | Nle | D | H | DNal(2′) | R | W | K | 1050±116 | 88 | 3660±410 | 95 | 244±31 | 90 | 91±9 | 86 |
| 14 | Nle | D | H | DNal(2′) | R | W | K | 4.9±0.9 | 90 | 65±11 | 90 | 507±120 | 90 | 35.2±5 | 90 |
| 15 | Nle | D | H | DNal(2′) | R | W | K | 4.0±0.7 | 90 | 15.3±2.8 | 80 | 98±4.1 | 90 | 6.33±0.9 | 90 |
| 16 | Nle | D | H | DNal(2′) | R | W | K | 2520±320 | 86 | 547±70 | 80 | 1700±340 | 90 | 1670±300 | 90 |
| 17 | Nle | D | H | DNal(2′) | R | W | K | 246±45 | 80 | 9.7±1.2 | 90 | 203±7 | 80 | 6.66±1 | 90 |
| 18 | Nle | D | H | DNal(2′) | R | W | K | 246±24 | 80 | 147±23 | 90 | 549±150 | 90 | 592±70 | 80 |
| 19 | Nle | D | H | DNal(2′) | R | W | K | 242±32 | 74 | 106±13 | 90 | 100±8 | 79 | 268±31 | 91 |
| 20 | Nle | D | H | DNal(2′) | R | W | K | 256±35 | 84 | 8±11 | 86 | 722±81 | 81 | 39.7±5 | 90 |
| 21 | Nle | D | H | DNal(2′) | R | W | K | 16.7±2.2 | 9 | 2.5±06 | 94 | 3.96±1 | 76 | 2.82±0.8 | 90 |
| 22 | Nle | D | H | DNal(2′) | R | W | K | 824±1 00 | 68 | 13.7±2 | 38 | 8740±841 | 59 | 388±41 | 70 |
| 23 | Nle | D | H | DNal(2′) | R | W | K | 1330±200 | 70 | 2220±242 | 61 | 99.9±10 | 36 | 159±19 | 67 |
| 24 | Nle | D | H | DNal(2′) | R | W | K | 9.4±1.2 | 86 | 51±9 | 78 | 1070±110 | 76 | 871±91 | 84 |
| 25 | Nle | D | H | DNal(2′) | R | W | K | 1740±210 | 68 | 988±110 | 90 | 505±111 | 67 | 571.5±61 | 93 |
| 26 | Nle | D | H | DNal(2′) | R | W | K | 1070±189 | 93 | 336±42 | 82 | 787±82 | 68 | 115±13 | 92 |
| 27 | Nle | D | H | DNal(2′) | R | W | K | 1220±123 | 94 | 1270±210 | 92 | 188±21 | 37 | 1510±159 | 32 |
| 28 | Nle | D | H | DNal(2′) | R | W | K | 9.9±1.2 | 93 | 20±3 | 88 | 0.5±0.02 | 64 | 8.1±1 | 89 |
| 29 | Nle | D | H | DNal(2′) | R | W | K | 1460±163 | 42 | 1670±193 | 97 | 304±35 | 27 | 4290±521 | 64 |
| 30 | Nle | D | H | DNal(2′) | R | W | K | 1380±150 | 87 | 6050±985 | 93 | 1790±190 | 24 | 1050±120 | 24 |
| 31 | Nle | D | H | DNal(2′) | R | W | K | 181 0±210 | 89 | 657±76 | 94 | 949±85 | 8 | 177±21 | 75 |
| 32 | Nle | D | H | DNal(2′) | R | W | K | 68.4±8 | 91 | 5820±200 | 84 | 3140±270 | 19 | 257±34 | 37 |
Amino acids with N-methyl groups are highlighted in gray. IC50, concentration of peptide at 50% specific binding (N = 4). NB, 0% 125I-NDP-α- MSH displacement observed at 10 µM. Percent binding efficiency, maximal % of 125I-NDP-α-MSH displacement observed at 10 µM.
The first group of peptides are mono-N-methylated derivatives of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 (peptides 2–6; Figures 1 and 2a). The competitive binding assays show that mono-N-methylation at each position of the pharmacophore does not have a remarkable influence on the binding affinity toward hMCRs except for the hMC4R for N-methylation at position 7 (NMe-DNal(2′)7, peptide 5), implying that NMe-DNal(2′)7 causes a drastic loss of binding affinity at hMC4R. cAMP assays show that peptide 3, with NMe-Trp9, leads to a greater loss of functional activity (cAMP activity) at hMC1R compared to that with the NMe-DNal(2′)7 analogue 5, except at hMC5R. Therefore, the single N-methylated derivatives of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 at the Trp9 position (peptide 3) lead to a selective agonist for hMC5R.
The second group of peptides are di-N-methylated derivatives in the core sequence of Ac-Nle4-c[Asp5,d-Nal-(2′)7,Lys10]-NH2 (peptides 7–16; Figure 1 and 2a). The first subgroup is NMe-Lys10 + NMeXaa (peptides 7–10; Xaa refers to the rest of the amino acids in the cyclic structure of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 considered for N-methylation). The results show that any site of N-methylation combined with NMe-Lys10 (peptides 7–10, Figure 2) retains the same binding behavior as that of the first group for hMC1R, hMC3R, and hMC5R and that a loss of binding affinity by 1 to 2 orders of magnitude occurs at hMC4R. The exception is peptide 9, which lost binding affinity for all subtypes of melanocortin receptors, possibly due to NMe-DNal(2′)7. It is interesting to note that when both of the positively charged amino acids (Arg8 and Lys10) are N-methylated (peptide 8) the binding affinity for hMC4R decreases by 3 orders of magnitude but increases at hMC3R. Therefore, peptide 8 becomes a potent selective antagonist of hMC3R (with pA2 value 9.1) and a potent selective agonist for hMC1R. This observation agrees with the previous studies using chimeric human melanocortin 4 receptor and truncation studies demonstrating that Arg8 is critical for binding at hMC4R.42 The N-methylation of Arg8 changes Φ and Ψ space and reduces the flexibility of the charged side chain group, which, in turn, reduces the binding affinity toward the receptor. When NMe-DNal(2′)7 is included (peptide 9), binding affinity is drastically lost at hMC1R, hMC3R, and hMC4R. Hence, peptide 9 is a very selective potential antagonist for hMC5R. The second subgroup of di-N-methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 analogues is NMe-Trp9 + NMeXaa (peptides 7 and 11–13). Peptide 7 retains the same binding affinities and functional activities as those of the mono-N-methylated derivatives of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2. Peptides 11–13 all show potential antagonist activity at hMC1R, hM3R, and hMC4R based on the EC50 and Act% values. In the third subgroup, consisting of NMe-Arg8 + NMeXaa (peptides 8, 11, 14, and 15), the combination with NMe-Arg8 shows no significant changes in the binding behavior compared to that of the mono-N-methylated compounds, except for hMC4R, as mentioned above. cAMP agonist activity at hMC5R is lost when it is combined with NMe-His. Interestingly, when NMe-Arg8 is combined with NMe-DNal(2′)7 (peptide 14), partial agonist activity is observed at hMC1R, hMC3R, and hMC4R. Among the fourth subgroup of NMe-DNal(2′)7 + NMeXaa peptides (peptides 9, 12, 14, and 16), the combination with NMe-DNal(2′)7 showed a significant loss of binding affinity at all receptor subtypes. The fifth and last of the di-N-methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 compounds is NMe-His6 + NMeXaa (peptides 10, 13, 15, and 16). When NMe-His6 is involved in di-N-methylated analogues, the agonist activity at hMC5R is diminished compared to that of the first subgroup. When the di-N-methylated combination of NMe-His6 and NMe-Trp9 is used, the binding affinity is often reduced by 1 order of magnitude and the functional cAMP activities are lost at the hMC1R, as was observed with the mono-N-methylated compounds (peptides 7, 12, and 13). When two N-methylated aromatic residues (NMe-His6 and NMe-DNal(2′)7, peptide 16) are used in combination, all of the agonist activities are lost and most of the binding affinities and cAMP activities are greatly diminished. However, the combination of NMe-His6 with positive charged residues NMe-Lys10 and NMe-Arg6 (peptides 10 and 15) shows increased, selective agonist activities at hMC1R. Peptide 15 is a selective agonist for hMC1R, with possible antagonist activities for the rest of the hMCRs. It has a 6-fold selective, potent antagonist activity at hMC3R with a pA2 value of 8.2 compared to that at hMC4R. Peptide 16, with binding affinity and binding efficiency toward all hMCRs, has possible universal antagonist activity for all subtypes of melanocortin receptors. To our knowledge, this is the first melanotropin peptide that has possible universal antagonist activity for all hMCRs. The fifth subgroup also presented moderate, selective antagonist for hMC5R (peptide 13). Generally, when the N-methylated aromatic amino acids are involved in dimethylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 analogues (peptide 16), most of the binding affinities and cAMP activities are greatly diminished.
The third group of peptides consists of tri-N-methylated derivatives in the core sequence of Ac-Nle4-c[Asp5,d-Nal-(2′)7,Lys10]-NH2 (peptides 17–26; Figure 1 and 2a). Except for a few cases of dramatically increased selectivity, this group of peptides in general shows significantly reduced binding affinities, which might be due to hindrance caused by the multiple N-methylations that are involved. However, in a few cases, there was a dramatic increase in selectivity. Peptide 17, with consecutive N-methylations at Arg8-Trp9-Lys10, leads to a selective partial agonist ligand at hMC4R. Compared to hMC4R, its binding affinity toward hMC3R is more potent than toward hMC4R, with binding affinities of 9.7 and 203 nM, respectively. The pA2 value for peptide 17 at hMC3R is 8.5. The combination of NMe-DNal(2′)7, NMe-Trp9, and NMe-Lys10 (peptide 18) shows full agonist activity at hMC1R and partial agonist activity at hMC4R. The combination of NMe-His6, NMe-Trp9, and NMe-Lys10 (peptide 19) shows good binding to all hMCRs but no activity to any hMCR. The pA2 values for peptide 19 at hMC1R, hMC3R, hMC4R, and hMC5R are 7.5, 8, 8, 7.4, respectively. Hence, in peptide 19, a potent, universal antagonist is discovered. Peptide 20 also shows potential antagonist activity at all 4 hMCRS (with a very weak activity at hMC3R). For the combination of NMe-DNal(2′)7 with positively charged residues NMe-His6 and NMe-Lys10 (peptide 22) or NMe-Arg8 and NMe-Trp9 (peptide 23), all functional activities are lost. The combination of NMe-His6, NMe-Trp9, and NMe-Arg8 (peptide 24) shows selective partial agonist activity at hMC1R and hMC5R. This observation is consistent with our earlier Ac-Nle4-c[Asp5,dPhe7, Lys10]-α-MSH(4–10)-NH2 N-methylation studies.41 In this group, peptide 19 is an antagonist toward all hMCRs with reduced binding affinities compared to those of the mono-N-methylated group of peptides (peptide 2–6), whereas peptide 21 retains agonist activity at hMC5R. It is noteworthy that when NMe-DNal(2′)7 is involved nearly all of the binding affinities are reduced (peptides 20, 22, 23, 25, and 26).
The fourth group of peptides has five different tetra-N-methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 derivatives (peptides 27–31; Figure 1 and 2a). The studies show that the sites of N-methylation when combined with NMe-DNal(2′)7 reduce the binding affinity at all hMCRs. Only compound 28 retains nanomolar binding at all hMCRs, whereas cAMP activity for this peptide shows agonist activity at hMC1R and hMC5R and potential antagonist activity at hMC3R and hMC4R, much like Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2.
The final penta-N-methylated compound 32 shows potent binding at hMC1R, with an IC50 of 68 nM (91% binding efficiency), poor binding efficiency at hMC5R (37%), and micromolar binding at hMC3R and hMC4R. Therefore, on the basis of its binding affinity, it has potent and selective binding to hMC1R, with a pA2 value of 8.8, consistent with its binding affinity. In the cAMP assay, the analogue shows much lower activation on peptide stimulation (lower Act% shown in Table 3); it was inactive at all hMCRs.
Comparison of N-Methylation on Ac-Nle4-c[Asp5,dPhe7, Lys10]-α-MSH(4–10)-NH2 (MT-II) and Ac-Nle4-c-[Asp5,d-Nal(2′)7,Lys10]-NH2 (SHU9119)
Our earlier N-methylation studies on Ac-Nle4-c[Asp5,d-Phe7, Lys10]-α-MSH(4–10)-NH2 demonstrated that N-methylation of cyclized melanotropins increased the selectivities for the hMCRs. For example, N-methylation of Ac-Nle4-c[Asp5,dPhe7, Lys10]-α-MSH(4–10)-NH2 led to the most selective agonist for hMC1R, which is also a specific biomarker for melanoma cells.41 The present results showed that N-methylation of Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 also leads to selective agonists and antagonists toward various hMCRs, but it does so with a much broader spectrum of selectivity compared to that for N-methylated Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2. In Figure 2, the red spots demonstrate increases and decreases in agonist activity at all subtypes of melanocortin receptors. In this study, we have found selective hMC1R agonists (peptide 8, 10, 15) and the most selective highly potent hMC1R antagonist (peptide 32), with a pA2 value of 8.8. In addition to possible selective antagonists for hMC3R (peptides 8, 15, 17), with pA2 values of 9.1, 8.2, and 8.5, respectively, a highly selective hMC5R agonist (peptide 3) and antagonists (peptides 9 and 13) were found. Finally, through this study, we discovered a unique universal antagonist (peptide 19) for all four hMCRs, which is an important advancement for examining the biological roles of the endogenous agonist MSH system. Direct comparison of binding and activity values for the same methylation pattern shows that N-methylation influences structural changes of Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH-(4–10)-NH2 (MT-II) and Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 (SHU9119) in different ways. It should not be expected that influencing both compounds in a parallel manner will lead to similar changes in binding and activity properties for each methylation pattern because the difference between Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II) and Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 (SHU9119) involves substitution of d-Phe7 by the bulky DNal(2′)7. Since this amino acid is presented in Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 but not in Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2, any N-methylation on these aromatic amino acids and combination with other N-methylated amino acids might induce different conformational changes that can dramatically change binding and functional selectivity properties. Introducing N-methylation on amide NHs of aromatic amino acids can be expected to reorient the backbone conformation and side chain stacking (aromatic groups), all of which can be important factors for melanocortin receptor subtype selectivity.
Binding Assays of Selective hMC1R Ligands at Human Melanoma Cells
The above-discussed SAR studies revealed that peptide 15 is a quite good, selective agonist of hMC1R. We have therefore further investigated binding and cAMP activities in human melanoma cells (A375). As shown in Table 4, these peptides have nanomolar binding affinities for A375 melanoma cells. The cAMP activities of these two peptides in melanoma cells are different from those for HEK 293 cells expressing hMC1R. This is due to the different expression level of hMC1R between these two cell lines. Another possible reason could be the different signaling system in these two cell lines. Nevertheless, these two peptides can be used to target melanoma cells.
Table 4.
Binding and cAMP Activities of Selected Peptides at Human Melanoma Cells (A375)
| Peptide | Sequence | IC50 (nM) | Binding Efficiency | EC50 (nM) | Act% |
|---|---|---|---|---|---|
| 15 | Ac-Nle-c[Asp-(NMe)His-DNal(2′)-(NMe)Arg-Trp-Lys]-NH2 | 14.8 ± 2 | 100 ± 10 | 22 ± 2.5 | NA |
| 28 | Ac-Nle-c[Asp-(NMe)His-DNal(2′)-(NMe)Arg-(NMe)Trp-(NMe)Lys]-NH2 | 8.7 ± 1 | 100 ± 10 | 20 ±2 | NA |
| MT-II | Ac-Nle-c[Asp-His-d-Phe-Arg-Trp-Lys]-NH2 | 6.7 ± 2.9 | 100 ± 10 | 110 ± 0.6 | 100 |
| 1 | Ac-Nle-c[Asp-His-DNal(2′)-Arg-Trp-Lys]-NH2 | 100 ± 96 | 80 ± 34 | 3.3 ± 2.6 | NA |
CONCLUSIONS
A lack of selectivity and potency in the agonist or antagonist properties of melantroptin ligands for the melanocortin receptor subtypes is still the most difficult hurdle for their application in medicine. To ameliorate the biological properties of melanocortin ligands and to achieve selective agonists and antagonists for melanocortin receptors via the conformational modulation of the peptide backbone that is imparted by N-methyl steric constraints, mono-and multiply N-methylated analogues of the Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 peptide were investigated. It was found that the activity and selectivity profile induced by N-methylation is very different for Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 compared to those resulting from a similar approach using Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2. This is striking as both stem peptides differ only in the exchange of one amino acid (DNal(2′)7 instead of d-Phe7 in Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2).
The systematic N-methylation of Ac-Nle4-c[Asp5,d-Nal-(2′)7,Lys10]-NH2 leads to a highly selective antagonist of hMC1R (peptide 32) based on binding affinity. This strategy still retains several hMC1R agonists (peptides 8, 10, and 15) as previously described.41 As anticipated, we have found a selective agonist for hMC5R (peptide 3). The selective antagonist for hMC3R (peptide 17) discovered in this study can be potentially used to treat obesity and diabetes. Apart from this, the newly discovered peptide 19 is a universal antagonist of the hMCR system, which could be important to modulate the endogenous agonist MSH in the melanocortin system. These results strengthen the role of our earlier discovered d-Trp8-γ-MSH as a selective agonist of hMC3R. Further conformational and topographical characterization of all of the important peptides discussed here to obtain a more detailed correlation between melanocortin ligand conformation and the observed receptor discrimination is in progress.
The results obtained from multiply the N-methylated Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 peptide along with those from multiply N-methylated Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II) will help to understand the finer details of the functional properties of melanocortin receptors and the bioactive conformational preferences of the ligands required by them. The studies further highlight the synthetic simplicity and impact of the N-methylation strategy on finetuning the conformational preferences of the ligands to achieve the desired biological effects. The studies, in general, are an encouragement toward a future for peptide chemistry for the development of peptide therapeutic agents.
EXPERIMENTAL SECTION
Materials
Nα-Fmoc-amino acids, peptide coupling reagents, Rink amide MBHA resin, and solvents were reagent grade and used without further purification unless otherwise specified. These chemicals were obtained from Aldrich, Novabiochem, Iris Biotech GmbH, Merck, NeoMPS, ORPEGEN Pharma, and GLS. The following amino acids were used: Fmoc-Lys(Alloc)-OH, Fmoc-Trp(Boc)-OH, Fmoc-Arg-(Pbf)-OH, Fmoc-DNal(2′)-OH, Fmoc-His(Trt)-OH, Fmoc-Asp-(OAllyl)-OH, and Fmoc-Nle-OH. The polypropylene reaction vessels (syringes with frits) were purchased from B. Braun Melsungen AG. The purity of the peptides was checked by analytical reverse-phase HPLC using an Amersham Pharmacia Biotech Äkta Basic 10F with a ODS-A C18 (120 Å, 5 µm, 250 mm × 4.6 mm) column (Omnicrom YMC Europe GmbH) monitored at 220 and 254 nm and by high-resolution mass spectral analysis
Synthesis
The Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2 analogues were synthesized manually by Fmoc-SPPS and characterized using methods described in Doedens et al.41 All physicochemical data for each new peptide is available in Table 1. All peptides were HPLC-purified using a ODS-A C18 (120 A, 5 µm, 250 mm × 20 mm) column (Omnicrom YMC Europe GmbH). For purification, a linear gradient of acetonitrile and water containing 0.1% TFA was used. All peptides when purified exhibited ≥95% purity, as analyzed by analytical HPLC and HPLC-MS.
Biological Activity Assays
Receptor Binding Assay
Competition binding experiments were carried out using both HEK 293 and A375 melanoma (ATCC) cell lines. HEK 293 cells stably expressed human MC1, MC3, MC4, and MC5 receptors as described before. HEK 293 cells transfected with hMCRs were seeded on 96-well plates 48 h before the assay was performed (50 000 cells/well). For the assay, the cell culture medium was aspirated and the cells were washed once with freshly prepared minimum essential medium (MEM) containing 100% minimum essential medium with Earle’s salt (MEM, GIBCO) and 25 mM sodium bicarbonate. Next, the cells were incubated for 40 min at 37 °C with different concentrations of unlabeled peptide and labeled [125I]-[Nle4,d-Phe7]-α-MSH (PerkinElmer Life Science, 20 000 cpm/well, 33.06 pM) diluted in 125 µL of freshly prepared binding buffer containing 100% MEM, 25 mM HEPES (pH 7.4), 0.2% bovine serum albumin, 1 mM 1,10-phenanthrolone, 0.5 mg/L leupeptin, and 200 mg/L bacitracin. The assay medium was subsequently removed, and the cells were washed once with basic medium and then lysed by the addition of 100 µL of 0.1 M NaOH and 100 µL of 1% Triton X-100. The total labeled [125I]-[Nle4,d-Phe7]-α-MSH detected in lysed cells was measured by a Wallac Micro β-TriLux 1450 LSC and luminescence counter (PerkinElmer Life Science, Boston, MA) (Table 2).
The binding studies with melanoma cells were done using the same methodology, and the results are presented in Table 4.
Adenylate Cyclase Assay
HEK 293 cells transfected with human melanocortin receptors were grown to confluence in MEM medium (GIBCO) containing 10% fetal bovine serum, 100 units/mL penicillin and streptomycin, and 1 mM sodium pyruvate. The cells were seeded on 96-well plates 48 h before the assay was performed (50 000 cells/well). For the assay, the cell culture medium was removed and the cells were rinsed with 100 µL of MEM buffer (GIBCO). An aliquot (100 µL) of Earle’s balanced salt solution with 5 nM isobutylmethylxanthine (IBMX) was placed in each well for 1 min at 37 °C. Next, aliquots (25 µL) of melanotropin peptides of varying concentration were added, and the cells were incubated for 3 min at 37 °C. The reaction was stopped by aspirating the assay buffer and adding 60 µL of ice-cold Tris/EDTA buffer to each well followed by placing the plates in a boiling water bath for 7 min. The cell lysates were then centrifuged for 10 min at 2300g. A 50 µL aliquot of the supernatant was transferred to another 96-well plate, and placed, along with 50 µL of [3H] cAMP and 100 µL of protein kinase A (PKA) buffer, in an ice bath for 2–3 h. The PKA buffer consisted of Tris/EDTA buffer with 60 µg/mL PKA and 0.1% bovine serum albumin by weight. The incubation mixture was filtered through 1.0 µm glass fiber filters in MultiScreen-FB 96-well plates (Millipore, Billerica, MA). The total [3H] cAMP was measured by a Wallac Micro β-TriLux 1450 LSC and luminescence counter (PerkinElmer Life Science, Boston, MA). The cAMP accumulation data for each peptide analogue was determined with the help of a cAMP standard curve generated by the same method as described above. The maximal cAMP produced at 10 µM of each ligand was compared to the amount of cAMP produced at 10 µM of the standard agonist, Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II), and is expressed as a percentage (% max effect) in Table 3. The antagonist properties of the lead compounds were evaluated by their ability to competitively displace the Ac-Nle4-c[Asp5,d-Phe7,Lys10]-α-MSH(4–10)-NH2 (MT-II) agonist in a dose-dependent manner at concentrations up to 10 µM.
The cAMP assays for the melanoma cell line were done using the same methods as above, and the results are given in Table 4.
Data Analysis
IC50 and EC50 values represent the mean of two experiments performed in triplicate. IC50 and EC50 estimates and their associated standard errors were determined by fitting the data using a nonlinear least-squares analysis with GraphPad Prism 5 (GraphPad Software, San Diego, CA). pA2 analysis was done with a Schild plot followed by the cAMP assay.
ACKNOWLEDGMENTS
These studies were supported in part by grants from the U.S. Public Health Service, National Institutes of Health, DK017420, GM 108040, and DA06284. We thank IGSSE (International Graduate School of Science and Engineering), the Bund der Freunde der TU München e.V., CompInt (Materials Science of Complex Interfaces) of the Elite Network of Bavaria for funding, IAS (Institute for Advanced Study) of Technische Universität München, CIPSM (Center for Integrated Protein Science Munich), KAU (Saudi Arabia), and Deutsche Forschungsgemeinschaft for a Koselleck grant.
ABBREVIATIONS USED
Abbreviations used for amino acids and designation of peptides follow the rules of the IUPAC-IUB Commission of Biochemical Nomenclature, J. Biol. Chem., 1972, 247, 977–983;
- Boc
tert-butyloxycarbonyl
- Fmoc
fluorenylmethoxycarbonyl
- CH3CN
acetonitrile
- Cl-HOBt
1-hydroxy-6-chlorobenzotriazole
- DCM
dichloromethane
- DIPEA
diisopropylethylamine
- DMF
N,N-dimethylformamide
- DIC
diisopropyl carbodiimide
- HOBt
N-hydroxybenzotriazole
- hMCR
human melanocortin receptor
- MSH
melanocyte-stimulating hormone
- Nal(2’)
2′-naphthylalanine
- Pbf
2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl
- TFA
trifluoroacetic acid
- SPPS
solid-phase peptide synthesis
- RP-HPLC
reverse-phase high-performance liquid chromatography
- Ac-Nle4-c[Asp5,d-Phe7,Lys10]-NH2
Ac-Nle-c[Asp-His-d-Phe-Arg-Trp-Lys]-NH2 (cyclic analogue of α-MSH)
- Ac-Nle4-c[Asp5,d-Nal(2′)7,Lys10]-NH2
Ac-Nle-c[Asp-His-d-Nal(2′)-Arg-Trp-Lys]-NH2 (cyclic analogue of α-MSH)
- GPCR
G protein coupled receptor
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Gantz I, Fong TM. The melanocortin system. Am. J. Physiol. Endocrinol. Metab. 2003;284:468–474. doi: 10.1152/ajpendo.00434.2002. [DOI] [PubMed] [Google Scholar]
- 2.Cone RD, editor. The Melanocortin System. Vol. 994. New York: New York Academy of Sciences; 2003. pp. 1–383. [Google Scholar]
- 3.Cone RD. Studies on the physiological functions of the melanocortin system. Endocr. Rev. 2006;27:736–749. doi: 10.1210/er.2006-0034. [DOI] [PubMed] [Google Scholar]
- 4.Getting SJ. Targeting melanocortin receptors as potential novel therapeutics. Pharmacol. Ther. 2006;111:1–15. doi: 10.1016/j.pharmthera.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 5.Hadley ME, Dorr RT. Melanocortin peptide therapeutics: Historical milestones, clinical studies and commercialization. Peptides. 2006;27:921–930. doi: 10.1016/j.peptides.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 6.Wikberg JE, Mutulis F. Targeting melanocortin receptors: an approach to treat weight disorders and sexual dysfunction. Nat. Rev. Drug Discovery. 2008;7:307–323. doi: 10.1038/nrd2331. [DOI] [PubMed] [Google Scholar]
- 7.Cai M, Nyberg J, Hruby VJ. Melanotropins as drugs for the treatment of obesity and other feeding disorders: potential and problems. Curr. Top. Med. Chem. 2009;9:554–563. doi: 10.2174/156802609788897817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadley ME. The Melanotropic Peptides. I–III. Boca Raton, FL: CRC Press; 1988. [Google Scholar]
- 9.Cone RD, editor. The Melanocortin Receptors. Totowa, NJ: Humana Press; 2000. [Google Scholar]
- 10.Brzoska T, Böhm M, Lüering A, Loser K, Luger TA. Terminal signal: anti-inflammatory effects of α-melanocyte-stimulating hormone related peptides beyond the pharmacophore. Adv. Exp. Med. Biol. 2010;681:107–116. doi: 10.1007/978-1-4419-6354-3_8. [DOI] [PubMed] [Google Scholar]
- 11.Muceniece R, Dambrova M. Melanocortins in brain inflammation: the role of melanocortin receptor subtypes. Adv. Exp. Med. Biol. 2010;681:61–70. doi: 10.1007/978-1-4419-6354-3_5. [DOI] [PubMed] [Google Scholar]
- 12.Wessells H, Fuciarelli K, Hansen J, Hadley ME, Hruby VJ, Dorr R, Levine N. Synthetic melanotropic peptide initiates erections in men with psychogenic erectile dysfunction: double-blind, placebo controlled crossover study. J. Urol. 1998;160:389–393. [PubMed] [Google Scholar]
- 13.King SH, Mayorov AV, Balse-Srinivasan P, Hruby VJ, Vanderah TW, Wessells H. Melanocortin receptors, melanotropic peptides and penile erection. Curr. Top. Med. Chem. 2007;7:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- 14.Ni XP, Butler AA, Cone RD, Humphreys MH. Central receptors mediating the cardiovascular actions of melanocyte stimulating hormones. J. Hypertens. 2006;24:2239–2246. doi: 10.1097/01.hjh.0000249702.49854.fa. [DOI] [PubMed] [Google Scholar]
- 15.Li SJ, Varga K, Archer P, Hruby VJ, Sharma SD, Kesterson RA, Cone RD, Kunos G. Melanocortin antagonists define two distinct pathways of cardiovascular control by alpha- and gamma-melanocyte-stimulating hormones. J. Neurosci. 1996;16:5182–5188. doi: 10.1523/JNEUROSCI.16-16-05182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature. 1997;385:165–168. doi: 10.1038/385165a0. [DOI] [PubMed] [Google Scholar]
- 17.Vergoni AV, Poggioli R, Bertolini A. Corticotropin inhibits food intake in rats. Neuropeptides. 1986;7:153–158. doi: 10.1016/0143-4179(86)90091-0. [DOI] [PubMed] [Google Scholar]
- 18.Vergoni AV, Poggioli R, Marrama D, Bertolini A. Inhibition of feeding by ACTH-(1–24): behavioral and pharmacological aspects. Eur. J. Pharmacol. 1990;179:347–355. doi: 10.1016/0014-2999(90)90175-6. [DOI] [PubMed] [Google Scholar]
- 19.Ellacott KLJ, Halatchev IG, Cone RD. Interactions between gut peptides and the central melanocortin system in the regulation of energy homeostasis. Peptides. 2006;27:340–349. doi: 10.1016/j.peptides.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 20.Butler AA. The melanocortin system and energy balance. Peptides. 2006;27:281–290. doi: 10.1016/j.peptides.2005.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang YK, Harmon CM. Recent developments in our understanding of melanocortin system in the regulation of food intake. Obes. Rev. 2003;4:239–248. doi: 10.1046/j.1467-789x.2003.00104.x. [DOI] [PubMed] [Google Scholar]
- 22.Morgan C, Thomas RE, Cone RD. Melanocortin-5 receptor deficiency promotes defensive behavior in male mice. Horm. Behav. 2004;45:58–63. doi: 10.1016/j.yhbeh.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Morgan C, Thomas RE, Ma W, Novotny MV, Cone RD. Melanocortin-5 receptor deficiency reduces a pheromonal signal for aggression in male mice. Chem. Senses. 2004;29:111–115. doi: 10.1093/chemse/bjh011. [DOI] [PubMed] [Google Scholar]
- 24.Mogil JS, Wilson SG, Chesler EJ, Rankin AL, Nemmani KV, Lariviere WR, Groce MK, Wallace MR, Kaplan L, Staud R, Ness TJ, Glover TL, Stankova M, Mayorov A, Hruby VJ, Grisel JE, Fillingim RB. The melanocortin-1 receptor gene mediates female-specific mechanisms of analgesia in mice and humans. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4867–4872. doi: 10.1073/pnas.0730053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalange AS, Kokare DM, Singru PS, Upadhya MA, Chopde CT, Subhedar NK. Central administration of selective melanocortin 4 receptor antagonist HS014 prevents morphine tolerance and withdrawal hyperalgesia. Brain Res. 2007;1181:10–20. doi: 10.1016/j.brainres.2007.08.054. [DOI] [PubMed] [Google Scholar]
- 26.Vrinten DH, Kalkman CJ, Adan RA, Gispen WH. Neuropathic pain: a possible role for the melanocortin system? Eur. J. Pharmacol. 2001;429:61–69. doi: 10.1016/s0014-2999(01)01306-1. [DOI] [PubMed] [Google Scholar]
- 27.Van der Ploeg LH, Martin WJ, Howard AD, Nargund RP, Austin CP, Guan X, Drisko J, Cashen D, Sebhat I, Patchett AA, Figueroa DJ, DiLella AG, Connolly BM, Weinberg DH, Tan CP, Palyha OC, Pong SS, MacNeil T, Rosenblum C, Vongs A, Tang R, Yu H, Sailer AW, Fong TM, Huang C, Tota MR, Chang RS, Stearns R, Tamvakopoulos C, Christ G, Drazen DL, Spar BD, Nelson RJ, MacIntyre DE. A role for the melanocortin 4 receptor in sexual function. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11381–11386. doi: 10.1073/pnas.172378699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellacott KLJ, Cone RD. The central melanocortin system and the integration of short- and long-term regulators of energy homeostasis. Recent Prog. Horm. Res. 2004;59:395–408. doi: 10.1210/rp.59.1.395. [DOI] [PubMed] [Google Scholar]
- 29.Marks DL, Hruby VJ, Brookhart G, Cone RD. The regulation of food intake by selective stimulation of the type 3 melanocortin receptor (MC3R) Peptides. 2006;27:259–264. doi: 10.1016/j.peptides.2005.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowley MA, Smart JL, Rubinstein M, Cerdán MG, Diano S, Horvath TL, Cone RD, Low MJ. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 31.Chen W, Kelly MA, Opitz Araya X, Thomas RE, Low MJ, Cone RD. Exocrine gland dysfunction in MC5-R-deficient mice: evidence for coordinated regulation of exocrine gland function by melanocortin peptides. Cell. 1997;91:789–798. doi: 10.1016/s0092-8674(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y. Structure, function and regulation of the melanocortin receptors. Eur. J. Pharmacol. 2011;660:125–130. doi: 10.1016/j.ejphar.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooray SN, Clark AJ. Melanocortin receptors and their accessory proteins. Mol. Cell. Endocrinol. 2011;331:215–221. doi: 10.1016/j.mce.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee J, Gilon C, Hoffman A, Kessler H. N-methylation of peptides: a new perspective in medicinal chemistry. Acc. Chem. Res. 2008;41:1331–1342. doi: 10.1021/ar8000603. [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee J, Laufer B, Kessler H. Synthesis of N-methylated cyclic peptides. Nat. Protoc. 2012;7:432–444. doi: 10.1038/nprot.2011.450. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee J, Rechenmacher F, Kessler H. N-methylation of peptides and proteins: an important element for modulating biological functions. Angew. Chem. Int. Ed. 2013;52:254–269. doi: 10.1002/anie.201205674. [DOI] [PubMed] [Google Scholar]
- 37.Biron E, Chatterjee J, Ovadia O, Langenegger D, Brueggen J, Hoyer D, Schmid H, Jelinek R, Gilon C, Hoffman A, Kessler H. Improving oral bioavailability of peptides by multiple N-methylation: somatostatin analogues. Angew. Chem. Int. Ed. 2008;47:2595–2599. doi: 10.1002/anie.200705797. [DOI] [PubMed] [Google Scholar]
- 38.White TR, Renzelman CM, Rand AC, Rezai T, McEwen CM, Gelev VM, Turner RA, Linington RG, Leung SS, Kalgutkar AS, Bauman JN, Zhang Y, Liras S, Price DA, Mathiowetz AM, Jacobson MP, Lokey RS. On-resin N-methylation of cyclic peptides for discovery of orally bioavailable scaffolds. Nat. Chem. Biol. 2011;7:810–817. doi: 10.1038/nchembio.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ovadia O, Greenberg S, Chatterjee J, Laufer B, Opperer F, Kessler H, Gilon C, Hoffman A. The effect of multiple N-methylation on intestinal permeability of cyclic hexapeptides. Mol. Pharmaceutics. 2011;8:479–487. doi: 10.1021/mp1003306. [DOI] [PubMed] [Google Scholar]
- 40.Beck JG, Chatterjee J, Laufer B, Kiran MU, Frank AO, Neubauer S, Ovadia O, Greenberg S, Gilon C, Hoffman A, Kessler H. Intestinal permeability of cyclic peptides: common key backbone motifs identified. J. Am. Chem. Soc. 2012;134:12125–12133. doi: 10.1021/ja303200d. [DOI] [PubMed] [Google Scholar]
- 41.Doedens L, Opperer F, Cai M, Beck JG, Dedek M, Palmer E, Hruby VJ, Kessler H. Multiple N-methylation of MT-II backbone amide bonds leads to melanocortin receptor subtype hMC1R selectivity: pharmacological and conformational studies. J. Am. Chem. Soc. 2010;132:8115–8128. doi: 10.1021/ja101428m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Obeidi F, Hruby VJ, de Lauro Castrucci A-M, Hadley ME. Design of potent linear α-melantropin 4–10 analogues modified in positions 5 and 10. J. Med. Chem. 1989;32:174–179. doi: 10.1021/jm00121a032. [DOI] [PubMed] [Google Scholar]
- 43.Hruby VJ, Lu D, Sharma SD, de L, Castrucci A, Kesterson RA, al-Obeidi FA, Hadley ME, Cone RD. Cyclic lactam α-melanotropin analogues of Ac-Nle4-c[Asp4,D-Phe7, Lys10] α-MSH(4–10)-NH2with bulky aromatic amino acids at position 7 show high antagonist potency and selectivity at specific melanocortin receptors. J. Med. Chem. 1995;38:3454–3461. doi: 10.1021/jm00018a005. [DOI] [PubMed] [Google Scholar]