Abstract
TfoX (Sxy) and CRP are two important competence activators. The link between tfoX and CRP has been shown in H. influenza but lacking evidence of direct interaction. Recently a Sxy-dependent CRP (CRP-S) site autoregulating Sxy was reported in E. coli. Here, we show that the cAMP-CRP complex transcriptionally regulates tfoX expression through multiple canonical CRP (CRP-N) sites in Vibrios. This conclusion is supported by an analysis of the tfoX mRNA levels and tfoX transcriptional reporter fusions. The reduced expression of tfoXVC was restored by trans-complementation of crp in ∆crp and by exogenous cAMP in ∆cya. A promoter deletion analysis and the site-directed mutagenesis of the putative CRP-N sites revealed the presence of two functional CRP-N sites. The direct binding of cAMP-CRP to the tfoXVCpromoter was demonstrated by EMSA assays. Additionally, the transcriptional start site (TSS) of tfoXVF in V. fluvialis was determined, and −10/−35 regions were predicted. Further comparison of the tfoX promoter in Vibrios revealed the existence of similar −10 motifs and putative CRP-N sites, indicating the conserved mechanism of CRP regulation on tfoX. Our study demonstrates the direct binding of the cAMP-CRP complex to tfoX promoter, and broadens the understanding of the molecular mechanism regulating tfoX in Vibrios.
Many bacterial species can take up environmental DNA under natural conditions, most of which is degraded in the cytoplasm for reuse. However, the DNA that escapes degradation might be incorporated into the host chromosomes by RecA-dependent homologous recombination1,2,3,4,5,6. This is a very complicated phenomenon with regard to its biological function, machinery components and the regulated and regulatory genes involved. In general, the process requires certain cellular structures, like type IV pili or related pseudopili, and many competence-related proteins, and interferes with other regulation systems, such as carbon catabolite repression (CCR), quorum sensing and nucleotide scavenging systems7,8,9. Although many advances have been made in this field, a clear picture of the molecular details of natural competence remains to be revealed.
Most of our understanding of the genetics and molecular mechanisms of competence originally came from studies of H. influenzae3,10. Nutrient starvation and the depletion of nucleotide pools induce natural competence/transformation in these bacteria3,8. The competence regulon of H. influenzae is characterized by a promoter-associated 22 bp competence regulatory element (CRE, 5′-T4G5C6G7A8–(N6)–T15C16G17C18A19-3′) that is closely related to the cAMP receptor protein (CRP) binding consensus (5′-T4G5T6G7A8–(N6)–T15C16A17C18A19-3′)10. The sequence has C and G instead of T and A at the highly conserved symmetrical positions 6 and 17, which are positions where CRP bends DNA11. The presence of cAMP and Sxy allows CRP to bind at CRE sites to activate the transcription of competence genes, and the affinity of these sites for CRP binding was experimentally demonstrated8,10,12. The terms CRP-S and CRP-N were respectively introduced to designate the new Sxy-dependent and canonical (Sxy-independent) CRP sites12.
In contrast to the naturally competent H. influenzae, Vibrio cholerae, the causative agent of cholera epidemics, was not known to be competent until Meibom et al. published their study in 2005 5. That study first demonstrated that chitin, a polymer of β-1,4-linked N-acetylglucosamine (GlcNAc), induces natural competence in V. cholerae5. In V. cholerae, competence development is triggered by a chitin-induced transcription regulator, TfoxVC, an ortholog of Sxy in H. influenzae, and a quorum-sensing regulator, HapR5. V. cholerae have counterparts of most H. influenzae Sxy-dependent, CRP-regulated genes12, and its competence phenotype is subject to catabolite repression5, which is positively regulated by two environmental signals, nutritional stress5 and nucleoside scavenging, similar to H. influenzae7. A natural competence phenotype was subsequently demonstrated to be a shared trait in other Vibrio species, such as V. fischeri13, V. vulnificus14,15, and V. parahaemolyticus16.
Sxy (TfoX) and CRP are two major shared activators that control the development of competence in the families Pasteurellaceae, Enterobacteriaceae, and Vibrionaceae12. Both sxy (tfoX) or crp/cya knockout mutants are unable to become competent17,18,19. Due to its vital role in regulating natural competence, the regulatory mechanism involving sxy (tfoX) has attracted increasing attentions from researchers. The secondary structure of sxy mRNA greatly increases its translational efficiency and promotes competence development in H. influenzae20. Besides chitin, a polymer of β-1,4-linked N-acetylglucosamine (GlcNAc), chitin disaccharide (GlcNAc)2 was also shown to be a minimum inducer of tfoX expression at both the transcriptional and translational levels in V. cholerae, and the corresponding cis-acting elements were previously identified21. A small regulator RNA, tfoR, activates the translation of tfoX in response to the presence of (GlcNAc)2 or chitin22.
CRP is one of major players in carbon catabolite repression (CCR)19, which allows bacteria to quickly respond to environmental changes by repressing genes responsible for the uptake, metabolism and assimilation of less favorable carbon sources when rapidly metabolizable carbohydrates, such as glucose, are present19,23,24. It has been shown that CRP plays a crucial role in the V. cholerae life cycle by regulating the expression of virulence factors (i.e., cholera toxin and toxin co-regulated pilus)25, affecting exopolysaccharide biosynthesis and rugose colonial morphology26, quorum sensing, motility, and multiple genes required for the survival of V. cholerae in the human host and the environment27,28.
The necessity of CRP and cAMP for chitin-induced competence was proven based on their interactions in the three interlinked aspects: they are required for the efficient colonization of the chitin surface, contribute to chitin degradation and utilization, and increase the expression of competence genes19. Our previous global gene expression profile based on a microarray analysis revealed that tfoX was down-regulated in a ∆crp mutant (Supplemental data)27, but this was not confirmed. In the present study, we add new information about the positive regulation of CRP on the development of competence by demonstrating the positive transcriptional regulation and direct binding of the cAMP-CRP complex on the tfoX promoter region in both V. cholerae and V. fluvialis, the latter of which is an emerging foodborne pathogen implicated in outbreaks and sporadic cases of cholera-like bloody diarrhea, and causes increasing public health concerns29,30. Many aspects of the ecology, epidemiology and pathogenesis of V. fluvialis remain to be investigated.
Results
Effect of CRP on tfoX gene expression
Yamamoto’s works greatly improved the knowledge of the regulation of tfoXVC in V. cholerae by identifying chitin disaccharide (GlcNAc)2 as the minimum inducer of chitin-dependent competence, by mapping the TSS, and the transcriptional and translational cis-acting elements, and by distinguishing the sRNA tfoR regulation21,22. Using this knowledge, combined with the prior clues provided by microarray data27, we reexamined the promoter region of tfoXVC and found a potential CRP-N binding site centered at −84.5 relative to the TSS, which contained a perfect TGTGA half-site and a 4/5 matched TCTCA half-site for the DNA binding domains of the active CRP dimer. These findings strongly suggested the possibility that CRP regulates tfoXVC expression.
To determine whether CRP was involved in the expression of tfoXVC, we examined the tfoXVC mRNA level in wild type and isogenic mutant crp via qRT-PCR (Fig. 1A). Deletion of the crp gene resulted in much lower expression of tfoXVC, which confirmed our microarray data. For further confirmation, we performed a complementation test by introducing plasmid pBADCRP7, which expresses biologically active CRP protein from the araBAD promoter. As shown in Fig. 1A, the tfoXVC expression was restored to a much higher level than the WT level in the pBADCRP7-complemented crp mutant. As expected, inclusion of the control vector did not restore the tfoXVC expression.
Figure 1. Effects of CRP on the tfoX gene expression.
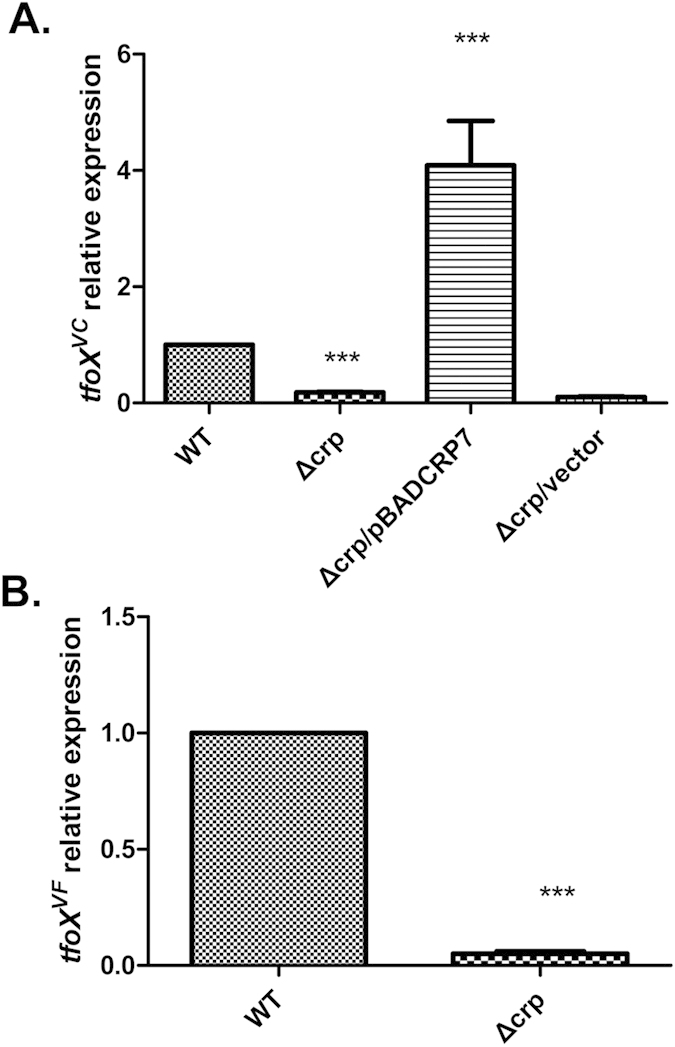
(A) V. cholerae strains C7258∆lacZ (WT), WL7258∆lacZ (∆crp), and WL7258∆lacZ containing the wild-type crp gene in trans and the control vector were grown in LB medium to late-log phase. (B) V. fluvialis strains 85003 (WT) and 85003∆crp (∆crp) were grown in LB medium, and cells were collected at the late-log phase. The tfoXVC and tfoXVF mRNA abundances were measured by qRT-PCR. The “WT” bar was set to 1 and used as a reference to calculate subsequent expression values. Error bars indicate the standard deviations of three independent cultures. ***Significantly different from the wild-type strain (t-test, P < 0.05).
To verify and expand the above finding that CRP was required for the expression of tfoX, we further examined the tfoX mRNA level in an emerging foodborne pathogen, V. fluvialis, and its isogenic crp mutant. The crp mutant was constructed by allelic exchange as described in the Materials and Methods section. Compared to the wild type pathogen, the tfoXVF mRNA level in the corresponding crp deletion mutant was significantly reduced (Fig. 1B). Taken together, these results indicate that the requirement of CRP for the expression of tfoX is a common feature in Vibrios. However, it was unclear whether this requirement is dependent on a direct interaction, or whether it may be a pleiotropic (or secondary) effect, because CRP is a global regulator and affects numerous genes.
Effects of cya mutation on the tfoX gene expression
To determine if the tfoXVC dependency on CRP can be fully accounted for by the cAMP binding to CRP, we measured the tfoXVC expression in WL7259 and WL7259 supplemented with 1.5 or 2.5 mM cAMP in the culture medium. As shown in Fig. 2, there was significantly lower expression of tfoXVC in the WL7259 lacking adenylate cyclase compared to the wildtype strain. Furthermore, the expression of tfoXVC was fully restored by supplementing the medium with exogenous cAMP. The activity of CRP is determined by the intracellular concentration of its allosteric activator, cAMP31. Consistent with this, the tfoXVC level in the strain with extra cAMP supplementation was more than 10-fold higher than that of the wild type strain. These results suggest that the expression of tfoXVC is under positive regulation by the cAMP-CRP complex. However, it was still unclear whether the dependence on both CRP and cAMP indicates the presence of a direct effect, and further evidence was required to clarify this point.
Figure 2. Effects of cya mutation on the tfoX gene expression.
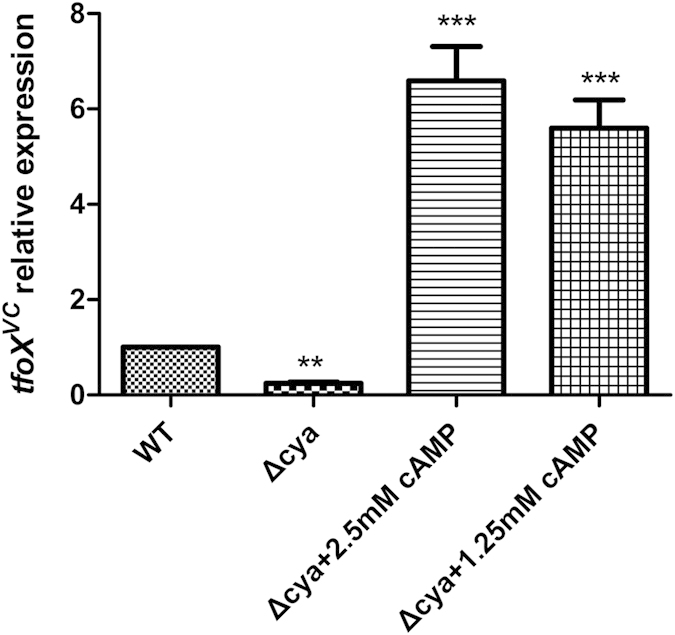
Overnight cultures of WL7259 (∆cya) were diluted 100-fold in LB and grown to OD600 0.5. Each culture was divided into three portions, and exogenous cAMP (Sigma Chemical Co.) was added to a final concentration of zero (control), 1.25, or 2.5 mM. The cultures were incubated at 37 °C for 1 h and the tfoXVC mRNA abundance was measured by qRT-PCR. The “WT” bar was set to 1 and used as a reference to calculate subsequent expression values. Error bars indicate the standard deviations of three independent cultures. **Significantly different from the wild-type strain (t-test, P < 0.05). ***Significantly different from the wild-type strain (t-test, P < 0.001).
CRP activates the transcription of tfoX
To determine if the CRP-mediated regulation of tfoX occurs at the level of transcription, we constructed transcriptional reporter plasmids by fusing the promoter regions of tfoXVC and tfoXVFto the reporter genes lacZ and luxCDABE, respectively. The −10 and −35 motifs of tfoXVC promoter have been determined in Yamamoto’s work21. The promoter of tfoXVF was predicted with sequence analogy of the tfoXVC promoter and TTS determined in our study (below part). In order to retain the full promoter activity, the sequences extending 408 bp and 381 bp upstream of tfoXVC and tfoXVF translational start sites were respectively amplified as promoter regions. The reporter constructs were transferred into the corresponding wild-type strains and the isogenic crp mutants. Significant differences in β-galactosidase expression (Fig. 3A) and luminescence activity (Fig. 3B) were detected between the WT and ∆crp containing the reporter constructs on both the V. cholerae and V. fluvialis backgrounds. Combined with the results about the tfoX mRNA expression level, these results strongly demonstrated that the promoter activity of tfoX is dependent on CRP in Vibrio species, i.e. CRP is directly required for tfoX expression.
Figure 3. Transcriptional fusion analysis of the cAMP–CRP regulation of tfoX expression.
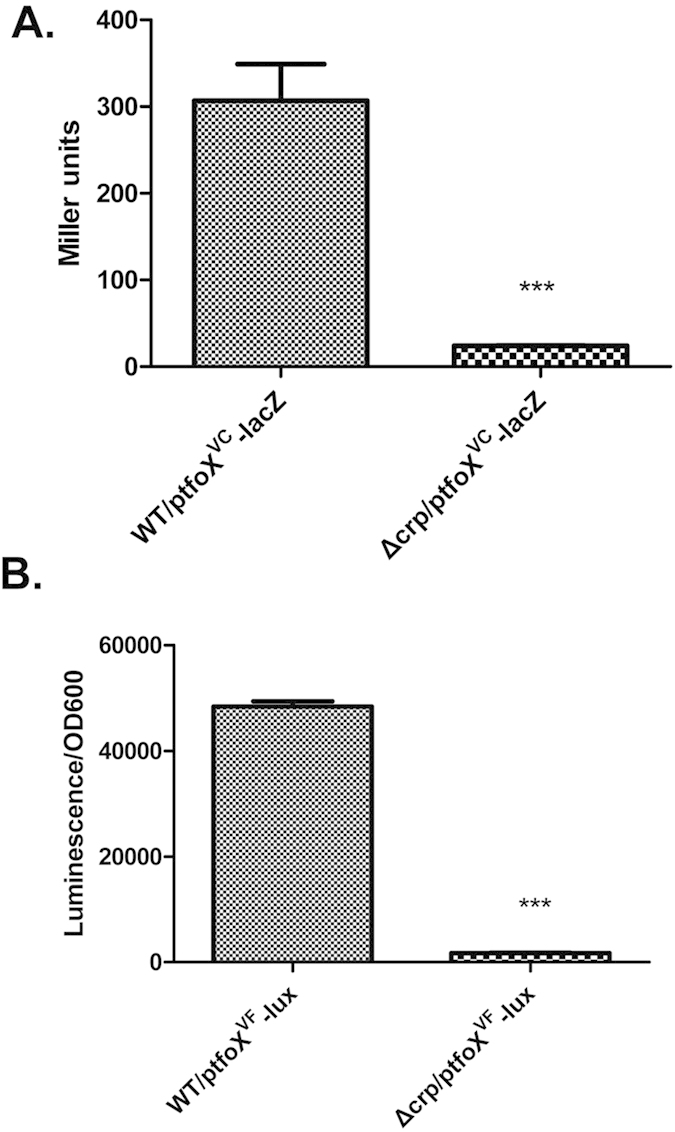
(A) The β-galactosidase expression in V. cholerae strains C7258∆lacZ (WT) and WL7258∆lacZ (∆crp) containing a ptfoXVC–lacZ transcriptional fusion. (B) The luminescence activity in V. fluvialis strains 85003 (WT) and 85003∆crp (∆crp) containing a ptfoXVF–lux transcriptional fusion. All strains were grown at 37 °C with shaking to the mid-log phase. The β-Galactosidase and bioluminescence activity were measured as described in the Methods. Error bars indicate the standard deviations of three independent cultures. ***Significantly different from the wild-type strain (t-test, P < 0.005).
Deletion analysis of the tfoX VC promoter region
To further delineate the cis DNA sequences in the tfoX promoter region required for CRP activation, we constructed additional transcriptional fusions containing 5′ deletions of the tfoXVC promoter (Fig. 4A). Plasmid p2tfoXVC-lacZ contained a DNA sequence extending from −74 bp upstream of the TSS, lacking the previously predicted CRP-binding sequence (TGTGA-N6-TCTCA); while p3tfoXVC-lacZ contained an even shorter sequence starting from the putative −35 element. As shown in Fig. 4B, p3tfoXVC-lacZ exhibited the lowest β-galactosidase activity, and no difference was observed between the WT and ∆crp. Surprisingly, plasmid p2tfoXVC-lacZ retained substantially high promoter activity, and a significant difference in β-galactosidase activity was still retained for the WT and ∆crp, indicating that the encompassed promoter region contains DNA sequences necessary for CRP activation.
Figure 4. Effects of putative CRP binding sites on the tfoXVC expression.
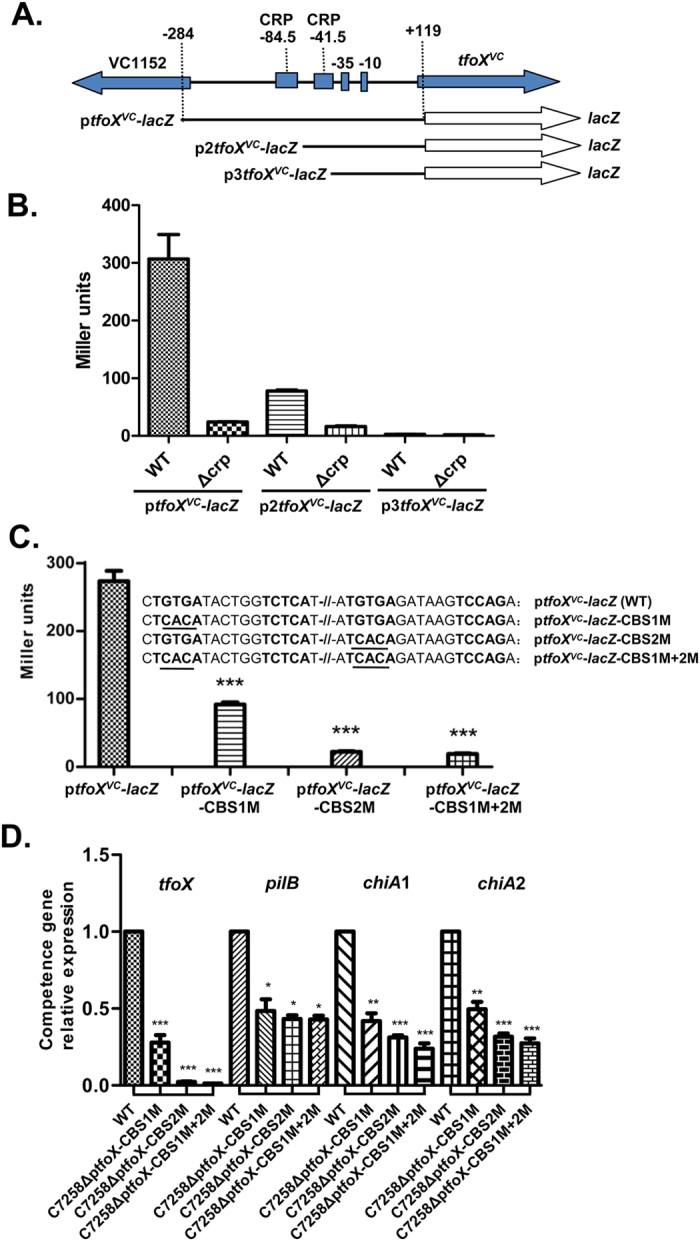
(A) The structural organizations of the tfoXVC promoter and transcriptional fusions. Two putative CRP binding sites were found in the upstream region of tfoXVC centered at positions −84.5 and −41.5. (B) A deletion analysis of the tfoXVC promoter. V. cholerae strains C7258∆lacZ (WT) and WL7258∆lacZ (∆crp) containing the p2tfoXVC–lacZ and p3tfoXVC–lacZ fusion, respectively, were grown at 37 °C to mid-log phase. (C) The promoter activities of wild-type and CBS mutated fusions. C7258∆lacZ containing ptfoXVC–lacZ, ptfoXVC-lacZ-CBS1M, ptfoXVC-lacZ-CBS2M, or ptfoXVC-lacZ-CBS1M+2M were grown at 37 °C to the mid-log phase. The β-galactosidase activity was measured as described in the Methods. The mutated bases in fusions were constructed by site-directed mutagenesis and underlined. (D) V. cholerae strains C7258 (WT), C7258∆ptfoX-CBS1M, C7258∆ptfoX-CBS2M and C7258∆ptfoX-CBS1M+2M were grown in LB medium to late-log phase. The tfoXVC and pilB, chiA-1 and chiA-2 mRNA abundances were measured by qRT-PCR. Error bars indicate the standard deviations of three independent cultures. The “WT” bar was set to 1 and used as a reference to calculate subsequent expression values. ***Significantly different from the wild-type strain (t-test, P < 0.0003). *Significantly different from the wild-type strain (t-test, P < 0.05).
Further inspection of the promoter region revealed another suboptimal CRP binding site (TGTGAGATAAGTCCAG), with three mismatches from the consensus motif on the one half-site (TCACA). This binding site is centered at −41.5, encompasses the predicted −35 hexamer, and thus extends into the region generally bound by the RNA polymerase (RNAP). Based on these results, we inferred that the tfoXVCpromoter resembles or belongs to the Class III CRP-dependent promoter, which requires multiple activator molecules for full transcriptional activation. In this case, it requires two CRP molecules. Specifically, one binding site is centered at position −84.5 as in the class I CRP-dependent promoters; the other is centered at −41.5, exactly like the class II CRP-dependent galP1 promoter32. The tandem binding of CRP causes synergistic effects on transcriptional activation, with the best effects resulting from the binding to a site situated 40 or 50 bp upstream33. The 43-bp spacer of the two binding sites (center-to-center distance) of tfoXVCseems to be an optimal distance that produces the best synergistic effects.
Site-directed mutagenesis of the CRP-binding site at the promoter of tfoX VC
The ptfoXVC-lacZ reporter fusion contains the full length functional tfoX promoter region so that it encompasses two putative CRP-binding sites which were designated as CBS1 and CBS2, respectively. To further verify and distinguish the roles of each CRP-binding site in tfoXVC transcription, we substituted the consensus GTG for ACA in the half-site of CBS1, CBS2, and both by site-directed mutagenesis (Fig. 4C). The promoter activities of the resultant mutant fusions ptfoXVC-lacZ-CBS1M, ptfoXVC-lacZ-CBS2M and ptfoXVC-lacZ-CBS1M+2M, were determined. As shown in Fig. 4C, the mutation of CBS1 resulted in a major decrease in the β-galactosidase activity of the ptfoXVC-lacZ-CBS1M fusion compared to the wild-type ptfoXVC-lacZ, which is consistent with the results of the p2tfoXVC-lacZ construct. As expected, ptfoXVC-lacZ-CBS1M+2M showed the lowest β-galactosidase activity due to the mutations of both CBS1 and CBS2 in the promoter region. Unexpectedly, mutation of CBS2 also results in a very low level of β-galactosidase activity in ptfoXVC-lacZ-CBS2M, similar to that observed for the double mutant fusion ptfoXVC-lacZ-CBS1M+2M. This result indicates that the CBS1 centered at −84.5 cannot efficiently initiate the transcription of the tfoXVCpromoter alone, and CBS2 is required for the synergistic activation of the tfoXVCpromoter by CBS1.
To further determine the mutation of either CBS or both in the tfoX promoter has an effect in competence development or on natural transformation in vivo, We subsequently introduced the site specific mutations to CBS1, CBS2 and both on the V. cholerae chromosomal tfoX promoter, and measured the mRNA levels of tfoX, and tfoX-induced genes, pilB, chiA1 and chiA2, which are all required for competence in V. cholerae5. As shown in Fig. 4D, the mRNA levels of tfoX, pilB, chiA1 and chiA2 in C7258∆ptfoX-CBS1M, C7258∆ptfoX-CBS2M and C7258∆ptfoX-CBS1M+2M were significantly reduced compare to the wildtype strain. These results coincide with the above results of ptfoXVC-lacZ-CBS mutant fusions and indicate the regulation of TfoX-dependent natural transformation by CRP through the CBS sites in vivo.
CRP activates transcription by directly interacting with RNAP and/or acting upon DNA to change its structure to facilitate RNAP binding34. At the class I promoter, CRP binds upstream of the promoter and increases the rate of initial binding of RNAP to form a closed promoter complex (RPc). At the class II promoter, CRP binds within the promoter to increase the rate of transition from the closed to open promoter complex (RPo), in which the DNA duplex becomes unwound around the −10 region and short RNA products are synthesized35. We speculate that the loss of the CBS2 site might cause a substantial decrease in the −35 and −10 motif binding by RNAP in the tfoXVC promoter and/or may decrease the efficiency of the transition from RPc to RPo. This will need to be investigated in the future studies. Another possibility is that an inverted repeat sequence (termed IR1), which was predicted to be a potential transcriptional operator21, overlaps the CBS2 site, so the site-directed mutagenesis of the consensus GTG to ACA in the CBS2 site could change the structure of IR1, thus affecting the transcription of tfoXVC. However, the present results demonstrate that the two putative CRP binding sites both play vital roles in the activation of tfoXVCexpression.
Determination of the TSS of tfoX VF and a comparative analysis of the tfoX promoter from different Vibrio species
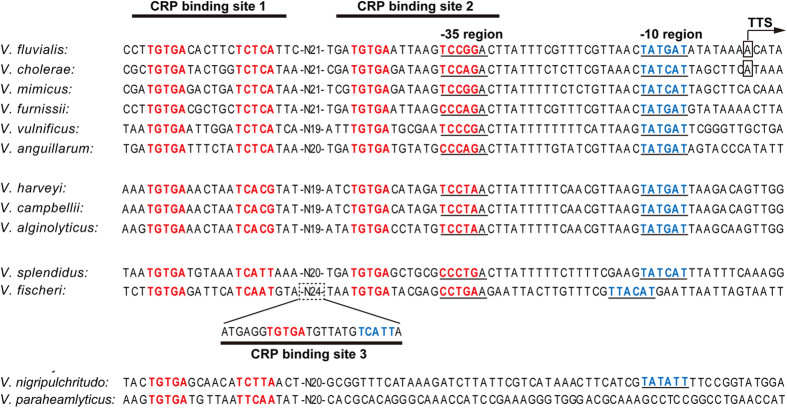
The molecular structural features, such as the −10 and −35 motifs, transcriptional and translational cis-acting elements, secondary structure of the tfoXVC mRNA, and TSS have already been determined21,22. The original annotated open reading frame (ORF) of tfoXVCwas 609 bp long, with GUG as the start codon and GGGA as the predicted Shine-Dalgarno (SD) sequence. Through a series of mutational analyses of potential start codons based on fusion-plasmid constructions, Yamamoto revised the start codon and SD sequence of tfoXVC to be ATG and GAAG, respectively, which are located 21 and 14 nucleotides downstream of the original sequences21,22. To assess the conservation of the molecular structural and regulatory characteristics of tfoX in different Vibrio species, we determined the TSS of tfoXVF and compared the tfoX promoters in different Vibrio species whose genome sequences are available from the NCBI database and for which the homologs of tfoX have been annotated.
The open reading frame (ORF) of tfoXVF shared 74.32% nucleotide identity with tfoXVC, but the intergenic regions showed much lower homology (55.87%). Using 5′ RACE, we identified a transcriptional start site 126 nucleotides upstream of the putative ATG start codon predicted according to the similarity to tfoXVC. Putative σ70- specific −10 (TATGAT) and −35 (TCCGGA) motifs separated by 19 bp were found in the upstream region of the TSS (Fig. 5), which coincides with the fact that competence genes are regulated by σ70 type promoters36. The −10 sequence has only one mismatch from the −10 motifs of E. coli (TATAAT) and tfoXVC(TATCAT). The −35 sequence has four mismatches from the E. coli consensus sequence TTGACA, but was a match at five of six nucleotides in the −35 region of tfoXVC(TCCAGA).
Figure 5. Structural organization of the tfoX promoter region.
The tfoX promoter-proximate sequences from V. fluvialis, V. cholerae, V. mimicus, V. furnissii, V. vulnificus, V. anguillarum, V. harveyi, V. campbellii, V. alginolyticus, V. splendidus, V. fischeri, V. nigripulchritudo and V. paraheamlyticus were compared. The CRP binding sites are indicated with horizontal lines and the invert repeats in the CRP binding box are in boldface type. The putative −10 and/or −35 elements are underlined. The TTS of V. fluvialis and V. cholerae is designated with a box. The putative third CRP binding site located in the 24 bp spacing sequences in V. fischeri is also shown.
As shown in Fig. 5, conserved −10 motifs and two putative CRP-binding sites were found around similar locations in the tfoX promoter in V. fluvialis, V. cholerae, V. mimicus, V. furnissii, V. vulnificus, V. anguillarum, V. harveyi, V. campbellii, V. alginolyticus, and V. splendidus. The −35 regions were less conserved. This was not unexpected, because no CRP-dependent promoter has a good −35 sequence, and some even lack good −10 sequences37. The center-to-center distances between the two putative CRP binding sites were 43, 42 or 41 bp in different species. Three putative CRP-binding sites were discernible in the promoter region of tfoX in V. fischeri. Notably, the spacer sequence between the two half-sites of CRP binding site 3 in V. fischeri was seven bases, rather than the conventional six bases present in standard CRP half-sites. In general, the distal CRP-binding sites are more conserved and can roughly be classified into four classes based on the sequence homology, although the corresponding regions in V. nigripulchritudo and V. paraheamlyticus display higher variation. It is difficult to predict the putative −35 and/or −10 elements in similar locations in these species compared with the above species, and only one potential CRP binding site was found. The different numbers of binding sites and the variations in the binding sequences may contribute to the fine-tuning of tfoX expression in response to the changes in the environment, and may reflect dynamic evolutionary histories of the acquisition and or loss of CRP-binding sites.
CRP binds directly to the tfoX promoter in vitro
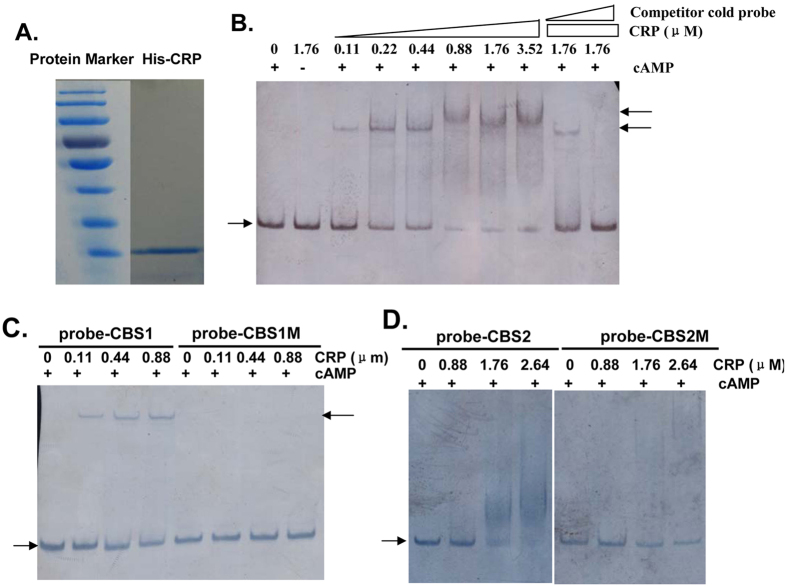
All of the above-described results suggest that tfoX expression is regulated at the transcriptional level via cAMP-CRP complex-mediated activation, and there are two cAMP-CRP binding sites at the tfoX promoter region. To verify whether there is cAMP-CRP binding to the putative sites, we measured the binding of purified CRP to a 152-bp DNA fragment of tfoXVC encompassing the two CRP binding sites. The fragment was labeled with biotin at the 5′ end and used as a probe. As shown in Fig. 6B, the addition of CRP (0.11 μM) resulted in a shift of the 152-bp DNA fragment to slower mobility. When the amount of CRP was increased (>0.88 μM), the shifted band slowed to an even higher position. The binding was abolished when CRP or cAMP was excluded from the reaction mixture. Inclusion of the same, but unlabelled, 152-bp DNA fragment greatly competed with the labeled probe for the binding sites in a dose-dependent manner. The addition of 100-fold of the competitor cold probe changed the higher-shifted band to the lower one, while the addition of 300-fold of the cold probe completely abolished the retarded-band, and the labeled probe in the reaction mixture was released as the free probe. On one hand, these results confirmed the specific binding of CRP to the tfoXVCpromoter, and on the other hand, suggested that one binding site exhibits lower-affinity binding. This is in agreement with the fact that the proximal CBS2 differs from the consensus sequence in three conserved positions, while CBS1 differs from the consensus sequence in only one conserved position, thus resulting in different binding affinity.
Figure 6. Binding of the cAMP-CRP complex to the promoter region of tfoXVC.
EMSA were performed as described in the Methods section to determine whether there was a direct interaction between cAMP-CRP and the promoter region of tfoXVC. The arrow on the left side indicates the unbound free probe, whereas the arrow on the right side indicates the probe bound with CRP. (A) The purity of His-CRP analyzed with SDS-PAGE. (B) A biotin-labeled 152-bp DNA probe containing two CRP binding sites (10 ng) was incubated with increasing amounts of CRP in the presence of cAMP (0.1 mM). For the competition analysis, the same, but unlabeled, 152-bp DNA was included as 100-fold and 300-fold concentrations relative to the labeled probes. (C) The biotin-labeled 75-bp DNA fragments containing the original distal CRP binding site (probe-CBS1) or the corresponding mutagenized binding site (probe-CBS1M) were used as probes in a gel shift assay. (D) The biotin-labeled 97-bp DNA fragments containing the original proximal CRP binding site (probe-CBS2) or the corresponding mutagenized binding site (probe-CBS2M) were used as probes in the gel shift assay.
For further confirmation, we performed EMSA with the fragments containing single CRP binding sites. Consistent with the more conserved features of CBS1 and the EMSA results described above, the 75 bp fragment with CBS1 was shifted to a single band with slower mobility following the addition of 0.11 μM of CRP, while mutagenesis of the conserved GTG completely abolished the binding (Fig. 6C). While the 97 bp fragment with CBS2 was not shifted even when up to 0.88 μM of CRP was added, smeared bands were formed when more than 1.76 μM CRP were added in the assay mixture, which may indicate the instability of the DNA-CRP complex formed in vitro due to lower affinity of binding (Fig. 6D). Although the EMSA assays indicated that CBS2 is a low affinity binding site, the site-directed mutagenesis analysis demonstrated that this binding site also plays a vital role in the initiation of the transcription of tfoXVC. It should also be kept in mind that it is impossible to rule out the possibility that the low affinity binding of CBS2 was due to the in vitro binding conditions in the EMSA assay, which do not exactly mimic the in vivo condition.
With V. cholerae CRP, which has a high amino acid identity (99.05%) to the CRP in V. fluvialis, we also found the direct and specific binding of CRP to the tfoXVF promoter in an EMSA assay (data not shown), further demonstrating the conserved direct regulatory effects of the cAMP-CRP complex on the competence regulator tfoX in Vibrio species.
Discussion
Competence development for DNA uptake by microorganisms is tightly regulated, and cAMP-CRP plays a central role in competence regulation in response to nutritional stress. The direct binding of CRP to the CRE site on the promoter region of competence-associated genes was predicted8 and experimentally confirmed10. The second essential regulator of competence, TfoX, was proposed to act cooperatively with CRP by directing it to CRE sites to allow maximal expression of the competence genes10, although it lacks the helix-turn-helix DNA binding motif12 and remains recalcitrant to overexpression and purification8. Recently Jaskolska and coworker demonstrated that TfoX is unstable and degraded by Lon protease in E.coli38. Previously, Zulty & Barcak39 and Cameron et al.20 showed a strong induction of sxy expression after the addition of 1 mM cAMP to H. influenzae, although evidence for direct binding is not yet available. Additionally, the opposite result was also reported, i.e., cya mutation has no effect on sxy expression10. While we were in the preparation of the manuscript, Jaskolska et al. published their data showing that in E. coli, Sxy is positively autoregulated at the transcriptional level by a mechanism requiring cAMP-CRP and the CRP-S site in the sxy promoter38. It is interesting that Vibrio species possess the CRP-N sites instead of CRP-S sites in the tfoX promoter regions. Experimental evidences have demonstrated that the CRP-N sites are higher affinity sites than the CRP-S sites10,12,36. The presence of CRP-N sites in tfoX could start higher expression of TfoX and in turn more efficiently induce the development of competence, thus to favor the survival of Vibrio species in the aquatic system which is normally nutrition-limited. This finding further indicates that though the members of gamma-proteobacteria families, such as Enterobacteriaceae, Pseudomonadaceae and Vibrionaceae share a common regulatory mechanism in competence development10,12, the divergence or variation in details exists which may facilitate to refine the regulation in different genetic backgrounds and or survival environments. We reasoned that this may due to the long-term evolution pressure selection between the different native habitant environments.
In V. cholerae, chitin induces natural competence5, and cAMP and CRP are required for efficient chitin colonization, degradation and increased competence gene expression19. The regulation of tfoX by CRP, particularly in Vibrio species, had not been established or investigated. In the present study, we experimentally demonstrated that cAMP and CRP positively regulate the expression of tfoX in V. cholerae and V. fluvialis. A set of promoter deletion fusions and site-directed mutagenesis analyses confirmed the functional existence of two CRP binding sites in the tfoXVC promoter region, and direct binding was further demonstrated by an EMSA in vitro.
A sequence comparison of the tfoX promoter region revealed the existence of optimal and suboptimal CRP-N binding sites in other Vibrio species homologs, suggesting that the transcriptional regulation of tfoX by CRP is a common feature in Vibrionaceae. The ecologies and natural hosts of Vibrio species vary, but free-living in sea water and attachment to zooplanktons are shared life stages. In general, sea water is a nutritionally-limited environment, where chitin is the most abundant nutrition alternative. The chitin utilization pathway has been known to be conserved in the Vibrionaceae40. Nutritional stress induces elevation of cAMP-CRP, and the availability of chitin efficiently activates tfoX expression. TfoX, as the early activated competence regulator, synergistically directs cAMP-CRP to the CRP-S sites on the competence regulon to further promote the competence development, thus allowing the bacteria to take up free DNA from the environment. The uptaken DNA can be used as either an energy source or to repair damaged DNA, or may be used to acquire new alleles/genes, which accounts for the intensive genetic diversity and the mosaic genome structure in Vibrio species revealed by recent genomic sequencing efforts41. It was thought that the reuse of nucleotides from DNA degradation in the cytoplasm may be more significant than other genetic benefits, at least in the short term6.
We also noticed that the degree of CRP binding sequence conservation varies and can roughly be classified into four different groups (Fig. 5). Generally, the distal CBS1 site is more conserved, with a match of at least eight of 10 nucleotides with the consensus binding site. In constrast, the half-site of the proximal CBS2 site obviously displays more sequence diversity, which may result in transient CRP-DNA interactions. It is tempting and logical to speculate that the regulatory features (such as the intensity and duration of binding) are distinct due to the differential binding affinity of CRP caused by sequence variations in different Vibrio species and between different binding sites. For example, V. fischeri possesses three regions that approximate the CRP consensus sequence. Even though V. fischeri has three CRP binding sites, the transformation efficiency has been reported to be 100-fold lower compared to that of V. cholerae13, indicating that the physiological significance of more binding sites needs to be established. The length of the spacer between the two CRP binding half-sites is usually 6 bp or 8 bp42,43. As mentioned before, the spacer region between the half-sites of the third CRP binding site in V. fischeri is 7 bp, which could potentially lead to a dramatic reduction in the binding affinity. The study by Pyles and Lee demonstrated that changing the spacer length from 7 bp to 8 bp increased the binding affinity of CRP for DNA44. Of course, considering the complexity of the competence-associated machinery components and their regulation, it cannot be stated this is the only reason for the 100-fold lower transformation.
In conclusion, our study provided a definitive analysis of the role of the cAMP-CRP in tfoX expression by experimentally demonstrating that the cAMP-CRP complex directly activates its transcriptional expression. These results, together with previous data5,19, demonstrate that the natural competence of Vibrios is subject to catabolite repression by the global transcriptional regulator, CRP, and this regulation is effected through direct control of both the vital competence regulator gene and the competence component genes. In addition, CRP indirectly regulates competence through quorum-sensing by activating primary autoinducer synthesis gene, cqsA27,28. The multiple forms of regulation at different layers or pathways mediated by CRP may maximize the input of simple environmental signals to induce or promote the development of competence for the efficient uptake of extracellular DNA as a nutritional supply or for other purposes, thus greatly favoring the survival and environmental fitness of the organism.
Methods
Strains and media
The V. cholerae and V. fluvialis mutants used in this study were constructed using the El Tor biotype strain, C7258 (Peru′ isolate, 1991), and the clinical strain, 85003 45, as wild-type precursors, respectively. The construction of strains WL7258 (C7258∆crp), WL7259 (C7258∆cya), C7258∆lacZ and WL7258∆lacZ has been described previously27,28,46. pBADCRP7 was introduced into WL7258∆lacZ by electroporation47. Escherichia coli DH5αλpir and S17-1λpir were used for cloning purposes. The V. cholerae and V. fluvialis strains were grown in LB broth containing 1% NaCl at 37 °C with agitation (250 rpm). Culture media were supplemented with ampicillin (Amp, 100 μg/ml), kanamycin (Km, 75 μg/ml), chloramphenicol (Cm, 5 μg/ml), streptomycin (Sm, 100 μg/ml), or polymyxin B (PolB, 100 units/ml) as required. All strains, plasmids and primers used in this study are listed in Table 1.
Table 1. Strains, plasmids and primers used in this study.
| Strain, plasmid or primer | Characteristics or sequence |
|---|---|
| Strains | |
| C7258 | V. cholerae, wild type, El Tor biotype (Peru´ isolate,1991) |
| WL7258 | C7258, ∆crp27 |
| WL7259 | C7258, ∆cya28 |
| C7258∆lacZ | C7258, ∆lacZ46 |
| WL7258∆lacZ | C7258, ∆crp, ∆lacZ46 |
| S17-1λpir | thi thr leu tonA lacY supE recA: :RP4-2Tc : :Mu (λpirR6K) (Laboratory stock) |
| 85003 | V. fluvialis, wild type, SmR 45 |
| 85003∆crp | 85003, ∆crp (This study) |
| C7258∆ptfoX-CBS1M | C7258 containing the site-specific mutagenesis of CRP binding site 1 on the chromosomal tfoXVC promoter region (This study) |
| C7258∆ptfoX-CBS2M | C7258 containing the site-specific mutagenesis of CRP binding site 2 on the chromosomal tfoXVC promoter region (This study) |
| C7258∆ptfoX-CBS1M+2M | C7258 containing the site-specific mutagenesis of CRP binding site 1 and 2 on the chromosomal tfoXVC promoter region (This study) |
| Plasmids | |
| pWM91 | Suicide vector containing R6K ori, sacB, lacZα; AmpR (Laboratory stock) |
| pCVD442 | Suicide vector containing R6K ori, sacB, AmpR (Laboratory stock) |
| pWM-VF∆crp | 1 kb SalI-SacI ∆crp fragment of V. fluvialis in pWM91 (This study) |
| pTT3 | rrnBT1T2 transcription terminator in pUC19; AmpR 48 |
| pTT-tfoXVC | 430 bp PstI- HindIII fragment of tfoXVC promoter region in pTT3 (This study) |
| phaplac7 | hapR promoter cloned in pKRZ1 in front of promoterless lacZ gene; AmpR 48 |
| ptfoXVC-lacZ | HindIII-KpnI fragment containing rrnBT1T2 and tfoXVC promoter from pTT-tfoXVC in phaplac7 (This study) |
| p2TTtfoXVC | 220 bp PstI- HindIII fragment of tfoXVC promoter region in ptfoXVC-lacZ (This study) |
| P3TTtfoXVC | 185 bp PstI- HindIII fragment of tfoXVC promoter region in ptfoXVC-lacZ (This study) |
| pBBRlux | bioluminescence based reporter plasmid containing a promoterless luxCDABE operon; CmR 49 |
| ptfoxVF-lux | 384 bp SacI-BamHI fragment of tfoXVFpromoter region in pBBRlux (This study) |
| ptfoXVC-lacZ-CBS1M | ptfoXVC-lacZ with the mutagenized sequence for the putative CRP binding site 1 (This study) |
| ptfoXVC-lacZ-CBS2M | ptfoXVC-lacZ with the mutagenized sequence for the putative CRP binding site 2 (This study) |
| ptfoXVC-lacZ-CBS1+2M | ptfoXVC-lacZ with the mutagenized sequence for the putative CRP binding site 1 and 2 (This study) |
| pBADCRP7 | V. cholerae crp ORF cloned in pBADHisB50 |
| pCVD-∆ptfoX-kan | 3.0 kb SalI-SphI fragment containing the flanking sequence of tfoXVCpromoter region and kan gene in pCVD442 (This study) |
| pCVD-∆ptfoX-CBS1M | 2.158 kb SalI-SphI fragment containing the tfoXVC promoter region with the mutagenized sequence for the putative CRP binding site 1 and the flanking sequence in pCVD442 (This study) |
| pCVD-∆ptfoX-CBS2M | 2.158 kb SalI-SphI fragment containing the tfoXVC promoter region with the mutagenized sequence for the putative CRP binding site 2 and the flanking sequence in pCVD442 (This study) |
| pCVD-∆ptfoX-CBS1M+2M | 2.158 kb SalI-SphI fragment containing the tfoXVC promoter region with the mutagenized sequence for both the putative CRP binding sites and the flanking sequence in pCVD442 (This study) |
| Primers | |
| VF-CRP-F1-up-SalI | 5′-AACGTCGACTACCCTTACCTGC-3′ |
| VF-CRP-F1-dn | 5′-GTGACGATTACAAAGTCTCTGCTTTTTCG-3′ |
| VF-CRP-F2-up | 5′-AGAGACTTTGTAATCGTCACCGAGACAGAA-3′ |
| VF-CRP-F2-dn-SacI | 5′-CGAGCTCCGTCTGTGGGATCTGAG-3′ |
| pVC1153-up-PstI | 5′-ACCTGCAGCGGGTAACCAGTAAAAAG-3′ |
| pVC1153-dn-HindIII | 5′-CCCAAGCTTCGAAAAACTGTTGCTCAT-3′ |
| VF-sxy-prom-up-SacI | 5′-CGAGCTCCATTGTTTATCATTGTTAGT-3′ |
| VF-sxy-prom-dn-BamHI | 5′-CGGGATCCCATATCCATTGATCCTTTAA-3′ |
| VF-sxy-qPCR-up | 5′-CGTTCTATGTTTGGTGGTATTG-3′ |
| VF-sxy-qPCR-dn | 5′-GCCGTTGTCTGCTTCTTC-3′ |
| VF-recA-qPCR-up | 5′-ACCGAGTCAACGACGATAAC-3′ |
| VF-recA-qPCR-dn | 5′-TGATGAACTGCTGGTGTCTC-3′ |
| VC1153-up | 5′-ACGCTCGATGTTTGGTGGTATTGG-3′ |
| VC1153-dn | 5′-ATTGGGTAAATCCCGTAGACGACG-3′ |
| VF-sxy-race | 5′-TCTGCTTCTTCACATGACGG-3′ |
| Shift-up1976/1996 | 5′-GGTTTAAGTATAGAGGAGCA-3′(5′ biotin-labeled) |
| Shift-dn2107/2127 | 5′-GTTCGTATGAGCTTGCTTGT-3′(5′ biotin-labeled) |
| VC1153-p1shift-dn | 5′-ATGCAATACTTTTGCGCCAG-3′(5′ biotin-labeled) |
| VC1153-p2shift-up | 5′-CTGGCGCAAAAGTATTGCAT-3′(5′ biotin-labeled) |
| VC1153CBS1M-for | 5′-TTGACGCTCACATACTGGTCTCATAATCTGG-3′ |
| VC1153CBS1M-rev | 5′-GACCAGTATGTGAGCGTCAATTTTTTGTTGC-3′ |
| VC1153CBS2M-for | 5′-TGCATCGATCACAGATAAGTCCAGACTTATTTCTC-3′ |
| VC1153CBS2M-rev | 5′-GACTTATCTGTGATCGATGCAATACTTTTGCGCCA-3′ |
| F1-up-SalI: | 5′-GCGTCGACATCACTTAGCTTGTTGTT-3′ |
| F1-dn-PstI: | 5′-GCTCATCTGCAGCTTTTTACTGGTTACCCG-3′ |
| F2-up: | 5′-GTAACCAGTAAAAAGATAGGTGACTCATAA-3′ |
| F2-dn: | 5′-ACTGTTGCTCATTCATATCCATTGATCCTT-3′ |
| F3-up-PstI: | 5′-AAAAGCTGCAGATGAGCAACAGTTTTTCGA-3′ |
| F3-dn-SphI: | 5′-CATGCATGCATTGGTATTCGTCAGTGG-3′ |
| pilB-real-u | 5′-ATGCTCACCAACCTTGTT-3′ |
| pilB-real-d | 5′-TGTAACCACCGCTTGTTC-3′ |
| chiA1-real-u | 5′-TGGCATAACTTCGTCAAT-3′ |
| chiA1-real-d | 5′-AAGGCAATATCAATCACATC-3′ |
| chiA2-real-u | 5′-GCTATCGGTGTTGGTCAT-3′ |
| chiA2-real-d | 5′-CGTAGAAGTCATAAGTCATTGC-3′ |
Construction of mutants
The V. fluvialis deletion mutant was constructed from clinical strain 85003 by allelic exchange45. Primers were designed based on the genome sequence of the V. fluvialis 85003 deposited in the Sequence Read Archive under accession no. SRX397301 45. To construct the ∆crp mutant, 85003∆crp, the upstream and downstream DNA fragments flanking the crp open reading frame (ORF) were amplified from 85003 genomic DNA using primer pairs VF-CRP-F1-up-SalI /VF-CRP-F1-dn and VF-CRP-F2-up/VF-CRP-F2-dn-SacI, respectively. The amplicons were mixed in equimolar concentration and used as the template to amplify the chromosomal fragment containing the crp deletion using primer pair VF-CRP-F1-up-SalI/VF-CRP-F2-dn-SacI. The resulting fragment was cloned into suicide plasmid pWM91 to generate pWM-VF∆crp. pWM-VF∆crp was constructed in E. coli S17-1λpir and transferred to strain 85003 by conjugation. Exconjugants were selected in LB medium containing Amp and Sm, and were streaked on LB agar containing 15% (w/v) sucrose. Sucrose-resistant colonies were tested for Amp sensitivity, and the deletion of crp was confirmed by DNA sequencing.
Construction of transcriptional reporter plasmids
To construct the ptfoXVC–lacZ transcriptional fusion, we cloned a 430-bp fragment containing the tfoXVC promoter region amplified with primers pVC1153-up-PstI/pVC1153-dn-HindIII into pTT3 48 downstream of the strong rrnBT1T2 transcription terminator to generate pTT-tfoXVC. Next, a 900 bp KpnI-HindIII fragment containing the rrnBT1T2 terminator and the tfoXVC promoter from pTT-tfoXVC was ligated into the big HindIII-KpnI fragment of phaplac7, which contains a promoterless lacZ gene, the pBR322 origin of replication, and the bla gene48. The resultant plasmid, ptfoXVC-lacZ, was introduced into C7258∆lacZ and WL7258∆lacZ by electroporation47. Additional DNA fragments harboring 5′ deletions of the tfoXVC promoter were amplified with primer pairs of pVC1153-up2-PstI/pVC1153-dn-HindIII and pVC1153-up3-PstI /pVC1153-dn-HindIII, and transcriptional fusions p2TTtfoXVC and p3TTtfoXVC were constructed in a similar manner. To construct the bioluminescence based ptfoxVF-lux fusion, we cloned a 384 bp fragment containing the tfoXVF promoter region amplified with primers VF-sxy-prom-up-SacI/VF-sxy-prom-dn-BamHI into reporter plasmid pBBRlux, which contains a promoterless luxCDABE operon49. ptfoxVF-lux was constructed in DH5αλpir and transferred into S17-1λpir, and then mobilized into V. fluvialis strains 85003 and 85003∆crp by conjugation.
Quantitative reverse transcription PCR (qRT-PCR)
The V. cholerae and V. fluvialis strains were grown in LB medium to the late-log phase. Cells were collected by centrifugation at 4 °C and immediately subjected to RNA extraction. Total RNA was extracted using the Trizol reagent (Ambion), followed by treatment with a TURBO DNA-freeTM kit (Ambion) to remove chromosomal DNA contamination. The purity and integrity of RNA samples was verified by UV spectrophotometry and agarose gel electrophoresis. Using 1 μg of total RNA per sample, cDNA was synthesized using random hexamer primers with SuperScriptTM III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. qRT-PCR was performed using SYBR Green (TaKaRa) on the Bio-Rad CFX96 Real-Time PCR detection system. The relative expression values (R) using recA mRNA as a reference were calculated using the equation R = 2−(ΔCq target −ΔCq reference), where Cq is the fractional threshold cycle. The following primer combinations were used: VC1153-up and VC1153-dn for tfoxVC; recA578 and recA863 for recAVC 27; VF-sxy-qPCR-up and VF-sxy-qPCR-dn for tfoxVF; VF-recA-qPCR-up and VF-recA-qPCR-dn for recAVF; pilB-real-u and pilB-real-d for pilB; chiA1-real-u and chiA1-real-d for chiA-1; chiA2-real-u and chiA2-real-d for chiA-2. A control mixture with total RNA as a template was used in each reaction to exclude the effects of chromosomal DNA contamination.
Determination of the 5′-end of tfoX VF mRNA
We used 5′ RACE (rapid amplification of cDNA ends) to determine the TSS of tfoXVF. V. fluvialis strain 85003 was grown in LB containing 1% NaCl at 37 °C with agitation to OD600 1.5. Total RNA was extracted as described in the preceding section. The cDNA was generated using the SMARTer™ RACE cDNA Amplification Kit (Clontech) according to the manufacturer’s instructions. The cDNA was then amplified by PCR using a kit provided with UPM (universal primer) and the gene specific primer, VF-sxy-race. The PCR product obtained was gel-purified and cloned into the pMD®18-T vector. Ten clones were sequenced using the primers M13-R and M13-F to determine the TSS.
Site-directed mutagenesis of the putative CRP binding sites of the tfoX VC promoter
The two putative CRP binding sites in the promoter region of tfoXVCwere mutated using the Hieff MutTM site-directed mutagenesis kit (Shanghai YEASEN Biotechnology Co., Ltd.) according to the manufacturer’s instructions. The above constructed ptfoXVC-lacZ plasmid containing the full-length functional tfoX promoter region was used as the template, and mutagenesis was induced using primers carrying the substituted nucleotides. Primer pairs VC1153CBS1M-for/VC1153CBS1M-rev and VC1153CBS2M-for/VC1153CBS2M-rev were used to substitute three bases in the half-site of putative CRP binding sites 1 and 2, respectively. To simultaneously mutate the two CRP binding sites, primer combinations VC1153CBS1M-for/VC1153CBS2M-rev and VC1153CBS1M-rev/ VC1153CBS2M-for were used. The resultant plasmids, ptfoXVC-lacZ-CBS1M, ptfoXVC-lacZ-CBS2M and ptfoXVC-lacZ-CBS1+2M, were sequenced to confirm the mutations. Subsequently, the obtained plasmid was introduced into V. cholerae strain C7258∆lacZ by electroporation.
Mutations in CBS1M, CBS2M and CBS1M+2M were further introduced into the chromosomal promoter region of tfoX of C7258 through suicide plasmid-mediated allelic exchange. For this purpose, the promoter region of tfoX was first replaced with a kanamycin gene, yielding strain C7258∆ptfoX-kan. Then, C7258∆ptfoX-kan was used as precursor to introduce CBS1M, CBS2M and CBS1M+2M mutations on the chromosome. To construct C7258∆ptfoX-kan, the flanking sequence of the tfoX promoter region was amplified with primer pairs F1-up-SalI/ F1-dn-PstI and F3-up-PstI/ F3-dn-SphI, and was sequentially cloned into pCVD442, then the kan gene was inserted at the PstI site. The resulting plasmid, pCVD-∆ptfoX-kan, was mobilized into C7258 by conjugation, and sucrose selection was applied to isolate segregants containing the kan replacement of the tfoX promoter region.
Subsequently, three recombinant suicide plasmids, pCVD-∆ptfoX-CBS1M, pCVD-∆ptfoX-CBS2M and pCVD-∆ptfoX-CBS1M+2M were generated. Primer pairs F1-up-SalI/F1-dn and F3-up/F3-dn-SphI were used to amplify the flanking sequence. Plasmids ptfoXVC-lacZ-CBS1M, ptfoXVC-lacZ-CBS2M and ptfoXVC-lacZ-CBS1+2M were used as a template to amplify the tfoX promoter region containing the CBS1M, CBS2M and CBS1M+2M mutations with the primer pair F2-up/F2-dn. Bridge PCR was then applied to join the three fragments, which were further cloned as SalI/SphI fragments into pCVD442. The resulting plasmids pCVD-∆ptfoX-CBS1M, pCVD-∆ptfoX-CBS2M and pCVD-∆ptfoX-CBS1M+2M were transferred into C7258∆ptfoX-kan. Exconjugants were selected in LB medium containing Amp and PolB and sucrose selection was applied to isolate segregants retaining the CBS1M, CBS2M and CBS1M+2M alleles. The corresponding mutants were named C7258∆ptfoX-CBS1M, C7258∆ptfoX-CBS2M and C7258∆ptfoX-CBS1M+2M, respectively.
Expression and purification of His-CRP
E. coli TOP10 harboring the recombinant plasmid pBADCRP750 were cultured at 37 °C in LB medium. The 6xHis-tagged CRP was induced by 0.2% (w/v) arabinose and purified by Ni-IDA affinity chromatography (Novagen) under native conditions according to the manufacturer’s instructions. The purity was analyzed by SDS-PAGE (Fig. 6A), and the concentration was determined with a Pierce BCA Protein Assay kit.
Electrophoretic mobility shift assays (EMSA)
EMSA was performed as described previously51. The 152 bp, 75 bp and 97 bp fragments of the tfoXVC promoter regions were amplified with the biotin-labeled primer pairs Shift-up1976-96/Shift-dn2107-27, Shift-up1976-96/VC1153-p1shift-dn, and VC1153-p2shift-up/Shift-dn2107-27 respectively, and were used as probes. The 152 bp probe covers fragments ranging from −127 to 24 relative to the reported TSS21 and encompasses two putative CRP binding sites. The same fragments without a biotin label were used as competing cold probes and were added in 100–300-fold excess of the labeled probes. The 75 bp and 97 bp probes extend from residues −127 to −53 and −74 to 24, respectively, and each contains a single CRP binding site. Binding reactions were performed by mixing each biotin-labeled probe with increasing quantities of purified CRP in 10 μl of reaction volume with binding buffer (50 mM Tris-HCl [pH7.8], 250 mM KCl, 5 mM MgCl2, 2.5 mM EDTA, 0.5 mM MnCl2, 2.5 mM DTT, 1 μg Poly (dI.dC) and 100 μM cAMP). The reaction mixture was incubated at room temperature for 30 min, and then separated on a 6% (for 152 bp probe) or 10% (for 75 bp and 97 bp probes) native polyacrylamide gel after adding loading buffer. The separated DNA and DNA-protein samples were transferred onto nylon membranes using the Mini Trans-Blot Electrophoretic Transfer cell (Bio-Rad), and were detected with the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) following the manufacturer’s instructions.
β-Galactosidase assays and bioluminescence assays
Overnight cultures of V. cholerae strains C7258∆lacZ and WL7258∆lacZ containing tfoXVC-lacZ fusion plasmids were diluted 100-fold into fresh LB, and were grown to mid-exponential phase. The specific activities of β-galactosidase are expressed in Miller units [1000 (OD420/t v OD600)], where t is the reaction time and v is the volume of enzyme extract per reaction52. Similarly, V. fluvialis strains 85003 and VF∆crp with ptfoxVF-lux were cultured to the mid-exponential phase. The absorbance and bioluminescence were quantified thereafter. The bioluminescence was measured on opaque-wall 96-well microtiter plates (ostar 3917) with a Tecan Infinite M200 Pro luminometer. The promoter activity is expressed as Lux/OD600.
Additional Information
How to cite this article: Wu, R. et al. Direct regulation of the natural competence regulator gene tfoX by cyclic AMP (cAMP) and cAMP receptor protein (CRP) in Vibrios. Sci. Rep. 5, 14921; doi: 10.1038/srep14921 (2015).
Acknowledgments
We thank Mr. Pengcheng Du and Dr. Jingyun Zhang for helps in preparing figures. This study was supported by Projects of the National Natural Science Foundation of China (81071410, 81171640), the Priority Project on Infectious Disease Control and Prevention (2012ZX10004215) and a project from Beijing Municipal Science & Technology Commission (D131100005313016).
Footnotes
Author Contributions Conceived and designed the experiments: W.L. and B.K. Performed the experiments: R.W., M.Z. and J.L. Analyzed the data: W.L., H.G., R.W. and M.Z. Contributed reagents/materials/analysis tools: W.L. and B.K. Wrote the paper: W.L. and R.W. All authors read and approved the final manuscript.
References
- Dubnau D.. DNA uptake in bacteria. Annu Rev Microbiol 53, 217–244 (1999). [DOI] [PubMed] [Google Scholar]
- Kruger N. J. & Stingl K.. Two steps away from novelty–principles of bacterial DNA uptake. Mol Microbiol 80, 860–867 (2011). [DOI] [PubMed] [Google Scholar]
- Sinha S., Mell J. C. & Redfield R. J.. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J Bacteriol 194, 5245–5254 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. H. & Azam F.. Microbiology. Chitin, cholera, and competence. Science 310, 1775–1777 (2005). [DOI] [PubMed] [Google Scholar]
- Meibom K. L., Blokesch M., Dolganov N. A., Wu C. Y. & Schoolnik G. K.. Chitin induces natural competence in Vibrio cholerae. Science 310, 1824–1827 (2005). [DOI] [PubMed] [Google Scholar]
- Redfield R. J.. Do bacteria have sex? Nat Rev Genet 2, 634–639 (2001). [DOI] [PubMed] [Google Scholar]
- Antonova E. S., Bernardy E. E. & Hammer B. K.. Natural competence in Vibrio cholerae is controlled by a nucleoside scavenging response that requires CytR-dependent anti-activation. Mol Microbiol 86, 1215–1231 (2012). [DOI] [PubMed] [Google Scholar]
- Macfadyen L. P.. Regulation of competence development in Haemophilus influenzae:Proposed Competence Regulatory Elements are CRP-Binding Sites. J Theor Biol 207, 349–359 (2000). [DOI] [PubMed] [Google Scholar]
- Chen I. & Dubnau D.. DNA uptake during bacterial transformation. Nat Rev Microbiol 2, 241–249 (2004). [DOI] [PubMed] [Google Scholar]
- Redfield R. J. et al. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J Mol Biol 347, 735–747 (2005). [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C. & Steitz T. A.. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253, 1001–1007 (1991). [DOI] [PubMed] [Google Scholar]
- Cameron A. D. & Redfield R. J.. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res 34, 6001–6014 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack-Berti A., Wollenberg M. S. & Ruby E. G.. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol 12, 2302–2311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P. A., Tucker M. S., Thiaville P. C., Joseph J. L. & Brown R. N.. USER friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl Environ Microbiol 75, 4936–4949 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman J., Guo Y. & Rowe-Magnus D. A.. Chitin-induced carbotype conversion in Vibrio vulnificus. Infect Immun 79, 3195–3203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Dai J., Morris J. G. Jr. & Johnson J. A.. Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol 10, 274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. S.. The gene encoding cAMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc Natl Acad Sci USA 89, 1626–1630 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorocicz I. R., Williams P. M. & Redfield R. J.. The Haemophilus influenzae adenylate cyclase gene: cloning, sequence, and essential role in competence. J Bacteriol 175, 7142–7149 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M.. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol 14, 1898–1912 (2012). [DOI] [PubMed] [Google Scholar]
- Cameron A. D., Volar M., Bannister L. A. & Redfield R. J.. RNA secondary structure regulates the translation of sxy and competence development in Haemophilus influenzae. Nucleic Acids Res 36, 10–20 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Morita M., Izumiya H. & Watanabe H.. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene tfoXVC. Gene 457, 42–49 (2010). [DOI] [PubMed] [Google Scholar]
- Yamamoto S. et al. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol 193, 1953–1965 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulke J. & Hillen W.. Carbon catabolite repression in bacteria. Curr Opin Microbiol 2, 195–201 (1999). [DOI] [PubMed] [Google Scholar]
- Bruckner R. & Titgemeyer F.. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett 209, 141–148 (2002). [DOI] [PubMed] [Google Scholar]
- Skorupski K. & Taylor R. K.. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA 94, 265–270 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Silva A. J. & Benitez J. A.. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl Environ Microbiol 73, 7482–7487 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Pascual-Montano A., Silva A. J. & Benitez J. A.. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153, 2964–2975 (2007). [DOI] [PubMed] [Google Scholar]
- Liang W., Sultan S. Z., Silva A. J. & Benitez J. A.. Cyclic AMP post-transcriptionally regulates the biosynthesis of a major bacterial autoinducer to modulate the cell density required to activate quorum sensing. FEBS Lett 582, 3744–3750 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa E. O. & Okoh A. I.. Vibrio fluvialis: an unusual enteric pathogen of increasing public health concern. Int J Environ Res Public Health 7, 3628–3643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Cui X., Du X., Kan B. & Liang W.. The virulence phenotypes and molecular epidemiological characteristics of Vibrio fluvialis in China. Gut Pathog 5, 6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Francke C. & Postma P. W.. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70, 939–1031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S. & Ebright R. H.. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293, 199–213 (1999). [DOI] [PubMed] [Google Scholar]
- Busby S. et al. Transcription activation by the Escherichia coli cyclic AMP receptor protein. Receptors bound in tandem at promoters can interact synergistically. J Mol Biol 241, 341–352 (1994). [DOI] [PubMed] [Google Scholar]
- Kolb A., Busby S., Buc H., Garges S. & Adhya S.. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem 62, 749–795 (1993). [DOI] [PubMed] [Google Scholar]
- Borukhov S. & Lee J.. RNA polymerase structure and function at lac operon. C R Biol 328, 576–587 (2005). [DOI] [PubMed] [Google Scholar]
- Cameron A. D. & Redfield R. J.. CRP binding and transcription activation at CRP-S sites. J Mol Biol 383, 313–323 (2008). [DOI] [PubMed] [Google Scholar]
- Fic E. et al. cAMP receptor protein from escherichia coli as a model of signal transduction in proteins–a review. J Mol Microbiol Biotechnol 17, 1–11 (2009). [DOI] [PubMed] [Google Scholar]
- Jaskolska M. & Gerdes K.. CRP-dependent Positive Autoregulation and Proteolytic Degradation Regulate Competence Activator Sxy of Escherichia coli. Mol Microbiol 95, 833–845 (2015). [DOI] [PubMed] [Google Scholar]
- Zulty J. J. & Barcak G. J.. Identification of a DNA transformation gene required for com101A+ expression and supertransformer phenotype in Haemophilus influenzae. Proc Natl Acad Sci USA 92, 3616–3620 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D. E., Gevers D., Vahora N. M. & Polz M. F.. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74, 44–51 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J. et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA 106, 15442–15447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber A. M., Zhurkin V. B. & Adhya S.. CRP-binding sites: evidence for two structural classes with 6-bp and 8-bp spacers. Gene 130, 1–8 (1993). [DOI] [PubMed] [Google Scholar]
- Barber A. M. & Zhurkin V. B.. CAP binding sites reveal pyrimidine-purine pattern characteristic of DNA bending. J Biomol Struct Dyn 8, 213–232 (1990). [DOI] [PubMed] [Google Scholar]
- Pyles E. A. & Lee J. C.. Mode of selectivity in cyclic AMP receptor protein-dependent promoters in Escherichia coli. Biochemistry 35, 1162–1172 (1996). [DOI] [PubMed] [Google Scholar]
- Lu X. et al. Identification of genetic bases of vibrio fluvialis species-specific biochemical pathways and potential virulence factors by comparative genomic analysis. Appl Environ Microbiol 80, 2029–2037 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J., Sultan S. Z., Liang W. & Benitez. J. A. Role of the histone-like nucleoid structuring protein in the regulation of rpoS and RpoS-dependent genes in Vibrio cholerae. J Bacteriol 190, 7335–7345 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus H., Ketley J. M., Kaper J. B. & Holmes R. K.. Effects of DNase production, plasmid size, and restriction barriers on transformation of Vibrio cholerae by electroporation and osmotic shock. FEMS Microbiol Lett 56, 149–154 (1990). [DOI] [PubMed] [Google Scholar]
- Silva A. J., Pham K. & Benitez J. A.. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149, 1883–1891 (2003). [DOI] [PubMed] [Google Scholar]
- Zhou Y. Y. et al. Plasticity of regulation of mannitol phosphotransferase system operon by CRP-cAMP complex in Vibrio cholerae. Biomed Environ Sci 26, 831–840 (2013). [DOI] [PubMed] [Google Scholar]
- Silva A. J. & Benitez J. A.. Transcriptional regulation of Vibrio cholerae hemagglutinin/protease by the cyclic AMP receptor protein and RpoS. J Bacteriol 186, 6374–6382 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Liang W., Wu R., Liang P. & Kan B.. Phenotype microarray screening of carbon sources used by Vibrio cholerae identifies genes regulated by the cAMP receptor protein. Can J Microbiol 59, 472–478 (2013). [DOI] [PubMed] [Google Scholar]
- Miller J. H.. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (1972). [Google Scholar]