Abstract
Rationale: Genetic variation plays a significant role in the etiology of sarcoidosis. However, only a small fraction of its heritability has been explained so far.
Objectives: To define further genetic risk loci for sarcoidosis, we used the Immunochip for a candidate gene association study of immune-associated loci.
Methods: Altogether the study population comprised over 19,000 individuals. In a two-stage design, 1,726 German sarcoidosis cases and 5,482 control subjects were genotyped for 128,705 single-nucleotide polymorphisms using the Illumina Immunochip for the screening step. The remaining 3,955 cases, 7,514 control subjects, and 684 parents of affected offspring were used for validation and replication of 44 candidate and two established risk single-nucleotide polymorphisms.
Measurements and Main Results: Four novel susceptibility loci were identified with genome-wide significance in the European case-control populations, located on chromosomes 12q24.12 (rs653178; ATXN2/SH2B3), 5q33.3 (rs4921492; IL12B), 4q24 (rs223498; MANBA/NFKB1), and 2q33.2 (rs6748088; FAM117B). We further defined three independent association signals in the HLA region with genome-wide significance, peaking in the BTNL2 promoter region (rs5007259), at HLA-B (rs4143332/HLA-B*0801) and at HLA-DPB1 (rs9277542), and found another novel independent signal near IL23R (rs12069782) on chromosome 1p31.3.
Conclusions: Functional predictions and protein network analyses suggest a prominent role of the drug-targetable IL23/Th17 signaling pathway in the genetic etiology of sarcoidosis. Our findings reveal a substantial genetic overlap of sarcoidosis with diverse immune-mediated inflammatory disorders, which could be of relevance for the clinical application of modern therapeutics
Keywords: Immunochip, HLA, IL23, BTNL2, association
At a Glance Commentary
Scientific Knowledge on the Subject
Sarcoidosis is a complex granulomatous disease with unknown etiology. There is a considerable genetic contribution; however, only a small fraction of the estimated heritability is explained, with variants in HLA region conferring the largest effects.
What This Study Adds to the Field
The study successfully identified four novel genetic risk loci for sarcoidosis and refined the association signal in the HLA region to three independent signals. Our results support the hypothesis that sarcoidosis partly shares a genetic background with other immune-related diseases. Thus, our findings may inspire a revised classification of clinical disease manifestations and subphenotypes. Our results further implicate a role of genetic factors in many aspects of sarcoidosis pathogenesis, especially in IL23/Th17 signaling, and may provide hypotheses on novel therapeutic targets.
Sarcoidosis is a complex inflammatory disease of mainly the lung with an unknown etiology that affects mostly young adults (20–40 yr) (1). It is characterized by heterogeneous clinical manifestations with diverse organ involvement and disease course. Previous genome-wide association studies (GWAS) and candidate gene-driven investigations identified several genetic risk loci for sarcoidosis, such as BTNL2 (2–4); ANXA11 (5–8); a locus on chromosome 11q13.1 (9); and, most consistently, several loci in the HLA region on chromosome 6p21 (10). Some risk loci are shared for sarcoidosis and other inflammatory or autoimmune conditions. For example, allelic variation of single-nucleotide polymorphism (SNP) rs11209026 (Arg381Gln) in the IL23R gene locus predisposes, among others, to sarcoidosis, inflammatory bowel disease, psoriasis, and ankylosing spondylitis (11–15).
Given the large overlap of the genetic risk maps of complex inflammatory and autoimmune diseases, the Immunochip array was designed for genotyping a comprehensive set of known immune-associated variants and for fine-mapping a subset of 186 selected risk loci (16, 17). This array was successfully used before to identify novel and shared risk loci for various complex immune-related diseases (18–24). We used the Immunochip to screen 1,726 German patients with sarcoidosis and 5,482 healthy control subjects for novel sarcoidosis risk loci, followed by replication in four independent European collections (4,605 cases and 12,673 control subjects) and another three populations for replication and subphenotype analysis (Table 1).
Table 1.
Study Populations
| Panel | Descent | Cases |
Control Subjects |
||||
|---|---|---|---|---|---|---|---|
| Number* | Male (%)* | Mean (SD) Age in 2014 (yr) | Number* | Male (%)* | Mean (SD) Age in 2014 (yr) | ||
| A | European (Germany) | 1,726 (1,869) | 39 (40) | 62.9 (11.6) | 5,482 (5,600) | 51 (51) | 57.2 (12.6) |
| B-I | European (Germany) | 573 (585) | 49 (49) | 55.7 (12.6) | 3,327 (3,500) | 47 (47) | 48.7 (14.9) |
| B-II | European (Germany) | 266 (307) | 46 (45) | 61.0 (13.7) | 266 (285) | 50 (50) | 58.3 (17.7) |
| C-I | European (Czech Republic) | 256 (267) | 45 (46) | 56.2 (13.1) | 305 (330) | 42 (43) | 45.6 (9.6) |
| C-II | European (Sweden) | 817 (1,121) | 57 (58) | 54.1 (12.4) | 2,040 | 28 | 53.6 (11.2) |
| D | African American (USA) | 781 | 26 | 47.9 (15.2) | 876 | 22 | 38.9 (10.2) |
| E | European (Germany) | 342† | 43 | 52.5 (7.2)‡ | — | — | — |
| F | European (Serbia) | 920 (920) | 47 | 53.3 (11.4) | — | — | — |
Data are given after quality control, with data before quality control in parentheses.
Number of trios (two parents with one affected offspring).
Given for the affected offspring.
Methods
Patients and Control Subjects
The study sample comprised 5,681 sarcoidosis cases, all of which were diagnosed according to international standards (25), 12,996 control subjects, and 684 parents of affected offspring. Analyses were performed in eight independent panels (A, B-I, B-II, C-I, C-II, D, E, and F) (Table 1). A detailed description and definition of the study sample including sarcoidosis subphenotypes is provided in the online supplement.
Immunochip Genotyping and Quality Control
DNA samples of panel A were genotyped using the Immunochip, comprising a total of 196,524 SNP assays (17). Quality control excluded 67,819 SNPs and 143 individuals from analysis. Principal component analysis revealed no population stratification in the remaining samples and no population outliers were detected. Unless described differently, genotype data of panels B, C, E, and F were generated using Sequenom Mass-ARRAY iPlex (Sequenom, Inc., San Diego, CA) (26) and Taqman technology (Applied Biosystems, Foster City, CA). In every analysis step, SNPs that had more than 5% missing data (call rate ≥ 95%), a minor allele frequency less than 1%, and exact Hardy-Weinberg equilibrium P less than 10−4 were excluded. The control subjects of panel C-III were genotyped using the Illumina Immunochip custom array (The Genome Institute, Singapore). Genotyping for panel D was performed at the Oklahoma Medical Research Foundation using the Illumina Immunochip. The online supplement provides further details.
Statistical Analysis and SNP Selection
Data filtering and any statistical analysis of genotype data was performed using PLINK v.1.07, applying logistic regression model throughout all case-control analysis, including conditional analysis using an allelic model for genotype coding (27). Most promising SNPs (excluding ANXA11 and HLA SNPs) were selected for follow-up and joint analysis on ranking top with their P value (P < 10−4) in the association analysis and a positive visual inspection of regional plots. The study design is visualized in the online supplement. HLA-haplotypes were imputed using HLA*IMP:02 with default parameters (28). All statistical tests used in this study and methods applied for in silico analyses are described in the online supplement.
Results
After applying conservative and established quality filters to panel A, 1,726 sarcoidosis cases, 5,482 control subjects, and 128,705 SNPs were included in the analysis of this dataset (Table 1; see the Methods section) and analyzed by a logistic regression analysis. The 44 most promising SNPs were selected based on ranked P values as described in the Methods section and investigated in a joint analysis of all available European case-control panels A, B-I, B-II, C-I, and C-II (total of 4,605 cases and 12,673 control subjects) (Table 1). This approach led to the identification of four novel susceptibility loci for sarcoidosis in Europeans with genome-wide significant association signals (Table 2, Figure 1). For the respective lead variants, the effect sizes did not differ significantly between the investigated populations as assessed by the Breslow-Day test.
Table 2.
Association Results for Newly Discovered Sarcoidosis Risk Variants
| dbSNP ID | rs653178 | rs4921492 | rs223498 | rs6748088 |
|---|---|---|---|---|
| Chromosome | 12 | 5 | 4 | 2 |
| Position | 112,007,756 | 158,832,277 | 103,651,962 | 203,264,771 |
| Candidate genes | ATXN2, SH2B3 | IL12B | NFKB, MANBA | FAM117B |
| A1 | G | A | C | C |
| A2 | A | C | A | T |
| Panel A | ||||
| AFcases | 0.55 | 0.39 | 0.51 | 0.34 |
| AFcontrols | 0.5 | 0.34 | 0.47 | 0.31 |
| P value | 1.36 × 10−6 | 7.56 × 10−7 | 6.68 × 10−6 | 4.30 × 10−4 |
| OR (95% CI) | 1.21 (1.12–1.30) | 1.22 (1.13–1.32) | 1.19 (1.10–1.29) | 1.16 (1.07–1.26) |
| Panel B-I | ||||
| AFcases | 0.54 | 0.39 | 0.52 | 0.35 |
| AFcontrols | 0.49 | 0.35 | 0.48 | 0.30 |
| P value | 8.06 × 10−3 | 0.019 | 0.012 | 1.34 × 10−4 |
| OR (95% CI) | 1.19 (1.05–1.34) | 1.17 (1.03–1.33) | 1.18 (1.04–1.33) | 1.29 (1.13–1.48) |
| Panel B-II | ||||
| AFcases | 0.54 | 0.4 | 0.5 | 0.37 |
| AFcontrols | 0.51 | 0.33 | 0.48 | 0.31 |
| P value | ns | 0.039 | ns | 0.048 |
| OR (95% CI) | 1.13 (0.89–1.44) | 1.33 (1.04–1.71) | 1.08 (0.85–1.38) | 1.29 (1.00–1.67) |
| Panel C-I | ||||
| AFcases | 0.58 | 0.38 | 0.44 | 0.36 |
| AFcontrols | 0.48 | 0.36 | 0.49 | 0.32 |
| P value | 8.94 × 10−4 | ns | ns | ns |
| OR (95% CI) | 1.49 (1.18–1.89) | 1.11 (0.87–1.41) | 0.82 (0.64–1.03) | 1.20 (0.94–1.54) |
| Panel C-II | ||||
| AFcases | 0.50 | 0.38 | 0.54 | 0.32 |
| AFcontrols | 0.48 | 0.35 | 0.48 | 0.30 |
| P value | ns | ns | 1.08 × 10−5 | ns |
| OR (95% CI) | 1.12 (0.99–1.26) | 1.14 (0.94–1.39) | 1.30 (1.16–1.46) | 1.10 (0.95–1.27) |
| Joint analysis | ||||
| P CMH | 1.64 × 10−10 | 2.14 × 10−9 | 1.28 × 10−9 | 2.10 × 10−8 |
| OR (95% CI) | 1.19 (1.14–1.27) | 1.20 (1.13–1.27) | 1.19 (1.12–1.26) | 1.18 (1.11–1.25) |
Definition of abbreviations: AF = allele frequency; CI = confidence interval; CMH = Cochran-Mantel-Haenzel test; dbSNP = National Institutes of Health SNP database; ns = not significant; OR = odds ratio.
The position of the respective lead variant is given according to human genome build 19. OR refers to allele 1 (A1), and P values are presented for the logistic regression model using an allelic model for genotype coding. The threshold for genome-wide significance (P < 5 × 10−8) in the CMH test (P CMH) was applied to define a true association.
Figure 1.
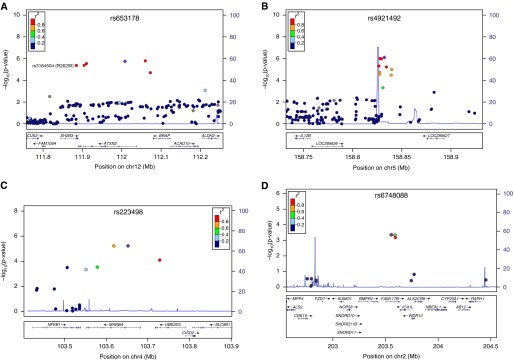
Association signals of newly discovered sarcoidosis risk regions panel A that showed genome-wide significance in the joint analysis of the European case-control panels A, B-I, B-II, C-I, and C-II on (A) chromosome 12q24.12 (lead single-nucleotide polymorphism [SNP] rs653178, nonsynonymous SNP rs3184504), (B) 5q33.3 (lead SNP rs4921492), (C) 4q24 (lead SNP rs223498), and (D) 2q33.2 (lead SNP rs6748088). The respective lead SNPs are marked by blue diamonds. The strength of linkage disequilibrium (r2) between the lead SNP and surrounding markers and the recombination rate is given according to 1,000 Genomes data (phase I, release March 2012) and is indicated by red to yellow coloring and the light blue line, respectively. Positions shown are according to National Center for Biotechnology Information’s build 37(hg19).
Novel Non-HLA Risk Loci with Genome-Wide Significance
SNP marker rs653178 on chromosome 12q24.12 was associated with the smallest P value in the joint analysis (P = 1.64 × 10−10; odds ratio [OR] [95% confidence interval (CI)], 1.19 [1.14–1.27]). The potentially causative nonsynonymous variant rs3184504 in the SH2B adaptor protein 3 (SH2B3) locus was in high linkage disequilibrium (LD) with the lead SNP rs653178 and strongly associated in panel A (r2 = 0.87; P = 3.42 × 10−6; OR [95% CI], 0.83 [0.77–0.90]). For rs653178, a subphenotype-differential effect was observed for Löfgren syndrome in panel B-II (P = 6.75 × 10−3; OR [95% CI], 1.97 [1.20–3.24]). Complete results for the subphenotype analysis are given in Table E1 in the online supplement.
Second, in the chromosome 5q33.3 region upstream of IL12B, SNP rs4921492 yielded a genome-wide significant result (P = 2.14 × 10−9; OR [95% CI], 1.20 [1.13–1.27]) for sarcoidosis. Concerning subphenotypes, this marker associated with nominal significance with the involvement of the central nervous system in panel F (P = 0.035; OR [95% CI], 1.51 [1.03–2.23]) (see Table E2 for complete subphenotype results). Besides the lead SNP rs4921492, five markers that are in high LD with it are strongly associated with sarcoidosis (see Table E3). One of those, namely SNP rs12651787 (P = 9.59 × 10−6; OR [95% CI], 1.20 [1.10–1.29]), is in high LD with rs1422877 (r2 = 0.97), which is predicted to reside in a nuclear factor-κB binding site and to confer allele-specific binding probabilities for the transcription factor (TF) RelA.
Third, SNP rs223498 located on chromosome 4q24 near NFKB1 showed a significant association in the joint analysis (P = 1.28 × 10−9; OR [95% CI], 1.19 [1.12–1.26]). In panel A, a neighboring SNP (rs227375) was associated with the same effect size (OR [95% CI], 1.19 [1.10–1.29]), but did not represent an independent signal as investigated by conditional regression analysis (data not shown). The region was sparsely covered with markers on the Immunochip (17 SNPs in a region of ±250 kb around the lead SNP). According to previous GWAS data (9), the association signal may extend beyond MANBA and NFKB1 (see Figure E1A). For rs223498, none of the investigated sarcoidosis subphenotypes was associated significantly (see Table E4).
Fourth, in the FAM117B gene region on chromosome 2q33.2, marker rs6748088 displayed a significant P value in the joint analysis (P = 2.10 × 10−8; OR [95% CI], 1.18 [1.11–1.25]). Only four markers were located in the ±500 kb-region around the lead SNP. Consulting previous GWAS data (9) did not allow the restriction of the association signal to a specific gene encoded in the region, but suggests a second independent signal in the genomic region (see Figure E1B). Conditional regression analysis in this dataset suggests an independent signal, represented by rs17469010 (P = 0.002; OR [95% CI], 1.68 [1.21–2.33]). Subphenotype analysis did not reveal any significant differential effects for this marker (see Table E5).
The complete results for the 44 selected SNPs, including P values for the Breslow-Day test, are listed in Tables E6 and E7. None of the four novel risk variants showed a significant association in the African American sample (panel D). Power calculations revealed limited power to replicate the findings in panel D (rs653178, 14.6%; rs4921492, 42.7%; rs223498, 26.1%; rs6748088, 21.4%). In the German trio panel E, one of the newly identified risk SNPs (rs4921492) showed nominal significance (P = 0.039; OR [95% CI], 1.27 [1.01–1.59]) (see Table E8). The statistical power to detect the three remaining association signals in panel E was low with 17.6% (rs653178) and 14.0% (rs223498 and rs6748088), respectively.
Candidates and Secondary Signals
One SNP in the TYK2 gene region, rs34536443, was associated with nearly genome-wide significance in the joint analysis of the European case-control panels (P = 5.48 × 10−8; OR [95% CI], 0.64 [0.55–0.76]) (see Table E3) and showed the same trend in the German trio panel E (P = 0.086; OR, 0.55 [0.27–1.10]) (see Table E4). It causes an amino acid change in the TYK2 protein (Pro1104Ala) and has therefore a high potential to be functionally relevant. No data were available for this SNP for the African American sample (panel D). Subphenotype analysis in panel F suggests an association with fibrosis compared with patients with no fibrosis (P = 0.01; OR [95% CI], 4.59 [1.33–15.76]; allele frequencyfibrotic = 0.06; allele frequencynonfibrotic = 0.01).
Furthermore, two SNPs in the known sarcoidosis risk locus IL23R on chromosome 1p31.3 were associated with genome-wide significance in panel A (rs12069782: P = 4.20 × 10−8; OR [95% CI], 1.30 [1.18–1.43] and rs12090164: P = 4.51 × 10−8; OR [95% CI], 1.30 [1.18–1.42]) (see Tables E6 and E7), of which rs12090164 was omitted from follow-up because of complete LD (r2 = 1). Subphenotype analysis did not reveal any subphenotype-differential effects (see Table E9). Conditional regression analysis showed that the association signal of rs12069782 was independent of the association previously reported (11, 29). The online supplement provides more details. The lead SNP rs12069782 was subjected to replication in the European case-control populations (panels B-I, B-II, C-I, and C-II). In this analysis the ORs ranged from 1.03 to 1.46, with P = 3.07 × 10−10 (OR [95% CI], 1.24 [1.16–1.33]) in the joint analysis of panels A, B, and C (see Tables E6 and E7). However, the association was not confirmed in the African American panel D (power, 68.3%) or the German trio panel E (power, 14.7%) (see Table E8).
Risk Variants in the HLA Region
The combined analysis of single markers in the HLA region and imputed classical HLA alleles in panel A revealed a total of 1,172 associated markers of the extended HLA region (chr6: 25–35 Mb; P < 5.0 × 10−8), comprising three independent association signals (see online supplement for complete results).
The BTNL2 Region
The strongest association in the HLA region was represented by marker rs5007259 in the BTNL2 promoter region (P = 1.55 × 10−35; OR [95% CI], 0.60 [0.55–0.65]) (Figure 2). In a recently published association study comprising panel D (samples of African American [AA] origin) and additional sarcoidosis patients and control subjects of European American (EA) origin (30), marker rs5007259 was associated with nominal significance in both subsets (pAA = 3.80 × 10−3; pEA = 7.05 × 10−7). Table E10 provides complete results on overlapping signals in the HLA region. Subphenotype analysis of rs5007259 revealed a nominally significant association with Löfgren syndrome, with OR ranging from 0.37 to 0.52. Complete results for the subphenotype analysis of this marker are given in Table E11.
Figure 2.
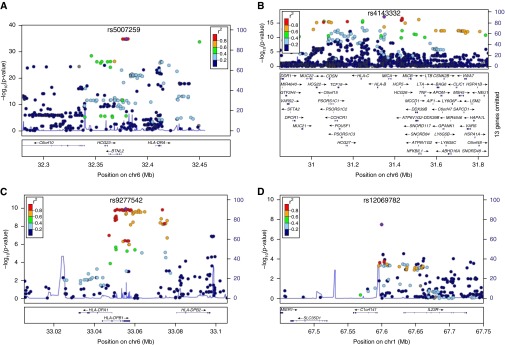
Association signals with genome-wide significance in the screening panel A. (A) BTNL2 (rs5007259), (B) HLA-B (rs4143332), (C) HLA-DPB1 (rs9277542), and (D) IL23R (rs12069782). For B and C, the depicted P values are derived from the sequential conditional regression analysis, conditioned on single-nucleotide polymorphism rs5007259 (B) and on rs5007259 and rs4143332 (C). See Figure 1 for descriptions.
The BTNL2 region was densely covered by the Immunochip, allowing fine-mapping of the major signal to the putative promoter region of BTNL2 (see Figure E2). The previously reported functional splice site variant rs2076530 was significantly associated (panel A; P = 9.64 × 10−25; OR [95% CI], 0.65 [0.60–0.70]), however no longer significant after including the allelic dosage for rs5007259 as a covariate (i.e., conditioning on rs5007259) (Pcond = 0.097). In contrast, the association of rs5007259 remained highly significant (Pcond = 5.00 × 10−13). According to FuncPred, the most strongly associated variants in the promoter region are predicted to reside in TF binding sites.
Analysis of HLA-haplotypes using logistic regression analysis revealed a significant association of HLA-DRB1-haplotypes (*0101: P = 1.42 × 10−18; OR [95% CI], 0.48 [0.41–0.57] and *0301: P = 4.99 × 10−19; OR [95% CI], 1.65 [1.48–1.84]) with sarcoidosis, a finding that has most consistently been reported (reviewed in Reference 31). HLA-DRB1*0301 was associated also in the small sample of patients with Löfgren syndrome (P = 2.25 × 10−13; OR [95% CI], 4.20 [2.86–6.86]), confirming previous findings (32). The BTNL2 SNPs rs5007259 remained significantly associated after conditioning on HLA-DRB1*0101 (P = 1.63 × 10−23; OR [95% CI], 0.65 [0.60–0.71]) and HLA-DRB1*0301 (P = 5.63 × 10−25; OR [95% CI], 0.64 [0.59–0.70]) in our study population. Vice versa, the association of both haplotypes was diminished by conditioning on rs5007259 (P = 2.29 × 10−8; OR [95% CI], 0.61 [0.52–0.73] and P = 5.43 × 10−7; OR [95% CI], 1.34 [1.20–1.51]).
HLA-B and HLA-DPB1
When conditioning all markers and the imputed haplotypes in the HLA region on rs5007259, marker rs4143332, which is located upstream of HLA-B, yielded the smallest P value (Pcond = 7.73 × 10−18; OR [95% CI], 1.64 [1.47–1.84]) (Figure 2). The most significantly associated haplotype HLA-B*0801 showed an association signal of similar strength (Pcond = 2.56 × 10−17; OR [95% CI], 1.63 [1.45–1.82]). Subphenotype analysis of rs4143332 revealed a significant difference in allele frequency between patients affected by acute and chronic sarcoidosis (OR, ≤2.21), and an even stronger differential effect for Löfgren syndrome (OR, ≤2.85). Table E12 provides complete results of the subphenotype analysis for this marker.
Conditioning on both rs5007259 and rs4143332 revealed a third independent association signal with genome-wide significance in the HLA region that is represented by SNP rs9277542 (Pcond = 1.13 × 10−10; OR [95% CI], 1.32 [1.21–1.44]) (Figure 2). In panel F, this marker displayed a differential effect for skin involvement (P = 7.13 × 10−3; OR [95% CI], 0.62 [0.44–0.88]). The complete results for the subphenotype association analysis are given in Table E13. Marker rs9277542 is located in the 3′-UTR of HLA-DPB1. None of the surrounding highly associated markers confer an amino acid change in the encoded protein, but functional predictions suggest a role in expression regulation via allele-specific miRNA binding (see Table E14) and alternative splicing according to SNPinfo (33). The online supplement and Data File E1 provide results of imputed HLA-haplotype analysis, subphenotype analysis, and complete information on the association of HLA haplotypes. Association signals of established sarcoidosis risk variants are given in the online supplement.
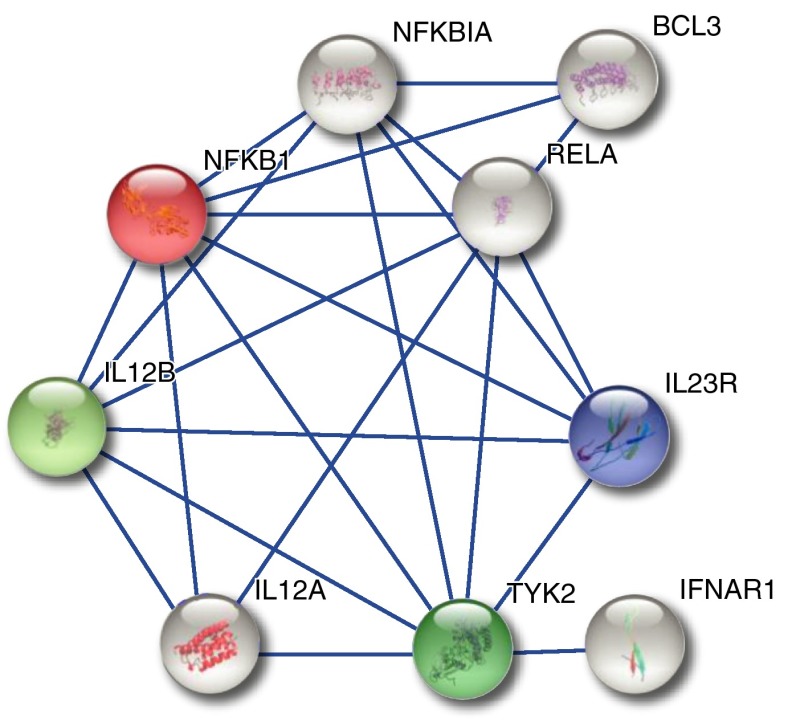
Protein Network Analysis
A network analysis of potentially affected gene products was performed using STRING (34). Proteins that were included in the analysis were selected based on the association and in silico analysis presented in this manuscript (i.e., BTNL2, HLA-DPB1, HLA-B, MICA, ANXA11, IL23R, SH2B3, CRIP1, IL12B, MANBA, NFKB1, and FAM117B) or were previously described or hypothesized as potentially affected (i.e., TNF, HLA-DRB1, HLA-DQB1, OS9, CYP27B1, KCNK4, CCDC88B, and RAB23). The analysis identified one prominent protein network, comprising key molecules of the IL12/IL23 signaling pathways (Figure 3).
Figure 3.
Protein interaction network, which was generated using STRING based on (1) coexpression of the respective transcripts, (2) experiments, and (3) information from (in parts) curated pathway databases, applying a confidence score >0.7. Colored nodes represent proteins that are or may be affected by sarcoidosis risk variants.
Sharing of Risk Loci and Cumulative Heritability
Our study demonstrated a considerable sharing pattern of sarcoidosis susceptibility factors, especially of the IL23/IL12 signaling pathway with other disorders and phenotypes, such as ankylosing spondylitis, inflammatory bowel disease, and psoriasis (IL23R, IL12B, and TYK2); rheumatoid arthritis (IL23R and TYK2); and multiple sclerosis (IL12B and TYK2; reviewed in Reference 17). However, the exact association patterns are not identical for these conditions as illustrated in a comparison of the IL12B and IL23R signal for sarcoidosis, psoriasis, and Crohn disease (35) (see Figure E3). Sequential conditional logistic regression analysis, calculation of LD, and test for epistasis proved the independence of the 11 sarcoidosis risk variants located in the BTNL2, HLA-B, HLA-DPB1, ANXA11, IL23R, ATXN2, IL12B, MANBA, FAM117B, chromosome 11q13.1, and RAB23 regions (see Figure E4). However, assuming a prevalence of 40 per 100,000, the cumulative heritability of the respective 11 lead variants was estimated to be only 2.78% (see Table E15).
Discussion
In our study, we successfully screened the largest sarcoidosis case-control study population to date for novel risk factors and identified chromosomes 12q24.12 (ATXN2/SH2B3), 5q33.3 (near IL12B), 4q24 (MANBA/NFKB1), and 2q33.2 (FAM117B) as susceptibility regions for sarcoidosis in the investigated European samples. We further found a novel, independent association signal on chromosome 1p31.3 (IL23R). Because of the extraordinary sample size and the applied level of significance, our findings are highly reliable to represent true genetic risk factors for sarcoidosis in Europeans. However, the analysis of the associated markers in an African American sarcoidosis population was not successful, which could have power (<68% throughout all tests), genetic, and functional reasons. This discrepancy points out the need for explicit transancestry association studies to get insight into the genetic architecture of sarcoidosis across populations of diverse ancestry. Analysis of sarcoidosis subphenotypes revealed several findings with nominal significance. However, because of the large numbers of tests, most of these results would not remain significant after correction for multiple testing and thus have to be investigated in independent samples for validation. Therefore these findings are not discussed in detail here.
The identified sarcoidosis susceptibility loci harbor genetic variants that may be functionally relevant in the context of sarcoidosis pathogenesis, affecting either known or novel players. First, variant rs3184504 on chromosome 12q24.12 induces an amino acid change (R262W) in the SH2B3 protein, an intriguing candidate. SH2B3 is involved in B-cell proliferation (36) and the endothelial response to TNF (37) and has not yet been implicated in sarcoidosis pathogenesis. The rs3184504*A risk allele is associated with stronger activation of the NOD2 recognition pathway in response to lipopolysaccharide and muramyl dipeptide, suggesting SH2B3 may play a role in protection against bacterial infection (38). Second, among the associated markers in the chromosome 5q33.3 region, SNP rs1422877 confers allele-specific binding probabilities for the TF RelA, which could influence the expression of neighboring genes (e.g., of the IL12B gene that encodes the p40 subunit of the IL12 and the IL23 receptors). Third, the lead SNP in the chromosome 1p31.3 region is located in the putative promoter region of the IL23R gene and might therefore influence IL23R expression.
Fourth, the associated variants on chromosome 2q33.2 could affect the expression of many genes, among them BMPR2, which is involved in the regulation of inflammatory processes in the lung (39) and has been suggested as a core mediator in sarcoidosis pathogenesis in comparison with idiopathic pulmonary fibrosis (40). Fifth, allelic variation of the candidate risk SNP rs34536443 (TYK2 Pro1104Ala) is known to influence Th2 lymphocyte polarization (41). The risk allele “C” reduces TYK2 activity in T lymphocytes and shifts the cytokine secretion toward a Th2 cytokine profile. This shift is also observed in severe sarcoidosis with progressing pulmonary fibrosis (42). Strikingly, the rare rs34536443-C allele is strongly associated with fibrosis among patients with sarcoidosis in panel F. Because of the large effect size (OR, 4.6), this finding provides potential for clinical application as a marker for high-risk patients, given a successful independent replication. Functional hypotheses on risk variants in the remaining novel susceptibility regions are given in the online supplement. Because the identification of genetic defects and deranged pathways does not necessarily imply a direct link to immunopathogenesis or therapy, further experimental work is necessary to assess these previously mentioned hypotheses and to clarify the potential role of the associated variants in the specific molecular processes in sarcoidosis pathogenesis and whether they represent useful targets in a therapeutic approach.
A network analysis of the potentially affected gene products defined in this study identified one prominent protein network, comprising key molecules of the IL12/IL23 signaling pathway (reviewed in Reference 17). From these data, we hypothesize that the IL12/Th1 and IL23/Th17 signaling pathways could be affected by the described risk alleles. Both Th1 and Th17 cells are present in granulomas of patients with sarcoidosis and are known to play a crucial role in disease process (43, 44). This is in line with emerging reports on the role of Th17 T cells in sarcoidosis and confirms Th1 cells as well-established players in disease pathogenesis (44, 45).
Of note, the two loci IL12B and IL23R are shared risk loci for sarcoidosis, the skin disease psoriasis, and the granulomatous Crohn disease, yet with differing association patterns. Given a more detailed genetic investigation, this information could be of value, because targeting the IL12/IL23 signaling pathway using the p40 antibody ustekinumab is approved in psoriasis treatment and also seems promising in the therapy of severe Crohn disease (46, 47). However, for sarcoidosis no such positive effect was observed (48). Because genetic data were not incorporated in this ustekinumab trial in sarcoidosis, we hypothesize that genetic information might aid the identification of a subset of patients with sarcoidosis that may have a higher chance to respond to the treatment, alike reported for a variant in the TNF locus and anti-TNF treatment (49). Functional and clinical studies are necessary to evaluate the usefulness of ustekinumab in such a genetically defined subcohort.
The HLA region has long been known and is most consistently reported to be associated with sarcoidosis. Our group had reported the association of BTNL2 and suggested the splice variant rs2076530 as the underlying risk factor before (2). Since then, this association has been replicated in other European and non-European populations (4, 50–55). However, in this study the most strongly associated variants were located in the BTNL2 promoter region and are predicted to reside in TF binding sites. They may therefore influence expression of BTNL2 by altering TF binding. Thus, this result may point to a different underlying mechanism, although it confirms BTNL2 as a risk factor for sarcoidosis.
The second independent signal in the HLA region peaked near HLA-B with the common haplotype HLA-B*0801 showing the strongest effect, with lead SNP rs4143332 being significantly associated with acute sarcoidosis and Löfgren syndrome, which is in line with previous findings (56, 57). It is part of the so-called “8.1 ancestral haplotype” (HLA A*0101: Cw*0701: B*0801: DRB1*0301: DQA1*0501: DQB1*0201), which is common in Europeans and includes a large number of genes related to the immune system. The 8.1 ancestral haplotype has been repeatedly reported in the context of sarcoidosis susceptibility and progression (56, 57) and is associated with several other immune-related diseases (58), such as systemic lupus erythematosus and primary sclerosing cholangitis (21, 59).
The third signal in the HLA-region is located in the 5′-UTR region of HLA-DPB1. This gene encodes a subunit of the HLA-DP receptor that plays a central role in the immune system by presenting peptides derived from extracellular proteins. HLA-DPB1 genotypes had been linked to sarcoidosis before (60–63) and the Glu69 variant (rs1042140) is known to be associated with chronic beryllium disease, a phenocopy of sarcoidosis (64). In our study population, none of the Glu69-containing haplotypes were associated with genome-wide significance (data not shown). Instead, this is the first report that locates this association at HLA-DPB1 to the 5′-UTR region of the gene, where the associated markers are predicted to affect miRNA binding and alternative splicing. The online supplement provides information for further functional hypotheses on associated variants. Overall, the genetic findings reported here are highly reliable, setting a most solid ground for subsequent experimental validation.
Altogether based on our study and previous findings, we defined 11 sarcoidosis risk loci (BTNL2, HLA-B, HLA-DPB1, ANXA11, IL23R, SH2B3/ATXN2, IL12B, NFKB1/MANBA, FAM177B, chromosome 11q13.1, and RAB23). However, the cumulative heritability of the 11 lead variants was estimated to be only 2.78%. This means that additional genetic risk factors for sarcoidosis remain to be discovered, which may include common variants with even smaller effects or risk alleles with lower frequencies, which were not targeted with the SNP selection strategy applied in this study. The detected loci show a considerable sharing pattern with other disorders and phenotypes. This overlap is supportive of the hypothesis that sarcoidosis shares a common genetic background with other immune-related diseases. Thus, our findings may inspire a revised classification of clinical disease manifestations and subphenotypes and may provide hypotheses for novel therapeutic targets.
Acknowledgments
Acknowledgment
The authors thank all study participants, families, and physicians for their contribution, and the German Sarcoidosis Patients Organization (Deutsche Sarkoidose Vereinigung e.V.) and the Popgen biobank for their support. They further thank the laboratory team of the Institute of Clinical Molecular Biology (Kiel, Germany) and The Swedish Epidemiological Investigation of Rheumatoid Arthritis group for technical assistance.
Footnotes
Supported by the Federal Ministry for Education and Research in Germany through the National Genome Research Network, within the framework of the e:Med (sysINFLAME) research and funding concept (01ZX1306A), the Genome Research Network “MooDS,” and the PopGen 2.0 network (01EY1103); by the Deutsche Forschungsgemeinschaft through grant FI 1935/1-1, MU 692/8-1, and BR 1912/6-1 and through the Clusters of Excellence “Inflammation at Interfaces” and “ImmunoSensation”; by the Palacky University (IGA PU LF 2015_020); by the Swedish Heart-Lung Foundation, the Swedish Medical Research Council, through the regional agreement on medical training and clinical research between Stockholm County Council and the Karolinska Institutet; by the Heinz Nixdorf Foundation and the Else-Kröner-Fresenius-Stiftung (Else Kröner-Exzellenzstipendium 2010_EKES.32); and by the National Institutes of Health/NHLBI (grant 1RC2HL101499). The KORA research platform (KORA, Cooperative Research in the Region of Augsburg) was initiated and financed by the Helmholtz Zentrum München–German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences, Ludwig-Maximilians-Universität, as part of LMUinnovativ.
Author Contributions: Study design, data analysis, and manuscript draft, A. Fischer. Data analysis, D.E., M.N., H.B., and C.G.M. Samples or datasets, C.G.M., M.C.I., B.A.R., M.P., F.M., S.P., C. Grohé, J.G., M.R., A.E., L.P., V.M.-V., D.J., M. Sterclova, J.H., M.M.N., S. Herms, C. Gieger, K.S., J.W., B.O.B., S.B., C.B., M. Schürmann, E.E., W.L., J.M.-Q., A. Franke, and S.S. Study design, S. Hofmann, A.N., A. Franke, and S.S. Final manuscript, all authors.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201503-0418OC on June 6, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Valeyre D, Prasse A, Nunes H, Uzunhan, Brillet PY, Muller-Quernheim J. Sarcoidosis. Lancet. 2014;383:1155–1167. doi: 10.1016/S0140-6736(13)60680-7. [DOI] [PubMed] [Google Scholar]
- 2.Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Wollnik B, Pabst S, Lennarz M, Rohmann E, Gillissen A, Vetter H, Grohé C. BTNL2 gene variant and sarcoidosis. Thorax. 2006;61:273–274. doi: 10.1136/thx.2005.056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rybicki BA, Walewski JL, Maliarik MJ, Kian H, Iannuzzi MC ACCESS Research Group. The BTNL2 gene and sarcoidosis susceptibility in African Americans and whites. Am J Hum Genet. 2005;77:491–499. doi: 10.1086/444435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI, Schürmann M, Müller-Quernheim J, Krawczak M, Rosenstiel P, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet. 2008;40:1103–1106. doi: 10.1038/ng.198. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Pabst S, Kubisch C, Grohé C, Wollnik B. First independent replication study confirms the strong genetic association of ANXA11 with sarcoidosis. Thorax. 2010;65:939–940. doi: 10.1136/thx.2010.138743. [DOI] [PubMed] [Google Scholar]
- 7.Morais A, Lima B, Peixoto M, Melo N, Alves H, Marques JA, Delgado L. Annexin A11 gene polymorphism (R230C variant) and sarcoidosis in a Portuguese population. Tissue Antigens. 2013;82:186–191. doi: 10.1111/tan.12188. [DOI] [PubMed] [Google Scholar]
- 8.Levin AM, Iannuzzi MC, Montgomery CG, Trudeau S, Datta I, McKeigue P, Fischer A, Nebel A, Rybicki BA. Association of ANXA11 genetic variation with sarcoidosis in African Americans and European Americans. Genes Immun. 2013;14:13–18. doi: 10.1038/gene.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer A, Schmid B, Ellinghaus D, Nothnagel M, Gaede KI, Schürmann M, Lipinski S, Rosenstiel P, Zissel G, Höhne K, et al. A novel sarcoidosis risk locus for Europeans on chromosome 11q13.1. Am J Respir Crit Care Med. 2012;186:877–885. doi: 10.1164/rccm.201204-0708OC. [DOI] [PubMed] [Google Scholar]
- 10.Müller-Quernheim J, Schürmann M, Hofmann S, Gaede KI, Fischer A, Prasse A, Zissel G, Schreiber S. Genetics of sarcoidosis. Clin Chest Med. 2008;29:391–414, viii. doi: 10.1016/j.ccm.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Fischer A, Nothnagel M, Franke A, Jacobs G, Saadati HR, Gaede KI, Rosenstiel P, Schürmann M, Müller-Quernheim J, Schreiber S, et al. Association of inflammatory bowel disease risk loci with sarcoidosis, and its acute and chronic subphenotypes. Eur Respir J. 2011;37:610–616. doi: 10.1183/09031936.00049410. [DOI] [PubMed] [Google Scholar]
- 12.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair RP, Ruether A, Stuart PE, Jenisch S, Tejasvi T, Hiremagalore R, Schreiber S, Kabelitz D, Lim HW, Voorhees JJ, et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J Invest Dermatol. 2008;128:1653–1661. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rueda B, Orozco G, Raya E, Fernandez-Sueiro JL, Mulero J, Blanco FJ, Vilches C, González-Gay MA, Martin J. The IL23R Arg381Gln non-synonymous polymorphism confers susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2008;67:1451–1454. doi: 10.1136/ard.2007.080283. [DOI] [PubMed] [Google Scholar]
- 16.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 18.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, Bakker SF, Bardella MT, Bhaw-Rosun L, Castillejo G, et al. Spanish Consortium on the Genetics of Coeliac Disease (CEGEC); PreventCD Study Group; Wellcome Trust Case Control Consortium (WTCCC) Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, et al. Collaborative Association Study of Psoriasis (CASP); Genetic Analysis of Psoriasis Consortium; Psoriasis Association Genetics Extension; Wellcome Trust Case Control Consortium 2. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu JZ, Hov JR, Folseraas T, Ellinghaus E, Rushbrook SM, Doncheva NT, Andreassen OA, Weersma RK, Weismüller TJ, Eksteen B, et al. UK-PSCSC Consortium; International IBD Genetics Consortium; International PSC Study Group. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JZ, Almarri MA, Gaffney DJ, Mells GF, Jostins L, Cordell HJ, Ducker SJ, Day DB, Heneghan MA, Neuberger JM, et al. UK Primary Biliary Cirrhosis (PBC) Consortium; Wellcome Trust Case Control Consortium 3. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2012;44:1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodríguez E, Matanovic A, Marenholz I, Hübner N, Schaarschmidt H, Novak N, Michel S, et al. High-density genotyping study identifies four new susceptibility loci for atopic dermatitis. Nat Genet. 2013;45:808–812. doi: 10.1038/ng.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, Goris A, et al. International Multiple Sclerosis Genetics Consortium (IMSGC); Wellcome Trust Case Control Consortium 2 (WTCCC2); International IBD Genetics Consortium (IIBDGC) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Statement on sarcoidosis. Joint Statement of the American Thoracic Society (ATS), the European Respiratory Society (ERS) and the World Association of Sarcoidosis and Other Granulomatous Disorders (WASOG) adopted by the ATS Board of Directors and by the ERS Executive Committee, February 1999. Am J Respir Crit Care Med. 1999;160:736–755. doi: 10.1164/ajrccm.160.2.ats4-99. [DOI] [PubMed] [Google Scholar]
- 26.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;60:2.12.1–2.12.18. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–972. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HS, Choi D, Lim LL, Allada G, Smith JR, Austin CR, Doyle TM, Goodwin KA, Rosenbaum JT, Martin TM. Association of interleukin 23 receptor gene with sarcoidosis. Dis Markers. 2011;31:17–24. doi: 10.3233/DMA-2011-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, Parker R, Adler A, Kelly JA, Kaufman KM, Lessard CJ, et al. Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS One. 2012;7:e43907. doi: 10.1371/journal.pone.0043907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer A, Grunewald J, Spagnolo P, Nebel A, Schreiber S, Müller-Quernheim J. Genetics of sarcoidosis. Semin Respir Crit Care Med. 2014;35:296–306. doi: 10.1055/s-0034-1376860. [DOI] [PubMed] [Google Scholar]
- 32.Grunewald J, Brynedal B, Darlington P, Nisell M, Cederlund K, Hillert J, Eklund A. Different HLA-DRB1 allele distributions in distinct clinical subgroups of sarcoidosis patients. Respir Res. 2010;11:25. doi: 10.1186/1465-9921-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellinghaus D, Ellinghaus E, Nair RP, Stuart PE, Esko T, Metspalu A, Debrus S, Raelson JV, Tejasvi T, Belouchi M, et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90:636–647. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaki S, Sauer K, Iritani BM, Chien S, Ebihara Y, Tsuji K, Takatsu K, Perlmutter RM. Control of B cell production by the adaptor protein lnk. Definition of a conserved family of signal-modulating proteins. Immunity. 2000;13:599–609. doi: 10.1016/s1074-7613(00)00060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitau J, Boulday G, Coulon F, Quillard T, Charreau B. The adaptor molecule Lnk negatively regulates tumor necrosis factor-alpha-dependent VCAM-1 expression in endothelial cells through inhibition of the ERK1 and -2 pathways. J Biol Chem. 2006;281:20148–20159. doi: 10.1074/jbc.M510997200. [DOI] [PubMed] [Google Scholar]
- 38.Zhernakova A, Elbers CC, Ferwerda B, Romanos J, Trynka G, Dubois PC, de Kovel CG, Franke L, Oosting M, Barisani D, et al. Finnish Celiac Disease Study Group. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 2010;86:970–977. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talati M, West J, Zaynagetdinov R, Hong CC, Han W, Blackwell T, Robinson L, Blackwell TS, Lane K. BMP pathway regulation of and by macrophages. PLoS One. 2014;9:e94119. doi: 10.1371/journal.pone.0094119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leng D, Huan C, Xie T, Liang J, Wang J, Dai H, Wang C, Jiang D. Meta-analysis of genetic programs between idiopathic pulmonary fibrosis and sarcoidosis. PLoS One. 2013;8:e71059. doi: 10.1371/journal.pone.0071059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Couturier N, Bucciarelli F, Nurtdinov RN, Debouverie M, Lebrun-Frenay C, Defer G, Moreau T, Confavreux C, Vukusic S, Cournu-Rebeix I, et al. Tyrosine kinase 2 variant influences T lymphocyte polarization and multiple sclerosis susceptibility. Brain. 2011;134:693–703. doi: 10.1093/brain/awr010. [DOI] [PubMed] [Google Scholar]
- 42.Patterson KC, Franek BS, Müller-Quernheim J, Sperling AI, Sweiss NJ, Niewold TB. Circulating cytokines in sarcoidosis: phenotype-specific alterations for fibrotic and non-fibrotic pulmonary disease. Cytokine. 2013;61:906–911. doi: 10.1016/j.cyto.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357:2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- 44.Facco M, Cabrelle A, Teramo A, Olivieri V, Gnoato M, Teolato S, Ave E, Gattazzo C, Fadini GP, Calabrese F, et al. Sarcoidosis is a Th1/Th17 multisystem disorder. Thorax. 2011;66:144–150. doi: 10.1136/thx.2010.140319. [DOI] [PubMed] [Google Scholar]
- 45.Ten Berge B, Paats MS, Bergen IM, van den Blink B, Hoogsteden HC, Lambrecht BN, Hendriks RW, Kleinjan A. Increased IL-17A expression in granulomas and in circulating memory T cells in sarcoidosis. Rheumatology (Oxford) 2012;51:37–46. doi: 10.1093/rheumatology/ker316. [DOI] [PubMed] [Google Scholar]
- 46.Sandborn WJ, Gasink C, Gao LL, Blank MA, Johanns J, Guzzo C, Sands BE, Hanauer SB, Targan S, Rutgeerts P, et al. CERTIFI Study Group. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 47.Kopylov U, Afif W, Cohen A, Bitton A, Wild G, Bessissow T, Wyse J, Al-Taweel T, Szilagyi A, Seidman E. Subcutaneous ustekinumab for the treatment of anti-TNF resistant Crohn’s disease—the McGill experience. J Crohn's Colitis. 2014;8:1516–1522. doi: 10.1016/j.crohns.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Judson MA, Baughman RP, Costabel U, Drent M, Gibson KF, Raghu G, Shigemitsu H, Barney JB, Culver DA, Hamzeh NY, et al. Safety and efficacy of ustekinumab or golimumab in patients with chronic sarcoidosis. Eur Respir J. 2014;44:1296–1307. doi: 10.1183/09031936.00000914. [DOI] [PubMed] [Google Scholar]
- 49.Wijnen PA, Cremers JP, Nelemans PJ, Erckens RJ, Hoitsma E, Jansen TL, Bekers O, Drent M. Association of the TNF-α G-308A polymorphism with TNF-inhibitor response in sarcoidosis. Eur Respir J. 2014;43:1730–1739. doi: 10.1183/09031936.00169413. [DOI] [PubMed] [Google Scholar]
- 50.Spagnolo P, Sato H, Grutters JC, Renzoni EA, Marshall SE, Ruven HJ, Wells AU, Tzouvelekis A, van Moorsel CH, van den Bosch JM, et al. Analysis of BTNL2 genetic polymorphisms in British and Dutch patients with sarcoidosis. Tissue Antigens. 2007;70:219–227. doi: 10.1111/j.1399-0039.2007.00879.x. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki H, Ota M, Meguro A, Katsuyama Y, Kawagoe T, Ishihara M, Asukata Y, Takeuchi M, Ito N, Shibuya E, et al. Genetic characterization and susceptibility for sarcoidosis in Japanese patients: risk factors of BTNL2 gene polymorphisms and HLA class II alleles. Invest Ophthalmol Vis Sci. 2012;53:7109–7115. doi: 10.1167/iovs.12-10491. [DOI] [PubMed] [Google Scholar]
- 52.Cozier Y, Ruiz-Narvaez E, McKinnon C, Berman J, Rosenberg L, Palmer J. Replication of genetic loci for sarcoidosis in US black women: data from the Black Women’s Health Study. Hum Genet. 2013;132:803–810. doi: 10.1007/s00439-013-1292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morais A, Lima B, Peixoto MJ, Alves H, Marques A, Delgado L. BTNL2 gene polymorphism associations with susceptibility and phenotype expression in sarcoidosis. Respir Med. 2012;106:1771–1777. doi: 10.1016/j.rmed.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Milman N, Svendsen CB, Nielsen FC, van Overeem Hansen T. The BTNL2 A allele variant is frequent in Danish patients with sarcoidosis. Clin Respir J. 2011;5:105–111. doi: 10.1111/j.1752-699X.2010.00206.x. [DOI] [PubMed] [Google Scholar]
- 55.Wennerström A, Pietinalho A, Lasota J, Salli K, Surakka I, Seppänen M, Selroos O, Lokki ML. Major histocompatibility complex class II and BTNL2 associations in sarcoidosis. Eur Respir J. 2013;42:550–553. doi: 10.1183/09031936.00035213. [DOI] [PubMed] [Google Scholar]
- 56.Idali F, Wikén M, Wahlström J, Mellstedt H, Eklund A, Rabbani H, Grunewald J. Reduced Th1 response in the lungs of HLA-DRB1*0301 patients with pulmonary sarcoidosis. Eur Respir J. 2006;27:451–459. doi: 10.1183/09031936.06.00067105. [DOI] [PubMed] [Google Scholar]
- 57.Grunewald J, Eklund A, Olerup O. Human leukocyte antigen class I alleles and the disease course in sarcoidosis patients. Am J Respir Crit Care Med. 2004;169:696–702. doi: 10.1164/rccm.200303-459OC. [DOI] [PubMed] [Google Scholar]
- 58.Price P, Witt C, Allcock R, Sayer D, Garlepp M, Kok CC, French M, Mallal S, Christiansen F. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 59.Morris DL, Taylor KE, Fernando MM, Nititham J, Alarcón-Riquelme ME, Barcellos LF, Behrens TW, Cotsapas C, Gaffney PM, Graham RR, et al. International MHC and Autoimmunity Genetics Network; Systemic Lupus Erythematosus Genetics Consortium. Unraveling multiple MHC gene associations with systemic lupus erythematosus: model choice indicates a role for HLA alleles and non-HLA genes in Europeans. Am J Hum Genet. 2012;91:778–793. doi: 10.1016/j.ajhg.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lympany PA, Petrek M, Southcott AM, Newman Taylor AJ, Welsh KI, du Bois RM. HLA-DPB polymorphisms: Glu 69 association with sarcoidosis. Eur J Immunogenet. 1996;23:353–359. doi: 10.1111/j.1744-313x.1996.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 61.Maliarik MJ, Chen KM, Major ML, Sheffer RG, Popovich J, Jr, Rybicki BA, Iannuzzi MC. Analysis of HLA-DPB1 polymorphisms in African-Americans with sarcoidosis. Am J Respir Crit Care Med. 1998;158:111–114. doi: 10.1164/ajrccm.158.1.9708111. [DOI] [PubMed] [Google Scholar]
- 62.Schürmann M, Bein G, Kirsten D, Schlaak M, Müller-Quernheim J, Schwinger E. HLA-DQB1 and HLA-DPB1 genotypes in familial sarcoidosis. Respir Med. 1998;92:649–652. doi: 10.1016/s0954-6111(98)90512-1. [DOI] [PubMed] [Google Scholar]
- 63.Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA, Pandey JP, Newman LS, Magira E, Beznik-Cizman B, et al. ACCESS Group. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet. 2003;73:720–735. doi: 10.1086/378097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 1993;262:242–244. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]