To the Editor:
Three meta-analyses of genome-wide association studies (GWAS) in healthy subjects have revealed multiple loci for FEV1 and FEV1/FVC (1–3), but only one study included even a small proportion of children (∼7%) (3). A separate GWAS of lung function in children and young adults (including subjects with and without respiratory diseases) did not yield significant results but replicated prior findings in adults (4), and a study in Hutterites identified variants in the 15q23 locus related to lung function (5). The only GWAS of lung function in subjects with asthma (aged 12 years and older, mostly adults) did not show novel results but replicated findings in healthy subjects (6). No prior GWAS has examined post-bronchodilator (post-BD) FEV1 and FEV1/FVC, which may best reflect true lung function (and not solely disease severity) in subjects with asthma.
Even though Puerto Ricans share a disproportionate burden from asthma in the United States (7), there has been no GWAS of lung function in this ethnic group. GWAS of minority populations at risk are important from a public health perspective and can identify novel susceptibility variants for well-characterized phenotypes (8). Moreover, loci identified in patients without asthma likely influence lung function in patients with asthma (9), but unidentified variants may also influence lung function in subjects with asthma (10). We conducted a GWAS of understudied lung function phenotypes (post-BD FEV1 and FEV1/FVC) in a unique high-risk population (Puerto Rican children with asthma) and attempted to replicate our top findings in independent cohorts. We also examined whether previous findings for GWAS of pre-BD lung function were replicated for post-BD lung function.
From September 2003 to June 2010, 618 children with asthma (physician-diagnosed asthma and one or more episode of wheeze in the previous year) were recruited in Hartford, Connecticut (n = 267), and San Juan, Puerto Rico (n = 351), as described elsewhere (11). At both study sites, eligibility criteria included age 6–14 years and having four Puerto Rican grandparents. All participants completed a protocol that included questionnaires, blood sample collection, and spirometry (conducted with an EasyOne spirometer; NDD Medical Technologies, Andover, MA), following American Thoracic Society recommendations for children (12). After baseline spirometry, subjects were given 200 μg of an albuterol metered-dose inhaler, and spirometry was repeated after 15 minutes. The best FEV1 and FVC from each test were selected for analysis. Of the 618 participants, 560 had blood samples and sufficient DNA for genome-wide genotyping, conducted using the HumanOmni2.5 BeadChip platform (Illumina Inc., San Diego, CA), as previously described (11). After excluding subjects with low marker call rate (n = 31), sex mismatch (n = 6), or missing post-BD phenotypes (n = 76), 447 subjects remained in the analysis. Written parental consent was obtained for participating children, from whom written assent was also obtained. The study was approved by the institutional review boards of the University of Puerto Rico (San Juan, PR), Brigham and Women’s Hospital (Boston, MA), and the University of Pittsburgh (Pittsburgh, PA).
Replication was attempted in three cohorts of children with asthma: the Genetics of Asthma in Costa Rica Study (13), the Childhood Asthma Management Program (14), and the Genes-environments and Admixture in Latino Americans study (15). The Genotype-Tissue Expression Biorepository (16) was then used to assess the association between our top-replicated single nucleotide polymorphisms (SNPs) and expression of nearby genes in human tissues. Bronchial epithelial brushings were obtained from healthy nonsmokers: three males and one female (median age, 27 yr; range, 24–40 yr). Expression data were generated using Illumina TrueSeq RNA sequencing. RNAseq data were mapped to the human genome (build 37.2), and relative expression level was assessed by fragments per kilobase of transcript per million mapped reads (17).
For the GWAS, we built linear regression models under an additive genetic model, adjusting for age, sex, principal components, and height. The top 20 SNPs were then tested using the same genetic model in the replication cohorts. The results for the replication cohorts were combined in a meta-analysis with weights proportional to the inverse variance of the beta.
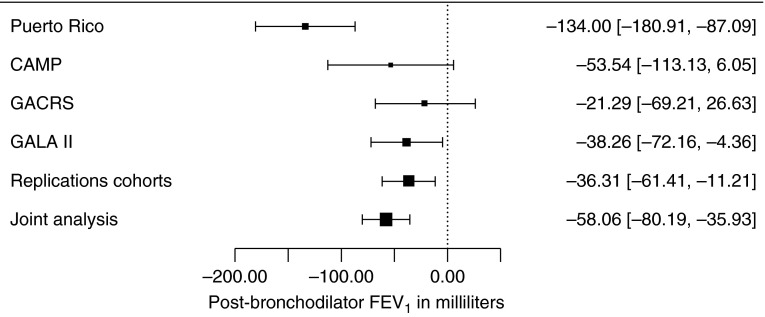
The main characteristics of study participants are shown in Table 1. Subjects at both study sites had similar age and lung function, but there was a slightly higher proportion of female subjects in Hartford than in San Juan. The GWAS results for both phenotypes did not show significant evidence of population stratification based on genomic inflation factors (see Figure E1 in the online supplement). Eight SNPs were associated with post-BD FEV1 at P < 9 × 10−7 in Puerto Ricans (Table E1 and Figure E2). Of these eight SNPs, two (rs7946574 and rs2984842) were associated with post-BD FEV1, at P < 5 × 10−8 (Figure E3). Results for rs7946574 were not significantly replicated, but findings for rs2984842 were replicated in the same direction of effect in all cohorts (combined P for the analysis in replication cohorts = 0.005) (Figure 1). This SNP, an intronic variant of the gene for fibroblast growth factor 14 (FGF14), is associated with increased expression of FGF14 in subcutaneous adipose tissue (P = 0.01) and lung (P = 0.10) (16). FGF14 was expressed in bronchial brushes from healthy nonsmokers at a level ∼2.5-fold greater than that for the cystic fibrosis transmembrane conductance regulator gene (CFTR) and 7-fold greater than that for the gene for hypoxanthine phosphoribosyltransferase 1 (HPRT1).
Table 1.
Baseline Characteristics of Participating Children
| Puerto Ricans (N = 447) | CAMP (N = 568) | GACRS (N = 556) | GALA II (N = 1,858) | |
|---|---|---|---|---|
| Age, yr, mean (SD) | 10 (2.7) | 8.9 (2.1) | 9.0 (2.0) | 12.5 (3.2) |
| Female sex | 225 (46%) | 228 (40%) | 230 (41%) | 835 (44.9%) |
| FEV1, L, mean (SD) | ||||
| Prebronchodilator | 1.9 (0.7) | 1.7 (0.5) | 1.7 (0.5) | 2.4 (0.8) |
| Post-bronchodilator | 2 (0.7) | 1.8 (0.5) | 1.8 (0.6) | 2.6 (0.9) |
| FEV1/FVC ratio, %, mean (SD) | ||||
| Prebronchodilator | 82 (9) | 79 (8) | 81 (13) | 84 (8) |
| Post-bronchodilator | 84 (9) | 85 (6) | 83 (17) | 88 (6) |
| FVC, L, mean (SD) | ||||
| Prebronchodilator | 2.3 (0.8) | 2.1 (0.6) | 2.1 (0.6) | 2.9 (1.0) |
| Post-bronchodilator | 2.4 (0.8) | 2.2 (0.6) | 2.1 (0.7) | 3.0 (1.0) |
| Genotyping platform(s) | Illumina Omni 2.5 M | Illumina 550 + 610k + custom arrays | Illumina 550 + 610 quad + custom arrays | Axiom LAT1 array (World Array 4) |
| Average (mean) ancestry | ||||
| European | Hartford, 63% | 99.1% | 55.7% | 53% |
| San Juan, 60% | ||||
| African | Hartford, 34% | 0.3% | 0.5% | 17% |
| San Juan, 36% | ||||
| Native American | Hartford, 3% | 0.6% | 43.8% | 30% |
| San Juan, 4% |
Definition of abbreviations: CAMP = Childhood Asthma Management Program; GACRS = Genetics of Asthma in Costa Rica Study; GALA II = Genes-environments and Admixture in Latino Americans.
Figure 1.
Forest plot of effect sizes on post-bronchodilator FEV1 (in milliliters) and 95% confidence intervals for rs2984842 in discovery and replication cohorts. CAMP = Childhood Asthma Management Program; GACRS = Genetics of Asthma in Costa Rica Study; GALA II = Genes-environments and Admixture in Latino Americans.
Five SNPs had genome-wide significant (P < 5 × 10−8) associations with post-BD FEV1/FVC in Puerto Ricans (Table E2, and Figures E2 and E4) but were not replicated in other cohorts.
We then tested whether SNPs identified by GWAS for pre-BD FEV1 are associated with post-BD FEV1 in Puerto Rican children. We identified four SNPs (tagging eight published SNPs for pre-BD FEV1) that were associated with post-BD FEV1 at P < 0.05 in the 4q24 locus (Table E3). This region contains multiple genes, including GSTCD, INTS12, and NPNT.
Our top replicated association for post-BD FEV1 (for rs2984842) is in FGF14, a gene in the fibroblast growth factor family that is most highly expressed in brain tissue; it is also expressed in lung, pituitary, and adipose tissues. Although FGF14 has not been associated with lung phenotypes, it was highly expressed in bronchial brushes from healthy adults, and other members of the FGF family (with which it shows homology) have been associated with lung development or disease, including FGF10 (18) and FGF7 (19). We also replicated previously published findings for pre-BD FEV1 in the 4q24 locus, using the post-BD FEV1 phenotype. Lack of replication of some previously identified variants may reflect lack of statistical power because of the small size of our discovery cohort, but it may also be a result of differences in linkage disequilibrium resulting from racial ancestry.
To our knowledge, this is the first GWAS of post-BD lung function and the first GWAS of lung function in Puerto Ricans. We have identified a novel gene associated with post-BD FEV1 in children with asthma. Further studies are needed to identify functional mechanisms.
Footnotes
The authors are supported by grants HL079966 and HL117191 from the U.S. National Institutes of Health (NIH) and by the Heinz Endowments. The Genetics of Asthma in Costa Rica Study was supported by grants HL066289 and HL04370 from the NIH. The Childhood Asthma Management Program Genetics Ancillary Study was supported by grants U01 HL075419, U01 HL65899, P01 HL083069, and R01 HL086601 from the NIH. J.M.B. was supported by grant K08HL111201 from the NIH. The Genes-environments and Admixture in Latino Americans study was supported by grants from the NIH to E.G.B. from the NHLBI (HL088133, HL004464, and HL117004); the National Institute of Environmental Health Sciences (ES015794); the National Institute on Minority Health and Health Disparities (MD006902); and the National Institute of General Medical Sciences (GM007546). E.G.B. was also supported by the Robert Wood Johnson Foundation Amos Medical Faculty Development Award, the American Asthma Foundation, and the Sandler Foundation. M.L.S. was supported by the National Science Foundation Graduate Research Fellowship under grant 1144247. M.P.-Y. was funded by a postdoctoral fellowship from Fundación Ramón Areces.
Author Contributions: J.M.B. and J.C.C. conceived of the study, participated in its design and coordination, and helped draft the manuscript. S.M.T., D.C.C.-C., D.H., Q.Y., M.P.-Y., and M.L.S. performed statistical analysis. E.G.B., G.C., E.F., A.A.L., B.A.R, W.C., N.B., E.A.-P., L.A., S.T.W., M.S.-Q., J.K.K., S.E.W., and M.M.C. participated in the design and coordination of the study. All authors read and approved the final manuscript.
This letter has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, Zhao JH, Ramasamy A, Zhai G, Vitart V, et al. Wellcome Trust Case Control Consortium; NSHD Respiratory Study Team. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ong BA, Li J, McDonough JM, Wei Z, Kim C, Chiavacci R, Mentch F, Caboot JB, Spergel J, Allen JL, et al. Gene network analysis in a pediatric cohort identifies novel lung function genes. PLoS ONE. 2013;8:e72899. doi: 10.1371/journal.pone.0072899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao TC, Du G, Han L, Sun Y, Hu D, Yang JJ, Mathias R, Roth LA, Rafaels N, Thompson EE, et al. Genome-wide association study of lung function phenotypes in a founder population. J Allergy Clin Immunol. 2014;133:248–255. doi: 10.1016/j.jaci.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Hawkins GA, Ampleford EJ, Moore WC, Li H, Hastie AT, Howard TD, Boushey HA, Busse WW, Calhoun WJ, et al. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmaticpatients. J Allergy Clin Immunol. 2013;132:313–320. doi: 10.1016/j.jaci.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen RT, Canino GJ, Bird HR, Shen S, Rosner BA, Celedón JC. Area of residence, birthplace, and asthma in Puerto Rican children. Chest. 2007;131:1331–1338. doi: 10.1378/chest.06-1917. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Howard TD, Moore WC, Ampleford EJ, Li H, Busse WW, Calhoun WJ, Castro M, Chung KF, Erzurum SC, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127:1457–1465. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imboden M, Bouzigon E, Curjuric I, Ramasamy A, Kumar A, Hancock DB, Wilk JB, Vonk JM, Thun GA, Siroux V, et al. Genome-wide association study of lung function decline in adults with and without asthma. J Allergy Clin Immunol. 2012;129:1218–1228. doi: 10.1016/j.jaci.2012.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brehm JM, Acosta-Pérez E, Klei L, Roeder K, Barmada MM, Boutaoui N, Forno E, Cloutier MM, Datta S, Kelly R, et al. African ancestry and lung function in Puerto Rican children. J Allergy Clin Immunol. 2012;129:1484–1490. doi: 10.1016/j.jaci.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 13.Hunninghake GM, Soto-Quiros ME, Avila L, Ly NP, Liang C, Sylvia JS, Klanderman BJ, Silverman EK, Celedón JC. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol. 2007;119:654–661. doi: 10.1016/j.jaci.2006.12.609. [DOI] [PubMed] [Google Scholar]
- 14.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 15.Pino-Yanes M, Thakur N, Gignoux CR, Galanter JM, Roth LA, Eng C, Nishimura KK, Oh SS, Vora H, Huntsman S, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol. 2015;135:228–235. doi: 10.1016/j.jaci.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium GT GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brehm JM, Hagiwara K, Tesfaigzi Y, Bruse S, Mariani TJ, Bhattacharya S, Boutaoui N, Ziniti JP, Soto-Quiros ME, Avila L, et al. Identification of FGF7 as a novel susceptibility locus for chronic obstructive pulmonary disease. Thorax. 2011;66:1085–1090. doi: 10.1136/thoraxjnl-2011-200017. [DOI] [PMC free article] [PubMed] [Google Scholar]