Abstract
Rationale: Hospitalization is associated with microbiome perturbation (dysbiosis), and this perturbation is more severe in patients treated with antimicrobials.
Objectives: To evaluate whether hospitalizations known to be associated with periods of microbiome perturbation are associated with increased risk of severe sepsis after hospital discharge.
Methods: We studied participants in the U.S. Health and Retirement Study with linked Medicare claims (1998–2010). We measured whether three hospitalization types associated with increasing severity of probable dysbiosis (non–infection-related hospitalization, infection-related hospitalization, and hospitalization with Clostridium difficile infection [CDI]) were associated with increasing risk for severe sepsis in the 90 days after hospital discharge. We used two study designs: the first was a longitudinal design with between-person comparisons and the second was a self-controlled case series design using within-person comparison.
Measurements and Main Results: We identified 43,095 hospitalizations among 10,996 Health and Retirement Study–Medicare participants. In the 90 days following non–infection-related hospitalization, infection-related hospitalization, and hospitalization with CDI, adjusted probabilities of subsequent admission for severe sepsis were 4.1% (95% confidence interval [CI], 3.8–4.4%), 7.1% (95% CI, 6.6–7.6%), and 10.7% (95% CI, 7.7–13.8%), respectively. The incidence rate ratio (IRR) of severe sepsis was 3.3-fold greater during the 90 days after hospitalizations than during other observation periods. The IRR was 30% greater after an infection-related hospitalization versus a non–infection-related hospitalization. The IRR was 70% greater after a hospitalization with CDI than an infection-related hospitalization without CDI.
Conclusions: There is a strong dose–response relationship between events known to result in dysbiosis and subsequent severe sepsis hospitalization that is not present for rehospitalization for nonsepsis diagnoses.
Keywords: microbiota, humans, dysbiosis, patient readmission, self-controlled case series
At a Glance Summary
Scientific Knowledge on the Subject
Hospitalization is associated with microbiome perturbation, especially in the setting of antibiotic therapy. Microbiome perturbation is, in turn, implicated in the pathogenesis of many diseases, including the inability to resist sepsis in animal models.
What This Study Adds to the Field
This study finds that the rate of sepsis is increased in the 90 days after hospital discharge and that the degree of increased sepsis risk correlates with type of prior hospitalization. Hospitalizations with higher probable microbiome perturbation are associated with greater risk of subsequent severe sepsis. This study raises the intriguing possibility that hospitalization-associated microbiome perturbation may be a mediator and actionable target for the pathogenesis of subsequent sepsis.
The role of the microbiome in human health is increasingly recognized (1, 2). Disruption to the microbiome (dysbiosis) is associated with increased host inflammation and has been implicated in the pathogenesis of many chronic diseases, including asthma (3), rheumatoid arthritis (4), inflammatory bowel disease (5), obesity (6), and cancer (7).
Animal studies have shown that gut microbiota may play an important role in the resistance to sepsis (8). However, there are few human studies in this regard. In a recent human case report, antibiotics were unable to resolve septic shock in a 29-year-old woman but fecal microbiota transplant cured her acute illness (9). Furthermore, small studies suggest that distortion of the microbiome is associated with the development of sepsis in preterm infants (10, 11). With this evidence in mind, we hypothesized that periods of dysbiosis may impair a patient’s ability to resist pathogenic assault. The disrupted microbial homeostasis and elevated inflammatory response inherent to dysbiosis create an environment conducive to the overgrowth of a single pathogen coupled with an overwhelming host inflammatory response (12, 13): the condition of severe sepsis. Determining whether dysbiosis is a risk factor for severe sepsis is critical because the human microbiome can be adjusted through therapeutic manipulation (14–16).
Rather than measure microbiome composition directly, we studied three major events that result in increasing degrees of probable dysbiosis: (1) hospitalization without infection, (2) hospitalization with infection, and (3) hospitalization with Clostridium difficile infection (CDI) (17–19). We hypothesized that all hospitalized patients experience diet, sleep, and lifestyle disruptions that perturb microbiome homeostasis (20), as confirmed through direct measurement of fecal diversity in hospitalized patients (17, 18). Patients hospitalized with infection experience further microbiome disruption, both from the infection and antimicrobial therapy (17, 19). Finally, C. difficile exploits a disordered microbiome, and thus serves as the gold-standard marker for dysbiosis (15, 21).
We tested whether (1) hospitalizations with probable dysbiosis are associated with increased risk of subsequent severe sepsis, (2) there is a correlation between the probable severity of dysbiosis during hospitalization and the magnitude of severe sepsis risk, and (3) there is specificity in the dose–response effect of hospitalization type on severe sepsis risk. Although this study cannot serve as definitive proof that dysbiosis is the mediator of the associations we test, we suggest that this work may be hypothesis-generating and brings a novel population-based perspective to the potential clinical importance of these associations. We have previously reported some of these results in an abstract (22).
Methods
Data Source
The Health and Retirement Study (HRS) is an ongoing, nationally representative, cohort study of older Americans. Started in 1992, the HRS has enrolled more than 37,000 participants, of whom 19,722 have agreed to link their data with Medicare (23). The cohort is reinterviewed every 2 years, with a follow-up rate consistently over 90% (23). Patients provided informed consent on enrollment in the HRS and again for linkage to Medicare. This work was approved by the University of Michigan Institutional Review Board.
Subjects
We included all fee-for-service Medicare beneficiaries with a baseline HRS survey in 1998–2008 for whom there were claims-based data on at least one hospitalization without inpatient mortality during 1998–2010.
Primary Outcome: Severe Sepsis Hospitalization
We identified severe sepsis hospitalizations with a commonly used, claims-based definition of severe sepsis that requires International Classification of Diseases (ICD)-9-CM codes for infection and acute organ dysfunction within the same hospitalization (24). This operationalization of severe sepsis has been validated against physician chart-review and found to have higher sensitivity and similar specificity to other claims-based definitions (25).
Exposures: Hospitalization, Hospitalization with Infection, and Hospitalization with CDI
Our exposures of interest were three types of hospitalizations: (1) non–infection-related hospitalizations, (2) infection-related hospitalizations (without CDI), and (3) hospitalizations with CDI (17, 18). We selected these exposures because we hypothesized that they would be associated with increasing magnitude of dysbiosis. We identified hospitalizations with infection as those with an ICD-9-CM code for infection (the same diagnostic codes included in the severe sepsis definition [24]) in any diagnosis field of the hospital claim. We identified hospitalization with CDI as those with ICD-9-CM code 008.45 (“intestinal infection due to Clostridium difficile”) in any diagnosis field. This identification method has been validated against a gold standard definition of positive toxin assay and/or visualization of pseudomembranes on colonoscopy, and been found to have high sensitivity and excellent specificity (26).
We considered the duration of the resulting microbiome perturbation to be the 90 days after hospitalization, because several studies suggest that the human microbiome closely resembles its predisturbance state within a few weeks to months after antibiotic exposure (27, 28). Any misspecification of the duration of the exposure biases our results toward the null hypothesis by imprecisely delineating higher and lower risk periods.
Covariates
We calculated Elixhauser comorbidities (n = 31) from inpatient and outpatient claims data for the year before each hospitalization (29). We identified functional limitations in six activities of daily living and five instrumental activities of daily living from the HRS survey immediately before each hospitalization (30). We also determined self-reported race, ethnicity, education, and wealth (sum of all assets and debts standardized to 2013 U.S. dollars) from survey data (23). Wealth, activities of daily living limitations, and instrumental activities of daily living limitations were missing in 0.1%, 4.1%, and 0.2%, respectively, and these values were imputed. The remaining covariates were present for 100% of the study population.
Statistical Analyses
We present categorical data as numbers (percentages) and continuous data as means (SDs) or medians (interquartile ranges) depending on the distribution. All analyses were conducted with Stata software version 13 (StataCorp, College Station, TX). We used two-sided hypothesis testing and set statistical significance at P less than 0.05.
Retrospective Longitudinal Design
We used multiple logistic regression models to evaluate the independent association between the three hospitalization types and probability of readmission for severe sepsis in the 90 days following live hospital discharge. In the regression model, we included all covariates (listed previously). We used hospitalization as the unit of analysis, adjusting for the nonindependence of observations within patients with Stata’s vce(cluster) command (31). We estimated missing covariate values (functional limitations, wealth) with multiple imputation by chained equations and five imputations (32).
To confirm that the observed differences in probability of severe sepsis following the three exposures represent differences in severe sepsis risk (not merely differences in propensity for hospital readmission), we also measured the associations with 90-day readmission for diagnoses other than severe sepsis. In the online supplement, we present supplemental analyses that account for patients’ competing risk of death before hospital readmission.
Self-controlled Case Series
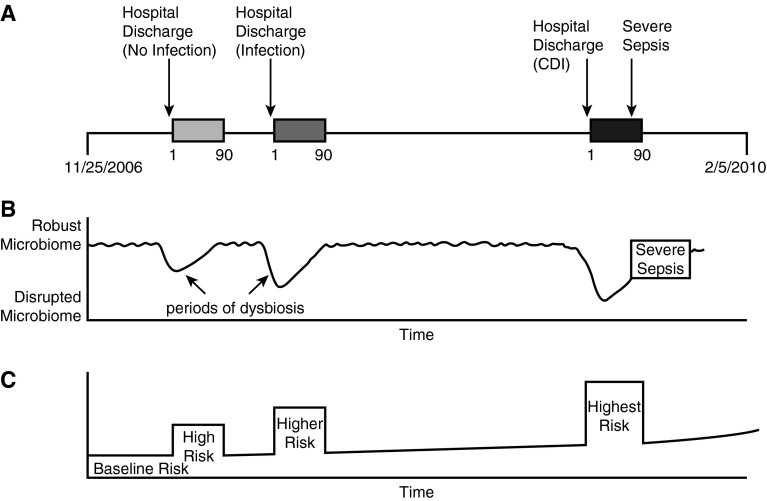
Because of the possibility for residual confounding with regression models, we also performed a self-controlled case series analysis (33). In the self-controlled case series method, each person serves as his or her own control, so that risk of severe sepsis in the 90 days after a hospitalization is compared with the patient’s own baseline risk of severe sepsis, before and after this 90-day period. As a result, temporally invariant covariates are controlled for implicitly. This method uses conditional fixed-effect Poisson regression to measure within-person differences in the rate of an outcome following different exposures (33). We modeled the marginal risk of severe sepsis (outcome) during four different time periods for each subject: (1) the 90-day window after hospitalization with CDI; (2) the 90-day window after infection-related hospitalization (non-CDI); (3) the 90-day window after non–infection-related hospitalization; and (4) all other times in the patient’s observation period, both before and after the 90-day windows. Figure 1 provides a conceptual diagram of the self-controlled case series analysis, linking the clinical history, microbiome health, and risk periods for a single hypothetical patient. Incidence rate ratios and 95% confidence intervals (CI) were generated to compare severe sepsis rates across different time periods.
Figure 1.
Conceptual diagram of the self-controlled case series analysis. (A) Hypothetical timeline for a patient with three exposures and one severe sepsis hospitalization. (B) Hypothetical shifts in microbiome health associated with the patient’s clinical history. Microbial diversity is in constant flux, with periods of disruption (dysbiosis) corresponding with hospitalization. (C) Classification of baseline and higher risk periods used to calculate the incidence risk ratios for severe sepsis following each of the three exposures. The baseline risk of sepsis increases over time as patients age, which is accounted for in the model. CDI = Clostridium difficile infection.
For each patient, we considered the start of his or her observation period to be the later of either the first date for which we had linked Medicare claims or the date when the patient was 65 years and 4 months old. We assumed that all Part A fee-for-service beneficiaries were enrolled by age 65 year and 4 months because this signifies the end of the standard enrollment, after which patients incur penalties for late enrollment (34). We considered the end of each patient’s observation to be the earlier of either the date of the patient’s death, determined from the National Death Index and confirmed by HRS interviewers and the Medicare Denominator File, or the date of the administrative censoring of the entire cohort at the end of the HRS–Medicare linkage on December 31, 2010. Because the incidence of severe sepsis rises precipitously with age (24), we controlled for age using a categorical age variable: 65–74, 75–79, 80–84, and greater than or equal to 85 years.
Results
We identified 43,095 exposure hospitalizations (28,465 hospitalizations without infection, 14,243 hospitalizations with non-CDI infection, and 387 hospitalizations with CDI) among 10,996 patients for inclusion in the longitudinal study (Table 1). Patients were predominantly female (58%), white persons (81%), with good baseline functional status, and mean age of 77 years.
Table 1.
Baseline Characteristics of Subjects in the Longitudinal Study
| Noninfection (n = 28,465) | Infection (n = 14,243) | CDI (n = 387) | |
|---|---|---|---|
| Age, yr, mean (SD) | 76.8 (9.0) | 78.3 (9.5) | 79.6 (8.4) |
| Male, n (%) | 12,244 (43.7%) | 5,695 (40.0%) | 159 (41.1%) |
| Self-reported race, n (%) | |||
| White | 23,307 (81.9%) | 11,470 (80.5%) | 312 (80.6%) |
| Black/African American | 4,562 (16.0%) | 2,447 (17.2%) | 69 (17.8%) |
| Self-reported Hispanic ethnicity, n (%) | 2,008 (7.1%) | 1,185 (8.3%) | 30 (7.8%) |
| Self-reported education, n (%) | |||
| No degree | 10,042 (35.3%) | 5,792 (40.7%) | 150 (38.8%) |
| General Education Development | 1,251 (4.4%) | 668 (4.7%) | 13 (3.4%) |
| High school diploma | 12,470 (43.8%) | 5,930 (41.6%) | 174 (45.0%) |
| College degree (2- or 4-yr) | 3,135 (11.0%) | 1,266 (8.9%) | 39 (10.1%) |
| Master or professional degree | 1,567 (5.5%) | 587 (4.1%) | 11 (2.8%) |
| Self-reported wealth, n (%) | |||
| Net negative or zero assets | 1,412 (5.0%) | 1,101 (7.7%) | 38 (9.8%) |
| Quartile 1 | 4,860 (17.1%) | 3,036 (21.3%) | 61 (15.8%) |
| Quartile 2 | 5,142 (18.1%) | 2,819 (19.8%) | 92 (23.8%) |
| Quartile 3 | 5,521 (19.4%) | 2,454 (17.2%) | 64 (16.5%) |
| Quartile 4 | 5,674 (19.9%) | 2,129 (15.0%) | 60 (15.5%) |
| Comorbidities, n (%) | |||
| Congestive heart failure | 9,889 (34.7%) | 6,338 (44.5%) | 193 (50.1%) |
| Kidney disease | 4,563 (16.0%) | 3,039 (21.3%) | 111 (28.7%) |
| Liver disease | 1,133 (4.0%) | 676 (4.8%) | 26 (6.7%) |
| Metastatic cancer | 1,352 (4.8%) | 742 (5.2%) | 20 (5.2%) |
| Diabetes with complication | 3,987 (14.0%) | 2,651 (18.6%) | 86 (22.2%) |
| Functional disability | |||
| Limitations of ADLs, median (IQR) | 0 (0–1) | 0 (0–2) | 0 (0–2) |
| Limitations of IADLs, median (IQR) | 0 (0–1) | 0 (0–2) | 0 (0–2) |
Definition of abbreviations: ADL = activities of daily living; CDI = Clostridium difficile infection; IADL = instrumental activities of daily living; IQR = interquartile range.
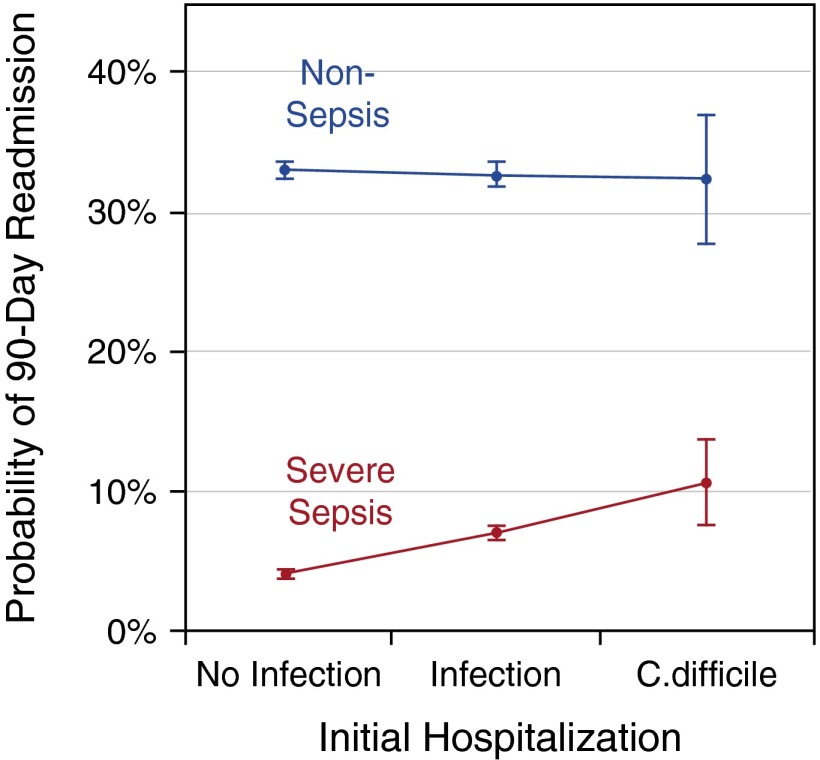
In unadjusted analyses, the probability of a 90-day readmission for severe sepsis was 3.7% (95% CI, 3.6–3.9%) following non–infection-related hospitalization; 8.4% (95% CI, 7.7–9.1%) following infection-related hospitalization, and 16.8% (95% CI, 12.2–21.4%) following hospitalization with CDI infection. After accounting for potential confounders, adjusted probabilities of 90-day readmission for severe sepsis were 4.1% (95% CI, 3.8–4.4%), 7.1% (95% CI, 6.6–7.6%), and 10.7% (95% CI, 7.7–13.8%), respectively, P less than 0.007 for each pairwise comparison (Figure 2, Table 2). Thus, risk of severe sepsis hospitalization was greater following infection-related hospitalization and still greater after a hospitalization with CDI, even after accounting for potential confounders.
Figure 2.
The probability of severe sepsis readmission correlates with probable microbiome disruption during index hospitalization. This figure depicts adjusted probabilities of 90-day hospital readmission for severe sepsis (red) and nonsepsis diagnoses (blue) following live discharge from hospitalization without infection, hospitalization with non–Clostridium difficile infection, and hospitalization with C. difficile infection. Probabilities are adjusted for age, sex, race, ethnicity, education, total wealth, limitations of activities and instrumental activities of daily living, and 31 Elixhauser comorbidities. Adjusted probabilities of rehospitalization for nonsepsis diagnoses are not different following the three exposures, whereas rehospitalization for severe sepsis is incrementally greater after exposures with increasing probable microbiome disruption.
Table 2.
Probabilities of 90-Day Readmissions for Severe Sepsis and Nonsepsis Diagnoses
| Index Hospitalization | Readmissions for Severe Sepsis |
Readmissions for Nonsepsis Diagnoses |
||
|---|---|---|---|---|
| Unadjusted Probability (95% CI) | Adjusted* Probability (95% CI)† | Unadjusted Probability (95% CI) | Adjusted* Probability (95% CI)‡ | |
| Noninfection-related hospitalization | 3.7% (3.6–3.9%) | 4.1% (3.8–4.4%) | 31.7% (31.0–32.5%) | 33.1% (32.4–33.7%) |
| Infection-related hospitalization | 8.4% (7.7–9.1%) | 7.1% (6.6–7.6%) | 34.7% (33.7–35.7%) | 32.7% (31.9–33.6%) |
| Hospitalization with CDI | 16.8% (12.2–21.4%) | 10.7% (7.7–13.8%) | 37.9% (32.7–43.3%) | 32.4% (27.8–37.0%) |
Definition of abbreviations: CDI = Clostridium difficile infection; CI = confidence interval.
Adjusted for age, sex, race, ethnicity, education, total wealth, limitations of activities and instrumental activities of daily living, and 31 Elixhauser comorbidities.
Adjusted comparisons: infection versus noninfection (P < 0.001); CDI versus infection (P = 0.007); CDI versus noninfection (P < 0.001).
Adjusted comparisons: infection versus noninfection (P = 0.51); CDI versus infection (P = 0.88); CDI versus noninfection (P = 0.78).
However, in contrast to severe sepsis, the probability of readmission for nonsepsis diagnoses did not differ following the three types of hospitalizations in adjusted analyses. The probability of a 90-day readmission for nonsepsis diagnoses was 33.1% (95% CI, 32.4–33.7%) after non–infection-related hospitalization, 32.7% (95% CI, 31.9–33.6%) after infection-related hospitalization, and 32.4% (95% CI, 27.8–37.0%) after hospitalization with CDI, P greater than 0.5 for each pairwise comparison (Figure 2, Table 2). Supplemental analyses accounting for the competing risk of death demonstrated a similar stepwise increase in the risk for severe sepsis rehospitalization across hospitalization types and no difference in risk for other readmissions (see Table E1 in the online supplement).
For the self-controlled case series analysis, we identified 1,983 patients with at least one severe sepsis and one exposure hospitalization. In total, the self-controlled case series analysis included 2,864 severe sepsis hospitalizations over 16,736 person-years (average 8.4-yr observation per patient).
Hospitalization, infection, and CDI were each associated with graduated increases in the rate of subsequent severe sepsis in the self-controlled case series, where each patient serves as his or her own control subject (Table 3). In the 90 days after hospitalization, the incidence rate ratio for severe sepsis was 3.3 (95% CI, 3.0–3.7; P < 0.001), indicating that, on average, the rate of severe sepsis was 3.3 times greater in the 90 days after hospitalization than during all times before or after this period. The rate of severe sepsis following hospitalization with non-CDI infection was 1.3 times (95% CI, 1.2–1.5; P < 0.001) greater than the rate following hospitalizations without infection. The rate of severe sepsis following hospitalization with CDI was 1.7 times (95% CI, 1.1–2.6; P = 0.015) greater than the rate following non-CDI infection.
Table 3.
Incidence Rate Ratios Comparing the Four Risk Periods*
| Comparison | IRR (95% CI) | P Value |
|---|---|---|
| 90 d after any hospitalization vs. not in the 90 d after hospitalization | 3.3 (3.0–3.7) | <0.001 |
| After infection-related hospitalization vs. after non–infection-related hospitalization | 1.3 (1.2–1.5) | <0.001 |
| After hospitalization with CDI vs. after infection-related hospitalization (non-CDI) | 1.7 (1.1–2.6) | 0.015 |
Definition of abbreviations: CDI = Clostridium difficile infection; CI = confidence interval; IRR = incidence rate ratio.
The four risk periods here are illustrated in Figure 1C: baseline risk, high risk (after non–infection-related hospitalization), higher risk (after infection-related hospitalization), and highest risk (after hospitalization with CDI).
Discussion
We have shown that the incidence of severe sepsis is elevated more than threefold in the 90 days following all-cause hospitalization among a nationally representative cohort of older adults. We have also demonstrated for the first time that the risk of severe sepsis is yet greater after hospitalizations with infection (by 30%), and greater still after hospitalizations with CDI (by 70%). Such a dose–response relationship is present for rehospitalizations with severe sepsis but not for rehospitalizations for other diagnoses. Furthermore, we show that these progressive increases in the rates of hospitalization are present with very tight control for extraneous factors, by using patients as their own control subjects. This accounts for the temporally invariant covariates, such as sex (a known microbiota variant [35]), medical history, and genetic predispositions to sepsis. In addition, we adjusted for age, thereby addressing the age-dependent incidence of severe sepsis.
Past studies have established that hospital readmission is frequent among Medicare beneficiaries (36), and that severe sepsis is a common reason for readmission (36, 37). Our findings advance the current understanding by demonstrating that severe sepsis readmission is not simply a reflection of patient’s underlying comorbidities before hospitalization. Rather, after careful adjustment for patient’s medical and sociodemographic factors, we found that the probability of severe sepsis readmission varies by hospitalization type. Hospitalizations with greater probable microbiome perturbation are associated with greater risk of subsequent severe sepsis, and this association seems to be temporally specific (to the 90 d following hospital discharge, relative to all times outside this 90-d period) and diagnosis specific (to the outcome of subsequent severe sepsis, but not hospitalizations for other diagnoses).
We hypothesize that hospitalization-associated dysbiosis may be an important mediator underlying the dose–response relationship between hospitalization type and risk for subsequent severe sepsis. The idea has strong face validity. Humans have evolved with resident bacteria and fungi, and these organisms serve varied functions, including bidirectional interactions with the immune system, and regulation of energy balance and metabolism (1, 38). Importantly, a diverse microbial community resists pathogen overgrowth and helps modulate both innate and adaptive immune responses (21). It is possible that dysbiosis may increase the likelihood of systemic inflammatory response syndrome and subsequent organ dysfunction by up-regulating the host inflammatory response (39, 40). Inflammasomes, which trigger the inflammatory cascade, are activated after gut microbial perturbations and are followed by a rapid sepsis-like death in the animal model (41).
Because we did not directly measure the microbiome in this study, we cannot definitively prove our hypothesis. Nevertheless, we maintain that hospitalization with C. difficile equates with intestinal perturbation of the microbiota. The arguments are (1) C. difficile, although ubiquitous in the environment, occurs almost exclusively in individuals receiving antibiotics, proton pump inhibitors, H2 receptor antagonists, and other drugs that disrupt the bacterial composition of the gastrointestinal tract (42, 43); (2) antibiotic stewardship programs decrease the incidence of CDI through reduction in antibiotic use (44); and, most importantly, (3) restoring the normal gut microbiome in persons with CDI cures the disease (15, 45). Although CDI may not be the only instance in which dysbiosis occurs, it is certainly a hallmark indication of intestinal perturbation.
Beyond microbiome perturbation, there are alternative mechanisms that may explain our findings. Our cohorts may have differed in the activation and lingering alteration of their immune response following their initial hospitalizations. Yende and coworkers (46, 47) have shown that inflammatory and coagulation markers are commonly elevated after pneumonia or sepsis and associated with worse overall and infection-related mortality. Meanwhile, compensatory antiinflammatory response or autoimmunosuppression following severe infection may also predispose patients to a second, and more severe, infection (48). It is likely that the human microbiome and immune response both contribute to the observed relationship because they are closely entwined. A healthy microbiome is critical to the development and function of the host immune response, and conversely, inflammatory mediators influence the microbiome composition (41, 49–52). Thus, we view alteration in host immunity as a complementary, rather than competing, hypothesis to explain our findings.
Understanding the association between transient dysbiosis and severe sepsis risk is especially important because of the potential for therapeutic manipulation of the human microbiome to reduce the incidence of severe sepsis during periods of heightened risk. Although attempts to modulate the immune system have failed to treat or prevent infection, the microbiome provides a promising therapeutic target. The human microbiome is amenable to such interventions as basic dietary modification and probiotic supplementation (53). Moreover, there have been some preliminary successes with respect to patient outcomes. Several studies indicate that microbiome modulation may reduce ventilator-associated pneumonia (54). Furthermore, microbiome replacement for recurrent CDI has demonstrated a 94% cure rate, better than that achieved with antibiotics (15). These studies suggest that treatment of dysbiosis can indeed result in improved health outcomes.
More detailed studies using longitudinal measurements of immune markers and microbial composition and function before, during, and after hospitalization are needed to further characterize these relationships. The microbiome composition could be assessed via 16S RNA sequencing of cheek swabs and/or stool samples, whereas microbial function could be assessed via metagenomic analysis (55). It is currently cost-prohibitive to complete serial analyses of this sort on large numbers of individuals, but may be feasible in the future as the cost of these technologies continues to decline. Functional genomics has been used to predict hospitalizations in psychiatric patients and may be feasible in select clinical populations, such as those recovering from a CDI (56). Going forward, it will also be important to understand the extent to which less severe stresses to the microbiome (e.g., minor infections not requiring hospitalization and common medications known to influence the microbiome [antibiotics, proton pump inhibitors (57), H2 receptor antagonists (58), and antidepressants (59)]) influence patients’ risk for developing severe sepsis, and whether these risks depend on the dosage and duration of medication. While we await these sorts of confirmatory studies, it may be possible to incorporate microbiome measurements into studies of selective oropharyngeal and/or gut decontamination and to monitor microbiome recovery and infections rates beyond hospital discharge.
If future studies do confirm an association between microbiome disruption and subsequent severe sepsis, then it will be important to test the extent to which preservation of the microbiome (through narrower-spectrum antibiotics, shorter treatment courses, elimination of other microbiome-altering medications, and so forth) or restoration of the microbiome (through diet, probiotic supplements, or fecal transplant) actually reduces risk for subsequent severe sepsis.
There are several potential limitations to our study. First, we performed a within-person analysis that limits the possibility of confounding by stable patient characteristics, but cannot fully rule out time-varying factors. Second, we used hospitalizations as proxies for the severity of dysbiosis rather than measuring microbial composition directly. The duration and severity of dysbiosis following antibiotic administration may vary across patients. Likewise, because we could not measure the microbiome recovery directly, we assumed that all patients experienced 90 days of dysbiosis. Although recovery of the human microbiome following a disturbance is incompletely understood (28, 60), several studies suggest that it closely resembles its predisturbance state within a few weeks to months after antibiotic exposure (27, 28). Any misspecification of the duration of the exposure biases our results toward the null hypothesis by imprecisely delineating higher and lower risk periods. Lastly, we rely on an implicit claims-based definition for identifying severe sepsis. Although this method performs favorably relatively to other claims-based definitions, it may result in some misclassification of patients as having severe sepsis versus another diagnosis (25). However, this potential for misclassification of readmission diagnosis should not differ by initial hospitalization type.
Despite these limitations, our study has several strengths. By using data from a nationally representative cohort with linked Medicare claims, we were able to study a large population of patients to assess whether proxies for dysbiosis are associated with increased risk of severe sepsis, the most costly cause of hospitalization in the United States (54). We examined these relationships using two designs: one a between-person longitudinal comparison and the other a within-person comparison of outcomes during different risk periods. We found a dose–response by severity of dysbiosis and demonstrated specificity of the association with severe sepsis hospitalizations.
Conclusions
Using a nationally representative sample of older Americans, we have shown that severe sepsis incidence is elevated threefold in the 90 days after hospitalization. Risk of severe sepsis is incrementally greater following hospitalizations with infection, and greater still following CDI. Although the cause of this relationship remains unproven, we hypothesize that the observed dose–response by hospitalization type may be caused by microbial perturbation. Because there are no current means to predict or prevent subsequent sepsis, the possibility of targeted restoration of the microbiome after a period of disruption holds the potential to reduce the incidence of subsequent severe sepsis and its associated morbidity and mortality.
Acknowledgments
Acknowledgment
The authors appreciate the expert programming of Ryan McCammon, M.S., and Vanessa Dickerman, M.S., at the University of Michigan.
Footnotes
Supported by grants T32 HL007749 (H.C.P.) and R01 AG030155 (K.M.L.) from the National Institutes of Health and IIR 11-109 (T.J.I.) from the U.S. Department of Veterans Affairs Health Services Research and Development Service. The Health and Retirement Study is funded by the National Institute on Aging (U01 AG009740) and performed at the Institute for Social Research, University of Michigan.
Author Contributions: H.C.P. designed the study, analyzed the data, interpreted the data, and drafted the manuscript. R.P.D. and M.A.M.R. interpreted the data and revised the manuscript critically for intellectual content. K.M.L. contributed to the design of the study, acquired the data, interpreted the data, and revised the manuscript critically for intellectual content. T.J.I. assisted with analysis of the data, interpreted the data, and revised the manuscript critically for intellectual content.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201503-0483OC on May 27, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, Curtis JL, Erb-Downward J, Lynch SV, Sethi S, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67:456–463. doi: 10.1136/thoraxjnl-2011-201183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7:569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, Oliver PM, Kolls JK, Weiser JN, Worthen GS. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Wang C, Tang C, He Q, Zhao X, Li N, Li J. Therapeutic modulation and reestablishment of the intestinal microbiota with fecal microbiota transplantation resolves sepsis and diarrhea in a patient. Am J Gastroenterol. 2014;109:1832–1834. doi: 10.1038/ajg.2014.299. [DOI] [PubMed] [Google Scholar]
- 10.Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, Shuster J, Sharma R, Hudak ML, Neu J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One. 2013;8:e52876. doi: 10.1371/journal.pone.0052876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuppala VS, Meinzen-Derr J, Morrow AL, Schibler KR. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J Pediatr. 2011;159:720–725. doi: 10.1016/j.jpeds.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham TA, Lawley TD. Emerging insights on intestinal dysbiosis during bacterial infections. Curr Opin Microbiol. 2014;17:67–74. doi: 10.1016/j.mib.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 16.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartosch S, Fite A, Macfarlane GT, McMurdo ME. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol. 2004;70:3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert AM, Rogers MA, Ring C, Mogle J, Petrosino JP, Young VB, Aronoff DM, Schloss PD. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;5:e01021-14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M, Tirrell M, Tiedje J, Gilbert JA, Zaborina O, et al. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio. 2014;5:e01361-14. doi: 10.1128/mBio.01361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Creditor MC. Hazards of hospitalization of the elderly. Ann Intern Med. 1993;118:219–223. doi: 10.7326/0003-4819-118-3-199302010-00011. [DOI] [PubMed] [Google Scholar]
- 21.Bien J, Palagani V, Bozko P. The intestinal microbiota dysbiosis and Clostridium difficile infection: is there a relationship with inflammatory bowel disease? Therap Adv Gastroenterol. 2013;6:53–68. doi: 10.1177/1756283X12454590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prescott HC, Dickson RP, Rogers MA, Langa KM, Iwashyna TJ. Is dysbiosis a mediator of sepsis? Studying a nationally representative patient sample. Am J Respir Crit Care Med. 2014;189:A2192. [Google Scholar]
- 23.Sonnega A, Faul JD, Ofstedal MB, Langa KM, Phillips JWR, Weir DR. Cohort profile: the Health and Retirement Study (HRS) Int J Epidemiol. 2014;43:576–585. doi: 10.1093/ije/dyu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Iwashyna TJ, Odden A, Rohde J, Bonham C, Kuhn L, Malani P, Chen L, Flanders S. Identifying patients with severe sepsis using administrative claims: patient-level validation of the angus implementation of the international consensus conference definition of severe sepsis. Med Care. 2014;52:e39–e43. doi: 10.1097/MLR.0b013e318268ac86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubberke ER, Reske KA, McDonald LC, Fraser VJ. ICD-9 codes and surveillance for Clostridium difficile-associated disease. Emerg Infect Dis. 2006;12:1576–1579. doi: 10.3201/eid1210.060016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elixhauser A, Steiner C, Harris DR, Coffey RM. Comorbidity measures for use with administrative data. Med Care. 1998;36:8–27. doi: 10.1097/00005650-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 31.Donner A, Eliasziw M. Sample size requirements for reliability studies. Stat Med. 1987;6:441–448. doi: 10.1002/sim.4780060404. [DOI] [PubMed] [Google Scholar]
- 32.Royston P. Multiple imputation of missing values. Stata J. 2004;4:227–241. [Google Scholar]
- 33.Whitaker HJ, Farrington CP, Spiessens B, Musonda P. Tutorial in biostatistics: the self-controlled case series method. Stat Med. 2006;25:1768–1797. doi: 10.1002/sim.2302. [DOI] [PubMed] [Google Scholar]
- 34.Medicare.govWhen can I sign up for Part A & Part B? [accessed 2013 Jan 24]. Available from: http://www.medicare.gov/sign-up-change-plans/get-parts-a-and-b/when-sign-up-parts-a-and-b/when-sign-up-parts-a-and-b.html
- 35.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jencks SF, Williams MV, Coleman EA. Rehospitalizations among patients in the Medicare fee-for-service program. N Engl J Med. 2009;360:1418–1428. doi: 10.1056/NEJMsa0803563. [DOI] [PubMed] [Google Scholar]
- 37.Dharmarajan K, Hsieh AF, Lin Z, Bueno H, Ross JS, Horwitz LI, Barreto-Filho JA, Kim N, Bernheim SM, Suter LG, et al. Diagnoses and timing of 30-day readmissions after hospitalization for heart failure, acute myocardial infarction, or pneumonia. JAMA. 2013;309:355–363. doi: 10.1001/jama.2012.216476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu GD. Diet, the gut microbiome and the metabolome in IBD. Nestle Nutrition Institute Workshop Series. 2014;79:73–82. doi: 10.1159/000360686. [DOI] [PubMed] [Google Scholar]
- 39.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal R, Coopersmith CM. Redefining the gut as the motor of critical illness. Trends Mol Med. 2014;20:214–223. doi: 10.1016/j.molmed.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slimings C, Riley TV. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother. 2014;69:881–891. doi: 10.1093/jac/dkt477. [DOI] [PubMed] [Google Scholar]
- 43.Deshpande A, Pasupuleti V, Thota P, Pant C, Rolston DD, Sferra TJ, Hernandez AV, Donskey CJ. Community-associated Clostridium difficile infection and antibiotics: a meta-analysis. J Antimicrob Chemother. 2013;68:1951–1961. doi: 10.1093/jac/dkt129. [DOI] [PubMed] [Google Scholar]
- 44.Feazel LM, Malhotra A, Perencevich EN, Kaboli P, Diekema DJ, Schweizer ML. Effect of antibiotic stewardship programmes on Clostridium difficile incidence: a systematic review and meta-analysis. J Antimicrob Chemother. 2014;69:1748–1754. doi: 10.1093/jac/dku046. [DOI] [PubMed] [Google Scholar]
- 45.Khan MA, Sofi AA, Ahmad U, Alaradi O, Khan AR, Hammad T, Pratt J, Sodeman T, Sodeman W, Kamal S, et al. Efficacy and safety of, and patient satisfaction with, colonoscopic-administered fecal microbiota transplantation in relapsing and refractory community- and hospital-acquired Clostridium difficile infection. Chin J Gastroenterol Hepatol. 2014;28:434–438. doi: 10.1155/2014/695029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yende S, D’Angelo G, Kellum JA, Weissfeld L, Fine J, Welch RD, Kong L, Carter M, Angus DC GenIMS Investigators. Inflammatory markers at hospital discharge predict subsequent mortality after pneumonia and sepsis. Am J Respir Crit Care Med. 2008;177:1242–1247. doi: 10.1164/rccm.200712-1777OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yende S, D’Angelo G, Mayr F, Kellum JA, Weissfeld L, Kaynar AM, Young T, Irani K, Angus DC GenIMS Investigators. Elevated hemostasis markers after pneumonia increases one-year risk of all-cause and cardiovascular deaths. PLoS One. 2011;6:e22847. doi: 10.1371/journal.pone.0022847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward NS, Casserly B, Ayala A. The compensatory anti-inflammatory response syndrome (CARS) in critically ill patients. Clin Chest Med. 2008;29:617–625, viii. doi: 10.1016/j.ccm.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamada N, Núñez G. Role of the gut microbiota in the development and function of lymphoid cells. J Immunol. 2013;190:1389–1395. doi: 10.4049/jimmunol.1203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 51.McDermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology. 2014;142:24–31. doi: 10.1111/imm.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morrow LE, Kollef MH, Casale TB. Probiotic prophylaxis of ventilator-associated pneumonia: a blinded, randomized, controlled trial. Am J Respir Crit Care Med. 2010;182:1058–1064. doi: 10.1164/rccm.200912-1853OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLOS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le-Niculescu H, Levey DF, Ayalew M, Palmer L, Gavrin LM, Jain N, Winiger E, Bhosrekar S, Shankar G, Radel M, et al. Discovery and validation of blood biomarkers for suicidality. Mol Psychiatry. 2013;18:1249–1264. doi: 10.1038/mp.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JW, Lee KL, Jeong JB, Kim BG, Shin S, Kim JS, Jung HC, Song IS. Proton pump inhibitors as a risk factor for recurrence of Clostridium-difficile-associated diarrhea. World J Gastroenterol. 2010;16:3573–3577. doi: 10.3748/wjg.v16.i28.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta RW, Tran L, Norori J, Ferris MJ, Eren AM, Taylor CM, Dowd SE, Penn D. Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J Pediatr Gastroenterol Nutr. 2013;56:397–400. doi: 10.1097/MPG.0b013e318282a8c2. [DOI] [PubMed] [Google Scholar]
- 59.Rogers MA, Greene MT, Young VB, Saint S, Langa KM, Kao JY, Aronoff DM. Depression, antidepressant medications, and risk of Clostridium difficile infection. BMC Med. 2013;11:121. doi: 10.1186/1741-7015-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;62:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]