Abstract
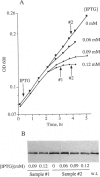
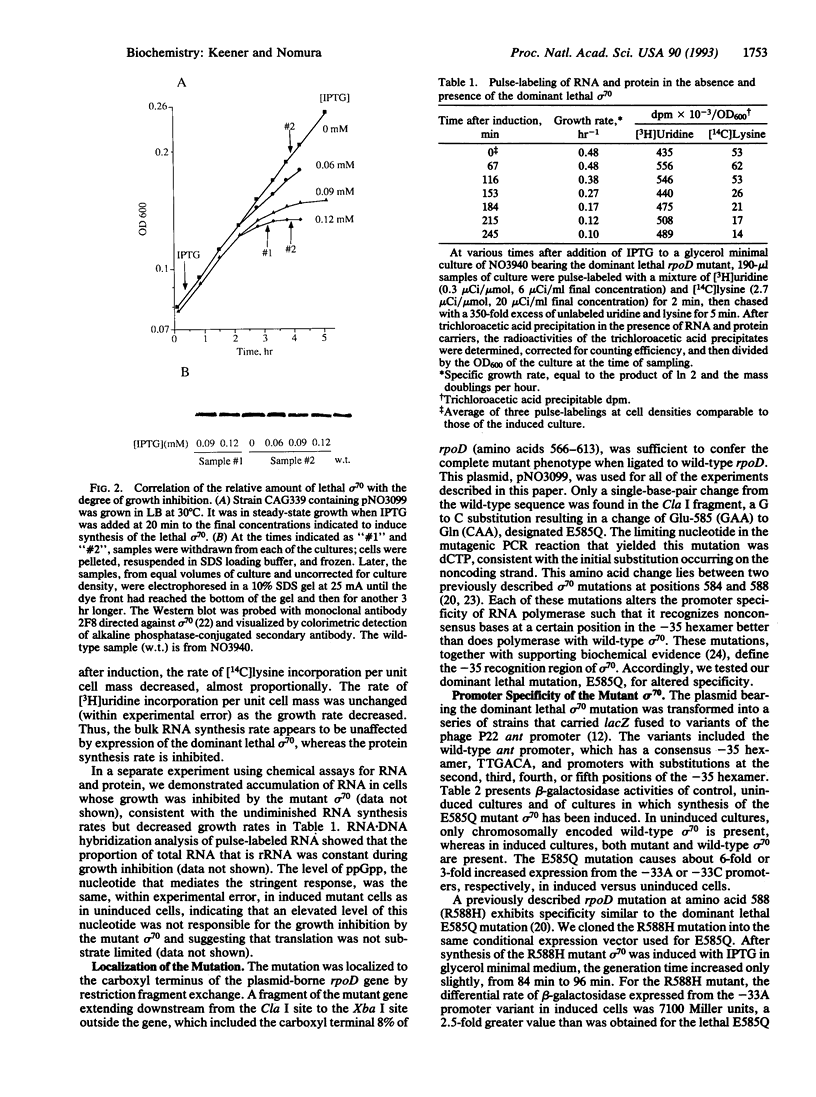
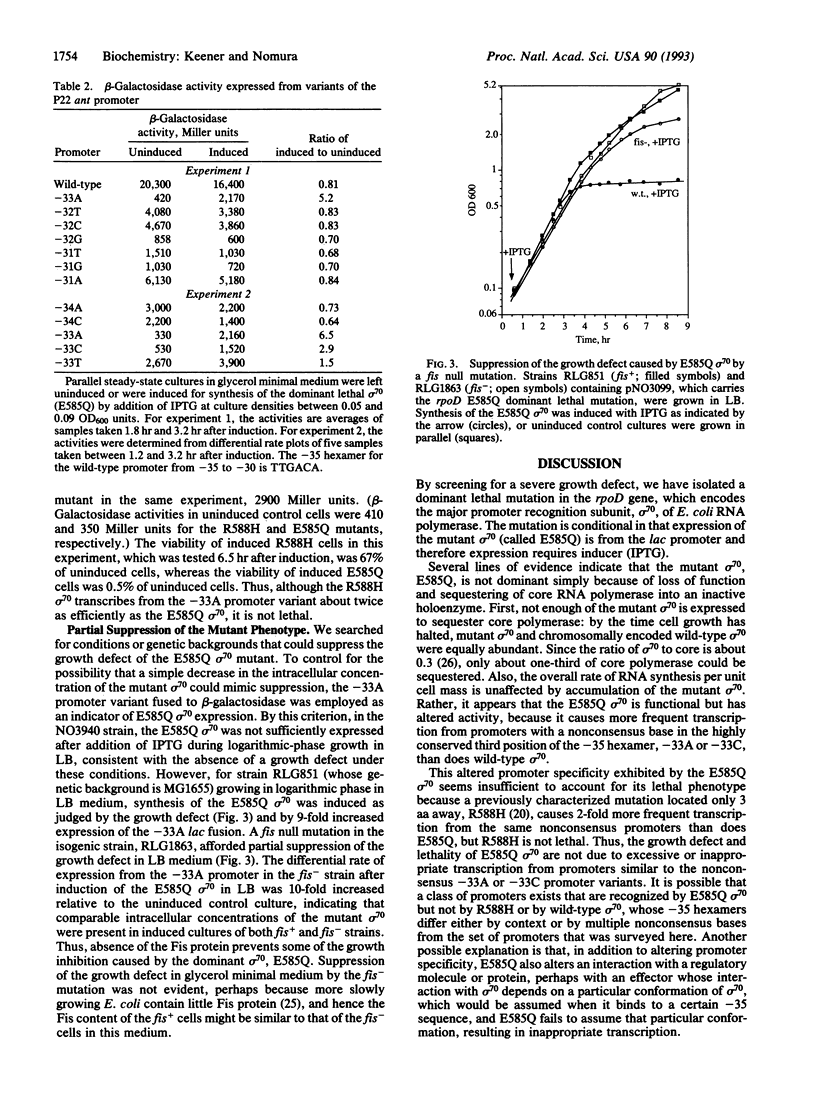
A dominant lethal mutation in the Escherichia coli rpoD gene, which encodes sigma 70, the promoter recognition subunit of RNA polymerase, was isolated after random mutagenesis. The lethal gene was maintained under control of the lac repressor on a low copy plasmid. An amount of lethal sigma 70 that was nearly equimolar with the chromosomally encoded sigma 70 was sufficient to cause cessation of growth. RNA synthesis per unit cell mass was unaffected, but protein synthesis was inhibited by the mutant sigma 70. The amino acid change (Glu-585 to Gln) was in a region of sigma 70 thought to bind the -35 hexamer of the promoter, and the mutant sigma 70 caused increased expression from promoters with nonconsensus bases in the third position of the -35 hexamer. A null mutation of the fis gene could partially suppress the mutant phenotype. These properties are consistent with those expected of a sigma 70 insensitive to growth rate control of rRNA and tRNA promoters.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burton Z., Burgess R. R., Lin J., Moore D., Holder S., Gross C. A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E coli K12. Nucleic Acids Res. 1981 Jun 25;9(12):2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. R., Gaal T., deBoer H. A., deHaseth P. L., Gourse R. L. Identification of promoter mutants defective in growth-rate-dependent regulation of rRNA transcription in Escherichia coli. J Bacteriol. 1989 Sep;171(9):4862–4870. doi: 10.1128/jb.171.9.4862-4870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski A. J., Walter W. A., Record M. T., Jr, Siegele D. A., Gross C. A. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992 Aug 7;70(3):501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- Gaal T., Barkei J., Dickson R. R., deBoer H. A., deHaseth P. L., Alavi H., Gourse R. L. Saturation mutagenesis of an Escherichia coli rRNA promoter and initial characterization of promoter variants. J Bacteriol. 1989 Sep;171(9):4852–4861. doi: 10.1128/jb.171.9.4852-4861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella T., Moyle H., Susskind M. M. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989 Apr 20;206(4):579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., de Boer H. A., Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986 Jan 17;44(1):197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- Hu J. C., Gross C. A. Marker rescue with plasmids bearing deletions in rpoD identifies a dispensable part of E. coli sigma factor. Mol Gen Genet. 1983;191(3):492–498. doi: 10.1007/BF00425768. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S., Gourse R. L., Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983 Jul;33(3):865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- Kiss A., Sain B., Venetianer P. The number of rRNA genes in Escherichia coli. FEBS Lett. 1977 Jul 1;79(1):77–79. doi: 10.1016/0014-5793(77)80354-2. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Moyle H., Waldburger C., Susskind M. M. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol. 1991 Mar;173(6):1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L., Verbeek H., Vijgenboom E., van Drunen C., Vanet A., Bosch L. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J Bacteriol. 1992 Feb;174(3):921–929. doi: 10.1128/jb.174.3.921-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris T. E., Koch A. L. Effect of growth rate on the relative rates of synthesis of messenger, ribosomal and transfer RNA in Escherichia coli. J Mol Biol. 1972 Mar 14;64(3):633–649. doi: 10.1016/0022-2836(72)90088-5. [DOI] [PubMed] [Google Scholar]
- Ross W., Thompson J. F., Newlands J. T., Gourse R. L. E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990 Nov;9(11):3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmientos P., Cashel M. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: differential control of dual promoters. Proc Natl Acad Sci U S A. 1983 Nov;80(22):7010–7013. doi: 10.1073/pnas.80.22.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shand R. F., Miercke L. J., Mitra A. K., Fong S. K., Stroud R. M., Betlach M. C. Wild-type and mutant bacterioopsins D85N, D96N, and R82Q: high-level expression in Escherichia coli. Biochemistry. 1991 Mar 26;30(12):3082–3088. doi: 10.1021/bi00226a015. [DOI] [PubMed] [Google Scholar]
- Siegele D. A., Hu J. C., Walter W. A., Gross C. A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989 Apr 20;206(4):591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Strickland M. S., Thompson N. E., Burgess R. R. Structure and function of the sigma-70 subunit of Escherichia coli RNA polymerase. Monoclonal antibodies: localization of epitopes by peptide mapping and effects on transcription. Biochemistry. 1988 Jul 26;27(15):5755–5762. doi: 10.1021/bi00415a054. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Albrechtsen B., Løve Larsen J. E. DNA-protein recognition: demonstration of three genetically separated operator elements that are required for repression of the Escherichia coli deoCABD promoters by the DeoR repressor. EMBO J. 1986 Aug;5(8):2015–2021. doi: 10.1002/j.1460-2075.1986.tb04458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek H., Nilsson L., Baliko G., Bosch L. Potential binding sites of the trans-activator FIS are present upstream of all rRNA operons and of many but not all tRNA operons. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):302–306. doi: 10.1016/0167-4781(90)90185-5. [DOI] [PubMed] [Google Scholar]
- Waldburger C., Gardella T., Wong R., Susskind M. M. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990 Sep 20;215(2):267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]