Abstract
In model organism Streptomyces coelicolor, γ-butyrolactones (GBLs) and antibiotics were recognized as signalling molecules playing fundamental roles in intra- and interspecies communications. To dissect the GBL and antibiotic signalling networks systematically, the in vivo targets of their respective receptors ScbR and ScbR2 were identified on a genome scale by ChIP-seq. These identified targets encompass many that are known to play important roles in diverse cellular processes (e.g. gap1, pyk2, afsK, nagE2, cdaR, cprA, cprB, absA1, actII-orf4, redZ, atrA, rpsL and sigR), and they formed regulatory cascades, sub-networks and feedforward loops to elaborately control key metabolite processes, including primary and secondary metabolism, morphological differentiation and stress response. Moreover, interplay among ScbR, ScbR2 and other regulators revealed intricate cross talks between signalling pathways triggered by GBLs, antibiotics, nutrient availability and stress. Our work provides a global view on the specific responses that could be triggered by GBL and antibiotic signals in S. coelicolor, among which the main echo was the change of production profile of endogenous antibiotics and antibiotic signals manifested a role to enhance bacterial stress tolerance as well, shedding new light on GBL and antibiotic signalling networks widespread among streptomycetes.
Microorganisms in the nature environment are overwhelmed with diverse chemicals and are engaged in extensive interactions with their community to modulate gene expression1. Chemical signalling has been shown to affect phenotypes significantly as diverse as differentiation, antibiotic production, biofilm formation and pathogenicity2,3,4. Streptomyces are important bacteria by producing a variety of active secondary metabolites and as one of the model systems for bacterial morphological development. Small molecule signals, such as γ-butyrolactones (GBLs) and antibiotics, have been reported to play vital roles in coordinating secondary metabolism and morphological development in Streptomyces species5,6. To understand these signalling pathways, many studies have been carried out by probing the functions of their signal receptors3,7.
In the model organism Streptomyces coelicolor A3(2), ScbR was characterized as a receptor of GBLs synthesised by the product of the scbA gene5, while ScbR2 was identified as a receptor of antibiotics6. The scbR, scbR2 and scbA genes are located in the cpk cluster, which determines production of the polyketide coelimycin P18. ScbR binds the scbR-scbA intergenic region to repress scbR while activating scbA in response to GBLs5. Another known target of ScbR is kasO, the pathway-specific activator gene of the cpk cluster9. Despite its high degree of homology (50%) with ScbR, ScbR2 does not bind GBL molecules, and was previously described as a “pseudo-GBL receptor”10. In fact, it binds and responds to endogenous antibiotics, actinorhodin (Act) and undecylprodigiosin (Red), as well as exogenous antibiotics, such as angucyclines6. Our previous work demonstrated that angucyclines affect the behavior of S. coelicolor by modulating the interaction of ScbR2 with adpA (encoding the master regulator of morphogenesis) and redD (the direct activator gene of the red cluster for Red production)6. Neither adpA nor redD is a target of ScbR. The scbR2 mutant displayed a complete loss of production of Act, Red and the calcium-dependent antibiotics (CDA), and showed precocious development of aerial hyphae6,10. These phenotypic effects are much more pronounced than those of the scbR mutant, which mainly showed delayed Red production5. Interestingly, ScbR2 also binds the same sites as ScbR in the scbR-scbA intergenic region to shut down GBL synthesis and in the promoter region of kasO to control the production of coelimycin P110,11. How do the signalling pathways mediated by ScbR and ScbR2 interplay and cooperate in eliciting specific physiological responses of S. coelicolor is largely unknown. Moreover, homologs of ScbR and ScbR2 are widespread among streptomycetes, but studies regarding to them were all focussed on the regulation of genes for antibiotic biosynthesis12,13. In order to gain a more comprehensive overview of the roles of such receptors, we have undertaken a genome-wide analysis of the regulatory targets of ScbR and ScbR2 in S. coelicolor.
In this work, targets of ScbR and ScbR2 were deciphered using chromatin immunoprecipitation followed by sequencing (ChIP-seq) and combined with transcriptomic expression analysis. These targets were engaged in diverse physiological processes, but a major role of them was to control secondary metabolism and elicit stress responses. Furthermore, ScbR and ScbR2 mediated regulatory cascades, feedforward loops (FFLs) and sub-networks were extracted to control Streptomyces phenotypes. The interplay among ScbR, ScbR2 and other regulators revealed intricate cross talks between signalling pathways triggered by GBLs, antibiotics, nutrient availability and stress.
Results
Mutational analysis and expression profiles of scbR and scbR2 in S. coelicolor
It was previously reported that an scbR mutant showed delayed Red and reduced Act production5. For further analysis, we constructed an in frame mutant of scbR in S. coelicolor M145. In ΔscbR, a delay of aerial development was noted after growing for 36 h on SMMS plates (Supplementary Fig. S1a). Red and Act in ΔscbR were produced almost synchronously with M145, but their production levels were greatly reduced (Supplementary Fig. S1bc). A similar pattern of production was observed for CDA: a plate-based bioassay of CDA production revealed a dramatic decrease of CDA level in ΔscbR (Supplementary Fig. S1d). Moreover, a yellow-pigmented secondary metabolite was observed in ΔscbR on SMMS plate, which was probably due to the production of coelimycin P1, as also reported for ΔscbR2 (also known as scbR2DM), in which coelimycin P1 was abundantly synthesised8,14 (Supplementary Fig. S1a). In comparison, ΔscbR2 showed a much more striking phenotype of complete loss of Act, Red and CDA production10 and precocious formation of aerial hyphae6. These observations demonstrated the pleiotropic effects of ScbR and ScbR2 on the physiology of S. coelicolor.
Before deciphering regulons of ScbR and ScbR2, their expression profiles at transcript and protein levels were monitored in SMM liquid culture. Our previous work established that transcription of scbR began at 24 h and peaked at 36 h11. In concert, ScbR protein was detected at 24 h by western blotting, and accumulated to the highest level at 36 h (Fig. 1). ScbR can be dissociated from its targets by SCB1, which is synthesized by ScbA, the mRNA for which also reached the highest level around 36 h11. Therefore, 30 h was chosen as the sampling time point for the ScbR ChIP experiment, when the level of ScbR protein was considerable but the transcript of scbA had not reached the highest level. In contrast, scbR2 transcript was not apparent until 36 h, peaked at 48 h and maintained a relatively high level until 72 h11. Also, ScbR2 protein could barely be detected at the beginning of growth (18 or 24 h), rose to the highest level at 42 h, and remained stable to 60 h (Fig. 1). ScbR2 can be dissociated from its targets by the endogenous antibiotics, Red and Act10. Act appeared in M145 after 48 h, while Red accumulated after 36 h (Supplementary Fig. S1bc). Therefore, 42 h was chosen as the sampling time point for the ScbR2 ChIP experiment, when ScbR2 protein was at the highest level and the production of Act and Red was still relatively low.
Figure 1. Protein levels of ScbR and ScbR2 during growth period.
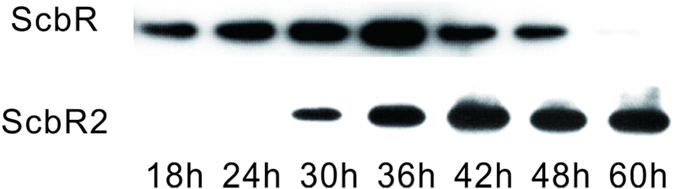
Western blotting was carried out to analyse the temporal expression of protein ScbR and ScbR2 during growth in liquid medium.
Overview of ChIP-seq and transcriptomic profiling results
ChIP-seq experiments of ScbR and ScbR2 were performed with purified antibodies in S. coelicolor M145 in liquid SMM culture as previously described6. As a control, sheared chromosome DNA (input DNA) was utilised to subtract background noise. On ChIP-seq maps, the fold change of peaks above 1.5 was fixed as the minimum cut-off value for ScbR and ScbR2 peak calling. On this basis, 144 ScbR peaks and 491 ScbR2 peaks were detected, distributed relatively even along the chromosome (Fig. 2a). Grouped together by the clustering algorithm MACS15, many of these significant peaks encompassed multiple genes and almost equally occurred in protein-coding and non-coding regions. The peaks generated included the majority of promoter regions known to be targeted by ScbR and ScbR2, i.e. scbR-scbA and kasO promoters were detected among the peaks of ScbR; scbR-scbA, kasO and adpA promoters were noted among peaks of ScbR2, suggesting the reliability of ChIP-seq.
Figure 2. Distribution and attributes of global targets of ScbR and ScbR2.
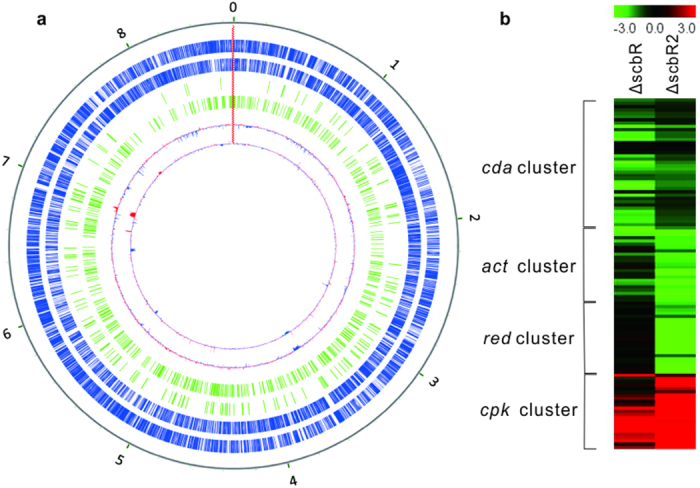
(a) A map of the S. coelicolor genome with ChIP-seq of ScbR and ScbR2 and transcriptome profiling data of their mutants. From the outmost to the innermost, coding sequences in the genome are shaded in blue as the two outer circles, the genomic distribution of ScbR and ScbR2 targets are shown in green. The transcriptome profiling of scbR and scbR2 mutants are indicated in the innermost two circles: the red lines represented genes up-regulated by mutation, while the blue indicated down-regulated genes. (b) Transcriptional effects of ScbR and ScbR2 on four antibiotic cluster genes. Red indicates activation; green indicates repression, while dark means no change.
To assess the transcriptional effects of ScbR and ScbR2 binding on targets genes, a genome-wide transcriptomic analysis of the parent strain M145 and the scbR or scbR2 mutants was performed. RNA samples were harvested at 30 h from ΔscbR and at 42 h from ΔscbR2, and they were analysed on an Agilent based microarray platform. 42.3% of genes demonstrated at least 20% difference in expression levels in ΔscbR, while 30.1% of genes showed greater than 20% change in ΔscbR2, underscoring the pleiotropic influence of the two regulators. Transcriptional changes of antibiotic synthesis gene clusters were consistent with phenotypes we observed (Supplementary Fig. S1). In ΔscbR, transcription of act and cda cluster genes was dramatically reduced, while cpk cluster genes were activated, but the mRNA level of red cluster genes decreased only slightly (Fig. 2b). Similarly, act, red and cda gene cluster mRNA was much less abundant in ΔscbR2, but cpk cluster mRNA was greatly enhanced.
In vitro confirmation of ScbR or ScbR2 binding events on selected promoter regions
To validate the results from ChIP-seq, the binding of ScbR and ScbR2 was analysed by electrophoretic mobility shift assays (EMSA). Peaks locating in the coding regions was first excluded, and attention was focused on regulation occurred in promoter regions (−600 ~ + 100 relative to the putative translational start point), especially those associated with genes that were well studied or vitally annotated were chosen for further validation (Supplementary Fig. S2). Thus, 23 targets of ScbR and 76 targets of ScbR2 (Supplementary Table S1 and S2) were chosen for evaluation. Recombinant ScbR or ScbR2 proteins were purified from E. coli and EMSA assays were performed with their selected promoter probes as described before11. In total, 7 out of 23 targets of ScbR, and 40 out of 76 targets of ScbR2 were newly confirmed to show direct binding (Table 1 and 2), among which five common targets could interact with both ScbR and ScbR2. According to our previous work11, ScbR and ScbR2 have identical binding sites; therefore we expected that ScbR and ScbR2 could also each bind newly identified targets of the other. EMSA experiments were then performed with ScbR protein and targets of ScbR2, and vice versa. It turned out 6 targets from ScbR2 were able to bind with ScbR, and the rest two targets of ScbR could also interact with ScbR2. In summary, 13 and 42 promoters were confirmed by EMSA to be bound by ScbR (Table 1) and ScbR2 (Table 2), respectively. Failure to detect those targets by ChIP was probably due to the competitive binding between ScbR and ScbR2, or to the use of different time points in the two ChIP experiments. Surprisingly, ScbR only bound a fraction of ScbR2 targets whereas ScbR2 bound to all ScbR targets. 69.2% of ScbR target genes and 58.1% of ScbR2 being transcriptionally activated or repressed (fold change > 1.2 or <0.85) (Table 1 and 2). Among those EMSA confirmed targets, we found the binding events did not necessarily lead to changes in expression of adjacent genes. This may be due to the need of assistant proteins to fulfil their roles, or to particular time points and culture conditions selected.
Table 1. Targets of ScbR confirmed by EMSA.
| Gene | Name | Fold change | Annotation | Possible binding site |
|---|---|---|---|---|
| sco1402-1403* | cvnA4 | 0.974 | Putative large secreted protein | AGTAGTAGGCTCGCGCCGTTTGTTG |
| 0.544 | Putative membrane protein | |||
| sco1947* | gap1 | 0.650 | Glyceraldehyde-3-phosphate dehydrogenase | ACGAACCGATCTCCTCGTTGGTAC |
| sco2373* | tcmA | 0.906 | Tetracenomycin C efflux protein | TTACTGACTCGTGAATTGGTTTGTCA |
| sco2907 | nagE2 | 1.043 | Putative PTS transmembrane component | TGGAAAAGACCGGATCCCCGCTTCTTT |
| sco3217 | cdaR | 0.420 | Putative transcriptional regulator | GCCGCACCGCTGCGCAGGTTTGG |
| sco3867-3868 | soyB1 | 0.611 | Putative ferredoxin | CGCACTCCAGCGGCGGTCGGTTTCGG |
| 1.398 | Putative uncharacterized protein | |||
| sco4423 | afsK | 1.405 | Serine/threonine-protein kinase AfsK | TCTTCCTGTCCGAGGCGCTGGTGGCGAACCT |
| sco4921* | accA2 | 1.650 | Putative acyl-CoA carboxylase A subunit | CGTGCTGCGGGCCACGCGGTTTCTTT |
| sco4947* | narG3 | 1.487 | Nitrate reductase alpha chain NarG3 | GCCGACGCCGCTGACCGGCTTCTGA |
| sco5423* | pyk2 | 1.017 | Pyruvate kinase | TTTTCGAACGGCGCATGGGTGCCATCCT ATCGGTTTGTTT |
| sco6268 | 1.874 | Putative histidine kinase | TTAACAAACCTCCTCGCCGGTTTTCAAT | |
| sco6312 | cprA | 0.969 | Transcriptional regulator | AAAAACAGGCACACGGTCTGTTG |
| sco6323-6324 | 1.875 | Putative tetR-family regulatory protein | ACTGAAAAGGGTTATTGCCTGTTTTGT | |
| 0.974 | Putative hydrolase |
*indicated targets obtained from ScbR2 targets. Divergent genes are listed in separated lines. Fold change value is the average of three biological duplicates.
Table 2. Targets of ScbR2 confirmed by EMSA.
| Gene | Name | Fold change | Annotation | Possible binding site |
|---|---|---|---|---|
| sco1346–1347 | fabG3 | 0.970 | Putative 3-oxoacyl-ACP reductase | CTAGAAGCCCTGGCACCCGGTGTCA |
| 1.128 | Putative secreted protein | |||
| sco1402–1403 | cvnA4 | 1.063 | Putative large secreted protein | AGTAGTAGGCTCGCGCCGTTTGTTG |
| 0.747 | Putative membrane protein | |||
| sco1505 | rpsD | 1.418 | 30 S ribosomal protein S4 | TGACAAGCCGGAAACCCAGAAAAGAGA |
| sco1570–1571 | argH | 1.161 | Argininosuccinate lyase | TGTCTAACGATTATGCGGGTGCGG |
| 1.178 | Putative uncharacterized protein | |||
| sco1697–1698 | soxR | 1.088 | Putative merR-family regulator | TCGCTCACCCGGTGCGCTCGTTTCTAAG |
| 1.637 | Putative uncharacterized protein | |||
| sco1947 | gap1 | 0.768 | Glyceraldehyde-3-phosphate dehydrogenase | ACGAACCGATCTCCTCGTTGGTAC |
| sco2373 | tcmA | 1.325 | Tetracenomycin C efflux protein | TTACTGACTCGTGAATTGGTTTGTCA |
| sco2528–2529 | leuA | 0.982 | 2-isopropylmalate synthase | GGTCACGCGGGTCCGTATCAGT |
| 1.004 | Putative metalloprotease | |||
| sco2615–2616 | valS | 0.806 | Folylpolyglutamate synthase | CCCGAAACG CGTTTCTTC |
| 1.455 | Putative membrane protein | |||
| sco2879 | cvnA12 | 1.366 | Putative uncharacterized protein | GGTCCTGGTAGTGGCTCAGTCGGTGT |
| sco2907 | nagE2 | 1.099 | Putative PTS transmembrane component | TGGAAAAGACCGGATCCCCGCTTCTTT |
| sco3067–3068 | 0.904 | Putative anti anti sigma factor | CGCACACCGCAGTGCACGTATTTG | |
| sig15 | 0.953 | RNA polymerase sigma factor | ||
| sco3217 | cdaR | 0.756 | Putative transcriptional regulator | GCCGCACCGCTGCGCAGGTTTGG |
| sco3224-3225 | 1.009 | Putative ABC transporter ATP-binding protein | CGACGAATCGAATCGCTTGTAC | |
| absA1 | 0.912 | Two component sensor kinase | ||
| sco3229–3230 | 0.410 | Putative 4-hydroxyphenylpyruvic acid dioxygenase | TTCGTTTTGCATTGTGAGGAGACAGGTGT | |
| cdaPSI | 0.341 | CDA peptide synthetase I | ||
| sco3249 | 0.379 | Putative acyl carrier protein | TTCGAACCTGCGACACCCGCTTTAGG | |
| sco3615–3616 | ask | 1.010 | Aspartokinase (EC 2.7.2.4) | GCTCCTCGCTCAATCCGTCTCTTT |
| 1.115 | Putative uncharacterized protein | |||
| sco3867–3868* | soyB1 | 0.613 | Putative ferredoxin | CGCACTCCAGCGGCGGTCGGTTTCGG |
| 2.278 | Putative uncharacterized protein | |||
| sco3961 | serS | 0.947 | Serine—tRNA ligase | AGGCCACCCTTCGTCCACCTGTTTCTTG |
| sco4035 | sigF | 1.077 | RNA polymerase sigma-F factor | TTGCACACAGTGGACATGTCTTGTGA |
| sco4118 | atrA | 0.563 | Putative tetR-family transcriptional regulator | ACGCACCCGGCGCTTGCGTTTGTCC |
| sco4423* | afsK | 1.110 | Serine/threonine-protein kinase AfsK | TCTTCCTGTCCGAGGCGCTGGTGGCGAACCT |
| sco4503 | 1.103 | Putative long-chain-fatty acid CoA ligase | CAGCGACAGCAGAAGCAGTGTCTTT | |
| sco4659 | rpsL | 1.871 | 30 S ribosomal protein S12 | TAGGCACTACTTCTCCGGTTTCTGT |
| sco4677 | 1.550 | Putative regulatory protein | GACGGACGCGGTGAGTTCGGTGGTGG | |
| sco4921 | accA2 | 2.464 | Putative acyl-CoA carboxylase complex A subunit | CGTGCTGCGGGCCACGCGGTTTCTTT |
| sco4947 | narG3 | 2.347 | Nitrate reductase alpha chain NarG3 | GCCGACGCCGCTGACCGGCTTCTGA |
| sco5085 | actII-orf4 | 0.250 | Actinorhodin operon activatory protein | ATAACAGGCCTACTCAACAGATTTCAAT |
| sco5216 | sigR | 1.620 | RNA polymerase sigma factor | AGTGAGACCGGTCTCGGTTTCACG |
| sco5423 | pyk2 | 0.783 | Pyruvate kinase | TTTTCGAACGGCGCATGGGTGCCATCCTATCGGTTTGTTT |
| sco5544–5545 | cvnA1 | 1.024 | Putative membrane protein | GGAATGATGCCTTCAGGTGTGCAA |
| 0.952 | Putative uncharacterized protein | |||
| sco5881 | redZ | 0.449 | Response regulator | GACGACCCGTGTCCTGGTGTGCTG |
| sco6060 | murC | 0.992 | UDP-N-acetylmuramate—L-alanine ligase | ACAAGGTCGGCGTGCCGGTCCTGAA |
| sco6071 | cprB | 0.777 | A-factor receptor homolog | ACTCAGAGCAGTTCGCTGGTCACTTG |
| sco6268 | 10.927 | Putative histidine kinase | TTAACAAACCTCCTCGCCGGTTTTCAAT | |
| sco6271–6272 | N/A | Putative acyl-CoA carboxylase complex A subunit | ACATTTCCTTCTCTCTTGTTCTCA | |
| 16.275 | Putative secreted FAD-binding protein | |||
| sco6275–6276 | 25.435 | Putative type I polyketide synthase | GACTGATCACCTACCCGGTGTTTCT | |
| 21.160 | Putative secreted protein | |||
| sco6282–6283 | 8.899 | Putative 3-oxoacyl-ACP reductase | CTGCAATTACCCTCGGCGGTATGACG | |
| 9.556 | Putative uncharacterized protein | |||
| sco6288 | 40.447 | Putative regulatory protein | GAAGAGACCGAGCGGTCCGTTTCATT | |
| sco6312 | cprA | 0.801 | Transcriptional regulator | AAAAACAGGCACACGGTCTGTTG |
| sco6323–6324 | 0.923 | Putative tetR-family regulatory protein | ACTGAAAAGGGTTATTGCCTGTTTTGT | |
| 1.082 | Putative hydrolase | |||
| sco7623 | 0.904 | NAD(P) transhydrogenase alpha subunit | CGAGAGACGGCCGTCGTTCTTG |
*indicated targets obtained from ScbR targets. Divergent genes are listed in separated lines. Fold change value is the average of three biological duplicates.
To gain further insights into the binding sites of ScbR or ScbR2, all binding sequences of ScbR or ScbR2 were submitted to the MEME algorithm16. A10-nt conserved motif 5′-MSGYTTSTTD-3′was derived for ScbR and a 5′-DYTYSTYSWS-3′ for ScbR2, respectively, as shown in Fig. 3a, resembling the consensus previously extracted from limited sequences9,11. The two motifs overlapped at a relatively conserved 5′-TTTSTT-3′ element, and overall, ScbR showed a higher specificity than ScbR2. The motif derived from exclusive targets of ScbR2 was almost the same as the motif derived from all target sequences of ScbR2 (data not shown), implying those ScbR2-specific binding sequences are more degenerate, and this degeneracy prevented recognition by ScbR that preferred more conserved binding sequences. Comparison of these two motifs could explain why ScbR2 is capable of binding to all ScbR targets while ScbR only binds a fraction of ScbR2 targets.
Figure 3. Conserved motifs and DNase I footprinting of ScbR and ScbR2 on sco6268 promoter.
(a) Conserved motifs of ScbR and ScbR2. All binding sequences of ScbR or ScbR2 were submitted to MEME algorithm for motif derivation. (b) Binding sites of ScbR and ScbR2 on sco6268 promoter. Footprints of ScbR and ScbR2 are shown between dashed lines, and the MEME predicted motifs were underlined.
To validate the MEME-predicted binding motifs, recombinant ScbR and ScbR2 were further used to conduct DNase I footprinting analysis as described before11, on the HEX dye labelled promoter region of sco6268, which was shown to bind with both ScbR and ScbR2 (Supplementary Fig. S3). Footprints of ScbR and ScbR2 on sco6268p covered the same 30 bp sequences, encompassing two mutually inverted 5′-TTTGG-3′ copies (Fig. 3b), resembling the conserved common element of ScbR and ScbR2 motifs (Fig. 3a). To predict the binding sites of ScbR and ScbR2 on those promoters more precisely, promoters with highly conserved palindromic motifs (scbA, kasO and sco6268 promoters), were then used as references for binding site extraction using the MEME algorithm, and the predicted binding sites were shown in Table 1 and 2. Most predicted binding sequences contained two copies of sequences similar to the 5′-TTTGG-3′ motif, but only one of the copies was highly conserved in some sequences. In some promoters, mainly those targeted by ScbR2, only half of the palindrome could be detected. This suggests a reliance on assistant proteins17 or a need for DNA configuration change to bring the halves closer.
Targets of ScbR and ScbR2 in secondary metabolism and development
When analysing the function of their targets, a predominant role of ScbR and ScbR2 was to regulate secondary metabolism both directly and via regulatory cascades and loops. By direct targeting, they both interacted with the promoter of sco6268 (which encodes a histidine kinase) in the cpk cluster, in addition to the promoter of kasO9,10, to repress coelimycin P1synthesis. As a result, most genes from the cpk cluster were transcriptionally activated in the scbR and scbR2 mutants (Fig. 2b). Since ScbR and ScbR2 were observed to be expressed during different time periods, single mutant of each regulator could exert a regulatory effect on cpk synthesis. Also, ScbR and ScbR2 influenced CDA synthesis by binding to the promoter of the activator gene, cdaR; as expected, most genes of the cda cluster were down-regulated in both ΔscbR and ΔscbR2 mutants (Fig. 2b). In this case, binding appears to activate, rather than repress, expression of the pathway activator gene. In addition, ScbR and ScbR2 may also exert an effect on reducing power supply for antibiotic production by targeting the expression of soyB1, which encodes a ferredoxin pivotal in transferring electrons to cytochrome P450 genes for secondary metabolism18; soyB1 was down-regulated in both ΔscbR and ΔscbR2 (0.611 and 0.613 fold, respectively).
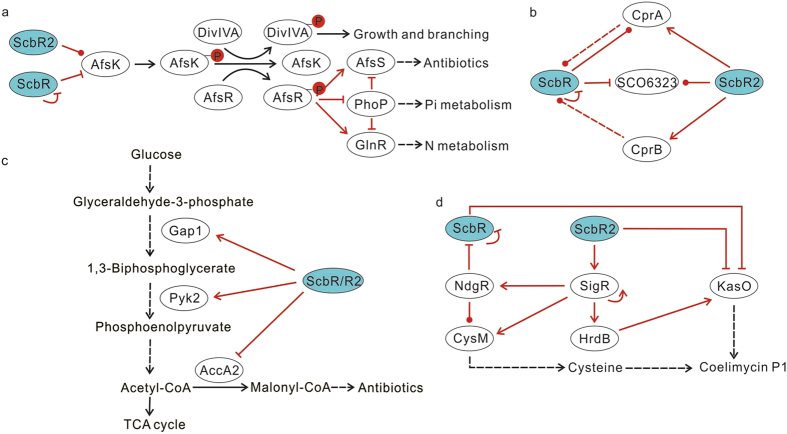
On the other hand, ScbR and ScbR2 made up the first step in some regulatory cascades (Fig. 4a). One case involved AfsS, target of the AfsKR two component system19 and was proposed to relate GBL signalling to the Act and Red production phenotypes20. Here, we further support this idea by identifying afsK as a target of ScbR and ScbR2. In ΔscbR, consistent with the diminished production of Act and Red, expression of afsS was greatly reduced (by 0.317 fold). Remarkably, AfsK is also implicated in polar growth and hyphal branching by phosphorylating DivIVA, and high AfsK activity could cause growth impediment21. This suggested a correlation between the phenotype of growth arrest (Supplementary Fig. S1) and the enhanced afsK expression (1.405 fold) in ΔscbR.
Figure 4. ScbR and ScbR2 mediated regulatory cascades and sub-networks.
(a) The involvement of AfsK in the regulatory cascades from ScbR or ScbR2 to growth or branching, antibiotic production and nutrition metabolism. (b) A complex regulatory network among GBL receptor homologues in S. coelicolor. (c) Control on the glycolysis and carbon flow by ScbR and ScbR2. (d) A sub-network stemming from the regulation of ScbR2 on sigma factor SigR. Regulatory interaction is indicated in red, while metabolite flow is indicated in black. Line with arrow represents activation, with bar represents repression, while with dot means the regulatory effect is unclear or dual function.
Thirdly, a sub-network involving ScbR and ScbR2 regulation was revealed. The promoter of cprA, which encodes another close homologue of ScbR, was bound by ScbR and ScbR2, and cprB, also encoding a close homologue of ScbR, was a ScbR2 target. Both CprA and CprB were reported to be involved in the regulation of antibiotic production and sporulation22, and recently we found they repress GBL synthesis by binding to the promoter of scbA (unpublished results). SCO6323, encoding another ScbR homologue in S. coelicolor, was also demonstrated here to be under the control of ScbR and ScbR2. Therefore, a complex regulatory network among GBL receptor homologues in S. coelicolor intervenes to control complex phenotypes (Fig. 4b).
Besides the common targets with ScbR, the antibiotic receptor ScbR2 appears to exert much more profound control on secondary metabolism. As a cluster-situated-regulator (CSR), ScbR2 bound more promoter regions within the cpk cluster. In addition to kasO and sco6268 promoters, it also bound at the intergenic regions of sco6271-sco6272, sco6275-6276, and sco6282-sco6283, and the promoter of another regulatory gene sco6288. Similarly, in the cda cluster, beside the CSR gene cdaR, it also targeted the promoters of structural genes sco3229-sco3230 and sco3249. Most strikingly, Act and Red production were completely abolished in ΔscbR2, a phenotype partly explained by the direct regulation by ScbR2 of the corresponding CSR genes actII-orf4, redD and redZ23,24. Transcription of them was greatly reduced in ΔscbR2 (Table 2). Furthermore, ScbR2 directly regulates the pleiotropic regulatory genes atrA25 and absA1/A226 to control antibiotic production. atrA was repressed (0.563 fold) and absA1/A2 was induced (Supplementary Fig. S4) in ΔscbR2. Overall, about one third of targets of ScbR2 were found to affect antibiotic biosynthesis, revealing a key role of ScbR2 in the control of antibiotic production phenotypes.
Targets of ScbR and ScbR2 involved in primary metabolism and other processes
The involvement of ScbR and ScbR2 in primary metabolism was mainly observed at three critical nodes in carbon flow, nitrate respiration and acetylglucosamine (GlcNAc) transport. Two genes encoding enzymes in glycolysis, gap1and pyk2 (the former encodes a glyceraldehyde-3-phosphate dehydrogenase responsible for the synthesis of 1,3-biphosphoglycerate from glyceraldehyde -3 phosphate; the latter encodes a pyruvate kinase controlling an irreversible reaction from phosphoenolpyruvate to pyruvate), were found as targets of ScbR and ScbR2 (Fig. 4c). Reduced transcription of gap1 in ΔscbR and ΔscbR2 (0.650 and 0.768 fold) suggests a slowdown of primary metabolism. Although, transcription of pyk2 was marginally changed in ΔscbR, it was reduced in ΔscbR2 (0.783 fold). Another important target gene of ScbR and ScbR2, sco4921 (accA2), encodes the A subunit of acetyl-CoA carboxylase, which is the key enzyme involved in converting acetyl-CoA to malonyl-CoA, thus providing precursors for antibiotic synthesis (notably polyketides such as Act and coelimycin P1) (Fig. 4c). Overexpression of accA2 in both ΔscbR and ΔscbR2 (1.650 and 2.464 fold) would direct more acyl-CoA flux toward malonyl-CoA for antibiotic production. NarG3, a component of respiratory nitrate reductase27, was also found as a target of ScbR and ScbR2, and its expression was induced in both ΔscbR and ΔscbR2 (1.487 and 2.347 fold). ScbR and ScbR2 were also involved in the regulation of nutrition utilization by targeting nagE2, which encodes the only permease for GlcNAc, a primary source of carbon and nitrogen for streptomycetes28,29. As mention above, afsK was under direct control from ScbR and ScbR2, downstream its response regulator AfsR was also closely involved with nitrogen and phosphate metabolism by binding with the promoters of the corresponding regulatory genes glnR and phoR/phoP30,31, and regulator PhoP could control GlnR and AfsS to correlate nutrition metabolism with antibiotic production31,32 (Fig. 4a). GBL and antibiotics are therefore involved in nutrition utilization as well.
Also ScbR2 controlled specific targets other than that shared with ScbR. The ribosome is a key node in cell associating with antibiotic-inducing responses and bacterial drug resistance33. Surprisingly, ScbR2 ChIP-seq binding peaks were associated with genes encoding ribosomal proteins, rpsD and rpsL, and were also present inside and upstream of ribosomal RNA genes (Supplementary Fig. S5), showing a role of ScbR2 in controlling ribosome assembly. The ability of a cell to synthesize proteins during stationary phase was thought the indication of its ability to produce secondary metabolite34. Accelerated protein synthesis displayed by overexpressed rpsL and rpsD in ΔscbR2 (1.418 and1.871 fold) could contribute to increased antibiotic production. In streptomycetes, mutants of σ factors implicated in stress-response are also perturbed in antibiotic production35. Among ScbR2 targets, we also identified genes for sigma factors. For example, SigR controls the response to thiol-oxidative stress36, and maintains the level and activity of the housekeeping sigma factor HrdB37. Interestingly, one of the targets of SigR, NdgR, can bind to the scbR-scbA intergenic region38, so the regulation on SigR resulted in two cascades from ScbR2 to KasO: ScbR2-SigR-NdgR-ScbR-KasO, and ScbR2-SigR-HrdB-KasO (the kasO promoter is recognised by the HrdB sigma factor39), as shown in Fig. 4d. Also, overexpressed sigR in ΔscbR2 (1.620 fold) was speculated to increase cysteine synthesis to meet the high demand of N-acetylcysteine in coelimycin P1 biosynthesis8 due to induction on cysteine synthesis gene cysM by SigR40. Three other sigma factors were found as targets of ScbR2: Sig15 was reported to play a role in osmotic stress response41; SigF controls late stages of spore development in Streptomyces42; and SCO4677, an anti-sigma factor (F), was found to repress antibiotic production and morphological differentiation43. Through regulation of sigma factors, ScbR2 also intensely relate with stress responses, further supporting a role of ScbR2 in eliciting survival responses in perception of antibiotic signals.
Refinement of a local regulatory network and FFL motifs
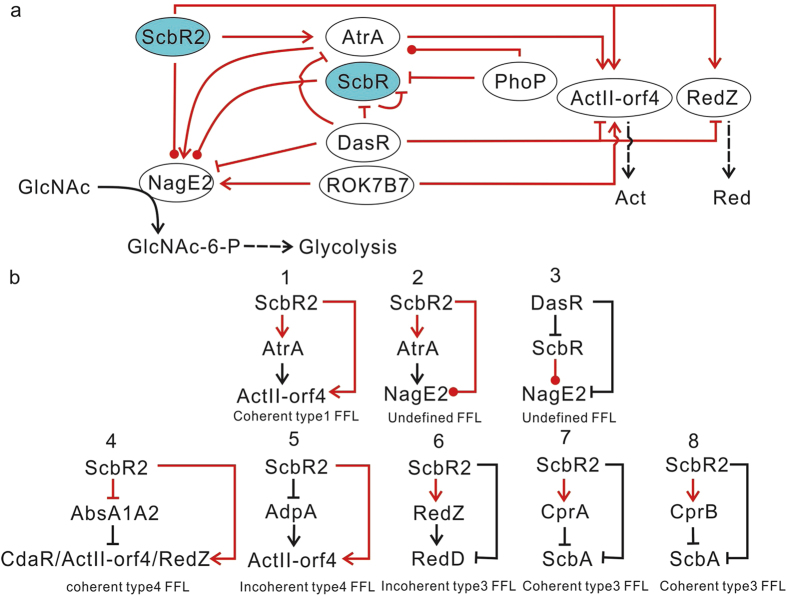
Besides ScbR and ScbR2 (Supplementary Fig. S3), nagE2 is also under multi-level regulation including repression by DasR (a master GlcNAc-responsive regulator)23,28, activation by AtrA (an activator of Act production)25, and activation from ROK7B7, which affects xylose utilization and carbon catabolite repression44,45. Recently, AtrA was reported to be regulated by DasR23, thus our work brought in more interplays among these regulators on the control of GlcNAc transport (Fig. 5a). Such interplay also occurred upstream of actII-orf4, a known target of DasR, AtrA and ROK7B725,28,46. Another antagonism happened between ScbR2 and DasR on the redZ promoter28, further indicating a close relation between GlcNAc transport and antibiotic production (Fig. 5a). Both actII-orf4 and atrA are shown here to be under the direct control of ScbR2 (Supplementary Fig. S3). Thus two FFLs are formed: ScbR2-AtrA-ActII-orf4 and ScbR2-AtrA-NagE2 (Fig. 5b, 1&2). The former is a type I coherent FFL47, in which both regulators X and Y activate the expression of gene Z while regulator X could also activate expression of regulator Y, but the latter FFL controlling NagE2 is an undefined type, since the regulatory effects of the binding of ScbR2 to NagE2 on the gene expression is statistically insignificant from microarray analysis. ScbR was reported to be down-regulated by DasR46, hence another FFL was formed by DasR-ScbR-NagE2 to control the expression of nagE2 (Fig. 5b 3). ScbR is also regulated by PhoP17, which could control the atrA expression17, indicating a complex interaction among GBL and antibiotic signalling with phosphate nutrition. Based on these interactions, comprehensive linkages were built between primary metabolism and secondary metabolism, nutritional signals (C, N and Pi sources) and signalling molecules (GBL and antibiotics) (Fig. 5a). This local regulatory network also provides a basic picture of regulatory networks and the underlying regulatory mechanisms.
Figure 5. The refinement of a comprehensive sub-network and 10 FFLs.
(a) A comprehensive sub-network involving control of GlcNAc transport and antibiotics production. Regulatory interaction is indicated in red, while metabolite flow is indicated in black. Lines with arrows represent activation; lines with bars represent repression, while lines with dots mean the regulatory effect is unclear. (b) 10 refined FFLs. Red lines represent newly found regulatory interaction.
Seven more FFL loops were extracted from the newly identified regulatory interactions (Fig. 5b 4–7). For example, actII-orf4 was found to be under control of two more FFL motifs via AbsA1A2 or AdpA as mediators26,48. When scbR2 is disrupted, AtrA is expected to be down-regulated, but AdpA and AbsA1A2 should be up-regulated (based on the Gus test shown in Supplementary Fig. S4), and the effects of reduced activation by AtrA and ScbR2, increased activation by AdpA, and increased repression from AbsA1A2 would then be integrated at the actII-orf4 promoter. So the abolishment of Act production in ΔscbR2 may mainly result from a more active AbsA1A2 repression. Likewise, multiple FFLs integration was also found to control expression of nagE2, cdaR, redD and scbA and their corresponding phenotypes. Therefore multi-FFLs were employed by S. coelicolor to control key cellular events.
Discussion
In this work, by ChIP-seq and transcriptome analysis, a global view of the specific responses triggered by GBL and antibiotic signalling and the regulatory networks downstream ScbR and ScbR2 were obtained. Unlike the well-studied GBL system of S. griseus, in which the GBL receptor ArpA mainly exerts its control by regulating the expression of AdpA that in turn binds and regulates multiple targets7, ScbR exerts its effects by directly binding to multiple targets and also binds to targets in primary metabolism that are not found in the GBL regulatory cascades in S. griseus. Therefore, the GBL signalling system of S. coelicolor is fundamentally different from that of S. griseus. ScbR2-mediated antibiotics signalling could provoke large scale physiological responses, including secondary metabolism change, ribosome assembly and induction of stress-related sigma factors. Such responses are beneficial for adaptation and could be vital to the survival of bacteria in their natural habitats. A major response mediated by ScbR and ScbR2 was the shift of endogenous antibiotics production, which could also serve as signals in intra- and interspecies communication or weapons in interspecies competition, implying a role of GBL and antibiotic signalling in streptomycetes ecology.
Interplays between ScbR, ScbR2 and other regulators allow the refinement of complex networks, among which several patterns of regulatory interconnection were extracted. By direct interaction, ScbR2 controls the sequent expression of multiple genes for coelimycin P1 synthesis according to the affinity of ScbR2 with their promoters, a common regulatory node in the metabolic pathway to perform a temporal regulation47. Also, ten FFLs involving ScbR and/or ScbR2 were defined in this work. FFLs are important building blocks of regulatory networks47. They can generate different phenotypes under different signal strength, as we observed previously with the incoherent FFL consisting of ScbR2-AdpA-RedD6, and could also serve other purposes, such as acceleration of signal response and noise filtration49,50. In the coherent type 1FFL ScbR2-AtrA-ActII-orf4 (Fig. 5b 1), both ScbR2 and AtrA can be deactivated from their targets by Act10,51. Therefore, when the concentration of Act is low, it would first disassociate the low affinity regulator, but the activation of actII-orf4 will be maintained by the other activator. Thus the FFL could function to delay and filter the turbulence in Act concentration on actII-orf4 expression. But when the Act concentration is high enough to disassociate both ScbR2 and AtrA from the actII-orf4 promoter, the FFL will accelerate the response to shut down the expression of actII-orf4 and Act production, forming a quick-responsive feedback inhibition mechanism (Fig. 5a). Interestingly, integration of multiple FFLs and utilization of feedback loop were discovered to control GlcNAc transport, antibiotics production, and SCB1 synthesis, which would benefit a stable gene expression52,53 and permit S. coelicolor to show robust adaptation to stimulus. Hence various strategies are used by S. coelicolor to adapt to chemical signals and to deal with fluctuating conditions in different environments.
Complex cross-talks between nutrient, stress, GBL and antibiotic signalling pathways were discovered in this work, involving interplays among ScbR, ScbR2 and many key regulators. Some same regulators were found to control both GlcNAc transport and antibiotic production, suggesting a close relation between GlcNAc transport and antibiotic production and the importance of this correlation in the physiology of S. coelicolor. GlcNAc is a major nutritional signal for streptomycetes to decide between growth and irreversible sporulation28. Therefore to guarantee the accuracy of the decision, multiple regulators, cross-talk between signalling pathways and FFLs formed by these regulators were employed to perform a delicate control on GlcNAc transport. Moreover, cross-talks with other regulators by ScbR or ScbR2, for example, with AdpA, AbsA2, AfsQ1,GlnR, DraR etc at the actII-orf4 promoter; AbsA2 and AfsQ1at the cdaR and redZ promoter; and DraR, AfsQ1 and PhoP at the kasO promoter20, allow the cells to integrate nutritional signals and signals from population growth and environment (manifested by the GBL and Act or Red signals) to adjust the activities of diverse processes, in order to maintain a nutrient homeostasis in natural condition54 and to make and support the important decision to sporulate and/or make antibiotics.
Methods
Bacterial strains, plasmids, oligonucleotides and growth conditions
Bacterial strains used in this study are listed in the Supplementary Table S3 and the oligonucleotide primers used are listed in Supplementary Dataset S1. S. coelicolor strains were incubated on MS solid medium for sporulation and Gus reporter assay. They were grown in liquid SMM medium at 30 °C for western blotting, antibiotic production, ChIP, and microarray experiments. SMMS plates were used for the observation of strain phenotypes. Antibiotic production was detected as described previously10. E. coli strains were grown in Luria–Bertani medium containing ampicillin (100 μg/mL), kanamycin (50 μg/mL), apramycin (50 μg/mL), hygromycin (50 μg/mL) or chloramphenicol (50 μg/mL) when necessary.
Western blotting
ScbR and ScbR2 monoclonal antibodies were prepared with recombinant proteins as antigens by CoWin Biotech Co. Ltd as described before6. Protein concentration was measured with Bradfold method. Total protein was extracted at 18, 24, 30, 36, 42, 48, 60 hour and 20 μg of each sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%). The primary antibodies were diluted at a ratio of 1:3000 from the concentration of 1 mg/ml, while goat anti-rabbit immunoglobulin G- horseradish peroxidase conjugate was used as a secondary antibody at a ratio of 1: 2000.
Construction of ΔscbR
To construct ΔscbR, a 1903 bp an a 1548 bp homologous arm were amplified from M145 genome with primers scbRLarmF/scbRLarmR and scbRRarmF/scbRRarmR, and digested with HindIII and BamHI, and BamHI and EcoRI, respectively. Digested fragments were then ligated with pKC1139 digested with HindIII and EcoRI to obtain the plasmid pKC1139-∆scbR, which was then conjugated into M145 to obtain the ΔscbR strain. Disruption of scbR was verified by PCR, showing that a 502 bp fragment internal to scbR gene was deleted.
ChIP-seq
ChIP experiments were carried out as described previously6. Samples of S. coelicolor M145 were grown in liquid SMM and harvested at 30 h and 42 h for ScbR and ScbR2 ChIP experiments, respectively. DNA obtained from ChIP experiments was then sonicated into shorter fragments and TruSeqTM DNA Sample Prep Kit-Set A was used to create a pair-end DNA library, which was subsequently amplified with TruSeq PE Cluster Kit and sequenced using Illumina Hiseq2500. MACS15 was used to identify peaks of ScbR and ScbR2 binding, and software CGview was used to create and view ChIP-seq maps55. Sequencing data were deposited in NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, number GSE64903).
Microarray transcriptional profiling
Spores of M145, ΔscbR and ΔscbR2 were inoculated in liquid SMM medium and grown at 30 °C. M145 and ΔscbR were harvested for RNA extraction at 30 h, M145 and ΔscbR2 were harvested at 42 h for RNA extraction. RNAs were then subjected to customized 8*60k Agilent microarray of S. coelicolor for hybridization according to the manufacturer’s published protocol (Agilent). Hybridization signals were extracted with Feature Extraction software to obtain raw data, which was introduced into GeneSpring GX software to set parameters and data obtained were then normalised. Six probes were designed for each gene and three biological replicates were analysed. Offset data of each gene were removed firstly based on criterion:  and average values were used to indicate transcriptional changes. Microarray data were deposited in NCBI (GEO number GSE64645).
and average values were used to indicate transcriptional changes. Microarray data were deposited in NCBI (GEO number GSE64645).
Construction of gus reporter plasmids
Promoters of absA1, actII-orf4 and cdaR were amplified from genomic DNA with primers absA1pGR/absA1pGF, actII-orf4pGF/actII-orf4pGR, and cdaRpGF/cdaRpGR, respectively. Plasmid backbone was amplified from plasmid pLC-gus6 with primers pLCgusF/pLCgusR. Promoters were then assembled with plasmid backbone by Gibsion assembly to construct plasmids pLC-absA1p-gus, pLC-actII-orf4p-gus and pLC-cdaRp-gus. Plasmids were sequenced for validation and transformed into WT and ΔscbR2 for coloration detection56.
Additional Information
How to cite this article: Li, X. et al. ScbR- and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci. Rep. 5, 14831; doi: 10.1038/srep14831 (2015).
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grant 31130001 and Ministry of Science and Technology of China Grant 2013CB734001. We are grateful to Professor Keith Chater for critical reading of this manuscript and Dr. Keqiang Fan for helpful comments, and Professor Qun He for ChIP technical support.
Footnotes
Author Contributions X.L., W.W. and K.Y. designed research; X.L. performed research; J.W. and J.J. contributed study materials; X.L. and S.L. analyzed data; X.L. and K.Y. wrote the paper. All authors reviewed the manuscript.
References
- Zhuang X. L., Gao J., Ma A. Z., Fu S. L. & Zhuang G. Q. Bioactive Molecules in Soil Ecosystems: Masters of the Underground. International Journal of Molecular Sciences 14, 8841–8868, 10.3390/Ijms14058841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. T. & Bassler B. L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harbor Perspectives in Medicine 2, a012427, 10.1101/cshperspect.a012427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front Biosci 7, d2045–2057 (2002). [DOI] [PubMed] [Google Scholar]
- Li Y.-H. & Tian X. Quorum Sensing and Bacterial Social Interactions in Biofilms. Sensors (Basel, Switzerland) 12, 2519–2538, 10.3390/s120302519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano E., Chakraburtty R., Nihira T., Yamada Y. & Calasso M. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 41, 1015–1028 (2001). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proceedings of the National Academy of Sciences of the United States of America 111, 5688–5693, 10.1073/pnas.1324253111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y., Yamazaki H., Kato J. Y., Tomono A. & Horinouchi S. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Bioscience, biotechnology, and biochemistry 69, 431–439, 10.1271/bbb.69.431 (2005). [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P. et al. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chemical Science 3, 2716–2720, 10.1039/C2SC20410J (2012). [DOI] [Google Scholar]
- Takano E. et al. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol Microbiol 56, 465–479, 10.1111/j.1365-2958.2005.04543.x (2005). [DOI] [PubMed] [Google Scholar]
- Xu G. et al. “Pseudo” gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem 285, 27440–27448, 10.1074/jbc.M110.143081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. A novel role of ‘pseudo’gamma-butyrolactone receptors in controlling gamma-butyrolactone biosynthesis in Streptomyces. Mol Microbiol 82, 236–250, 10.1111/j.1365-2958.2011.07811.x (2011). [DOI] [PubMed] [Google Scholar]
- Bunet R. et al. Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of Kinamycins. J Bacteriol 193, 1142–1153, 10.1128/jb.01269-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Najafabadi Z. et al. The gamma-butyrolactone receptors BulR1 and BulR2 of Streptomyces tsukubaensis: tacrolimus (FK506) and butyrolactone synthetases production control. Appl Microbiol Biotechnol 98, 4919–4936, 10.1007/s00253-014-5595-9 (2014). [DOI] [PubMed] [Google Scholar]
- Gottelt M., Kol S., Gomez-Escribano J. P., Bibb M. & Takano E. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156, 2343–2353, 10.1099/mic.0.038281-0 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137, 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research 37, W202–208, 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allenby N. E., Laing E., Bucca G., Kierzek A. M. & Smith C. P. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic acids research 40, 9543–9556, 10.1093/nar/gks766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang Y., Zhang L., Yang X. & Jiang X. Exploring the electron transfer pathway in the oxidation of avermectin by CYP107Z13 in Streptomyces ahygroscopicus ZB01. PLoS One 9, e98916, 10.1371/journal.pone.0098916 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. C., Umeyama T. & Horinouchi S. afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol Microbiol 43, 1413–1430 (2002). [DOI] [PubMed] [Google Scholar]
- Liu G., Chater K. F., Chandra G., Niu G. & Tan H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiology and molecular biology reviews: MMBR 77, 112–143, 10.1128/MMBR.00054-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel A. M. et al. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proceedings of the National Academy of Sciences of the United States of America 109, E2371–E2379, 10.1073/pnas.1207409109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H., Nakagawa T. & Horinouchi S. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol Microbiol 28, 743–753 (1998). [DOI] [PubMed] [Google Scholar]
- Swiatek-Polatynska M. A. et al. Genome-Wide Analysis of In Vivo Binding of the Master Regulator DasR in Streptomyces coelicolor Identifies Novel Non-Canonical Targets. PLoS One 10, e0122479, 10.1371/journal.pone.0122479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdeno A. M., Bibb M. J. & Challis G. L. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem Biol 8, 817–829 (2001). [DOI] [PubMed] [Google Scholar]
- Uguru G. C. et al. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol Microbiol 58, 131–150, 10.1111/j.1365-2958.2005.04817.x (2005). [DOI] [PubMed] [Google Scholar]
- McKenzie N. L. & Nodwell J. R. Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J Bacteriol 189, 5284–5292, 10.1128/jb.00305-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Alderson J., van Keulen G., White J. & Sawers R. G. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 156, 3166–3179, 10.1099/mic.0.042572-0 (2010). [DOI] [PubMed] [Google Scholar]
- Rigali S. et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Reports 9, 670–675, 10.1038/embor.2008.83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Biro S., Lee K. J., Palmer T. & Schrempf H. The complex extracellular biology of Streptomyces. FEMS microbiology reviews 34, 171–198, 10.1111/j.1574-6976.2009.00206.x (2010). [DOI] [PubMed] [Google Scholar]
- Santos-Beneit F., Rodríguez-García A. & Martín J. F. Overlapping binding of PhoP and AfsR to the promoter region of glnR in Streptomyces coelicolor. Microbiological Research 167, 532–535, 10.1016/j.micres.2012.02.010 (2012). [DOI] [PubMed] [Google Scholar]
- Santos-Beneit F., Rodriguez-Garcia A., Sola-Landa A. & Martin J. F. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol Microbiol 72, 53–68, 10.1111/j.1365-2958.2009.06624.x (2009). [DOI] [PubMed] [Google Scholar]
- Rodríguez-García A., Sola-Landa A., Apel K., Santos-Beneit F. & Martín J. F. Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Research 37, 3230–3242, 10.1093/nar/gkp162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Spiegelman G. B. & Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9, 445–453, 10.1016/j.mib.2006.08.006 (2006). [DOI] [PubMed] [Google Scholar]
- Hosaka T., Xu J. & Ochi K. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol Microbiol 61, 883–897, 10.1111/j.1365-2958.2006.05285.x (2006). [DOI] [PubMed] [Google Scholar]
- Lian W. et al. Genome-wide transcriptome analysis reveals that a pleiotropic antibiotic regulator, AfsS, modulates nutritional stress response in Streptomyces coelicolor A3(2). BMC genomics 9, 56, 10.1186/1471-2164-9-56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget M. S., Kang J. G., Roe J. H. & Buttner M. J. sigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). The EMBO journal 17, 5776–5782, 10.1093/emboj/17.19.5776 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallifidas D., Thomas D., Doughty P. & Paget M. S. The sigmaR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology 156, 1661–1672, 10.1099/mic.0.037804-0 (2010). [DOI] [PubMed] [Google Scholar]
- Yang Y. H. et al. NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Appl Microbiol Biotechnol 82, 501–511, 10.1007/s00253-008-1802-x (2009). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. An engineered strong promoter for streptomycetes. Appl Environ Microbiol 79, 4484–4492, 10.1128/AEM.00985-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget M. S. B., Molle V., Cohen G., Aharonowitz Y. & Buttner M. J. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Molecular Microbiology 42, 1007–1020, 10.1046/j.1365-2958.2001.02675.x (2001). [DOI] [PubMed] [Google Scholar]
- Homerova D., Sevcikova B., Rezuchova B. & Kormanec J. Regulation of an alternative sigma factor sigmaI by a partner switching mechanism with an anti-sigma factor PrsI and an anti-anti-sigma factor ArsI in Streptomyces coelicolor A3(2). Gene 492, 71–80, 10.1016/j.gene.2011.11.011 (2012). [DOI] [PubMed] [Google Scholar]
- Potuckova L. et al. A new RNA polymerase sigma factor, sigma F, is required for the late stages of morphological differentiation in Streptomyces spp. Mol Microbiol 17, 37–48 (1995). [DOI] [PubMed] [Google Scholar]
- Kim E. S., Song J. Y., Kim D. W., Chater K. F. & Lee K. J. A possible extended family of regulators of sigma factor activity in Streptomyces coelicolor. J Bacteriol 190, 7559–7566, 10.1128/jb.00470-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H. et al. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol 75, 1133–1144, 10.1111/j.1365-2958.2009.07020.x (2010). [DOI] [PubMed] [Google Scholar]
- Swiatek M. A. et al. The ROK family regulator Rok7B7 pleiotropically affects xylose utilization, carbon catabolite repression, and antibiotic production in streptomyces coelicolor. J Bacteriol 195, 1236–1248, 10.1128/jb.02191-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel G. P. & McDowall K. J. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Natural product reports 28, 1311–1333, 10.1039/c1np00003a (2011). [DOI] [PubMed] [Google Scholar]
- Shen-Orr S. S., Milo R., Mangan S. & Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature genetics 31, 64–68, 10.1038/ng881 (2002). [DOI] [PubMed] [Google Scholar]
- Park S. S. et al. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). Journal of industrial microbiology & biotechnology 36, 1073–1083, 10.1007/s10295-009-0591-2 (2009). [DOI] [PubMed] [Google Scholar]
- Mangan S., Itzkovitz S., Zaslaver A. & Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. Journal of molecular biology 356, 1073–1081, 10.1016/j.jmb.2005.12.003 (2006). [DOI] [PubMed] [Google Scholar]
- Mangan S., Zaslaver A. & Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. Journal of molecular biology 334, 197–204 (2003). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Binding of a biosynthetic intermediate to AtrA modulates the production of lidamycin by Streptomyces globisporus. Mol Microbiol, 10.1111/mmi.13004 (2015). [DOI] [PubMed] [Google Scholar]
- Ma W., Trusina A., El-Samad H., Lim W. A. & Tang C. Defining network topologies that can achieve biochemical adaptation. Cell 138, 760–773, 10.1016/j.cell.2009.06.013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselius L. et al. Modelling and analysis of a gene-regulatory feed-forward loop with basal expression of the second regulator. Journal of theoretical biology 363, 290–299, 10.1016/j.jtbi.2014.08.043 (2014). [DOI] [PubMed] [Google Scholar]
- Martin J. F. et al. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microbial biotechnology 4, 165–174, 10.1111/j.1751-7915.2010.00235.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. & Wishart D. S. Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539, 10.1093/bioinformatics/bti054 (2005). [DOI] [PubMed] [Google Scholar]
- Rudolph M. M., Vockenhuber M. P. & Suess B. Synthetic riboswitches for the conditional control of gene expression in Streptomyces coelicolor. Microbiology 159, 1416–1422, 10.1099/mic.0.067322-0 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.