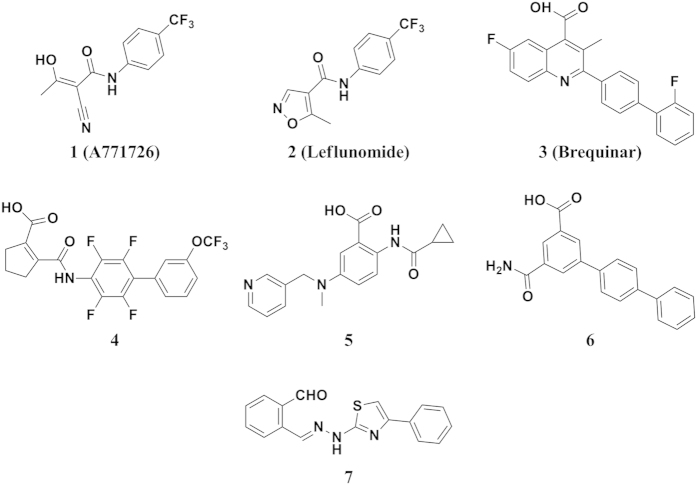
Abstract
Human dihydroorotate dehydrogenase (hDHODH) is an attractive therapeutic target for the treatment of rheumatoid arthritis, transplant rejection and other autoimmune diseases. Based on the X-ray structure of hDHODH in complex with lead compound 7, a series of benzylidenehydrazinyl-substituted thiazole derivatives as potent inhibitors of hDHODH were designed and synthesized, of which 19 and 30 were the most potent with IC50 values in the double-digit nanomolar range. Moreover, compound 19 displayed significant anti-arthritic effects and favorable pharmacokinetic profiles in vivo. Further X-ray structure and SAR analyses revealed that the potencies of the designed inhibitors were partly attributable to additional water-mediated hydrogen bond networks formed by an unexpected buried water between hDHODH and the 2-(2-methylenehydrazinyl)thiazole scaffold. This work not only elucidates promising scaffolds targeting hDHODH for the treatment of rheumatoid arthritis, but also demonstrates that the water-mediated hydrogen bond interaction is an important factor in molecular design and optimization.
Human dihydroorotate dehydrogenase (hDHODH) is a mitochondrial inner membrane anchored enzyme that belongs to the class 2 DHODH family of enzymes1. It is the fourth enzyme in the de novo pyrimidine nucleotide biosynthesis pathway. In humans, resting or fully differentiated cells fulfil their metabolic requirement for pyrimidines through a salvage pathway, whereas activated lymphocytes and other rapidly proliferating tumor cells primarily depend on de novo pyrimidine biosynthesis to support their enhanced growth rate2,3. Inhibition of hDHODH can block this de novo pyrimidine nucleotide biosynthesis pathway, and reducing the survival of these proliferating lymphocytes and tumor cells. Consequently, hDHODH is an attractive target for the treatment of cancer and autoimmune diseases such as rheumatoid arthritis, psoriasis and multiple sclerosis4,5,6.
Clardy et al. first solved the three-dimensional crystal structures of hDHODH in complex with A771726 (1), the active metabolite of leflunomide (2), and a brequinar analog, providing the first insights into the specific structural features of class 2 DHODH enzymes. hDHODH is composed of two domains connected by an extended loop: a large α/β barrel C-terminal domain and a small N-terminal domain containing two α-helices7. The C-terminal domain offers binding sites for FMN and DHO, while the N-terminal domain houses the ubiquinone-binding site8. Several anti-tumor and immunosuppressive molecules have been reported to target hDHODH at the ubiquinone-binding site1,9,10,11. The two best-studied examples (Fig. 1) are brequinar (3)12 and leflunomide (2)13,14,15. The latter, which is the prodrug of the active metabolite A771726 (1), has been approved by the FDA for the treatment of rheumatoid arthritis. Unfortunately, both compounds are associated with various side effects. Brequinar was used for the treatment of cancer and graft-versus-host disease on the basis of satisfactory efficacy in animal models but failed in clinical trials due to its narrow therapeutic window16. Leflunomide has a disadvantageously long half-life of approximately two weeks in patients and has many side effects, such as symptomatic liver damage and interstitial lung disease17. Consequently, the identification of novel potent inhibitors of hDHODH as therapeutic agents that inhibit pyrimidine biosynthesis for the treatment of cancer and autoimmune diseases remains of considerable interest1,9.
Figure 1. Selected structures of reported hDHODH inhibitors.
Compound 7 is the lead in this study identified in our previous work.
Water plays a crucial role in molecular recognition and molecular association18. A comprehensive study of water molecules at protein-ligand interfaces was performed by Wang et al. in 200719. A total of 1829 ligand-bound water molecules, of which more than half bridged ligand and protein atoms by polar interactions, were identified in the crystal structures of 392 high-resolution protein-ligand complexes. The formation of hydrogen bond networks is specific features of water molecules in active sites20. According to Essex et al., a tightly bound water molecule can form three or four hydrogen bonds with the protein and the ligand, contributing to a stronger negative binding free energy compared to a loosely bound water molecule, which makes less than three hydrogen bonds21. Water molecules have been confirmed to be essential mediators of binding affinity or binding specificity in several ligand-protein systems. Recently, Boxer and Levinson shed light on the significance of water molecules at the structural interface in inhibitor recognition by demonstrating that two structural water molecules in the active site assisted the clinical kinase inhibitor bosutinib in recognizing its binding targets22. Hangauer et al. suggested that water molecules could strengthen the cooperativity between the functional groups of the ligand in the thermolysin-binding pocket23. Three interfacial water molecules were identified by Christensen and coworkers as engaged in an extensive hydrogen bonding network between thapsigargin and the backbone of sarco/endoplasmic reticulum calcium ATPase (SERCA), which could influence the orientation of the thapsigargin molecule to bind to the active site24. In addition, Woods et al. addressed the impact of a conserved water molecule that mediates saccharide binding to legume lectin concanavalin A25. These studies have demonstrated that certain structural water molecules within protein active sites are relevant for ligand-protein recognition and contribute to binding affinity by modifying the active site geometry and generating water-mediated hydrogen bond networks, and thus should be considered in drug design18,21,26,27,28.
In this study, unexpected water-mediated hydrogen bond networks were identified in the interface between hDHODH and benzylidenehydrazinyl-substituted thiazole inhibitors in the analysis of the X-ray crystal structure of hDHODH in complex with the lead compound 7 (PDB ID: 4LS0), which was identified via virtual screening (Fig. 1)6. Three rounds of structure-guided lead optimization and X-ray crystallographic investigations revealed that the water-mediated binding modes were specific to inhibitors harboring benzylidenehydrazinyl-substituted thiazole skeletons. Compared to other published structures of hDHODH inhibitors, the benzylidenehydrazinyl-substituted thiazole analogs exhibited greater flexibility within the binding site due to the two rotatable bonds of the amphipathic methylenehydrazine linker connecting the hydrophilic and hydrophobic portions of the ligands. Water molecules located at the P3 and P4 positions acted as anchors to help maintaining the bioactive conformation of the inhibitors in this series. Therefore, the benzylidenehydrazinyl-substituted thiazole inhibitors developed in this study exhibited relatively high binding affinities against hDHODH. Of these inhibitors, compounds 19 and 30 were the most potent with the IC50 values in the double-digit nanomolar scale. Moreover, 19 exhibited remarkable anti-arthritic efficacy in vivo and markedly alleviated foot swelling in a dose-dependent manner. Compound 19 also displayed good pharmacokinetic profiles and a half-life of 9.69 h indicating its promise as a scaffold for further development as an anti-arthritic agent. Interrogating the distributions and binding patterns of all the bridging water molecules in the 35 X-ray crystal structures of hDHODH deposited in the Protein Data Bank29 suggested that the locations and binding modes of the binding-enhancing interfacial water molecules in the ubiquinone-binding site tend to be complementary with the polar moiety of the ligands and should be exploited rationally in the future drug design targeting hDHODH enzyme. In addition to providing novel chemical scaffolds for the treatment of rheumatoid arthritis, the present study also sheds light on the importance of water-mediated hydrogen bond networks for molecular design and optimization.
Results and Discussion
Lead Generation and X-ray Structure Analysis
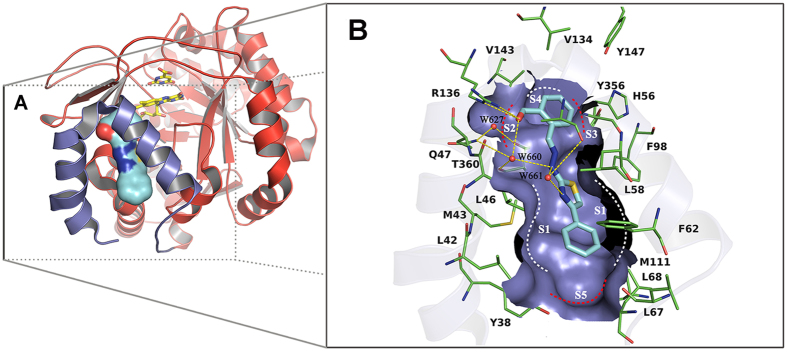
Compound 7, a benzylidenehydrazinyl-substituted thiazole derivative, was first identified in a hierarchical structure-based virtual screen by our group as a potent hDHODH inhibitors with an IC50 value of 0.365 μM6. To elucidate the binding mode of 7 and promote the structure-based development of hDHODH inhibitors of this series, compound 7 was co-crystallized with hDHODH, yielding X-ray structures that diffracted to 2.07 Å. The coordinates of the crystal structure have been deposited in the Protein Data Bank with PDB ID code 4LS0. The X-ray structure of 7 and hDHODH reveals that the formyl-substituted phenyl ring moiety of 7 is located at the inner region of the ubiquinone-binding pocket of hDHODH. A hydrogen bond is observed between the oxygen atom of the aldehyde group of 7 and the guanidine group of Arg136 at subsite S2 (Fig. 2). Two buried water molecules (W627 and W660) in this hydrophilic region form a complicated water-bridging hydrogen bond network with the ligand and residues Gln47, Arg136 and Thr360. More specifically, W627 forms hydrogen bonds with the sidechain nitrogen atom of Gln47 and the sidechain oxygen atom of Thr360, while W660 is more versatile and participates in the formation of four hydrogen bonds with the sidechain nitrogen atom of Gln47, the backbone oxygen atom of Thr360, the oxygen atom of the formyl group of 7, and the secondary amine nitrogen atom of the hydrazine group of 7. Another structural water molecule, W661, is located near the thiazole moiety and forms a bridge between the nitrogen atom of the thiazole group, the secondary amine of the hydrazine moiety and the carbonyl oxygen atom of residue Ala55. Thus, W661 not only enhances the intramolecular interactions within 7, but also participates in polar interactions between the ligand and hDHODH within the hydrophobic subsite S1. In addition to the hydrogen bond interactions in the hydrophilic region, the hydrophobic interactions between 7 and hDHODH near the entrance of the ubiquinone-binding site (S1, Fig. 2) are also of crucial importance. Because the 4-phenyl-thiazole moiety extends to the entrance of the pocket and fits the pocket shape well, hydrophobic effects and VDW interactions occur between the 4-phenyl-thiazol moiety and residues Leu42, Met43, Leu46, Leu58, Phe62, Leu68, Phe98 and Met111.
Figure 2. X-ray structure determination of hDHODH in complex with 7 (PDB ID: 4LS0).
(A) Overview of the structure of hDHODH in complex with compound 7. The receptor is shown in cartoon with the large C-terminal domain colored red and the small N-terminal domain colored purple. Flavin mononucleotide (FMN) and orotate (ORO) are displayed as yellow sticks. Compound 7 is rendered as cyan surface. (B) Detailed description of the ubiquinone-binding site. The hydrophilic pockets are highlighted with red dashed lines, and the hydrophobic ones are marked with white dashed lines. Critical residues are represented as thin green sticks. Oxygen atoms are colored red and nitrogen atoms blue. Water molecules are depicted as red balls. Hydrogen bonds are shown as yellow dashed lines.
Analysis of the binding mode of the lead compound in the X-ray structure highlighted three water molecules (W627, W660 and W661) embedded in the binding site. Compared with the water molecule W627, which consistently bridges Gln47 and Thr360 of the receptor and the polar groups (like carbonyl, carboxyl and cyano) of the ligands in subsite S2 (Fig. 2), W660 and W661 exhibit unexpected binding modes by engaging the 2-(2-methylenehydrazinyl)thiazole moiety of 7 in close contacts with the receptor. The methylenehydrazine linker that connects the hydrophilic and hydrophobic portions of the inhibitor provides two rotatable bonds and results in a more flexible structure. Therefore, the presences of W660 and W661 act as anchors to maintain the skeleton of 7 in its bioactive conformation, thereby enhancing the binding affinity between 7 and hDHODH.
To identify more potent hDHODH inhibitors and to determine if the three water molecules, particularly W660 and W661, are present in other X-ray structures of hDHODH in complex with the benzylidenehydrazinyl-substituted thiazole analogs, lead optimization was conducted in three phases: i) a substructure-based database search to diversify the scaffold of the lead compound and obtain beneficial information; ii) hydrophilic modification of the benzene ring located at subsite S2 to improve polar contacts and determine if variation of the hydrophilic substitutions will influence the presence of the buried water molecule located within the binding site; and iii) hydrophobic optimization at subsite S1 to strengthen nonpolar contacts and to determine if the appearance of the structural water molecule is affected by stronger hydrophobic interactions.
First-Round Optimization of hDHODH Inhibitors
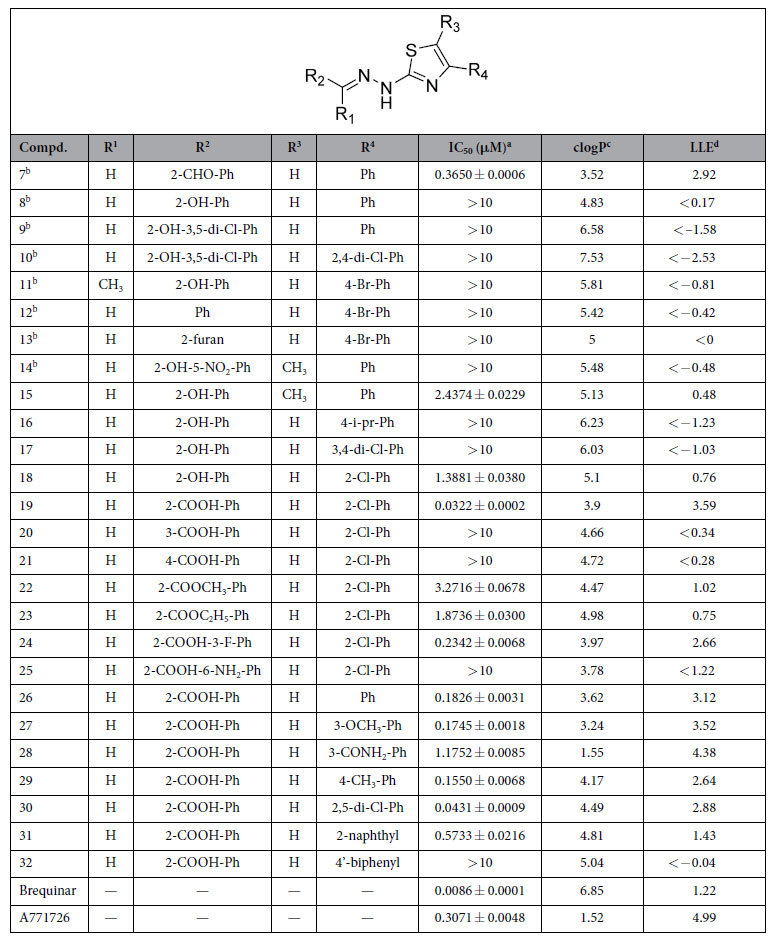
To detect what functional groups are more favorable for the binding at subsite S2, the substructure-based database searching approach was applied in the first round against the currently available databases (such as SPECS and MayBridge database) to rapidly obtain the derivatives. Using unsubstituted (E)-2-(2-benzylidenehydrazinyl)-4-phenylthiazole as the query scaffold, a focused library containing several thiazole derivatives was extracted from the SPECS database30, and all compounds in this focused library (8–14) were purchased from the vendor. HPLC analysis data confirmed the purity of these compounds to be ≥95% (Supplementary Table S1). However, none of the analogs were active (Table 1). Combined with the binding information of 7, we then analyzed the structure-activity relationship of compounds 8–14 and synthesized 15–18. Compounds 15 and 18 exhibited moderate inhibition of hDHODH with increased lipophilicity, resulting in sharply decreased ligand lipophilicity efficiency (LLE)31,32. In contrast to the formyl group of 7, compounds 8–18 contained a hydroxyl-substituted or unsubstituted aromatic ring in subsite S2 (Fig. 2), which undermined the hydrogen bond interactions between the compounds and residues Arg136 and Gln47. This led to a sharp decrease in inhibitory activity as well as a disfavored increase in ligand lipophilicity. By comparing the inhibitory activity of the compounds in the first-round, it was indicated from the opposite side that a carbonyl-substituted aromatic ring in R2 is more preferred than the hydroxyl-substituted or unsubstituted one by subsite S2. In addition, an o-chlorine atom in the benzene group of the 4-phenyl-thiazol moiety was tolerated (18). Accordingly, in the next round of optimization, the accepted o-chlorine atom located in hydrophobic subsite S1 was retained and more polar group was introduced at subsite S2.
Table 1. Chemical structure, ligand lipophilicity efficiency (LLE), and pharmacological activity of compounds 7–32.
Second-Round Optimization of hDHODH Inhibitors
Based on results of the first round, a carboxyl group was introduced into the phenyl to replace the hydroxy group. The resulting compound 19 had an IC50 value of 0.032 μM against hDHODH, a 43-fold increase compared to the hydroxy-substituted derivative 18. Coupled with a reduction in lipophilicity of more than one log unit lipophilicity, 19 exhibits a pronounced advantage over 18 in terms of LLE. Obviously, the greatly increased activity of 19 is attributable to the strong salt bridge interaction between the carboxyl group and Arg365. However, due to an inappropriate orientation and steric clashes, compounds with a meta-substitution (20) or para-substitution (21) of the carboxyl group were unable to directly interact with Arg136, thus leading to a loss of activity. Esterification of the ortho-substituted carboxyl group of 19 with methyl alcohol (22) or ethyl alcohol (23) affected the aforementioned salt bridge, resulting in decreases in potency of 101- and 58-fold, respectively, compared with 19. The esterification also increased the lipophilicity of 22 and 23 compared to 19, leading to an obvious decrease in LLE.
Compound 23 was slightly more potent than 22, which may be attributable to the stronger hydrophobic interactions between the ethyl group and the small hydrophobic pocket formed by residues Val134 and Val143 at subsite S4 (Fig. 2). To detect subsite S4, a fluorine atom was introduced (24). Unfortunately, 24 displayed a 7-fold decrease in inhibition activity compared with 19. This decrease is attributable to the effect of the electron-withdrawing nature of the fluorine atom impacting the orientations of the two oxygen atoms of the carboxyl group, which slightly weakens the salt bridge and the water-mediated hydrogen bond interaction network. In an attempt to establish polar contacts with residues Tyr356, His56 and Tyr147, an amino group was introduced at subsite S3 (25). 25 exhibited a sharp decrease in inhibitory activity, which was likely due to steric clashes. In summary, the second round of optimization revealed that the derivative with an ortho carboxyl-substituted group on the benzene ring at subsite S2 was strongly preferred.
Analysis of the X-ray Structure of 19 with hDHODH.
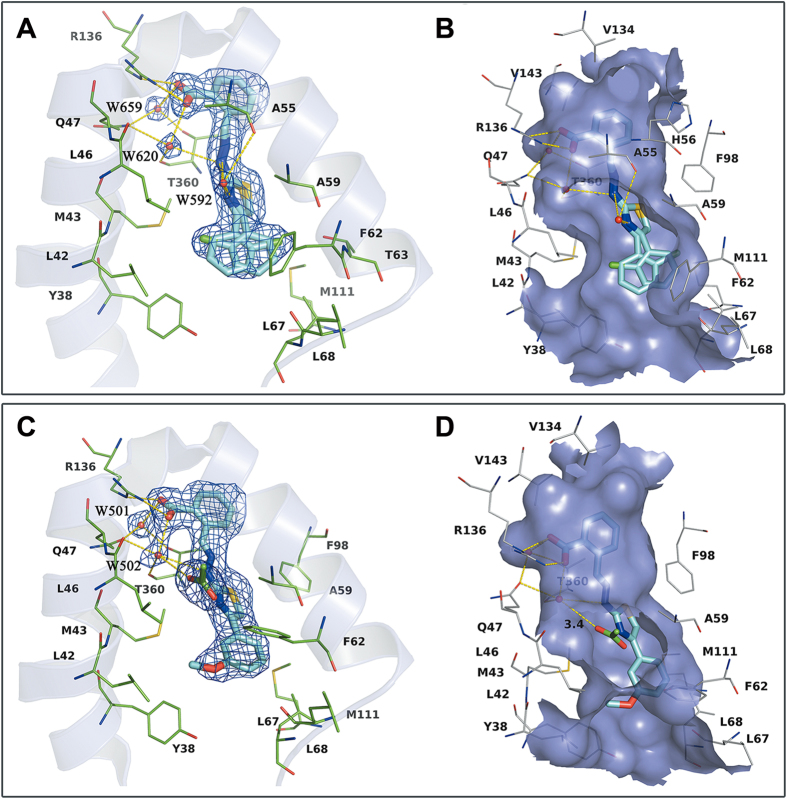
Using our established X-ray crystallographic protocols (see Experimental Section), we obtained a 2.20 Å X-ray structure of compound 19 in complex with hDHODH. The coordinates of the complex structure have been deposited in the Protein Data Bank as the entry 4LS1. The binding mode of compound 19 is presented in Fig. 3A,B. A similar water-bridging hydrogen bond network was observed around the benzylidenehydrazinyl-substituted thiazole portion of inhibitor 19. Crystallographic water molecules (W592, W620, and W659) were observed at nearly the same location and function in the same way as those depicted in Fig. 2. W659 mediates the interaction between Gln47, Thr360 and the carboxyl group. W620 is also tetra-coordinated with the ligand and the protein by simultaneously forming two hydrogen bonds with the receptor and another two with compound 19. A 1.1-Å deviation is apparent between W592 and W661 in Fig. 2. Compared to W661, W592 is closer to Ala55 and slightly further from the hydrazine moiety. However, the bridging functions of W592 are unaffected, and W592 is involved in three hydrogen bonds with the nitrogen atom of the thiazole group, the secondary amine of the hydrazine moiety and the carbonyl oxygen atom of residue Ala55. The novel binding modes of W592 and W620, coupled with the typical binding pattern of W659, result in the formation of a complicated hydrogen bond network that contributes to the stability of the bioactive conformation of the flexible scaffold provided by the benzylidenehydrazinyl-substituted thiazole moiety, which enables favorable polar interactions with hDHODH. Moreover, in subsite S2 of the ubiquinone-binding site, the carboxyl of the benzoic acid moiety forms a nearly perfectly oriented salt bridge with the side chain of Arg136. The combination of this favorable salt bridge and the strong water-bridging hydrogen bond network increase the potency of compound 19 compared to compound 18.
Figure 3. X-ray structure determination of hDHODH in complex with 19 (PDB ID: 4LS1) and 27 (PDB ID: 4LS2).
(A) 2Fo-Fc electron density (blue) for 19 contoured at 1σ. Critical residues are represented as thin green sticks. (B) Specific binding information of the tunnel-like binding site of 4LS1. The surface of the binding site is colored purple. Compound 19 is displayed as cyan sticks and important residues are represented as gray lines. (C) 2Fo-Fc electron density (blue) for 27 contoured at 1σ. (D) Specific binding information of the tunnel-like binding site of 4LS2. Compound 27 (cyan) and acetate molecule (green) are displayed in stick mode. Water molecules are displayed as red balls and hydrogen bonds are shown as yellow dashed lines.
Notably, the chlorophenyl group exhibits a dual binding mode at the entrance of the binding tunnel. The chlorophenyl moiety perfectly accommodates the concave surfaces formed by Met43 and Leu46 on the left side and Ala59, Phe62, and Met111 on the right side (Fig. 3B). In addition to the shape complementarity, the o-substitution of a chlorine atom on the benzene group presumably has the same favorable effects on binding affinity as the methyl group by stabilizing the bioactive conformation of compound 19. This conformation more closely matches the conformer observed in the X-ray structure of the hDHODH and 19, thus decreasing the conformational reordering required upon binding33,34,35. The amphipathic character, salt bridge, water mediation effect and conformational advantages combine to establish compound 19 as the most potent hDHODH inhibitor among the benzylidenehydrazinyl-substituted thiazole derivatives, with an IC50 of 32 nM.
Third-Round Optimization of hDHODH Inhibitors
A third round of modification was launched to further determine if the water-bridging hydrogen bond network would be destroyed when the R4 group (2-chlorine-Ph) of 19 was replaced by more diverse substituents. When there was no substitution on the phenyl moiety, the hydrophobic interactions were weakened, resulting in a slight decrease in activity (26). When another chlorine atom was introduced at the 5-position of the phenyl moiety, compound 30 (IC50 = 0.043 μM) did not exhibit any advantage over 19. Because 30 exhibited increased lipophilicity but similar potency, resulting in a decreased LLE value. Baumgartner et al.11 proposed a new, predominantly polar subsite S5 (Fig. 2) consisting of the hydroxy group of Tyr38 and the backbone carbonyl of Leu67 in the hydrophobic section. This subsite was probed by attaching a -OCH3 group to the 3-position of the phenyl moiety (27). Unfortunately, no obvious polar interactions were observed between the proposed subsite S5 and the -OCH3 moiety (Fig. 3C). However, 27 was equipotent with 19 in terms of LLE, as its reduced lipophilicity compensated for its decreased potency. Efforts to explore this subsite were continued, and 28 was synthesized. The lipophilicity of 28 was reduced by two log units at the expense of a sharp decrease in potency, making 28 less attractive than 19. Therefore, attempts to generate polar contacts with residues Tyr38 and Leu67 at subsite S5 by modifying the 3-position of the phenyl group were unsatisfactory. In addition, the binding mode of 27 in the crystal structure (Fig. 3C) also suggested that substitutions at the 4-position of the phenyl moiety could be accommodated. The biological activity data for 29 and 32 indicated that small hydrophobic groups such as -CH3 are favored at the 4-position, whereas larger groups such as phenyl, may collide with Leu67 and Leu68 and result in a loss of activity. Interestingly, when the biphenyl group was converted into a naphthalene group (31), the latter was capable of forming π-π stacking interactions with Phe62. Thus, compound 31 exhibited moderate inhibitory activity. None of the compounds in the third round performed better than 19, but this round revealed that small substituting groups such as -Cl, -CH3 and -OCH3 are favored at the entrance of the binding site, whereas substituents at the 4-position of the phenyl moiety are actually size-constrained. More importantly, the crystal structure of hDHODH in complex with 27 was solved (PDB ID: 4LS2), which confirmed the functions of the versatile water molecules observed in the other structures.
Analysis of the X-ray Structure of 27 with hDHODH
The X-ray structure of hDHODH with compound 27 was obtained, and the binding mode of 27 is shown in Fig. 3C,D. Interestingly, there are only two structural water molecules (W501 and W502) in the inner polar site of the tunnel-like binding site. These water molecules form nearly the same water-bridging hydrogen bond network observed in compounds 7 (W627 and W660) and 19 (W659 and W620). W501 mediates the interaction between Gln47, Thr360, and the polar carboxyl group of 27. The versatile tetra-coordinated W502 appears in the same place and mediates interactions in the same manner as W660 and W620, even though some modifications to the benzene motif stretching along the hydrophobic entrance of the binding site are apparent. Nevertheless, an acetate molecule instead of a water molecule (W661 in Fig. 2, W592 in Fig. 3A) is in close proximity to the thiazole moiety of the inhibitor, which came from the crystallization agent. The size of the final 2Fo-Fc electron density map at this site is too big for one water molecule but most suitable for one acetate molecule, and the acetate molecule happened to locate at the entrance of the inhibitor binding pocket and formed hydrogen bonds with compound 27 in the complex structure. In addition to the polar contacts with the nitrogen atom of the thiazole group and the secondary amine of the hydrazine moiety, this polar solvent molecule also forms hydrogen bonds with W502 at a distance of 3.4 Å. This structure indicates that the water molecules positioned at W501 and W502 (Fig. 3C) are stable and complement the binding of the scaffold of the benzylidenehydrazinyl-substituted thiazole inhibitor and hDHODH. However, the water molecule located around the thiazole portion of the ligand is loosely bound in the hDHODH-ligand system and is susceptible to be replaced by a solvent molecule. As shown in Fig. 3B,D, the hydrophobic entrance of the binding pocket is exposed to solvent, and its openness is largely influenced by the rotation of the flexible residue Phe62.
In addition to the water-bridging interactions, the salt bridge between residue Arg136 and the carboxyl moiety of 27 is also a valuable feature for binding affinity. The -OCH3 group at the 3-position of the phenyl moiety matches the concave surface perfectly, forming close hydrophobic interactions with the residues of Leu42, Met43 and Leu46 in subsite S1. Figure 3C also indicates that little space remains between the 4-position of the phenyl moiety and the receptor at the entrance of the binding site, and only small substitutions can be accommodated, as verified using compounds 29 and 32. This round of X-ray crystallographic exploration confirmed once again that the binding of the benzylidenehydrazinyl-substituted thiazole inhibitor to hDHODH is accompanied by two tightly bound water molecules and a loosely bound water molecule that is exposed to solvent.
Analysis of Structural Water Molecules in the Ubiquinone-Binding Site.
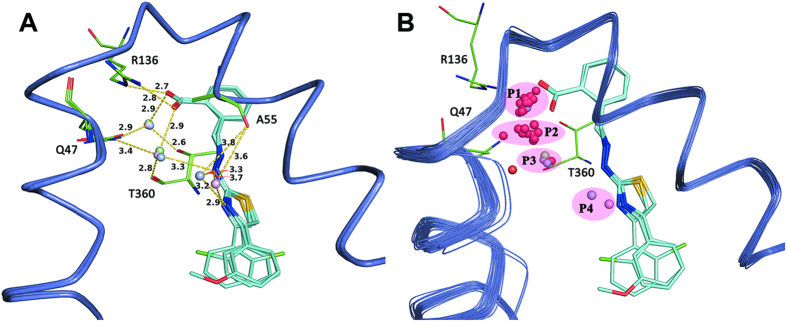
In each of the crystal structures of the three complexes derived in our study, an unexpected water bridging pattern was discovered (Fig. 4A). Figure 4B presents the alignment of all 32 reported protein-ligand complex structures of hDHODH deposited in the Protein Data Bank (Supplementary Information Table S2), with the addition of the 3 structures solved in this study. In total, there are 43 interfacial water molecules clustered into four positions in the ubiquinone-binding site of these 35 crystal structures, of which 58.1% are located in the P1 position and 23.3% are located in the P2 position. Of the five water molecules at the P3 position, three were identified in this study. In addition, two water molecules were observed at the P4 position for the first time in this study.
Figure 4. Analysis of the structural water molecules at the ubiquinone-binding site of hDHODH.
(A) Water-bridging hydrogen bond network. Three X-ray crystal structures solved in this study are aligned together (purple ribbon) and the corresponding ligand are displayed as thin cyan sticks. Interfacial water molecules of 4LS0 (lightblue), 4LS1 (lightpink) and 4LS2 (palegreen) are shown as round balls. Hydrogen bonds are rendered as yellow dashed lines. (B) Distributions of the buried water molecules. Besides the three structures, all the other 32 crystal structures of hDHODH deposited in the Protein Data Bank previously are aligned together, and their buried water molecules are colored red.
Apparently, more than half of the interfacial water molecules are located at the P1 position, where they perform the general functions of bridging Gln47, Thr360 and polar groups (like carbonyl, carboxyl and cyano) of the ligands. Therefore, the water molecule at the common P1 position is adequately considered in molecular docking or virtual screening work6. The appearance of a water molecule at the P2 position is always accompanied by polar groups such as that of A771726. In detail, the water molecule at the P2 position is involved in an intricate hydrogen bond network between residues Gln47, Arg136 and the carbonyl group of the amide moiety in the A771726-like ligand. This binding mode was observed in several X-ray structures, such as 1D3H, 3F1Q, 3FJL, 3G0U, 3G0X and 3U2O. By contrast, unlike the water molecule commonly observed near the P1 or P2 position, the water molecules located at the P3 and P4 positions were rarely observed in other previously published X-ray structures of hDHODH. Interestingly, the co-occurrences of water molecules at the P3 and P4 positions were found together with the scaffold of the benzylidenehydrazinyl-substituted thiazole inhibitors designed in this study. These water molecules cooperate with each other to form a complicated hydrogen bond network that facilitates the binding of the inhibitors in this series to the active site of hDHODH. The water molecule at P3 region was tetra-coordinated with the ligand and the protein through an unexpected bridging pattern in which the water molecule simultaneously forms two hydrogen bonds with the ligand (the benzylidenehydrazinyl portion) and an additional two hydrogen bonds with the protein (the residues Gln47 and Thr360). Moreover, three rounds of verification demonstrated that the water molecule at the P3 position is robust, tightly bound and not shifted by molecular decoration at the hydrophilic or hydrophobic portions of lead compound 7. The water molecule at P3 position appears to be a specialized binding partner that maintains the bioactive conformation of the benzylidenehydrazinyl-substituted thiazole inhibitor, which has a more flexible methylenehydrazine linker connecting the polar and nonpolar portions of the ligand in the binding site compared to other known inhibitors. The water molecule located at the P4 position can also bridge the nitrogen atom of the thiazole group, the secondary amine of the hydrazine moiety, and the carbonyl oxygen atom of residue Ala55. Thus, this water molecule enhances intramolecular interactions in addition to participating in polar interactions between the ligand and hDHODH in the hydrophobic subsite S1. Therefore, the water molecules at the P3 and P4 positions actively contribute to maintaining the bioactive conformations of the benzylidenehydrazinyl-substituted thiazole inhibitors in the ubiquinone-binding site of hDHODH. However, the water molecule at the P4 position is bound more loosely than the water molecule located at the P3 Position, and thus is susceptible to be replaced by the polar acetate molecule (Fig. 4) because the P4 position is exposed to solvent.
Analysis of the distributions and binding modes of the water molecules in the ubiquinone-binding site of hDHODH indicated that the hydrophilic pocket of the active site is water accessible and that the locations and binding patterns of the water molecules complement the polar moiety of the ligands that bind to hDHODH. Given the crucial role of water in protein-ligand association, water molecules should be exploited in the design of hDHODH inhibitors.
In vivo Pharmacokinetic Study of Compound 19
A pharmacokinetic study of the most potent inhibitor, 19, was performed by administering rats a 1 mg/kg intravenous (IV) dose and a 10 mg/kg oral dose (PO) of the compound. After IV dosing, 19 exhibited a terminal half-life of 9.69 h, a steady-state volume of distribution of 0.35 L/kg, and a low plasma clearance of 0.04 L/h/kg (Table 2). After oral administration, 19 exhibited an exposure (AUC0-∞) of 53047.73 μg/L*h, leading to an oral bioavailability of 22.75%. The maximum plasma concentration (Cmax) of 19 was 5310.50 μg/L, and the time to reach the maximum concentration (Tmax) of the inhibitor was 1.00 h. Compared with Vidofludimus (4SC-101), which targets hDHODH and is in phase II clinical trials for the treatment of rheumatoid arthritis and inflammatory bowel disease (with a terminal half-life of 2.3 to 3 hours in MRLlpr/lpr mice36,37), 19 likely has an advantage in terms of its terminal half-life (9.69 h). The oral bioavailability of 19 could be improved by administration in its salt formation and their drug-like properties along with pharmacokinetic characteristics will be optimized further in the future work.
Table 2. Pharmacokinetic parameters of 19 after intravenous (IV) and oral (PO) administration.
| Parametera | 19 |
|---|---|
| IV dose (mg/kg) | 1 |
| T1/2 (h) | 9.69 ± 3.15 |
| AUC0–∞ (μg/L*h) | 23318.81 ± 4501.24 |
| Cl (L/h/kg) | 0.04 ± 0.01 |
| Vdss (L/kg) | 0.35 ± 0.02 |
| PO dose (mg/kg) | 10 |
| T1/2 (h) | 8.20 ± 1.72 |
| AUC0–∞ (μg/L*h) | 53047.73 ± 11030.40 |
| Cmax (μg/L) | 5310.50 ± 1910.47 |
| Tmax (h) | 1.00 ± 0.87 |
| F (%) | 22.75 ± 4.73 |
aCompound was dosed to equal number of male Sprague-Dawley rats in IV and PO administration respectively (n = 3).
In vivo Anti-arthritic Efficacy of Compound 19.
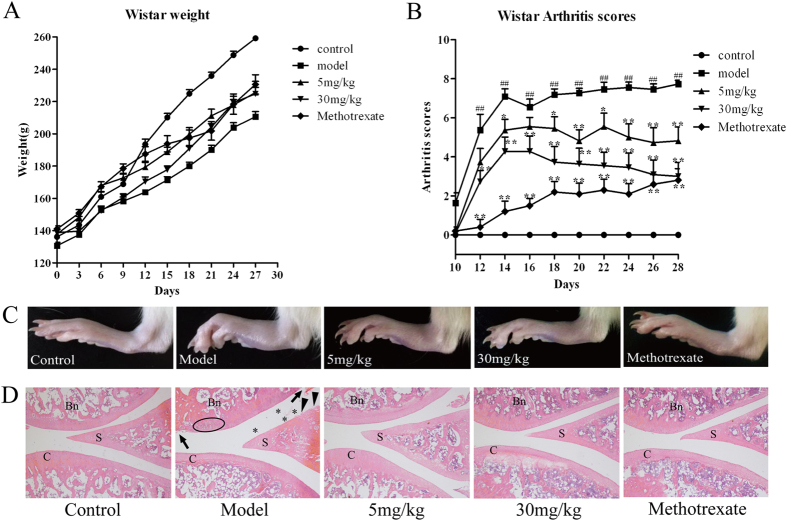
Compound 19 and methotrexate were injected intraperitoneally once per day for 28 days into the Wistar rats with collagen-induced arthritis (CIA). The swelling scores of the arthritis and morphological observations of the rats’ joint tissues were employed to examine the anti-arthritic effect of compound 19 (Fig. 5). With the onset of arthritis, the swelling scores were an indication of the disease states. The maximum score was 8, which is the sum of the scores from both hind paws of each rat and indicates a severe arthritis state. During the experiment, the control rats had normal eating patterns, shiny hair and continuously increasing body weight. By contrast, the rats in the model group exhibited a rough and dull hair at the beginning of the trial, and a slower increase in body weight relative to the normal group after day 9 (Fig. 5A). Although treatment with compound 19 or methotrexate had no obvious influence on increase in body weight, compound 19 displayed significant dose-dependent anti-arthritic effect (p < 0.05) and obviously alleviated foot swelling (Fig. 5B,C). The arthritis scores of the CIA rats reached their peak and stabilized (Fig. 5B) around 8 after day 18, presenting a severe arthritis state, while the arthritis scores of the compound 19 treated rats decreased and were around 5 (5 mg/kg) and 3 (30 mg/kg) at the day 28, which indicated that compound 19 can display substantial anti-inflammation effects and alleviate foot swelling in a dose-dependent manner (Fig. 5B,C).
Figure 5. In vivo effects of compound 19 on collagen-induced arthritis (CIA) rats.
Arthritis was induced in Wistar rats by twice immunization with type II collagen on day 0 and day 7. Compound 19 was administered i.p. daily at 5 mg/kg and 30 mg/kg until the end of this experiment. Methotrexate, as the positive control, was administered at the dose of 0.3 mg/kg. (A) Measurement of body weight was taken every three days. Treatment of compound 19 or methotrexate had no obvious effect on the growth of body weight versus model group. (B) The incidence and severity of arthritis in different groups were evaluated via observation of the changes of arthritis scores every two days. Compound 19 displayed significant anti-arthritic effect in vivo (p < 0.05) and obviously alleviated foot swelling in a dose-dependent manner. (C) The representative photomicrographs of knee joints in different treatments groups taken after 28 day’s trial. The arthritis swelling of the model group was the worst. The swelling hind paw was obviously relieved in the group treated with compound 19. (D) Representative H & E stained sections of joint tissue of each groups. Compared with model group and compound 19 treated groups, the latter could remarkably alleviate the histological changes of severe arthritis. Asterisk indicates the infiltration of the inflammatory cells; solid triangles indicate areas with synovial degradation; solid arrows indicate areas of cartilage degradation; and the oval highlights the inflammatory cells. S, synovium; C, cartilage; Bn, bone. Magnification = 50 ×. Data was expressed as mean ± SEM. ##P < 0.01 vs normal control, *P < 0.05, **P < 0.01 vs CIA group.
Besides, histological changes in the joint tissues of the normal, CIA, compound 19 and methotrexate treated rats were investigated by the hematoxylin and eosin (H&E) staining method. In the joint tissues of the CIA rats with serious arthritis (Fig. 5D), pathological features including the infiltration of the inflammatory cells (marked with asterisk), synovial degradation (marked with solid triangles), cartilage erosion in the joint tissues (marked with solid arrows) and inflammatory lesions of some cells (highlighted with an oval) were clearly observed. Besides, due to the serious arthritis, the articular cavity was full of exudate which was larger than the other compound treated groups. Compared to the CIA rats, the previously mentioned pathological changes were evidently alleviated in the joint tissues of the rats treated with compound 19 in both dosage, demonstrating that compound 19 displayed anti-arthritic effects in a dose-related manner. The significant in vivo anti-arthritic effects of 19 not only confirmed that hDHODH is an effective anti-arthritic target but also confirmed that this compound is a promising chemical scaffold for further development as an immunosuppressant and antiproliferative agent targeting hDHODH.
Conclusion
In summary, this work presented a series of benzylidenehydrazinyl-substituted thiazole inhibitors to suppress the bioactivity of hDHODH. Compound 19 displayed the lowest IC50 value among these compounds, with notable anti-arthritic efficacy and acceptable pharmacokinetic profiles in vivo. These results suggest the potential of 19 for application as an immunosuppressive agent targeting hDHODH. More importantly, water-bridged interactions between the ligands and hDHODH were identified in the three X-ray structures solved in this study. Two robust bridging water molecules co-occurred with the ligands to help maintain their bioactive conformations by generating a strong water-mediated hydrogen bond network between the flexible 2-(2-methylenehydrazinyl)thiazole linker and hDHODH. The locations and H-bonding patterns of the water molecules are highly complementary with the polar moieties of the ligands in the ubiquinone-binding site of hDHODH and should be considered in future drug design and optimization workflows targeting hDHODH.
Methods
In vitro Enzyme Assay
The plasmid coding for human DHODH was kindly provided by Prof. Jon Clardy (Harvard Medical School)7. hDHODH (Met30-Arg396) was expressed and purified in the same manner as described in our recently published work38. We determined the effect of benzylidenehydrazinyl-substituted thiazole derivatives to inhibit hDHODH by using a DCIP assay method38. The assay buffer contained 50 mM HEPES pH 8.0, 150 mM KCl, 0.1% Triton X-100, 100 μM UQ0 and 120 μM DCIP. The results were presented as the mean ± standard deviation of three independent experiments (Table 1).
Crystal Structure Determination
Co-crystallization of hDHODH with compound was performed as previously described38. X-ray diffraction data were collected at 100 K at the synchrotron beamline BL17U1 at SSRF (Shanghai, China). Statistics of data collection, processing, and refinement were summarized in Table 3. The data were processed with MOSFLM39, and scaled using the SCALA program from the CCP4 suite40. Structural elucidation and refinement were carried out using the CCP4 suite of programs40,41. The PDB entry 1D3G (stripped of ligands and water molecules) was used as a molecular replacement model for phasing of the X-ray data. Iterative model building and refinement were performed using Coot and Refmac5, respectively42,43. The molecular graphics package PyMOL (DeLano, 2002) was used to generate the figures.
Table 3. Data collection and refinement statistics for the three hDHODH X-ray structures.
| Inhibitor | 7 | 19 | 27 |
|---|---|---|---|
| Wavelengths (Å) | 0.97852 | 0.97852 | 0.97852 |
| Space group | P 32 2 1 | P 32 2 1 | P 32 2 1 |
| Cell dimensions (Å) | 90.530/90.530/123.820 | 90.860/90.860/122.940 | 90.390/90.390/122.900 |
| Resolution (Å) | 2.07 | 2.20 | 2.27 |
| Number of reflections | 36008 | 30357 | 27386 |
| Redundancy | 7.6(7.7) | 8.8(9.0) | 8.5(8.7) |
| Completeness (%) | 99.6(99.9) | 100.0(100.0) | 99.9(100.0) |
| Rmerge (%) | 11.0(29.0) | 14.9(36.7) | 10.6(33.0) |
| I/σ (I) | 14.8(8.3) | 12.2(6.3) | 18.9(9.4) |
| R/Rfree (%) | 15.1(17.2) | 15.1(18.5) | 15.1(19.0) |
| Bonds (Å) | 0.007 | 0.007 | 0.007 |
| Angles (deg.) | 1.226 | 1.297 | 1.206 |
| PDB ID code | 4LS0 | 4LS1 | 4LS2 |
In vivo Pharmacokinetic Assay
Compound 19, the most potent inhibitor, was selected to perform the single dose pharmacokinetic studies. Male Sprague-Dawley rats were raised according to the Guide for the Care and Use of Laboratory Animals. Blood samples of the rats were collected at the intervals of 5 min, 15 min, 30 min, 1 h, 2 h, 4 h, 8 h and 24 h. Heparin sodium was applied as the anticoagulant of the blood samples. Based on the LC/MS/MS quantitative data, pharmacokinetic parameters were analyzed using WinNonlin5.2 with noncompartment model.
In vivo Efficacy Study
Adult male Wistar rats (120–140 g) were purchased from Shanghai SLRC Laboratory animal company and housed in standard pathogen-free cages. The environment was climate-controlled (20–25 °C, 50–60% relative humidity) with a 12-hour light and 12-hour dark cycle as described previously38. Sterilized food and water were provided naturally. All procedures involving animals conform to the Chinese government guidelines for animal experiments, and were conducted with approval of the Institutional Animal Care and Use Committee of East China University of Science & Technology. Bovine type II collagen (CII, Chondrex, USA) was emulsified in the same manner as that conducted previously to get the CIA model44. Specifically, 49 rats were randomly assigned into five groups: normal control (solvent, i.p., n = 6), CIA model (solvent, i.p., n = 11), Methotrexate-treated (0.3 mg/kg, i.p., n = 10), compound 19-treated (5 mg/kg, i.p., n = 11) and compound 19-treated (30 mg/kg, i.p., n = 11) groups. Methotrexate, reported to has a therapeutic effect on rheumatoid arthritis45, was used as the positive control in this study. The main workflows of this experiment were consistent with that reported recently38. During the 28 days of the experiment, compound 19 (5 mg/kg and 30 mg/kg) and Methotrexate (0.3 mg/kg) were injected intraperitoneally once every day. At the same time, the normal control group and model group were administered the same amount of solution. The solvent of 19 was made up of 5% DMSO, 45% saline and 50% PEG400.
In this study, the clinical-mimic arthritis was observed and the severity was assessed by the same criteria as that applied in reference 65: 0, normal, without any swelling; 1, mild swelling; 2, swelling; 3, significant swelling; 4, severe swelling. The maximum score was 8, which was the sum of the scores from both hind paws of each rat. When arthritic signs appeared, the incidence and severity of the arthritis in rats with different treatments were evaluated via arthritis scores recorded every 2 days under blinded conditions. Measurement of body weight was taken every 3 days. All Wistar rats were killed after 28 day’s trails. Pathological detections against the whole knee joints of the rats were conducted using the same method as described previously38. Mean ± standard deviation (mean ± SEM) was applied to present the data in this study. SPSS was used to evaluate the statistical significance of differences among the groups. P ≤ 0.05 was the threshold to be statistically significant46.
Chemistry
A series of benzylidenehydrazinyl-substituted thiazole derivatives were designed and synthesized as potential hDHODH inhibitors. These analogs were primarily prepared via the cyclization of thiosemicarbazone with 2-bromo-1-phenylethanones47, and the thiosemicarbazones were obtained from the condensation reaction of commercially available substituted benzaldehydes and thiosemicarbazide47. The preparation of 2-bromo-1-phenylethanones began with commercially available acetophenones with diverse substituents48. Following this design route, we successfully acquired compounds 15–18 and 20–21 (as shown in Supplementary Scheme S1). When the substituted benzaldehyde was transformed to the o-carboxybenzaldehyde, further intramolecular cyclization between the carboxyl group and the second nitrogen atom in the hydrazine group occurred in addition to the anticipated reaction (as shown in Supplementary Scheme S2). To obtain the target compounds, extensive efforts were made to regulate the hydrolysis of the intramolecular amide bond. Fortuitously, this problem was resolved by referring to an article in which NaOH was used in dry THF49. However, several substituted benzaldehydes, such as 24a and 25a in Supplementary Scheme S2, were commercially unavailable. Therefore, a five-step workflow comprising esterification, benzylic bromination, lactonization, benzylic bromination and hydrolysis was designed to obtain their crucial o-carboxybenzaldehyde intermediates (24V and 25V). For this workflow, 2-methyl benzoic acid derivatives were used as the starting products (Supplementary Scheme S3)50,51,52,53,54,55,56,57.
The preparations of the esterified derivatives (22, 23) are shown in Supplementary Scheme S4. For these compounds, the esterification reactions of 2-formylbenzoic acid were firstly performed to obtain the methyl ester and ethyl esters without affecting the aldehyde group58. Thiazole hydrazine was obtained via the condensation of thiosemicarbazide and 2-bromo-1-phenylethanones in 1,4-dioxane at room temperature. Finally, the target products, the benzylidenehydrazinyl-substituted thiazole analogs, were obtained via a dehydration reaction of methyl or ethyl 2-formylbenzoate and thiazole hydrazine59. In general, all reaction conditions were mild, and no dangerous or toxic materials were used in the reactions. The structures of all products were verified by 1H NMR, 13C NMR and HRMS (ESI).
General methods
All chemical reagents and solvents were obtained from commercial sources and used without further purification. Thin-layer chromatography (TLC) was carried out to monitor the process of reactions. Purification of compounds was achieved by column chromatography with silica gel (HaiYang, Qingdao) 200–300 mesh. 1H NMR and 13C NMR spectra were recorded on a Bruker AM-400 spectrometer with chemical shifts expressed as ppm (in CDCl3 or DMSO-d6, Me4Si as internal standard). Melting points were recorded on a WRS-1B-digital melting point apparatus. The mass spectra were measured at The Institute of Fine Chemistry of ECUST. The MS data were obtained by Agilent 5975C GC-MS instrument and Agilent LC-MS 1200 and 6120 system. Analytical HPLC was performed on a Hewlett-Packard 1100 chromatography system equipped with photodiode array detector and a Zorbax XDB-C18 column (250 mm × 4.6 mm) was used to determine the purity of the products. The mobile phase A was water with 1‰ TFA and mobile phase B was acetonitrile. A gradient of 30–100% B over 20 minutes was run at a flow rate of 1.0 mL/min. Compounds synthesized in our laboratory were generally varied from 95% to 99% in purity, and the biological experiments were employed on compound whose purity is at least 95%.
methyl 2-(bromomethyl)-6-fluorobenzoate (24III)
1H NMR (400 MHz, CDCl3): δ 7.39 (m, 1H), 7.22 (d, J = 7.6 Hz, 1H), 7.09 (t, J = 8.4 Hz, 1H), 4.66 (s, 2H), 3.99 (s, 3H). GC-MS (EI), 246.1.
methyl 2-(bromomethyl)-3-nitrobenzoate (25III)
1H NMR (400 MHz, CDCl3): δ 8.12 (d, J = 8.0 Hz, 1H), 7.97 (d, J = 8.0 Hz, 1H), 7.56 (t, J = 8.0 Hz, 1H), 5.17 (s, 2H), 4.01 (s, 3H). GC-MS (EI), 273.0.
7-fluoroisobenzofuran-1(3H)-one (24IV)
1H NMR (400 MHz, CDCl3): δ 7.69 (m, 1H), 7.29 (d, J = 7.6 Hz, 1H), 7.17 (t, J = 8.4 Hz, 1H), 5.33 (s, 2H). GC-MS (EI), 152.2. The 1H NMR data are matched with the data in the literature60.
4-nitroisobenzofuran-1(3H)-one (25IV)
1H NMR (400 MHz, CDCl3): δ 8.55 (d, J = 8.0 Hz, 1H), 8.29 (d, J = 7.6 Hz, 1H), 7.83 (t, J = 8.0 Hz, 1H), 5.78 (s, 2H). GC-MS (EI), 179.0. The 1H NMR data are matched with the data in the literature61.
2-fluoro-6-formylbenzoic acid (24V)
1H NMR (400 MHz, DMSO-d6): δ 8.27 (s, 1H), 7.84 (m, 1H), 7.50 (m, 2H), 6.66 (s, 1H). MS (ESI) m/z 167.1 [M − H]−.
2-formyl-3-nitrobenzoic acid (25V)
1H NMR (400 MHz, DMSO-d6): δ 8.54 (d, J = 8.4 Hz, 1H), 8.51 (d, J = 8.0 Hz, 1H), 8.27 (d, J = 7.6 Hz, 1H), 7.97 (t, J = 8.0 Hz, 1H), 7.08 (d, J = 7.6 Hz, 1H). MS (ESI) m/z 194.1 [M − H]−.
2-(2-hydroxybenzylidene)hydrazinecarbothioamide (15a)
1H NMR (400 MHz, DMSO-d6): δ 11.37 (s, 1H), 9.99 (s, 1H), 8.37 (s, 1H), 8.09 (s, 1H), 7.92 (d, J = 8.4 Hz, 1H), 7.91 (s, 1H), 7.21 (t, J = 7.6 Hz, 1H), 6.91 (d, J = 8.4 Hz, 1H), 6.82 (t, J = 7.6 Hz, 1H). MS (ESI) m/z 194.0 [M − H]−.
2-((2-carbamothioylhydrazono)methyl)benzoic acid (19a)
1H NMR (400 MHz, DMSO-d6): δ 13.28 (s, 1H), 11.59 (s, 1H), 8.79 (s, 1H), 8.25 (d, J = 7.2 Hz, 1H), 8.22 (s, 1H), 7.96 (s, 1H), 7.83 (d, J = 8.0 Hz, 1H), 7.57 (t, J = 7.6 Hz, 1H), 7.48 (t, J = 7.6 Hz, 1H). MS (ESI) m/z 222.0 [M − H]−.
3-((2-carbamothioylhydrazono)methyl)benzoic acid (20a)
1H NMR (400 MHz, DMSO-d6): δ 13.11 (s, 1H), 11.48 (s, 1H), 8.23 (s, 1H), 8.21(s, 1H), 8.13 (d, J = 6.0 Hz, 1H), 8.12 (s, 1H), 8.09 (s, 1H), 7.95 (d, J = 8.0 Hz, 1H), 7.54 (t, J = 7.6 Hz, 1H). MS (ESI) m/z 222.0 [M − H]−.
4-((2-carbamothioylhydrazono)methyl)benzoic acid (21a)
1H NMR (400 MHz, DMSO-d6): δ 13.04 (s, 1H), 11.57 (s, 1H), 8.30 (s, 1H), 8.11(s, 1H), 8.10 (s, 1H), 7.96–7.91 (m, 4H). MS (ESI) m/z 222.0 [M − H]−.
2-((2-carbamothioylhydrazono)methyl)-6-fluorobenzoic acid (24a)
1H NMR (400 MHz, DMSO-d6): δ 13.86 (s, 1H), 11.68 (s, 1H), 8.40 (s, 1H), 8.20 (s, 1H), 7.93 (d, J = 8.0 Hz, 1H), 7.76 (s, 1H), 7.55–7.49 (m, 1H), 7.34 (t, J = 7.6 Hz, 1H). MS (ESI) m/z 240.0 [M − H]−.
2-((2-carbamothioylhydrazono)methyl)-3-nitrobenzoic acid (25a)
1H NMR (400 MHz, DMSO-d6): δ 13.80 (s, 1H), 11.80 (s, 1H), 8.53 (s, 1H), 8.37 (s, 1H), 8.14 (d, J = 8.0 Hz, 1H), 8.09 (d, J = 8.0 Hz, 1H), 7.74 (t, J = 8.0 Hz, 1H), 7.08 (s, 1H). MS (ESI) m/z 267.0 [M − H]−.
(E)-2-((2-(5-methyl-4-phenylthiazol-2-yl)hydrazono)methyl)phenol (15)
mp 227.5–229.0 °C. 1H NMR (400 MHz, DMSO-d6): δ 11.90 (s, 1H), 10.20 (s, 1H), 8.30 (s, 1H), 7.62 (d, J = 7.6 Hz, 2H), 7.59 (d, J = 8.4 Hz, 1H), 7.44 (t, J = 7.6 Hz, 2H), 7.33 (t, J = 7.6 Hz, 1H), 7.22 (t, J = 8.0 Hz, 1H), 6.91 (d, J = 7.6 Hz, 1H), 6.88 (t, J = 7.6 Hz, 1H), 2.42 (s, 3H). 13C NMR (100 MHz, DMSO-d6): 164.2, 156.0, 140.3, 134.8, 131.5, 130.3, 128.6, 128.3, 127.9, 127.1, 126.8, 120.0, 119.4, 116.1, 12.3. HRMS (ESI) calcd for C17H16N3OS [M + H]+ 310.1014, found 310.1020. Purity: 99.51% (tR 11.82 min).
(E)-2-((2-(4-(4-isopropylphenyl)thiazol-2-yl)hydrazono)methyl)phenol (16)
mp 158.0–160.0 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.16 (s, 1H), 10.11 (s, 1H), 8.32 (s, 1H), 7.77 (d, J = 8.0 Hz, 2H), 7.63 (d, J = 7.2 Hz, 1H), 7.28 (d, J = 8.4 Hz, 2H), 7.23 (t, J = 8.0 Hz, 2H), 6.91 (d, J = 8.0 Hz, 1H), 6.89 (t, J = 7.2 Hz, 1H), 2.91 (m, 1H), 1.23 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, DMSO-d6): 169.4, 157.3, 150.9, 149.6, 146.5, 131.9, 130.8, 130.4, 127.2, 126.7, 119.4, 117.6, 116.6, 102.2, 33.9, 23.9. HRMS (ESI) calcd for C19H20N3OS [M + H]+ 338.1327, found 338.1331. Purity: 95.76% (tR 21.79 min).
(E)-2-((2-(4-(3,4-dichlorophenyl)thiazol-2-yl)hydrazono)methyl)phenol (17)
mp 226.9–228.0 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.10 (s, 1H), 10.10 (s, 1H), 8.36 (s, 1H), 8.08 (d, J = 2.0 Hz, 1H), 7.84 (dd, J1 = 8.4 Hz, J2 = 2.0 Hz, 1H), 7.66 (d, J = 8.0 Hz, 1H), 7.64 (d, J = 6.8 Hz, 1H), 7.54 (s, 1H), 7.24 (t, J = 8.0 Hz, 1H), 6.92 (d, J = 8.4 Hz, 1H), 6.89 (t, J = 7.6 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 168.2, 156.0, 147.9, 134.0, 135.1, 131.4, 130.8, 130.6, 129.7, 127.2, 126.4, 125.5, 120.0, 119.5, 116.1, 105.6. HRMS (ESI) calcd for C16H12N3OSCl2 [M + H]+ 364.0078, found 364.0071. Purity: 99.89% (tR 21.16 min).
(E)-2-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)phenol (18)
mp 204.5–205.1 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.15 (s, 1H), 10.13 (s, 1H), 8.36 (s, 1H), 7.88 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.65 (d, J = 8.0 Hz, 1H), 7.53 (d, J = 7.6 Hz, 1H), 7.42 (t, J = 7.6 Hz, 1H), 7.36 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.33 (s, 1H), 7.24 (t, J = 7.6Hz, 1H), 6.93 (d, J = 8.4 Hz, 1H), 6.90 (t, J = 7.6 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 167.1, 156.0, 147.0, 140.0, 133.2, 131.0, 130.8, 130.5, 130.3, 129.00, 127.2, 126.5, 120.8, 119.5, 116.1, 108.2. HRMS (ESI) calcd for C16H13N3OSCl [M + H]+ 330.0468, found 330.0472. Purity: 99.43% (tR 12.44 min).
(E)-3-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (20)
mp 274.6–275.2 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.48 (s, 1H), 8.24 (s, 1H), 8.11 (s, 1H), 7.94 (d, J = 8.0 Hz, 1H), 7.89 (d, J = 6.4 Hz, 1H), 7.87 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.57 (t, J = 7.6 Hz, 1H), 7.54 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.42 (t, J = 7.6 Hz, 1H), 7.36 (s, 1H), 7.35 (t, J = 8.0 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 167.7, 167.5, 147.6, 140.9, 135.3, 133.7, 131.9, 131.5, 131.2, 131.0, 130.9, 130.3, 129.7, 129.5, 127.7, 127.2, 109.3. HRMS (ESI) calcd for C17H11N3O2SCl [M − H]− 356.0261, found 356.0260. Purity: 96.55% (tR 12.89 min).
(E)-4-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (21)
mp 258.9–259.5 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.41 (s, 1H), 8.09 (s, 1H), 7.90 (d, J = 8.4 Hz, 2H), 7.87 (dd, J1 = 7.6 Hz, J2 = 2.0 Hz, 1H), 7.77 (d, J1 = 8.4 Hz, 2H), 7.54 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.42 (t, J = 7.6 Hz, 1H), 7.39 (s, 1H), 7.36 (t, J = 7.6 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 167.6, 167.4, 147.4, 140.7, 138.9, 133.6, 131.5, 131.4, 131.3, 130.9, 130.3, 129.6, 127.7, 126.7, 109.5. HRMS (ESI) calcd for C17H11N3O2SCl [M − H]− 356.0261, found 356.0264. Purity: 98.38% (tR 12.74 min).
2-(4-(2-chlorophenyl)thiazol-2-yl)phthalazin-1(2H)-one (19c)
1H NMR (400 MHz, CDCl3): δ 8.83 (s, 1H), 8.44 (d, J = 8.0 Hz, 1H), 8.10 (t, J = 7.6 Hz, 2H), 8.04 (s, 1H), 8.01 (d, J = 8.0 Hz, 1H), 7.98 (d, J = 7.2 Hz, 1H), 7.60 (d, J = 8.0 Hz, 1H), 7.49 (t, J = 7.6 Hz, 1H), 7.43 (t, J = 7.6 Hz, 1H). 13C NMR (100 MHz, CDCl3): 158.4, 156.7, 146.8, 141.5, 135.3, 133.5, 133.3, 132.0, 131.5, 130.8, 130.1, 129.0, 128.1, 127.9, 127.0, 118.0. HRMS (ESI) calcd for C17H10N3OSClNa [M + Na]+ 362.0131, found 362.0129.
2-(4-(2-chlorophenyl)thiazol-2-yl)-8-fluorophthalazin-1(2H)-one (24c)
1H NMR (400 MHz, CDCl3): δ 8.51(d, J = 2.4 Hz, 1H), 7.96 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.93–7.87 (m, 1H), 7.73 (s, 1H), 7.63 (d, J = 7.6 Hz, 1H), 7.56–7.52 (m, 1H), 7.47 (dd, J1 = 8.0 Hz, J2 = 1.6 Hz, 1H), 7.35 (t, J = 7.6 Hz, 1H), 7.31–7.26 (m, 1H). 13C NMR (100 MHz, CDCl3): 163.1, 156.0, 155.4, 147.8, 139.7, 136.0, 133.4, 132.3, 131.8, 130.8, 130.2, 129.1, 127.0, 122.9, 120.1, 116.9, 115.9. HRMS (ESI) calcd for C17H9N3ONaSClF [M + Na]+ 380.0037, found 380.0037.
2-(4-(2-chlorophenyl)thiazol-2-yl)-5-nitrophthalazin-1(2H)-one (25c)
1H NMR (400 MHz, DMSO-d6): δ 9.14 (s, 1H), 8.82 (d, J = 8.0 Hz, 1H), 8.73 (dd, J1 = 8.0 Hz, J2 = 0.8 Hz, 1H), 8.17 (t, J = 8.0 Hz, 1H), 8.10 (s, 1H), 7.96 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.61 (dd, J1 = 8.0 Hz, J2 = 1.6 Hz, 1H), 7.49 (t, J = 7.6 Hz, 1H), 7.43 (t, J = 7.6 Hz, 1H). HRMS (ESI) calcd for C17H9N4O3NaSCl [M + Na]+ 406.9982, found 406.9986.
2-(4-phenylthiazol-2-yl)phthalazin-1(2H)-one (26c)
1H NMR (400 MHz, DMSO-d6): δ 8.85(s, 1H), 8.45 (d, J = 7.6 Hz, 1H), 8.11–8.01 (m, 6H), 7.49 (t, J = 7.6 Hz, 2H), 7.38 (t, J = 7.2 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 158.4, 157.5, 150.3, 141.4, 135.2, 134.6, 133.5, 129.2, 128.9, 128.5, 128.0, 127.0, 127.0, 126.4, 113.1. HRMS (ESI) calcd for C17H11N3ONaS [M + Na]+ 328.0521, found 328.0524.
2-(4-(2-methoxyphenyl)thiazol-2-yl)phthalazin-1(2H)-one (27c)
1H NMR (400 MHz, DMSO-d6): δ 8.84 (s, 1H), 8.43 (d, J = 8.0 Hz, 1H), 8.11–8.04 (m, 3H), 7.99 (t, J = 7.6 Hz, 1H), 7.61 (d, J = 7.6 Hz, 1H), 7.56 (s, 1H), 7.39 (t, J = 8.0 Hz, 1H), 6.95 (d, J = 8.4 Hz, 1H), 3.84 (s, 1H). 13C NMR (100 MHz, CDCl3): 159.9, 158.3, 157.4, 151.3, 140.6, 135.7, 134.4, 132.7, 129.7, 128.6, 127.4, 127.3, 126.8, 119.2, 114.1, 111.9, 111.9, 55.4. HRMS (ESI) calcd for C18H14N3O2S [M + H]+ 336.0807, found 336.0809.
2-(2-(1-oxophthalazin-2(1H)-yl)thiazol-4-yl)benzamide (28c)
1H NMR (400 MHz, DMSO-d6): δ 8.84 (s, 1H), 8.52 (s, 1H), 8.43 (d, J = 7.2 Hz, 1H), 8.16–7.98 (m, 6H), 7.86 (d, J = 6.0 Hz, 1H), 7.55 (s, 1H), 7.46 (s, 1H). 13C NMR (100 MHz, DMSO-d6): 168.3, 158.4, 157.7, 149.8, 141.5, 135.4, 135.3, 134.6, 133.5, 129.2, 129.0, 128.9, 128.1, 127.4, 127.0, 125.7, 113.8. HRMS (ESI) calcd for C18H12N4O2SNa [M + Na]+ 371.0579, found 371.0575.
2-(4-(p-tolyl)thiazol-2-yl)phthalazin-1(2H)-one (29c)
1HNMR (400 MHz, DMSO-d6): δ 8.82 (s, 1H), 8.42 (d, J = 8.0 Hz, 1H), 8.08 (t, J = 7.6 Hz, 1H), 8.05 (t, J = 7.6 Hz, 1H), 7.99 (d, J = 7.6 Hz, 1H), 7.95 (s, 1H), 7.90 (d, J = 8.0 Hz, 2H), 7.27 (d, J = 7.6 Hz, 2H), 2.35 (s, 3H). 13C NMR (100 MHz, DMSO-d6): 158.3, 157.3, 151.5, 140.6, 138.0, 134.3, 132.7, 131.6, 129.3, 128.6, 127.4, 127.3, 126.8, 126.4, 110.8. HRMS (ESI) calcd for C18H14N3OS [M + H]+ 320.0858, found 320.0862.
2-(4-(2,5-dichlorophenyl)thiazol-2-yl)phthalazin-1(2H)-one (30c)
1H NMR (400 MHz, DMSO-d6): δ 8.84 (s, 1H), 8.44 (d, J = 7.6 Hz, 1H), 8.17 (s, 1H), 8.12–7.98 (m, 4H), 7.64 (d, J = 7.6 Hz, 1H), 7.51 (d, J = 8.4 Hz, 1H). 13C NMR (100 MHz, CDCl3): 158.4, 156.7, 146.5, 140.9, 134.7, 134.5, 132.9, 132.9, 131.5, 131.3, 130.4, 129.0, 128.7, 127.5, 127.2, 126.9, 117.4. HRMS (ESI) calcd for C17H9N3 NaSCl2 [M + Na]+ 395.9741, found 395.9745.
2-(4-(naphthalen-2-yl)thiazol-2-yl)phthalazin-1(2H)-one (31c)
1H NMR (400 MHz, DMSO-d6): δ 8.89 (s, 1H), 8.59 (s, 1H), 8.46 (d, J = 8.0 Hz, 1H), 8.20–8.02 (m, 7H), 7.96 (d, J = 7.2 Hz, 1H), 7.59–7.53 (m, 2H). 13C NMR (100 MHz, CDCl3): 158.4, 157.6, 151.4, 140.8, 134.4, 133.6, 133.2, 132.8, 131.6, 128.7, 128.6, 128.3, 127.7, 127.5, 127.3, 126.9, 126.3, 126.1, 125.7, 124.4, 112.0. HRMS (ESI) calcd for C21H13N3OSNa [M + Na]+ 378.0677, found 378.0674.
2-(4-([1,1’-biphenyl]-4-yl)thiazol-2-yl)phthalazin-1(2H)-one (32c)
1H NMR (400 MHz, CDCl3): δ 8.57 (s, 1H), 8.56 (d, J = 6.8 Hz, 1H), 8.05 (d, J = 7.6 Hz, 2H), 7.91 (t, J = 7.2 Hz, 1H), 7.86 (t, J = 7.6 Hz, 1H), 7.82 (d, J = 7.6 Hz, 1H), 7.67 (d, J = 8.4 Hz, 2H), 7.65 (d, J = 7.2 Hz, 2H), 7.48 (s, 1H), 7.45 (t, J = 8.0 Hz, 2H), 7.35 (t, J = 7.6 Hz, 1H). 13C NMR (100 MHz, CDCl3): 158.4, 157.6, 151.1, 140.8, 140.8, 140.7, 134.4, 133.4, 132.8, 128.8, 128.7, 127.5, 127.4, 127.4, 127.1, 127.0, 126.9, 111.6. HRMS (ESI) calcd for C23H16N3OS [M + H]+ 382.1014, found 382.1017.
5-amino-2-(4-(2-chlorophenyl)thiazol-2-yl)phthalazin-1(2H)-one (25d)
1H NMR (400 MHz, DMSO-d6): δ 8.90 (s, 1H), 7.99 (s, 1H), 7.97 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.59 (t, J = 8.0 Hz, 1H), 7.59 (dd, J1 = 8.0 Hz, J2 = 1.6 Hz, 1H), 7.53 (d, J = 7.2 Hz, 1H), 7.48 (t, J = 7.6 Hz, 1H), 7.42 (t, J = 8.0 Hz, 1H). 7.14 (dd, J1 = 8.0 Hz, J2 = 1.2 Hz, 1H), 6.66 (s, 2H). 13C NMR (100 MHz, DMSO-d6): 158.7, 156.9, 147.6, 146.8, 137.7, 134.2, 133.5, 132.0, 131.5, 130.8, 130.0, 127.9, 127.7, 119.3, 117.7, 113.6, 113.5. HRMS (ESI) calcd for C17H11N4OSClNa [M + Na]+ 377.0240, found 377.0240.
(E)-2-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (19)
mp 183.9–185.8 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.80 (s, 1H), 8.84 (s, 1H), 8.01 (d, J = 7.6 Hz, 1H), 7.88 (t, J = 7.2 Hz, 2H), 7.63 (t, J = 7.2 Hz, 1H), 7.53 (dd, J1 = 7.6 Hz, J2 = 1.2 Hz, 1H), 7.48 (t, J = 7.2 Hz, 1H), 7.43 (t, J = 7.2 Hz, 1H), 7.41(t, J = 7.2 Hz, 1H ), 7.41 (s, 1H).13C NMR (100 MHz, DMSO-d6): 168.6, 167.8, 147.6, 140.8, 135.1, 133.7, 132.4, 131.6, 131.2, 130.8, 130.3, 129.5, 129.3, 127.7, 126.5, 109.3. HRMS (ESI) calcd for C17H11N3O2SCl [M − H]− 356.0261, found 356.0261. Purity: 99.11% (tR 13.58 min).
(E)-2-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)-6-fluorobenzoic acid (24)
mp 211.2–211.7 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.40 (s, 1H), 8.18 (s, 1H), 7.86 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.69 (d, J = 8.0 Hz, 1H), 7.55–7.50 (m, 2H), 7.41 (t, J = 7.6 Hz, 1H), 7.37 (s, 1H), 7.35 (t, J = 7.6 Hz, 1H), 7.29(t, J = 8.8 Hz, 1H ). 13C NMR (100 MHz, DMSO-d6): 167.6, 166.1, 160.5, 147.6, 138.2, 138.1, 133.9, 133.8, 133.7, 131.5, 131.2, 130.8, 129.5, 127.7, 122.0, 116.5, 109.6. HRMS (ESI) calcd for C17H10N3O2SClF [M − H]− 374.0166, found 374.0164. Purity: 97.09% (tR 12.05 min).
(E)-3-amino-2-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (25)
mp 220.9–221.6 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.8 (s, 1H), 11.1 (s, 1H), 7.93 (s, 1H), 7.89 (d, J = 7.2 Hz, 1H), 7.62 (d, J = 7.6 Hz, 1H), 7.52–7.43(m, 3H), 7.12 (d, J = 7.2 Hz, 1H), 7.08 (d, J = 8.4 Hz, 1H), 6.77 (s, 2H). 13C NMR (100 MHz, DMSO-d6): δ 165.8, 155.2, 154.3, 147.1, 146.3, 134.0, 132.7, 131.7, 131.5, 131.1, 130.3, 129.7, 128.1, 121.3, 114.5, 113.1, 111.6. Purity: 97.06% (tR 19.58 min).
(E)-2-((2-(4-phenylthiazol-2-yl)hydrazono)methyl)benzoic acid (26)
mp 175.4–176.4 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.80 (s, 1H), 8.84 (s, 1H), 7.99 (d, J = 7.6 Hz, 1H), 7.89–7.85 (m, 3H), 7.62 (t, J = 7.2 Hz, 1H), 7.47 (t, J = 7.6 Hz, 1H), 7.41 (t, J = 7.6 Hz, 2H), 7.33 (s, 1H), 7.30 (t, J = 7.6 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 168.6, 151.1, 140.8, 135.1, 135.0, 132.3, 130.8, 130.5, 129.3, 129.1, 128.0, 126.4, 126.0, 104.3. HRMS (ESI) calcd for C17H12N3O2S [M-H]- 322.0650, found 322.0653. Purity: 95.72% (tR 9.27 min).
(E)-2-((2-(4-(3-methoxyphenyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (27)
mp 167.3–167.9 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.80 (s, 1H), 8.82 (s, 1H), 8.00 (d, J = 8.0 Hz, 1H), 7.88 (d, J = 7.6 Hz, 1H), 7.62 (t, J = 7.6 Hz, 1H), 7.49–7.43 (m, 3H), 7.37 (s, 1H), 7.32 (t, J = 8.0 Hz, 1H), 6.88 (dd, J1 = 8.4 Hz, J 2 = 2.0 Hz, 1H), 3.80 (s, 3H). 13C NMR (100 MHz, DMSO-d6): 168.7, 168.5, 160.0, 150.9, 140.8, 136.5, 135.0, 132.4, 130.8, 130.4, 130.1, 129.3, 126.4, 118.4, 113.8, 111.3, 104.7, 55.5. HRMS (ESI) calcd for C18H14N3O3S [M − H]− 352.0756, found 352.0754. Purity: 95.56% (tR 7.94 min).
(E)-2-((2-(4-(3-carbamoylphenyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (28)
mp 297.9–298.4 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.20 (s, 1H), 8.83 (s, 1H), 8.39 (s, 1H), 8.05 (s, 1H), 8.00 (dd, J1 = 7.2 Hz, J2 = 3.2 Hz, 2H), 7.88 (d, J = 7.6 Hz, 1H), 7.80 (d, J = 7.6 Hz, 1H), 7.63 (t, J = 7.6 Hz, 1H), 7.51–7.46 (m, 2H), 7.41 (s, 2H). 13C NMR (100 MHz, DMSO-d6): 168.8, 168.7, 168.4, 150.6, 140.8, 135.2, 135.1, 135.0, 132.4, 130.8, 130.4, 129.3, 129.0, 128.6, 126.9, 126.5, 125.4, 105.0. HRMS (ESI) calcd for C18H15N4O3S [M + H]+ 367.0865, found 367.0863. Purity: 97.89% (tR 7.832 min).
(E)-2-((2-(4-(p-tolyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (29)
mp 225.5–226.1 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.80 (s, 1H), 8.83 (s, 1H), 8.00 (d, J = 7.2 Hz, 1H), 7.88 (d, J = 7.6 Hz, 1H), 7.75 (d, J = 8.0 Hz, 2H), 7.62 (t, J = 7.6 Hz, 1H), 7.47 (t, J = 7.6 Hz, 1H), 7.25 (s, 1H), 7.21 (t, J = 8.0 Hz, 2H), 2.32 (s, 3H). 13C NMR (100 MHz, DMSO-d6): 168.7, 168.6, 151.1, 140.7, 137.3, 135.0, 132.5, 132.3, 130.8, 130.6, 129.6, 129.2, 126.4, 126.0, 103.4, 21.3. HRMS (ESI) calcd for C18H14N3O2S [M − H]− 336.0807, found 336.0808. Purity: 96.31% (tR 13.17 min).
(E)-2-((2-(4-(2,5-dichlorophenyl)thiazol-2-yl)hydrazono)methyl)benzoic acid (30)
mp 205.0–206.0 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.20 (s, 1H), 8.84 (s, 1H), 7.99 (d, J = 7.2 Hz, 1H), 7.95 (d, J = 6.4 Hz, 1H), 7.88 (d, J = 8.0 Hz, 1H), 7.62 (t, J = 7.2 Hz, 1H), 7.57 (d, J = 8.4 Hz, 1H), 7.52 (s, 1H), 7.47 (t, J = 7.6 Hz, 1H), 7.41 (dd, J1 = 8.8 Hz, J2 = 2.8 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 168.7, 167.9, 146.1, 141.2, 135.0, 134.9, 132.7, 132.3, 132.3, 130.8, 130.7, 130.6, 129.7, 129.4, 129.0, 126.5, 110.6. HRMS (ESI) calcd for C17H10N3O2SCl2 [M − H]− 389.9871, found 389.9871. Purity: 97.66% (tR 9.88 min).
(E)-2-((2-(4-(naphthalen-2-yl)thiazol-2-yl)hydrazono)methyl)benzoic acid (31)
mp 238.1–239.4 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.80 (s, 1H), 8.87 (s, 1H), 8.40 (s, 1H), 8.04–8.01 (m, 2H), 7.96–7.89 (m, 4H), 7.64 (t, J = 7.6 Hz, 1H), 7.54–7.46 (m, 4H). 13C NMR (100 MHz, DMSO-d6): 168.8, 168.7, 151.0, 140.8, 135.1, 133.7, 132.9, 132.6, 132.4, 130.9, 130.4, 129.3, 128.6, 128.6, 128.1, 126.9, 126.5, 124.6, 124.4, 105.2. HRMS (ESI) calcd for C21H16N3O2S [M + H]+ 374.0963, found 374.0959. Purity: 96.39% (tR 19.11 min).
(E)-2-((2-(4-([1,1′-biphenyl]-4-yl)thiazol-2-yl)hydrazono)methyl)benzoic acid (32)
mp 227.1–228.1 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.35 (s, 1H), 8.83 (s, 1H), 8.01 (d, J = 6.0 Hz, 1H), 7.96 (d, J = 6.8 Hz, 2H), 7.88 (d, J = 6.0 Hz, 1H), 7.74–7.71 (m, 4H), 7.64 (t, J = 6.4 Hz, 1H), 7.48 (t, J = 6.0 Hz, 3H), 7.42 (s, 1H), 7.38 (t, J = 6.0 Hz, 1H). 13C NMR (100 MHz, DMSO-d6): 169.3, 169.3, 151.4, 141.4, 140.8, 140.2, 135.8, 134.9, 133.1, 131.5, 131.0, 130.1, 130.0, 128.6, 128.0, 127.6, 127.3, 127.1, 105.3. HRMS (ESI) calcd for C23H18N3O2S [M + H]+ 400.1120, found 400.1114. Purity: 97.60% (tR 9.16 min).
(E)-methyl 2-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)benzoate (22)
mp 145.8–147.7 °C. 1H NMR (400 MHz, CDCl3): δ 8.57 (s, 1H), 8.11 (d, J = 8.0 Hz, 1H), 7.95 (d, J = 7.6 Hz, 1H), 7.85 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.59 (t, J = 8.4 Hz, 1H), 7.43 (t, J = 7.6 Hz, 1H), 7.42 (t, J = 8.0 Hz, 1H), 7.28 (s, 1H), 7.27 (t, J = 8.0 Hz, 1H ), 7.19 (t, J = 8.0 Hz, 1H), 3.93 (s, 3H). 13C NMR (100 MHz, CDCl3): 167.9, 167.2, 146.4, 141.8, 134.8, 132.7, 132.2, 132.0, 131.0, 130.6, 130.5, 129.0, 128.9, 128.6, 127.2, 127.0, 108.7, 52.4. HRMS (ESI) calcd for C18H15N3O2SCl [M − H]− 372.0574, found 372.0575. Purity: 98.10% (tR 15.32 min).
(E)-ethyl 2-((2-(4-(2-chlorophenyl)thiazol-2-yl)hydrazono)methyl)benzoate (23)
mp 172.0–172.6 °C. 1H NMR (400 MHz, CDCl3): δ 8.47 (s, 1H), 8.08 (d, J = 8.0 Hz, 1H), 7.94 (d, J = 8.0 Hz, 1H), 7.84 (dd, J1 = 7.6 Hz, J2 = 1.6 Hz, 1H), 7.57 (t, J = 8.0 Hz, 1H), 7.44–7.39 (m, 2H), 7.24 (t, J = 7.6 Hz, 1H ), 7.20 (s, 1H), 7.14 (t, J = 8.0 Hz, 1H), 4.37 (q, J = 7.2 Hz, 2H ), 1.40 (t, J = 7.2Hz, 3H). 13C NMR (100 MHz, CDCl3): 167.9, 166.7, 147.3, 141.1, 135.0, 133.3, 132.0, 132.0, 131.1, 130.5, 130.4, 128.9, 128.7, 128.6, 127.1, 126.9, 108.9, 61.3, 14.2. HRMS (ESI) calcd for C19H17N3O2SCl [M − H]− 386.0730, found 386.0729. Purity: 99.78% (tR 20.22 min).
Additional Information
How to cite this article: Li, S. et al. Rational Design of Benzylidenehydrazinyl-Substituted Thiazole Derivatives as Potent Inhibitors of Human Dihydroorotate Dehydrogenase with in Vivo Anti-arthritic Activity. Sci. Rep. 5, 14836; doi: 10.1038/srep14836 (2015).
Supplementary Material
Acknowledgments
We thank the staff at Shanghai Synchrotron Radiation Facility for assistances with data collection. This work was supported by the Fundamental Research Funds for the Central Universities, the National Natural Science Foundation of China (grants 81102375, 21372078, 81302697, 81222046 and 81230076) (X.L., Z.Z., L.Z. and H.L.), the Shanghai Committee of Science and Technology (grant 14431902400) (H.L.), and the 863 Hi-Tech Program of China (grant 2012AA020308) (H.L.). Lili Zhu is also sponsored by the Shanghai Natural Science Fund for Youth Scholars (grant 12ZR1442800). Honglin Li is also sponsored by Shanghai Rising-Star Tracking Program (grant 13QH1401100), the Innovation Program of Shanghai Municipal Education Commission (grant 13SG32) and Fok Ying Tung Education Foundation (141035).
Footnotes
The authors declare no competing financial interests.
Author Contributions S.L., G.L., X.R. and J.Z. performed research and drafted the manuscript; M.X., W.S., L.X. and D.D. performed research and helped to draft the manuscript; Y.D., X.L., L.Z., R.W. and Z.Z. performed research and helped to draft the manuscript; Y.X. and H.L. designed and performed research, interpreted data and approved the final manuscript.
References
- Munier-Lehmann H., Vidalain P.-O., Tangy F. & Janin Y. L. On dihydroorotate dehydrogenases and their inhibitors and uses. J. Med. Chem. 56, 3148–3167 (2013). [DOI] [PubMed] [Google Scholar]
- Jones M. E. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 49, 253–279 (1980). [DOI] [PubMed] [Google Scholar]
- Rückemann K. et al. Leflunomide inhibits pyrimidine de novo synthesis in mitogen-stimulated T-lymphocytes from healthy humans. J. Biol. Chem. 273, 21682–21691 (1998). [DOI] [PubMed] [Google Scholar]
- Gummert J. F., Ikonen T. & Morris R. E. Newer immunosuppressive drugs a review. J. Am. Soc. Nephrol. 10, 1366–1380 (1999). [DOI] [PubMed] [Google Scholar]
- Löffler M., Fairbanks L. D., Zameitat E., Marinaki A. M. & Simmonds H. A. Pyrimidine pathways in health and disease. Trends Mol. Med. 11, 430–437 (2005). [DOI] [PubMed] [Google Scholar]
- Diao Y. Y. et al. Discovery of diverse human dihydroorotate dehydrogenase inhibitors as immunosuppressive agents by structure-based virtual screening. J. Med. Chem. 55, 8341–8349 (2012). [DOI] [PubMed] [Google Scholar]
- Liu S., Neidhardt E. A., Grossman T. H., Ocain T. & Clardy J. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure 8, 25–33 (2000). [DOI] [PubMed] [Google Scholar]
- Phillips M. A. & Rathod P. K. Plasmodium dihydroorotate dehydrogenase: a promising target for novel anti-malarial chemotherapy. Infect. Disord. Drug. Targets 10, 226–239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas V. K. & Ghate M. Recent developments in the medicinal chemistry and therapeutic potential of dihydroorotate dehydrogenase (DHODH) inhibitors. Mini. Rev. Med. Chem . 11, 1039–1055 (2011). [DOI] [PubMed] [Google Scholar]
- Deng X. Y. et al. Fluorine modulates species selectivity in the triazolopyrimidine class of plasmodium falciparum dihydroorotate dehydrogenase inhibitors. J. Med. Chem. 57, 5381–5394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner R. et al. Dual binding mode of a novel series of DHODH inhibitors. J. Med. Chem. 49, 1239–1247 (2006). [DOI] [PubMed] [Google Scholar]
- Maroun J. et al. Multicenter phase II study of brequinar sodium in patients with advanced lung cancer. Cancer Chemother. Pharmacol. 32, 64–66 (1993). [DOI] [PubMed] [Google Scholar]
- Alcorn N., Saunders S. & Madhok R. Benefit-risk assessment of leflunomide: an appraisal of leflunomide in rheumatoid arthritis 10 years after licensing. Drug Saf. 32, 1123–1134 (2009). [DOI] [PubMed] [Google Scholar]
- White R. M. et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva Junior H. T. & Morris R. E. Leflunomide and malononitrilamides. Am. J. Med. Sci. 313, 289–301 (1997). [DOI] [PubMed] [Google Scholar]
- Pally C. et al. Side effects of brequinar and brequinar analogues, in combination with cyclosporine, in the rat. Toxicology 127, 207–222 (1998). [DOI] [PubMed] [Google Scholar]
- Suissa S., Hudson M. & Ernst P. Leflunomide use and the risk of interstitial lung disease in rheumatoid arthritis. Arthritis Rheum. 54, 1435–1439 (2006). [DOI] [PubMed] [Google Scholar]
- Amadasi A. et al. Robust classification of “relevant” water molecules in putative protein binding sites. J. Med. Chem. 51, 1063–1067 (2008). [DOI] [PubMed] [Google Scholar]
- Lu Y., Wang R., Yang C. Y. & Wang S. Analysis of ligand-bound water molecules in high-resolution crystal structures of protein-ligand complexes. J. Chem. Inf. Model. 47, 668–675 (2007). [DOI] [PubMed] [Google Scholar]
- Sharrow S. D., Edmonds K. A., Goodman M. A., Novotny M. V. & Stone M. J. Thermodynamic consequences of disrupting a water-mediated hydrogen bond network in a protein:pheromone complex. Protein Sci. 14, 249–256 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillari C., Taylor J., Viner R. & Essex J. W. Classification of water molecules in protein binding sites. J. Am. Chem. Soc. 129, 2577–2587 (2007). [DOI] [PubMed] [Google Scholar]
- Levinson N. M. & Boxer S. G. A conserved water-mediated hydrogen bond network defines bosutinib’s kinase selectivity. Nat. Chem. Biol. 10, 127–132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasief N. N., Tan H. W., Kong J. & Hangauert D. Water mediated ligand functional group cooperativity: the contribution of a methyl group to binding affinity is enhanced by a COO- group through changes in the structure and thermodynamics of the hydration waters of ligand-thermolysin complexes. J. Med. Chem. 55, 8283–8302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen E. S. et al. Water-mediated interactions influence the binding of thapsigargin to sarco/endoplasmic reticulum calcium adenosinetriphosphatase. J. Med. Chem. 56, 3609–3619 (2013). [DOI] [PubMed] [Google Scholar]
- Kadirvelraj R., Foley B. L., Dyekjaer J. D. & Woods R. J. Involvement of water in carbohydrate-protein binding: concanavalin A revisited. J. Am. Chem. Soc. 130, 16933–16942 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homans S. W. Water, water everywhere—except where it matters? Drug Discov. Today 12, 534–539 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang B., Tan V. B., Lim K. M. & Tay T. E. Significance of water molecules in the inhibition of cylin-dependent kinase 2 and 5 complexes. J. Chem. Inf. Model. 47, 1877–1885 (2007). [DOI] [PubMed] [Google Scholar]
- Wei D., Wilson W. D. & Neidle S. Small-molecule binding to the DNA minor groove is mediated by a conserved water cluster. J. Am. Chem. Soc. 135, 1369–1377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specs: chemistry solutions for drug discovery. http://www.specs.net/ (accessed June 1, 2012).
- Hopkins A. L., Keseru G. M., Leeson P. D., Rees D. C. & Reynolds C. H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 13, 105–121 (2014). [DOI] [PubMed] [Google Scholar]
- Tarcsay A., Nyiri K. & Keseru G. M. Impact of lipophilic efficiency on compound quality. J. Med. Chem. 55, 1252–1260 (2012). [DOI] [PubMed] [Google Scholar]
- Schonherr H. & Cernak T. Profound methyl effects in drug discovery and a call for new ch methylation reactions. Angew. Chem. Int. Ed. Engl. 52, 12256–12267 (2013). [DOI] [PubMed] [Google Scholar]
- Angell R. et al. Biphenyl amide p38 kinase inhibitors 3: Improvement of cellular and in vivo activity. Bioorg. Med. Chem. Lett. 18, 4428–4432 (2008). [DOI] [PubMed] [Google Scholar]
- Leung C. S., Leung S. S., Tirado-Rives J. & Jorgensen W. L. Methyl effects on protein-ligand binding. J. Med. Chem. 55, 4489–4500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusai K. et al. Immunosuppression with 4SC-101, a novel inhibitor of dihydroorotate dehydrogenase, in a rat model of renal transplantation. Transplantation 93, 1101–1107 (2012). [DOI] [PubMed] [Google Scholar]
- Kulkarni O. P. et al. 4SC-101, a novel small molecule dihydroorotate dehydrogenase inhibitor, suppresses systemic lupus erythematosus in MRL-(Fas)lpr mice. Am. J. Pathol. 176, 2840–2847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. S. et al. Design, synthesis, X-ray crystallographic analysis, and biological evaluation of thiazole derivatives as potent and selective inhibitors of human dihydroorotate Dehydrogenase. J. Med. Chem. 58, 1123–1139 (2015). [DOI] [PubMed] [Google Scholar]
- Powell H. R. The rossmann fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D-Biol. Crystallogr . 55, 1690–1695 (1999). [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D-Biol. Crystallogr . 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- Potterton E., Briggs P., Turkenburg M. & Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. Sect. D-Biol. Crystallogr . 59, 1131–1137 (2003). [DOI] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D-Biol. Crystallogr . 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr. D-Biol. Crystallogr . 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue R. C. et al. Rapid-resolution liquid chromatography TOF-MS for urine metabolomic analysis of collagen-induced arthritis in rats and its applications. J. Ethnopharmacol. 145, 465–475 (2013). [DOI] [PubMed] [Google Scholar]
- Weinblatt M. E. Methotrexate in rheumatoid arthritis: a quarter century of development. Trans. Am. Clin. Climatol. Assoc. 124, 16–25 (2013). [PMC free article] [PubMed] [Google Scholar]
- Rosillo M. A. et al. Anti-inflammatory and joint protective effects of extra-virgin olive-oil polyphenol extract in experimental arthritis. J Nutr Biochem 25, 1275–1281 (2014). [DOI] [PubMed] [Google Scholar]
- Bharti S. K., Nath G., Tilak R. & Singh S. Synthesis, anti-bacterial and anti-fungal activities of some novel Schiff bases containing 2, 4-disubstituted thiazole ring. Eur. J. Med. Chem. 45, 651–660 (2010). [DOI] [PubMed] [Google Scholar]
- Sekimata K. et al. A specific and potent inhibitor of brassinosteroid biosynthesis possessing a dioxolane ring. J. Agric. Food Chem. 50, 3486–3490 (2002). [DOI] [PubMed] [Google Scholar]
- Furuta T. et al. Synthesis of axially chiral amino acid and amino alcohols via additive−ligand-free Pd-catalyzed domino coupling reaction and subsequent transformations of the product amidoaza [5] helicene. J. Org. Chem. 75, 7010–7013 (2010). [DOI] [PubMed] [Google Scholar]
- Silva N. M., Tributino J. L., Miranda A. L., Barreiro E. J. & Fraga C. A. New isoxazole derivatives designed as nicotinic acetylcholine receptor ligand candidates. Eur. J. Med. Chem. 37, 163–170 (2002). [DOI] [PubMed] [Google Scholar]
- Xu B. H. et al. O-Spiro C-aryl glucosides as novel sodium-dependent glucose co-transporter 2 (SGLT2) inhibitors. Bioorg. Med. Chem. Lett. 19, 5632–5635 (2009). [DOI] [PubMed] [Google Scholar]
- He Y. et al. Highly enantioselective synthesis of 2, 3-dihydro-1 H-imidazo [2, 1-a] isoindol-5 (9b H)-ones via catalytic asymmetric intramolecular cascade imidization-nucleophilic addition-lactamization. Org. Lett. 16, 6366–6369 (2014). [DOI] [PubMed] [Google Scholar]
- Mirabdolbaghi R. & Dudding T. An indium-mediated allylative/transesterification DFT-directed approach to chiral C (3)-functionalized phthalides. Org. Lett. 14, 3748–3751 (2012). [DOI] [PubMed] [Google Scholar]
- Koeberle S. C. et al. Design, synthesis, and biological evaluation of novel disubstituted dibenzosuberones as highly potent and selective inhibitors of p38 mitogen activated protein kinase. J. Med. Chem. 55, 5868–5877 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang S., Xu L. & Trudell M. L. Selective oxidation of benzylic alcohols and TBDMS ethers to carbonyl compounds with CrO3-H5IO6. Synthesis , 1757–1760 (2005). [Google Scholar]
- van den Heuvel M., van den Berg T. A., Kellogg R. M., Choma C. T. & Feringa B. L. Synthesis of a non-heme template for attaching four peptides: an approach to artificial iron (II)-containing peroxidases. J. Org. Chem. 69, 250–262 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang H. et al. Synthesis of chiral 3-substituted phthalides by a sequential organocatalytic enantioselective aldol-lactonization reaction. Three-step synthesis of (S)-(−)-3-butylphthalide. J. Org. Chem. 75, 368–374 (2009). [DOI] [PubMed] [Google Scholar]
- Kiselev E., Empey N., Agama K., Pommier Y. & Cushman M. Dibenzo [c, h][1, 5] naphthyridinediones as topoisomerase I inhibitors: design, synthesis, and biological evaluation. J. Org. Chem. 77, 5167–5172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcal W. et al. New trypanocidal hybrid compounds from the association of hydrazone moieties and benzofuroxan heterocycle. Bioorg. Med. Chem. 16, 6995–7004 (2008). [DOI] [PubMed] [Google Scholar]
- Soucy C., Favreau D. & Kayser M. The regioselectivity of metal hydride reductions of 3-substituted phthalic anhydrides. J. Org. Chem. 52, 129–134 (1987). [Google Scholar]
- Kayser M. M. & Morand P. Regioselectivity of metal hydride reductions of unsymmetrically substituted cyclic anhydrides. systems where” steric hindrance along the preferred reaction path” rationalization is not applicable. Can. J. Chem. 58, 2484–2490 (1980). [Google Scholar]
- QikProp, version 3.2 , Schrodinger, LLC, New York, NY, 2009. [Google Scholar]
- Leeson P. D. & Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 6, 881–890 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.