Abstract
The most economically important disease of cultivated grapevines worldwide is powdery mildew (PM) caused by the ascomycete fungus Erysiphe necator. The majority of grapevine cultivars used for wine, table grape, and dried fruit production are derived from the Eurasian grape species Vitis vinifera because of its superior aroma and flavor characteristics. However, this species has little genetic resistance against E. necator meaning that grape production is highly dependent on the frequent use of fungicides. The integration of effective genetic resistance into cultivated grapevines would lead to significant financial and environmental benefits and represents a major challenge for viticultural industries and researchers worldwide. This review will outline the strategies being used to increase our understanding of the molecular basis of V. vinifera susceptibility to this fungal pathogen. It will summarize our current knowledge of different resistance loci/genes that have evolved in wild grapevine species to restrict PM infection and assess the potential application of these defense genes in the generation of PM-resistant grapevine germplasm. Finally, it addresses future research priorities which will be important in the rapid identification, evaluation, and deployment of new PM resistance genes which are capable of conferring effective and durable resistance in the vineyard.
Introduction
Grapevine (Vitis spp.) has been cultivated for human consumption for over 7000 years. Few horticultural crops have had more historical, cultural, and social impacts than grapevine. Grapevines are estimated to be cultivated on over 7.6 million of hectares of land worldwide1. The majority of wine grape cultivars are derived from the species Vitis vinifera which originated in Eurasia but are highly susceptible to the pathogens and pests that are thought to have evolved on the wild grapevines native to North America. The ascomycete fungus, Erysiphe necator (syn. Uncinula necator), the causal agent of grapevine powdery mildew (PM) disease, was inadvertently introduced into Europe from North America in the 1850s and caused significant losses to viticultural production2. The fungus has subsequently spread to other grape-growing regions throughout the world and has changed the practice of viticulture by requiring the use of frequent and prophylactic spray programs. Indeed, a report on the use of fungicides in the European Union over the period 2001–2003 indicated that while viticulture only accounted for 3.3% of the agricultural area, a staggering 81,000 tonnes of active substance were applied annually to grapevines in European vineyards, which represented 67% of all fungicides applied to crops in the EU3. Not only does this translate into increased production costs for growers, but there is also the potential impact of these chemicals on the health of beneficial organisms in the vineyard4 and vineyard workers5, as well as increased carbon emissions generated from their frequent application. Thus, the integration of effective genetic resistance into grape cultivars would reduce the dependence of viticulture on chemical inputs, leading to significant financial, health, and environmental benefits.
Wild North American Vitis spp. including V. rotundifolia (syn. Muscadinia rotundifolia), V. rupestris, V. riparia, and V. aestivalis are more resistant to PM than European V. vinifera cultivars6. As early as the late 1800s, grape breeders began introgressing genetic resistance from the North American Vitis spp. into V. vinifera, resulting in the generation of many Vitis interspecific ‘French–American’ hybrids. However, commercial adoption of these new grape cultivars has been limited, due to the reduced quality of wine made from these resistant hybrids. Selected accessions of a number of wild Chinese Vitis species7,8 have also been reported to show strong resistance to PM, but apart from some specific examples described below, little information is available regarding the genetic basis of PM resistance or the quality of wine produced from Vitis interspecific ‘French–Chinese’ hybrids.
The biotrophic fungal pathogen – erysiphe necator
There is insufficient space in this short review to provide a detailed description of the biology, ecology, and epidemiology of grapevine PM. Instead, readers are directed to an excellent review published by Gadoury et al.9 However, a brief description of the infection process is presented here in order to understand the resistance strategies used by the grapevine host to restrict fungal invasion and colonization.
Erysiphe necator is an obligate biotrophic fungus that relies fully on a host cell in photosynthesis-active tissues to complete its life cycle. Once a conidiospore of E. necator lands on the epidermis of photosynthesis-active tissues, it germinates to form a lobed appressorium. Based on studies with other PMs, it is likely that germination involves the secretion of fungal lytic enzymes such as lipases, esterases, and cutinases10 which leads to the release of long-chain fatty acid derivatives which enhance fungal germination and development10,11. From the lower surface of the appressorium, a penetration peg emerges which penetrates the cell wall and invades the host epidermal cell to form a specialized intracellular structure called a haustorium. The haustorium is an interface between the fungus and the host cell that facilitates the dynamic exchange of molecules derived from both fungal and host cells. The fungus retrieves hexoses, amino acids, vitamins, and other nutrients from host cells, through the haustorium, while at the same time secreting proteins to suppress host defences. If the establishment of the haustorium and the uptake of nutrients is successful, the fungus continues to spread via hyphae across the surface, producing more appressoria and haustoria at regular intervals. After 5–25 days, sporulation occurs in the form of conidiophores perpendicular to the epidermis on which chains of asexual conidia are produced and spores are released to start a new cycle of infection12.
Plant defenses against biotrophic fungal pathogens
There are two main strategies that plants use to restrict the invasion and growth of biotrophic fungal pathogens: penetration resistance and programmed cell death (PCD)-mediated resistance (Figure 1). Penetration resistance blocks the breach of the cell wall and membrane by the germinated spore and thus prevents the formation of the haustorium. The PCD-mediated resistance is exerted inside the penetrated epidermal cell and induces the death of invaded cell, thereby terminating the supply of nutrients required by the biotrophic fungus for further growth and development.
Figure 1.
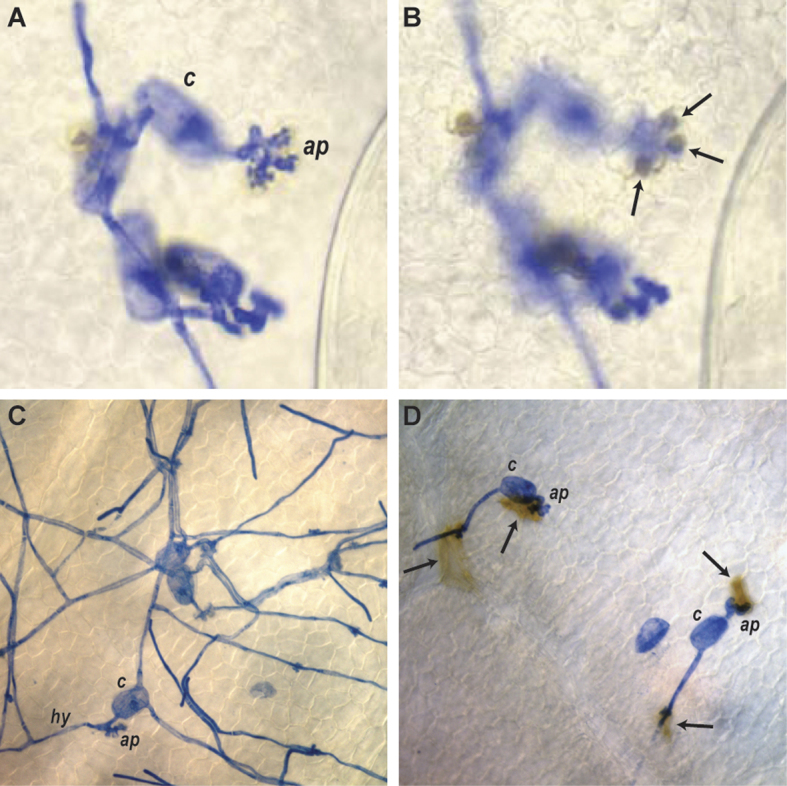
Mechanisms of grapevine defense against the biotrophic fungal pathogen powdery mildew (E. necator). Grapevine powdery mildew spores were inoculated onto detached leaves of M. rotundifolia (A & B), V. vinifera cv. Cabernet Sauvignon (C) and a V. vinifera backcross progeny plant containing the powdery mildew resistance gene MrRUN1 (D). Leaf samples were collected after 2 days and fixed and stained with Coomassie brilliant blue to visualize fungal structures. Panels A and B represent the same field of view but are focused at different levels to show the germinated conidium (c) and appressoria (ap) on the surface of the leaf (panel A) and the globular papillae (arrows) beneath the appressoria (panel B) which are blocking penetration and haustoria formation. Panel C shows normal growth of E. necator hyphae (hy) across the leaf surface of a susceptible grapevine cultivar. Panel D shows the induction of MrRUN1-mediated programmed cell death in penetrated epidermal cells (arrows) which effectively halts further growth of this biotrophic pathogen.
The innate immune responses in a plant cell happen, consecutively and are interconnected, in two basic forms: pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI)13. PTI is activated by the interaction of extracellular pattern-recognition receptors in the plasma membrane of the host cell and pathogen-specific molecules that are released from a pathogen14. The fungal PAMP chitin, which is a major constituent of fungal cell walls, is released during infection by PM and is detected by a LysM (lysin motif)-receptor-like kinase15. This, in turn, activates the mitogen-activated protein kinase cascade which triggers multiple defense responses, including the generation of reactive oxygen species, defense gene activation, biosynthesis/signaling of plant stress/defense hormones, phytoalexin biosynthesis, and cell wall strengthening16. PTI is the first line of defense and provides protection against the majority of microbes that plants face. However, through evolution, certain isolates have become ‘adapted’ to a specific host through the development of effector proteins that are secreted into the plant cell to suppress PTI and enable the pathogen to become virulent on the host13. Over time, selected plant species in which PTI had been compromised, acquired additional receptors (resistance (R) proteins) that specifically recognize these effectors, leading to ETI. R proteins interact with the effector directly, or indirectly through partner proteins, leading to the induction of defense responses that share overlapping pathways with PTI17. ETI is most commonly associated with PCD (observed as a hypersensitive response) which prevents biotrophic pathogens, including PM, from obtaining nutrients and completing its life cycle.
Pamp-triggered immunity against powdery mildew in grapevine
The cultivated grapevine, V. vinifera is resistant to species of PM that are not adapted to grapevine. For example, the ‘non-adapted’ PM species, Erysiphe cichoracearum that causes PM disease of cucurbits, shows much lower rates of penetration of grapevine epidermal cells than E. necator18 and, as such, is unable to establish a successful infection. Penetration resistance represents the major component of PTI against non-adapted PMs in most plant species and has been shown by forward genetic screens, in Arabidopsis and barley, to involve the combined action of at least three PENETRATION (PEN) genes: PEN1, PEN2, and PEN319–21. PEN1 is a member of the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) family which includes proteins which mediate membrane fusion events22 and is proposed to have a role in the trafficking of secretory vesicles to the plasma membrane that contain cargo required for penetration resistance against PM. PEN2 and PEN3 function in the same penetration resistance pathway in Arabidopsis which is independent of PEN120. PEN2 is a myrosinase involved in the biosynthesis of antimicrobial molecules that are delivered to the site of PM penetration via PEN3 which is an ATP-binding cassette transporter23,24.
As grapevine is a woody perennial and much more difficult to transform, it has not been possible to use similar forward genetic screens to identify basic components of PTI against E. necator. However, accumulating evidence from studies using other experimental approaches indicates that the PEN1- and PEN2/PEN3-like pathways are also important components of PTI in grapevine. The first piece of evidence comes from inhibitor studies. Penetration resistance of grapevine against the non-adapted PM species E. cichoracearum was shown to be compromised by the actin cytoskeleton inhibitor cytochalasin18 which was subsequently shown, in Arabidopsis, to inhibit the focal accumulation of PEN3 under the site of PM penetration23. This suggests a role for a PEN3-like protein in PTI against non-adapted PM in grapevine, but as yet, no orthologues of the Arabidopsis PEN3 protein have been identified in any other plant. Recently a PEN1 orthologue from grapevine (VvPEN1) was cloned and its functional complementation of the Arabidopsis pen1 mutant demonstrated25. A VvPEN1-GFP fusion protein was also shown to accumulate under the site of PM penetration as has been demonstrated for PEN1 in Arabidopsis and barley26. The accumulation of PEN1 and VvPEN1 under the site of attempted PM penetration is inhibited by the endomembrane trafficking inhibitors brefeldin A and wortmannin25,27. Feechan et al.18 also demonstrated that penetration resistance against the non-adapted PM species E. cichoracearum, in grapevine, was compromised by brefeldin A and wortmannin, suggesting the existence of a PEN1-mediated secretory pathway that is also an important component of PTI against PM in grapevine.
Effector-triggered immunity against powdery mildew in grapevine
The most important class of R-genes in plants are those encoding proteins with nucleotide binding (NB) site – leucine-rich repeat (LRR) domains28. These NB-LRR proteins specifically recognize the microbial effector molecules secreted during infection and initiate ETI which is highly effective against biotrophic pathogens such as PM. The genomes of perennial woody plants appear to possess a larger number of NB-LRR resistance genes than annual herbaceous plants which most probably reflects the more diverse range of pathogens that perennial plants have to deal with over their lifespan29.
The grapevine genome encodes a large family of NB-LRR genes that are clustered in tandem repeats in genomic regions30,31. Tandem repetitive and paralogous R-genes constitute a reserved genetic army that can be activated whenever there is a need to battle against invading pathogens. It is therefore reasonable to expect that a significant amount of genomic diversity exists within the large number of accessions of wild Vitis species. To date, nine loci have been identified from a range of different grape species native to North America, China, and Central Asia, which are thought to contain R-genes that confer strong resistance to E. necator (Table 1). The chromosomal location of these loci have been genetically mapped using molecular markers, although the exact position on the chromosome in question is only currently known for the Resistance to Uncinula necator (RUN1) locus which is co-located with a locus for Resistance to Plasmopora viticola (downy mildew) (RPV1) on chromosome 1232.
Table 1. Major resistance loci in grapevine species that confer resistance to the powdery mildew fungus Erysiphe necator.
| R-locus | Source of resistance | Origin of R-loci | Resistance mechanism | Chromosome | R-gene type | References |
|---|---|---|---|---|---|---|
| RUN1 | M. rotundifolia ‘Thomas’ | North America | PCD of penetrated cell (rapid) | 12 | TIR-NB-LRR gene (MrRUN1) functionally confirmed in transgenic susceptible cultivars | 32 |
| RUN2 | M. rotundifolia ‘Magnolia’/‘Trayshed’ | North America | PCD of penetrated cell (lower frequency compared to RUN1) | 18 | n.d. | 35,84 |
| REN1 | V. vinifera ‘Kishmish vatkana’ | Central Asia | PCD of penetrated cell (significantly slower and lower frequency than RUN1) – hyphal growth and sporulation restricted | 13 | Maps to a CC-NB-LRR gene cluster | 34,39,40 |
| REN2 | V. cinerea ‘Illinois 547-1’ | North America | PCD of penetrated cell (slower than RUN1) | 14 | n.d. | 35,85 |
| REN3 | Interspecific hybrid ‘Regent’ | North America | n.d. | 15 | n.d. | 86 |
| REN4 | V. romanetii | China | PCD of penetrated cell (rapid)/callose encasement of haustoriuma | 18 | n.d. | 37 |
| REN5 | M. rotundifolia ‘Regale’ | North America | Post-penetration but mechanism not reported | 14 | n.d. | 36 |
| REN6 | V. piasezkii | China | PCD of penetrated cell (very rapid)b | 9 | n.d. | Riaz S, 2015, unpublished data |
| REN7 | V. piasezkii | China | n.d. | 19 | n.d. | Riaz S, 2015, unpublished data |
n.d. – not determined.
Feechan A, 2015, unpublished data.
Dry I, 2015, unpublished data.
RUN1 is one of the three PM resistance loci that have been identified from different accessions of the wild North American grapevine species M. rotundifolia (Table 1) and is, to date, the only pathogen resistance locus that has been cloned from any grapevine species32. Sequencing of the RUN1/RPV1 locus revealed that it contains a family of seven putative Toll/interleukin-1 receptor (TIR)-NB-LRR-type R-genes. However, only one of the candidate TIR-NB-LRR genes at the locus (MrRGA10) was found to confer strong resistance to PM when transformed into a range of susceptible V. vinifera cultivars including Shiraz, Tempranillo, and Portan. This gene, designated MrRUN1, confers complete resistance against isolates from Australia, North America, and France by rapidly inducing PCD in penetrated epidermal cells32. However, a PM isolate (Musc4) collected from the southeastern region of North America33, to which M. rotundifolia is native, was found to be capable of breaking MrRUN1 resistance32 indicating that the effector recognized by the MrRUN1 protein has either been mutated or completely lost from the Musc4 isolate.
The other two PM resistance loci identified in M. rotundifolia are located on different chromosomes to RUN1 (Table 1). Allelic variants of the RUN2 locus, RUN2.1 and RUN2.2 on chromosome 18, have been identified in the M. rotundifolia cultivars ‘Magnolia’ and ‘Trayshed’, respectively34 whereas RUN1 is thought to have originated from the cultivar ‘Thomas’35. Interestingly, while RUN2.1-mediated PM resistance does not appear to be as strong as RUN1, it is not broken by the Musc4 isolate making it a potential candidate for pyramiding with RUN135. REN5 was derived from the M. rotundifolia cultivar ‘Regale’ and maps to the upper portion of chromosome 1436. The mechanism underlying the resistance mediated by REN5 is yet to be determined but appears to be initiated at the post-penetration phase36.
Recent research has demonstrated that wild Chinese Vitis species also represent an important source of major dominant R-genes for PM resistance. REN4 has been successfully introgressed into V. vinifera from the Chinese species V. romanetii and shown to segregate as a single dominant R-locus37. REN4 resistance was initially reported to be associated with high levels of penetration resistance and did not appear to be dependent on the induction of PCD37. However, more recent studies indicate that REN4-mediated resistance occurs post-penetration and involves two different mechanisms – penetrated epidermal cells either undergo PCD or the haustoria become encased in callose thereby effectively blocking nutrient uptake (Feechan A, 2015, unpubl. data). Interestingly, this dual resistance phenotype is reminiscent of the type of response mediated by the broad spectrum PM resistance gene RPW8.2 from Arabidopsis which has a unique structure in terms of other known R proteins38. Another important observation is that REN4 resistance is not broken by the Musc4 isolate35 suggesting that REN4 targets a different E. necator effector to that recognized by the RUN1 protein. This is most probably the result of REN4 co-evolving with different E. necator isolates in China to those in North America.
The wild Chinese grapevine species V. piasezkii also appears to contain at least two PM resistance loci, designated REN6 and REN7, on chromosomes 9 and 19 respectively (Riaz S, 2015, unpublished data). A comparison of the resistance responses of the REN6, REN4, and RUN1 loci, against the same Australian E. necator isolate, in the same genetic background, indicated that PCD initiation is most rapid in penetrated cells containing REN6, with less than 5% of appressoria producing a secondary hypha, compared to ∼15% and 30% in grapevines containing REN4 or RUN1 respectively (Dry I, 2015, unpublished data).
Finally, it is now clear that certain accessions of V. vinifera from Central Asia also contain a major R-gene that, while less effective than RUN1-mediated resistance, still significantly restricts PM growth and sporulation. Two V. vinifera cultivars, ‘Kishmish vatkana’ and ‘Dzhandzhal kara’, originating from Uzbekistan, were shown to induce PCD in penetrated epidermal cells at a higher frequency than susceptible vines, but the speed of the PCD induction was much slower than that observed in a genotype containing RUN139. As a result, more PM hyphal growth and sporulation is observed on REN1 plants than on RUN1 plants, but this is still much less than observed on susceptible V. vinifera cultivars. The REN1 locus has been mapped to a 1.4 Mbp region on chromosome 1340. The syntenous region in the PN40024 V. vinifera reference genome contains a cluster of CC-NB-LRR genes40, but no data have yet been published to indicate what candidate R-genes are present in this region in ‘Kishmish vatkana’ or ‘Dzhandzhal kara’. Riaz and co-workers34 subsequently identified an additional six V. vinifera and two V. vinifera subsp. sylvestris accessions from Central Asia that also contained a REN1-like locus. Based on genetic marker analysis, they concluded that the REN1-like resistance in V. vinifera subsp. sylvestris was most likely the progenitor of the resistance in the Central Asian V. vinifera accessions.
The existence of major R-gene resistance against E. necator in Vitis species native to China and Central Asia brings into question the current dogma that this pathogen is native to North America and was spread to all grape-growing regions from this one source. It seems more likely that E. necator isolates have been in existence in Central Asia and China for a much longer period than previously thought to explain the evolution of these R-genes in the wild grape species from these regions34.
Downstream genes implicated in resistance to powdery mildew
In addition to the isolate-specific R-genes that confer ETI in penetrated epidermal cells (Table 1), a number of other genes have also been implicated in PM resistance in certain wild Vitis species (Table 2). These genes have been identified because (i) they show increased transcription during PM infection and/or show differential expression levels between PM-resistant wild Vitis species and susceptible V. vinifera cultivars and (ii) they confer increased levels of resistance to PM when overexpressed transiently in grapevine leaves or stably transformed into wild-type or mutant lines of the model species A. thaliana41–45.
Table 2. Genes from wild grapevine species postulated to be involved in resistance to the powdery mildew fungus Erysiphe necator.
| Gene | Description | Vitis species | Function and phenotype | References |
|---|---|---|---|---|
| VaEDS1 | Enhanced Disease Susceptibility ortholog | V. aestivalis ‘Norton’ | Defense pathway regulator – complements Arabidopsis eds1 mutant. Constitutively high expression in V. aestivalis resistant genotype and regulated by SA and PM | 41,49 |
| VpPR10.1 | Pathogenesis-related protein 10 | V. pseudoreticulata ‘Baihe-35-1’ | Antifungal activity. Increases resistance to PM in agroinfiltrated grapevine leaves | 71 |
| VpALDH2B4 | Aldehyde dehydrogenase | V. pseudoreticulata ‘Baihe-35-1’ | Activation of SA signaling? Enhanced resistance to PM when overexpressed in Arabidopsis | 43 |
| VpWRKY1 | WRKY domain Transcription factor | V. pseudoreticulata ‘Baihe-35-1’ | Transcriptional activator of defense-related genes. Enhanced resistance to PM when overexpressed in Arabidopsis | 42 |
| VpRFP1 | C4C4-type RING finger protein | V. pseudoreticulata ‘Baihe-35-1’ | Transcriptional activator of defense-related genes? Enhanced resistance to PM when overexpressed in Arabidopsis | 45 |
| VpEIRP1 | E3 ubiquitin ligase Erysiphe necator-induced C3HC4 RING finger protein | V. pseudoreticulata ‘Baihe-35-1’ | Ubiquitination and degradation of a negative transcriptional regulator of defense? Enhanced resistance to PM when overexpressed in Arabidopsis | 44 |
One example of this is the cultivar Norton which is derived from the North American grapevine species V. aestivalis and which is highly resistant to E. necator in comparison to V. vinifera cv. Cabernet Sauvignon46. Investigation of the PM-responsive transcriptome of the two grapevine species revealed that an ortholog of the Arabidopsis Enhanced Disease Susceptibility 1 (EDS1) was differentially expressed in Norton and Cabernet Sauvignon. EDS1 transcription was induced in response to PM in Cabernet Sauvignon whereas its transcription levels were constitutively high in Norton and always exceeded the levels induced in Cabernet Sauvignon41. The level of salicylic acid was also found to be significantly higher in Norton than in Cabernet Sauvignon under non-PM-infected conditions46. EDS1 has been previously been shown to regulate resistance to host-adapted biotrophic pathogens in Arabidopsis in a SA-dependent manner47,48. Constitutively expressed VaEDS1 complemented the function of the mutant eds1 gene in Arabidopsis41 and rendered the pen2/eds1 mutant resistant to PM. The VaEDS1 promoter was also shown to be inducible by SA and PM49. These results strongly suggest that the constitutively high levels of SA and SA/PM-responsive EDS1 in Norton may account for the elevated resistance of this genotype to PM.
A large number of studies have also been carried out to determine the genetic basis of PM resistance in certain accessions of the wild Chinese grapevine V. pseudoreticulata. At least five different genes have been identified that may contribute to PM resistance in this wild species (Table 2). The transcription factor, VpWRKY1, was rapidly induced in V. pseudoreticulata within 12 h of inoculation with E. necator and the level of expression was found to be correlated with the level of resistance42. Furthermore, ectopic expression of VpWRKY1 in Arabidopsis enhanced resistance to E. cichoracearum. Two other genes that appear to be upregulated in the V. pseudoreticulata accession Baihe-35-1, in response to PM inoculation, and which both confer resistance to PM when ectopically expressed in Arabidopsis, belong to the Really Interesting New Gene (RING) finger protein gene family44,45. The RING finger domain has been shown to have E3 ligase activity, which is important in ubiquitin-dependent protein degradation50. Although plant cells contain hundreds of distinct E3 ligases involved in ubiquitination reactions, in a range of different biological processes, to date there are only limited reports of any RING-type E3 ligases involved in plant defense51,52.
Evidence for powdery mildew susceptibility genes in grapevine
As described above, adapted PM species are able to successfully penetrate their cognate host by secreting effector proteins that suppress host PTI. However, successful penetration by the adapted PM species has been shown to be dependent on the presence of a functional allele of the Mildew resistance Locus O (MLO) in a range of host species including barley53, Arabidopsis54, tomato55, and pea56.
MLO proteins belong to large gene families which are unique to plants and encode seven-transmembrane domain proteins of unknown biochemical activity localized in the plasma membrane57. Significantly, only specific MLO genes within the family are capable of acting as PM susceptibility genes and these encode proteins with conserved motifs within the cytoplasmic C-terminal domain of the MLO protein58. The mechanism by which MLO proteins act as PM susceptibility factors is unknown. One possibility is that adapted PM species are able to utilize these specific MLO proteins to suppress host PTI, perhaps through the secretion of an effector that targets MLO either directly or indirectly through another protein. Support for this hypothesis comes from the recent observation that the Arabidopsis PM susceptibility protein AtMLO2, also acts as a susceptibility factor for infection by the bacterial pathogen Pseudomonas syringae and that AtMLO2 is targeted by the P. syringae effector HopZ259.
Based on sequence homology, the presence of the C-terminal conserved motifs and expression kinetics following PM infection, Feechan et al.60 identified three VvMLOs (VvMLO3, VvMLO4, and VvMLO17) that may act as PM susceptibility genes in V. vinifera. VvMLO3 and VvMLO4, but not VvMLO17, were subsequently shown to partially rescue an Arabidopsis mlo2 mlo6 mlo12 triple mutant25. Furthermore, GFP fusions of both VvMLO3 and VvMLO4 were demonstrated to localize to the site of PM appressoria formation, in agreement with previous localization studies in barley with HvMLO26. These data strongly support a role for VvMLO3 and VvMLO4 as PM susceptibility factors in grapevine. However, despite the generation of numerous single and double VvMLO3/4 knockout mutants in V. vinifera using RNAi techniques, it has not yet been to recover transgenic grapevines with high rates of reduced PM penetration (Feechan A, 2015, unpubl. data). It is interesting to note that PM-resistant mlo mutants of barley, tomato, and pea have been identified in naturally occurring segregating populations in which mlo is in the homozygous state53,55,56. Therefore, an alternative strategy in grapevine, may be to employ techniques such as EcoTILLING61 to search for point mutations and/or small insertions/deletions in VvMLO3 and VvMLO4 in V. vinifera germplasm collections where the mutation, and thus the PM resistance phenotype, is masked by presence of the wild-type MLO allele.
Evidence for the presence of PM susceptibility gene(s) in grapevine also comes from a recent study which used genotype-by-sequencing to identify a QTL for PM susceptibility from Chardonnay named Sen1 (Susceptibility to Erysiphe necator 1)62. Isolation and analysis of genes located at the Sen1 locus will assist in helping us understand the ways that E. necator establishes and maintains a compatible biotrophic relationship with the grapevine host cell.
Developmental changes in grapevine resistance to powdery mildew
Age-related or ontogenic resistance has been observed in a number of plant species to viral, bacterial, oomycete, and fungal pathogens63. Both grapevine berries and leaves display ontogenic resistance to PM. This is particularly apparent in developing berries of V. vinifera cultivars which are highly susceptible to infection in the first 1–2 weeks after fruit set64 but then become increasingly resistant to PM penetration65,66. In contrast, berries of most North American Vitis species exhibit strong resistance at all stages of berry development67, a constitutive resistance that may have developed during the coevolution of wild Vitis species and E. necator populations.
While the genetic and mechanistic basis of ontogenic resistance to PM in grapevine is still unknown, it is important to note two things. First, that ontogenic resistance to PM is not unique to grapevine. For example, strawberry leaves and fruit also display ontogenic resistance to PM (Podosphaera aphanis)68. Second, the ontogenic resistance phenotype appears to be the result of an increase in PTI and does not appear to be associated with changes in the availability of nutrients during leaf or berry development. In experiments reported by Ficke et al.,65 conidial germination and appressorium formation were unaffected by berry development but the rate of penetration, formation of haustoria, and development of secondary hyphae was almost completely halted on older berries. Furthermore, the increased PM resistance could not be ascribed to any changes in cuticle or cell wall thickness in the developing berries66. Based on the evidence presented in the previous section, which shows that the successful penetration of epidermal cells, by an adapted PM species, is dependent on MLO-mediated suppression of host PTI, it is tempting to speculate that changes in host MLO expression or activity may be involved in this process.
Perspectives and challenges
With the ongoing development of cheaper and faster sequencing technologies it will be possible to undertake complete genome and transcriptome sequencing of an increasing number of wild Vitis species that display resistance to E. necator. This will facilitate comparative genomics studies leading to the identification of key components in PTI and ETI against E. necator in grapevines. Identification and characterization of more grapevine R-genes will unveil deeper insights into and shed more light onto the complex genetic mechanisms of grapevine disease resistance and also provide more molecular markers and genes for breeding resistant grapevine cultivars.
With the identification of an increasing number of R-gene candidates, it will be essential that techniques are available to functionally characterize these genes. Ideally, this would involve stable transformation of a susceptible V. vinifera cultivar with the R-gene candidates to challenge them with a range of E. necator isolates32. However, R-loci typically contain multiple R-gene candidates and stable grapevine transformation, despite technical improvements69, remains a long and technically challenging process. Alternative strategies are needed to facilitate rapid evaluation of these R-gene candidates. One possibility is the use of transient expression systems such as agroinoculation70. This approach has been used to demonstrate the anti-fungal activity of VpPR10.1 against E. necator71 (Table 2). It might also be feasible to test grapevine R-gene function by transforming into Arabidopsis mutants in which PTI has been compromised allowing E. necator to penetrate and form haustoria72. Figure 2 shows the results of an experiment in which the PM resistance gene MrRUN1, was transformed into the Arabidopsis pen1-1 mutant. Inoculation of these pen1-1 mutants with E. necator resulted in a significant induction of PCD in transgenic lines containing MrRUN1 but not in the pen1-1 control lines32. Furthermore, MrRUN1-mediated PCD in Arabidopsis was only induced in response to penetration by E. necator and not observed with E. cichoracearum demonstrating the response is specific to grapevine PM. Thus, this approach could be used to rapidly evaluate multiple R-gene candidates before selected genes are introduced into susceptible V. vinifera cultivars for final validation.
Figure 2.
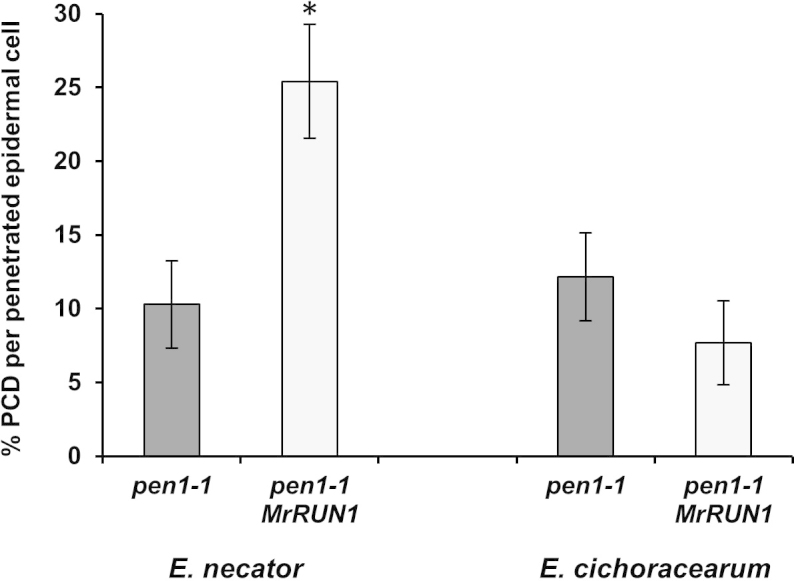
Use of the Arabidopsis pen1 mutant for rapid screening of candidate grapevine powdery mildew resistance genes. The pen1-1 mutant line that allows increased penetration of non-adapted powdery mildew species was transformed with the grapevine powdery mildew resistance gene MrRUN1 and inoculated with either grapevine powdery mildew (E. necator) or Arabidopsis powdery mildew (E. cichoracearum). Programmed cell death (PCD) was estimated by trypan blue staining of inoculated leaves at 2 dpi. Each data point is the mean of three independent experiments (±standard deviation). In each experiment, a minimum of 100 germinated conidia were scored on each of three leaves for each line. Asterisk indicates a statistically significant difference from pen1-1 (P < 0.05; Student's t-test).
Due to its mixed reproductive system (sexual and asexual) and large population size, PM is considered to be a pathogen with a high evolutionary potential and therefore a high risk to overcome genetic resistance73. This was demonstrated by the fact that PM resistance conferred by the apple R-gene Pl2 was found to have broken down in experimental orchards after only 6 years74. Jones et al.75 recently reported on the genome sequencing of five E. necator isolates collected from Californian vineyards that had regularly been treated with synthetic fungicides. Their results showed there to be a significant amount of structural variation in the genomes of the different E. necator isolates and, in particular, a variation in copy number of the EnCYP51 gene which is the target of the commonly used sterol demethylase fungicide75. This demonstrates that E. necator is able to readily respond to strong selection pressures in the field. This has important implications for the potential deployment of major dominant R-genes outlined in Table 1 and highlights the importance of using pyramiding strategies involving multiple R-genes to maintain durable PM resistance in the vineyard.
While it is generally accepted that pyramiding R-genes is an effective approach for increasing the durability of field resistance76, it is essential that the R-genes to be combined do not rely on recognition of the same E. necator effector protein to initiate PCD (defined as the avirulence (Avr) effector), because resistance conferred by both genes would be lost simultaneously should a new isolate evolve in which this effector has been lost or mutated. At present, we have only limited information about the range of PM isolates detected by some of the R-loci listed in Table 135 and breeding strategies are currently being employed that are based essentially on the availability of R-genes from different Vitis species, rather than on any empirical knowledge regarding the isolate specificity of these different R-genes. However, the assumption that R-genes from different Vitis species have different recognition specificities may not always be true. This was clearly demonstrated by a study on resistance to the oomycete pathogen Phytophthora infestans, which causes late blight in potato, showing that R-genes from three different Solanum species were functionally equivalent and recognized the same P. infestans effector77.
The simplest approach to determining Avr effector recognition specificity is to challenge grape genotypes containing different R-loci to PM, with a range of PM isolates with different Avr specificities35. However, we are currently limited by the paucity of E. necator isolate collections and by the fact that it is not always feasible to bring the grape genotypes and E. necator isolates together in the same country, region, or research facility because of quarantine issues and concerns associated with accidental release of the pathogen. Therefore alternative approaches will be required to enable Avr recognition specificity of PM R-genes from different grape species. One such approach is ‘Effectoromics’, a high-throughput functional genomics approach that uses the transient expression of Avr effectors to probe plant germplasm to detect and characterize R-genes78. However, this is only possible once the Avr effectors have been identified. This will be challenging given that sequencing of the E. necator genome has revealed it to contain at least 150 candidate-secreted effector proteins (CSEPs)75. Furthermore, unlike functionally characterized Avr effectors from oomycete pathogens, which have a conserved host cell targeting motif (RXLR) in the N-terminal region of their mature protein79, there are insufficient Avr effectors characterized from PM species, or for that matter, fungal biotrophic pathogens in general, to use a bioinformatics approach based on conserved domains to enable prediction of the potential Avr candidates from the E. necator effectorome. Thus, more targeted approaches may be required involving the use of a comparative genomics approach in which the effectoromes of isolates possessing different pathogenicity specificities are compared80 or a map-based cloning approach using PM populations segregating for the Avr effector gene81.
The identification of Avr effectors will not only provide us with tools to enable testing of the recognition specificity of new R-genes without the need to have access to the PM isolate, but will also provide us with a diagnostic test to be able to follow the appearance of resistance-breaking strains in the vineyard. Characterization of the E. necator effectorome will also facilitate the identification of the host targets of the effectors that suppress the grapevine PTI pathway as has been done with the Blumeria graminis f. sp. hordei – barley interaction82,83. This knowledge could possibly be used to modify the host targets to avoid PTI suppression and, in so doing, re-establish PTI against E. necator in grapevine.
Further work is also needed to properly characterize the genes and pathways underpinning the quantitative PM resistance displayed by certain wild Vitis species. Much of the evidence implicating a role for the genes listed in Table 2, in PM resistance, is based on correlations between an elevated level of gene expression (either constitutive or PM-induced) in the resistant genotype vs. the susceptible genotype and functional assays showing reduced pathogen infection when overexpressed in Arabidopsis. However, in order to prove conclusively that these genes do contribute to PM resistance in these wild grapevine species, it will be necessary to demonstrate that the segregation of resistance is genetically linked to the inheritance of these candidate genes. This will also test the degree to which these genes are able to function in different genetic backgrounds i.e. V. vinifera which is essential to know if they are being considered as part of any gene pyramiding strategy.
In summary, we see the following research priorities as critical in the ongoing development of durable and effective PM-resistant grapevine germplasm:
Genome and transcriptome sequencing of more PM-resistant wild grape species to identify new R-genes.
Genome and transcriptome sequencing of multiple virulent and avirulent grapevine powdery mildew isolates to facilitate construction of the E. necator effectorome and identify Avr effectors.
Functional characterization of R-genes from different Vitis species in terms of effector recognition specificity.
Whole genome association analysis of disease resistance using high-density SNPs to identify genes conferring partial resistance to PM for pyramiding with major R-genes.
Continued exploration of grapevine genes involved in PM susceptibility, including host targets of PTI suppression by E. necator effectors.
Further genetic analysis of ontogenic resistance in developing leaves and berries to identify the genes and pathways underlying this process.
Acknowledgments
The authors thank Dr Summaira Riaz and Dr Lance Cadle-Davidson for providing unpublished data. Researchers in IBD’s laboratory were supported by grants from the Australian Grape and Wine Authority. CSIRO Agriculture is part of the Wine Innovation Cluster. Research projects in WQ’s laboratory were sponsored by the USDA-NIFA grants, Missouri State University and the Missouri Wine and Grape Board.
The authors declare no conflict of interest.
References
- OIV. International Organisation of Vine and Wine. Statistical Report on World Vitiviniculture 2012. Available at http://www.oiv.int/oiv/files/0%20-%20Actualites/EN/Report.pdf (accessed 3rd March 2015).
- Campbell C. Phylloxera: how wine was saved for the world. Harper Collins London, 2004, p 368. [Google Scholar]
- EUROSTAT EC. The use of plant protection products in the European Union. Data 1992-2003. Luxembourg: Office for Official Publications of the European Communities, 2007. ISBN 92-79-03890-7. [Google Scholar]
- Gadino AN, Walton VM, Dreves AJ. Impact of vineyard pesticides on a beneficial arthropod, Typhlodromus pyri (Acari: Phytoseiidae), in laboratory bioassays. J Econ Entomol 2011; 104: 970–977. [DOI] [PubMed] [Google Scholar]
- Le Moal J, Rolland M, Goria S, Wagner V, De Crouy-Chanel P, Rigou A et al. Semen quality trends in French regions are consistent with a global change in environmental exposure. Reproduction 2014; 147: 567–574. [DOI] [PubMed] [Google Scholar]
- Cadle-Davidson L, Chicoine DR, Consolie NH. Variation within and among Vitis spp. for foliar resistance to the powdery mildew pathogen Erysiphe necator. Plant Dis 2011; 95: 202–211. [DOI] [PubMed] [Google Scholar]
- Wan Y, Schwaniniger H, He P, Wang Y. Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 2007; 46: 132–136. [Google Scholar]
- Wang Y, Liu Y, He P, Chen J, Lamikanra O, Lu J. Evaluation of foliar resistance to Uncinula necator in Chinese wild Vitis species. Vitis 1995; 34: 159–164. [Google Scholar]
- Gadoury DM, Cadle-Davidson L, Wilcox WF, Dry IB, Seem RC, Milgroom MG. Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol Plant Pathol 2012; 13: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wang F, Liu GS, Greenshields D, Shen WY, Kaminskyj S et al. Analysis of a Blumeria graminis-secreted lipase reveals the importance of host epicuticular wax components for fungal adhesion and development. Mol Plant Microbe Interact 2009; 22: 1601–1610. [DOI] [PubMed] [Google Scholar]
- Weidenbach D, Jansen M, Franke RB, Hensel G, Weissgerber W, Ulferts S et al. Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol 2014; 166: 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delp CJ. Effect of temperature and humidity on the grape powdery mildew fungus. Phytopathology 1954; 44: 615–626. [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science 2013; 341: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 2009; 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Wan JR, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008; 20: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XZ, Zhang SQ. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 2013; 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 2010; 13: 459–465. [DOI] [PubMed] [Google Scholar]
- Feechan A, Kabbara S, Dry IB. Mechanisms of powdery mildew resistance in the Vitaceae family. Mol Plant Pathol 2011; 12: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 2003; 425: 973–977. [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 2005; 310: 1180–1183. [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sanchez-Rodriguez C, Hou BH, Molina A, Schulze-Lefert P et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 2006; 18: 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli J, Sutter JU, Blatt MR. A new catch in the SNARE. Trends Plant Sci 2004; 9: 187–195. [DOI] [PubMed] [Google Scholar]
- Underwood W, Somerville SC. Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc Natl Acad Sci USA 2013; 110: 12492–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009; 323: 101–106. [DOI] [PubMed] [Google Scholar]
- Feechan A, Jermakow AM, Ivancevic A, Godfrey D, Pak H, Panstruga R et al. Host cell entry of powdery mildew is correlated with endosomal transport of antagonistically acting VvPEN1 and VvMLO to the papilla. Mol Plant Microbe Interact 2013; 26: 1138–1150. [DOI] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 2005; 102: 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen ME, Feechan A, Bohlenius H, Ueda T, Thordal-Christensen H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci USA 2012; 109: 11443–11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW. Plant disease resistance genes: current status and future directions. Physiol Mol Plant Pathol 2012; 78: 51–65. [Google Scholar]
- Tobias PA, Guest DI. Tree immunity: growing old without antibodies. Trends Plant Sci 2014; 19: 367–370. [DOI] [PubMed] [Google Scholar]
- Moroldo M, Paillard S, Marconi R, Fabrice L, Canaguier A, Cruaud C et al. A physical map of the heterozygous grapevine ‘Cabernet Sauvignon’ allows mapping candidate genes for disease resistance. BMC Plant Biol 2008; 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright D, Cestaro A, Pruss D et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2007; 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Anderson C, Torregrosa L, Jermakow A, Mestre P, Wiedemann-Merdinoglu S et al. Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant J 2013; 76: 661–674. [DOI] [PubMed] [Google Scholar]
- Brewer M, Milgroom M. Phylogeography and population structure of the grape powdery mildew fungus, Erysiphe necator, from diverse Vitis species. BMC Evol Biol 2010; 10: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz S, Boursiquot J-M, Dangl G, Lacombe T, Laucou V, Tenscher A et al. Identification of mildew resistance in wild and cultivated Central Asian grape germplasm. BMC Plant Biol 2013; 13: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Kocsis M, Riaz S, Zhang W, Gadoury DM, Walker MA et al. Strategies for RUN1 deployment using RUN2 and REN2 to manage grapevine powdery mildew informed by studies of race-specificity. Phytopathology 2015; in press. [DOI] [PubMed]
- Blanc S, Wiedemann-Merdinoglu S, Dumas V, Mestre P, Merdinoglu D. A reference genetic map of Muscadinia rotundifolia and identification of Ren5, a new major locus for resistance to grapevine powdery mildew. Theor Appl Genet 2012; 125: 1663–1675. [DOI] [PubMed] [Google Scholar]
- Ramming DW, Gabler F, Smilanick J, Cadle-Davidson M, Barba P, Mahanil S et al. A single dominant locus, Ren4, confers rapid non-race-specific resistance to grapevine powdery mildew. Phytopathology 2010; 101: 502–508. [DOI] [PubMed] [Google Scholar]
- Wang WM, Wen YQ, Berkey R, Xiao SY. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 2009; 21: 2898–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, Di Gaspero G, Kovács L, Howard S, Kiss E, Galbács Z et al. Resistance to Erysiphe necator in the grapevine ‘Kishmish vatkana’ is controlled by a single locus through restriction of hyphal growth. Theor Appl Genet 2008; 116: 427–438. [DOI] [PubMed] [Google Scholar]
- Coleman C, Copetti D, Cipriani G, Hoffmann S, Kozma P, Kovacs L et al. The powdery mildew resistance gene REN1 co-segregates with an NBS-LRR gene cluster in two Central Asian grapevines. BMC Genet 2009; 10: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Shu X, Ali M, Howard S, Li N, Winterhagen P et al. A functional EDS1 ortholog is differentially regulated in powdery mildew resistant and susceptible grapevines and complements an Arabidopsis eds1 mutant. Planta 2010; 231: 1037–1047. [DOI] [PubMed] [Google Scholar]
- Li H, Xu Y, Xiao Y, Zhu Z, Xie X, Zhao H et al. Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 2010; 232: 1325–1337. [DOI] [PubMed] [Google Scholar]
- Wen Y, Wang X, Xiao S, Wang Y. Ectopic expression of VpALDH2B4, a novel aldehyde dehydrogenase gene from Chinese wild grapevine (Vitis pseudoreticulata), enhances resistance to mildew pathogens and salt stress in Arabidopsis. Planta 2012; 236: 525–539. [DOI] [PubMed] [Google Scholar]
- Yu Y, Xu W, Wang J, Wang L, Yao W, Yang Y et al. The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol 2013; 200: 834–846. [DOI] [PubMed] [Google Scholar]
- Yu Y, Xu W, Wang S, Xu Y, Li He, Wang Y et al. VpRFP1, a novel C4C4-type RING finger protein gene from Chinese wild Vitis pseudoreticulata, functions as a transcriptional activator in defence response of grapevine. J Exp Bot 2011; 62: 5671–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung R, Gonzalo M, Fekete C, Kovacs L, He Y, Marsh E et al. Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol 2008; 146: 236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 2005; 8: 383–389. [DOI] [PubMed] [Google Scholar]
- Straus MR, Rietz S, van Themaat EVL, Bartsch M, Parker JE. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J 2010; 62: 628–640. [DOI] [PubMed] [Google Scholar]
- Gao F, Dai R, Pike S, Qiu W, Gassmann W. Functions of EDS1-like and PAD4 genes in grapevine defenses against powdery mildew. Plant Mol Biol 2014; 86: 381–393. [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 2004; 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Takai R, Matsuda N, Nakano A, Hasegawa K, Akimoto C, Shibuya N et al. EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J 2002; 30: 447–455. [DOI] [PubMed] [Google Scholar]
- Hondo D, Hase S, Kanayama Y, Yoshikawa N, Takenaka S, Takahashi H. The LeATL6-associated ubiquitin/proteasome system may contribute to fungal elicitor-activated defense response via the jasmonic acid-dependent signaling pathway in tomato. Mol Plant Microbe Interact 2007; 20: 72–81. [DOI] [PubMed] [Google Scholar]
- Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A et al. The barley mlo gene: a novel control element of plant pathogen resistance. Cell 1997; 88: 695–705. [DOI] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 2006; 38: 716–720. [DOI] [PubMed] [Google Scholar]
- Bai YL, Pavan S, Zheng Z, Zappel NF, Reinstadler A, Lotti C et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of Mlo function. Mol Plant Microbe Interact 2008; 21: 30–39. [DOI] [PubMed] [Google Scholar]
- Humphry M, Reinstadler A, Ivanov S, Bisseling T, Panstruga R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol 2011; 12: 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo-Garcia J, Kusch S, Panstruga R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol 2014; 204: 273–281. [DOI] [PubMed] [Google Scholar]
- Panstruga R. Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol Biol 2005; 59: 485–500. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Wan J, Ford R, Gong YC, Fung P, Nahal H et al. Quantitative interactor screening with next-generation sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics 2012; 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Jermakow AM, Torregrosa L, Panstruga R, Dry IB. Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct Plant Biol 2008; 35: 1255–1266. [DOI] [PubMed] [Google Scholar]
- Mejlhede N, Kyjovska Z, Backes G, Burhenne K, Rasmussen SK, Jahoor A. EcoTILLING for the identification of allelic variation in the powdery mildew resistance genes mlo and Mla of barley. Plant Breed 2006; 125: 461–467. [Google Scholar]
- Barba P, Cadle-Davidson L, Harriman J, Glaubitz J, Brooks S, Hyma K et al. Grapevine powdery mildew resistance and susceptibility loci identified on a high-resolution SNP map. Theor Appl Genet 2014; 127: 73–84. [DOI] [PubMed] [Google Scholar]
- Develey-Riviere MP, Galiana E. Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytol 2007; 175: 405–416. [DOI] [PubMed] [Google Scholar]
- Gadoury DM, Seem RC, Ficke A, Wilcox WF. Ontogenic resistance to powdery mildew in grape berries. Phytopathology 2003; 93: 547–555. [DOI] [PubMed] [Google Scholar]
- Ficke A, Gadoury DM, Seem RC, Dry IB. Effects of ontogenic resistance upon establishment and growth of Uncinula necator on grape berries. Phytopathology 2003; 93: 556–563. [DOI] [PubMed] [Google Scholar]
- Ficke A, Gadoury DM, Godfrey D, Dry IB. Host barriers and responses to Uncinula necator in developing grape berries. Phytopathology 2004; 94: 438–445. [DOI] [PubMed] [Google Scholar]
- Gee CT, Gadoury DM, Cadle-Davidson L. Ontogenic resistance to Uncinula necator varies by genotype and tissue type in a diverse collection of Vitis spp. Plant Dis 2008; 92: 1067–1073. [DOI] [PubMed] [Google Scholar]
- Asalf B, Gadoury DM, Tronsmo AM, Seem RC, Dobson A, Peres NA et al. Ontogenic resistance of leaves and fruit, and how leaf folding influences the distribution of powdery mildew on strawberry plants colonized by Podosphaera aphanis. Phytopathology 2014; 104: 954–963. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Dai LM, Cheng SY, He J, Wang D, Zhang JX et al. A circulatory system useful both for long-term somatic embryogenesis and genetic transformation in Vitis vinifera L. cv. Thompson Seedless. Plant Cell Tiss Org 2014; 118: 157–168. [Google Scholar]
- Santos-Rosa M, Poutaraud A, Merdinoglu D, Mestre P. Development of a transient expression system in grapevine via agro-infiltration. Plant Cell Rep 2008; 27: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Xu T-F, Zhao X-C, Jiao Y-T, Wei J-Y, Wang L, Xu Y. A Pathogenesis related protein, VpPR-10.1, from Vitis pseudoreticulata: an insight of its mode of antifungal activity. PLoS ONE 2014; 9: e95102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Jermakow AM, Anderson C, Bouquet A, Adam-Blondon A-F, Thomas MR et al. Exploiting a non-host Arabidopsis system to identify powdery mildew resistance genes in grapevine XIV International Congress on Molecular Plant-Microbe Interactions. Quebec City, Canada; 2009. [Google Scholar]
- McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 2002; 40: 349–379. [DOI] [PubMed] [Google Scholar]
- Caffier V, Laurens F. Breakdown of Pl2, a major gene of resistance to apple powdery mildew, in a French experimental orchard. Plant Pathol 2005; 54: 116–124. [Google Scholar]
- Jones L, Riaz S, Morales-Cruz A, Amrine KCH, McGuire B, Gubler WD et al. Adaptive genomic structural variation in the grape powdery mildew pathogen, Erysiphe necator. BMC Genomics 2014; 15: 1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt CC. Durable resistance: a key to sustainable management of pathogens and pests. Infect Genet Evol 2014; 27: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, Oh SK et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 2008; 3: e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Oliver RP. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Int 2014; 27: 196–206. [DOI] [PubMed] [Google Scholar]
- Petre B, Kamoun S. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol 2014; 12: e1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Yoshida K et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 2009; 21: 1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout CJ, Skamnioti P, Porritt O, Sacristan S, Jones JDG, Brown JKM. Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 2006; 18: 2402–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SM, Kuhn H, Micali C, Liller C, Kwaaitaal M, Panstruga R. Interaction of a Blumeria graminis f. sp hordei effector candidate with a barley ARF-GAP suggests that host vesicle trafficking is a fungal pathogenicity target. Mol Plant Pathol 2014; 15: 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Pedersen C, Kwaaitaal M, Gregersen PL, Morch SM, Hanisch S et al. Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol Plant Pathol 2012; 13: 1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz S, Tenscher AC, Ramming DW, Walker MA. Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor Appl Genet 2011; 122: 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbó MA, Ye GN, Weeden NF, Wilcox WF, Reisch BI. Marker-assisted selection for powdery mildew resistance in grapes. J Am Soc Hortic Sci 2001; 126: 83–89. [Google Scholar]
- Welter LJ, Gokturk-Baydar N, Akkurt M, Maul E, Eibach R, Topfer R et al. Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol Breed 2007; 20: 359–374. [Google Scholar]


