Abstract
Grapevine is a perennial crop often cultivated by grafting a scion cultivar on a suitable rootstock. Rootstocks influence scions, particularly with regard to water uptake and vigor. Therefore, one of the possibilities to adapt viticulture to the extended drought stress periods is to select rootstocks conferring increased tolerance to drought. However, the molecular mechanisms associated with the ability of rootstock/scion combination to influence grape berry metabolism under drought stress are still poorly understood. The transcriptomic changes induced by drought stress in grape berries (cv. Pinot noir) from vines grafted on either 110R (drought-tolerant) or 125AA (drought-sensitive) rootstock were compared. The experiments were conducted in the vineyard for two years and two grape berry developmental stages (50% and 100% veraison). The genome-wide microarray approach showed that water stress strongly impacts gene expression in the berries, through ontology categories that cover cell wall metabolism, primary and secondary metabolism, signaling, stress, and hormones, and that some of these effects strongly depend on the rootstock genotype. Indeed, under drought stress, berries from vines grafted on 110R displayed a different transcriptional response compared to 125AA-concerning genes related to jasmonate (JA), phenylpropanoid metabolism, and pathogenesis-related proteins. The data also suggest a link between JA and secondary metabolism in water-stressed berries. Overall, genes related to secondary metabolism and JA are more induced and/or less repressed by drought stress in the berries grafted on the drought-sensitive rootstock 125AA. These rootstock-dependent gene expression changes are relevant for berry composition and sensory properties.
Introduction
Grapevine is one of the most economically important fruit species worldwide.1 Grapevine development and ripening are sensitive to environmental factors’ and climate change can potentially influence yield, berry composition,2 and phenology,3 consequently affecting the viticulture suitability, especially in Mediterranean regions.4 Among the parameters affected by climate change, water availability drastically limits the distribution and the yield of grapes around the world.5,6
The progress made at molecular level concerning gene expression, transcriptional regulation, and signal transduction has improved our understanding of the regulatory networks that control the plant responses to water deficit.7 Considering grapes, genomic approaches have been used to promote large-scale mRNA expression profiling studies of water and salinity stress.5,8,9
Among the different abiotic stresses that affect grapevine, only water deficit, at a moderate level, have been used in a positive way to improve flavor and quality characteristics in grape berries.10 Significant biochemical changes occurring in berries subjected to water stress may influence berry flavor and composition.9,11 The total anthocyanin content of water-stressed berries increase9,11,12 probably due to the upregulation of genes encoding dihydroflavonol-4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT), and VvMybA1.13 In parallel with flavonoid induction, sugars also accumulate more under water stress conditions.9,11,12 Furthermore, both abscisic acid (ABA) and sugar signaling might affect berry ripening.11,13, 14, 15, 16 Concerning amino acid biosynthesis, the accumulation of numerous transcripts associated with glutamate and proline (PRO) biosynthesis is upregulated by water deficit.9 Scion cultivars differ in their response to water stress. For instance, under water stress, flavonol content and flavonol synthase transcripts at ripening stage were higher in Chardonnay than in Cabernet Sauvignon.9 The ABA metabolic pathway was affected for both cultivars, but interestingly, under water restriction, ABA concentration was also higher in Cabernet Sauvignon than in Chardonnay.
Currently, almost all vineyards over the world are grafted on rootstocks which are hybrids of three species: Vitis berlandieri, V. riparia, and V. rupestris. In addition to phylloxera resistance, grapevine rootstock genotypes are characterized by the vigor they confer to the scion, which may influence yield,17 grape berry quality,18 berry size, sugar content, anthocyanins, and organic acids19,20 under different irrigation treatments.12 The characterization of the hydraulic features of Vitis rootstocks showed a tight correlation between the vigor conferred by the rootstock and the expression of aquaporins (VvPIP2-1, VvPIP2-2) under well-watered conditions.21 Grafting induces massive transcriptional modifications in the shoot apical meristem of the grape scion.22
The selection and breeding of rootstocks improving water use efficiency is one of the key strategies to face climate change. Nevertheless, scion–rootstock interactions, especially under drought stress, are poorly documented at the molecular level. Although some pathways or enzymes affected by water deficit in grape berry have been characterized in the literature, to date no study aimed to evaluate the combined impact of rootstock and water stress on gene expression in the berries in field conditions. In our previous work, two different rootstock genotypes differing in their degree of tolerance to water stress, 110R (drought tolerant) and 125AA (drought sensitive) grafted with the same scion (cv. Pinot noir) were exposed to water limitation in the field, during three years (2009, 2010, and 2011).23 Due to yearly climate variations, the grapes only underwent a significant water stress in 2009 and 2010, and years 2009 and 2010 were therefore selected for detailed metabolite analysis. Under control conditions, predawn leaf water potential (ΨPD) did not differ between the two rootstocks, and was stable at about −0.10 MPa at harvest for both growing seasons. A weak to moderate water deficit for the two stressed rootstocks was observed during both seasons, with no significant differences of ΨPD between stressed rootstocks for both years.23 Water stress did not significantly affect berry mass, yield, sugar and organic acids concentration but increased anthocyanin concentration of the berries regardless of the rootstock. Furthermore, the data showed that the berry amino acid concentration at harvest depended on interactions between water supply and rootstocks.
In the present study, a genome-wide transcriptomic approach (microarray) was used to identify ontology categories and genes that were differentially expressed in grape berry grafted on 110R vs. 125AA under water stress, with a special focus on categories linked with grape berry quality.
Materials and methods
Field conditions and sampling
Grape berries from Pinot noir (Vitis vinifera) grafted on rootstocks Richter 110 (V. rupestris × V. berlandieri) and Kober 125AA (V. riparia × V. berlandieri), were sampled from field-grown vines at two different developmental stages (E-L 35 and E-L 36) according to the modified Eichhorn and Lorenz developmental scale,24 during two growing seasons (2009 and 2010). Vines were trained using a vertical shoot positioning system. The experimental vineyard belongs to the University of Geisenheim (49°59′N, 7°57′W, Germany). Twenty rows of Pinot noir were selected, with an average of 17 plants per row.
Two water conditions were established for the vines grafted on the two rootstocks: control (C) and water stress treatment (S). To impose water stress, half the rows of the vineyard chosen were covered with a removable plastic film from flowering onwards, covering completely the width of the inter-row on both sides of the plants during rainy days. Water stress was applied to the upper part of the vineyard (slope with 6–7% of inclination) and the control was situated at the lower part. Additional details about the training system, ripeness parameters (total soluble solids, titratable acidity, anthocyanins) were published previously.23 Three biological replicates (each including 85 berries from 17 plants per row) for each condition were collected in the vineyard. Sampling was carried out at two berry developmental stages, including 50% veraison when 50% of the berries of a cluster changed their color, and 100% veraison when all berries of a cluster colored. Berries were immediately frozen in liquid nitrogen and stored at −80 °C for further RNA extraction and gene expression analysis.
RNA extraction
Total RNA from berries of Pinot noir grafted onto 110R and 125AA was extracted as previously described.25 Seeds of each berry were removed before grinding in liquid nitrogen. Total RNA was subjected to DNA digestion with five units of RNase-free DNase I (Promega) for 1 h at 37 °C. RNA integrity was measured at 260 nm with a micro-spectrophotometer (NanoDrop 200C, NanoDrop products, Wilmington, USA) and visualized by electrophoresis on 1.2% agarose gels. RNA quality was also confirmed using a lab-on-chip (2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA). The RNA obtained was then used for quantitative polymerase chain reaction (qPCR) and microarray analysis.
Quantitative RT-PCR
Quantitative real-time PCR expression analysis was carried out using the CFX96 Real-Time PCR Detection system (Bio-Rad). For each sample, reverse transcription was performed with 2 µg of purified RNA using the Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer’s instructions. The cDNA obtained was diluted (1/20) in distilled water. Ten µL reaction mixes were prepared, that included 5 µL of iQTM SYBR Green Supermix (Bio-Rad), 0.2 µM of each primer and 2 µL of diluted berry cDNA. Gene transcripts were quantified upon normalization to VvGAPDH and VvEF1γ according to.26 Samples from 2009 and 2010 (12 samples for each treatment and rootstock combination) were tested with three technical replicates for each biological sample, and SD values of means were calculated using standard statistical methods. Specific oligonucleotides primer pairs were designed with Beacon Designer 7 software (Premier Biosoft International). Specific annealing of the oligonucleotides was controlled by dissociation kinetics performed at the end of each PCR run. The efficiency of each primer pair was measured on a PCR product serial dilution. Quantitative RT-PCR primer sequences are listed in Table S1.
Microarray experiment and analyses
The microarrays used were the grape whole-genome microarrays from NimbleGen microarrays 090818 Vitus exp HX12 (Roche, NimbleGen Inc., Madison, WI, USA), representing 29.549 genes based on the 12x genome assembly, using the grapevine V1 gene model predictions (http://genomes.cribi.unipd.it). The chip probe design is available at the following address, http://ddlab.sci.univr.it/FunctionalGenomics. The microarray contains 118.015 probes with an average of four probes per gene. The correspondence between probe identifiers and gene identifiers was obtained from http://genomes.cribi.unipd.it/. All the samples were hybridized following a complete randomized factorial design to take into account the variability through the glass-slide microarrays for broader statistical inference.27 Microarray hybridizations were done for 48 samples (two rootstock × two phenological stages × two years × two treatments × three biological replicates) by the Plateforme Biopuces (Institut National des Sciences Appliquées, Toulouse, France) according to the manufacturer’s instructions.
Microarray data were analyzed using the R28 and R/Bioconductor softwares.29 Quality control was done using the ArrayQualityMetrics package,30 and identified outliers were excluded from further analyses. Data were rma normalized31 and differentially expressed genes between stressed and control samples as well as stressed/control differentials between 110R and 125AA rootstocks were identified using the Limma package.32 Differentials with absolute fold changes above 1.5 and P-values below 0.05 were considered significant. Significantly affected gene categories were identified using a Wilcoxon rank sum test in R and the MapMan Ontology.33,34 Differentials for the different categories were visualized using PageMan.35 MapMan mappings for the Cribi 12X grapevine genome are based on closest homologs regarding to the Arabidopsis thaliana genome. Concerning results presented in the Tables and Supplemental Tables, each sequence corresponding to the ID indicated was manually blasted (blast n) against NCBI database. Microarray data have been deposited in the NCBI GEO database with accession number GSE66391.
Results and discussion
Differential gene expression triggered by water stress
The transcriptomic study was performed at two developmental stages that correspond to E-L 35 (50% veraison) and E-L 36 (100% veraison). Indeed, earlier studies reported that veraison corresponds to a key transcriptional reprogramming stage of cellular metabolism that determines most of the phenotypic traits of the ripe berry.36,37 RNA were extracted from berries sampled in 2009 and 2010 on 110R and 125AA plants grown under either control and water stress conditions.
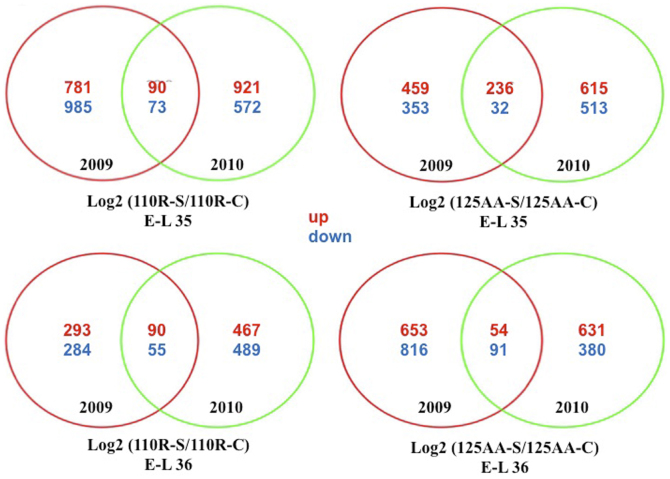
Venn diagrams (Figure 1) shows that at E-L 35, more genes were affected in berries grafted in 110R than in berries grafted on 125AA, while the reverse was true at E-L 36. The proportion of upregulated genes shared for both years ranged between 10 and 30% of the total number of upregulated genes, except for E-L 36 125AA, where this figure drops down to less than 10%. Among the genes differentially expressed for both years, more genes were upregulated than downregulated by water stress, except for E-L 36 125AA. The proportion of downregulated genes shared for both years is 10–20% (Figure 1).
Figure 1.
Two-way Venn diagram of the genes differentially expressed at E-L 35 and E-L 36 for the log2(110R-S/110R-C) and log2(125AA-S/125AA-C) conditions in 2009 and 2010.
Tables 1 and 2 detail the ontology categories that were identified by Wilcoxon rank sum test as more significantly affected by water stress than other categories at E-L 35 and E-L 36, respectively. For berries grafted on both rootstock genotypes and for both years studied, water stress affected mostly the same categories (‘cell wall,’ ‘lipid metabolism,’ ‘secondary metabolism,’ ‘aromatic amino acid metabolism,’ ‘stress,’ ‘regulation of transcription,’ ‘signaling,’ etc.). However, this analysis also underlines some specificity due to the rootstock. Indeed, at E-L 35, some categories were specifically affected for 125AA for both years. These included genes related to ‘major and minor carbohydrate metabolism,’ ‘TAG metabolism,’ ‘aspartate metabolism,’ ‘flavonoids dihydroflavonols,’ ‘jasmonate (JA) metabolism,’ ‘biotic stress,’ or ‘UDP glucosyltransferases’ (Table 1). In contrast, at the same stage, and for both years, transcript abundance of genes involved in ‘flavonoids flavonols,’ ‘polyamine synthesis,’ or ‘receptor kinases’ are more particularly affected in berries grafted on 110R (Table 1). At E-L 36, functional categories specifically affected in berries grafted on 125AA, encompass gene related to ‘cellulose synthesis’ and ‘receptor kinases LRR’ (Table 2). In contrast, the categories ‘PS light reactions,’ ‘fatty acid metabolism,’ ‘flavonols,’ ‘glutathione S transferases,’ and ‘ribosomal proteins’ were specifically affected in 110R only (Table 2). Altogether, the data show that water stress strongly impacts gene expression in the berries, through major categories that cover cell wall metabolism, secondary metabolism, signaling, stress, and hormones, and that some of these effects strongly dependent on the rootstock genotype.
Table 1. Significantly enriched ontology categories identified by Wilcoxon test at E-L 35 in 2009 and 2010 for Log2 110R-S/110R-C, and Log2 125AA-S/125AA-C respectively.
| Category name | Bin | 110R |
125AA |
||
|---|---|---|---|---|---|
| P-value | P-value | P-value | P-value | ||
| E-L 35 2009 | E-L 35 2010 | E-L 35 2009 | E-L 35 2010 | ||
| Major CHO metabolism | 2 | 4.0 × 10−2 | Not significant | 1.2 × 10−3 | 2.9 × 10−2 |
| Minor CHO metabolism.raffinose family | 3.1 | Not significant | Not significant | 6.9 × 10−3 | 1.1 × 10−2 |
| Cell wall.cellulose synthesis | 10.2 | Not significant | Not significant | 2.3 × 10−2 | 3.0 × 10−2 |
| Cell wall.degradation | 10.6 | 2.8 × 10−2 | 1.3 × 10−2 | 1.8 × 10−2 | Not significant |
| Lipid metabolism.TAG synthesis | 11.4 | Not significant | Not significant | 6.5 × 10−4 | 2.3 × 10−3 |
| Lipid metabolism.lipid degradation | 11.9 | 3.3 × 10−3 | 2.0 × 10−2 | 4.1 × 10−2 | 2.0 × 10−2 |
| Amino acid metabolism.synthesis.aspartate family | 13.1.3 | Not significant | Not significant | 8.8 × 10−3 | 9.7 × 10−3 |
| Amino acid metabolism.synthesis.aromatic aa | 13.1.6 | 3.7 × 10−2 | 1.1 × 10−2 | 4.8 × 10−2 | 2.3 × 10−5 |
| Secondary metabolism.isoprenoids | 16.1 | Not significant | 1.7 × 10−2 | 4.6 × 10−2 | 2.3 × 10−3 |
| Secondary metabolism.phenylpropanoids. lignin biosynthesis | 16.2.1 | 3.3 × 10−4 | 4.0 × 10−2 | 4.6 × 10−8 | 3.2 × 10−7 |
| Secondary metabolism.flavonoids. chalcones.naringenin-chalcone synthase | 16.8.2.1 | 2.9 × 10−13 | 1.8 × 10−5 | 0.0 | 0.0 |
| Secondary metabolism.flavonoids.di-hydroflavonols.flavonoid 3-monooxygenase | 16.8.3.3 | Not significant | Not significant | 4.0 × 10−2 | 1.3 × 10−2 |
| Secondary metabolism.flavonoids.flavonols | 16.8.4 | 2.4 × 10−2 | 7.5 × 10−6 | Not significant | Not significant |
| Hormone metabolism.jasmonate.synthesis-degradation | 17.7.1 | Not significant | Not significant | 6.9 × 10−3 | 1.9 × 10−2 |
| Stress.biotic | 20.1 | Not significant | 4.9 × 10−4 | 4.2 × 10−4 | 2.6 × 10−2 |
| Stress.abiotic | 20.2 | 6.5 × 10−9 | 3.0 × 10−4 | 1.6 × 10−3 | 1.2 × 10−4 |
| Polyamine metabolism.synthesis | 22.1 | 3.1 × 10−2 | 5.2 × 10−3 | Not significant | Not significant |
| Misc. UDP glucosyl and glucoronyl transferases | 26.2 | Not significant | Not significant | 2.7 × 10−2 | 3.6 × 10−4 |
| RNA.regulation of transcription | 27.3 | 2.4 × 10−2 | 4.2 × 10−2 | 2.1 × 10−3 | 3.2 × 10−3 |
| Protein.synthesis.ribosomal protein | 29.2.1 | Not significant | 0.0 | 9.7 × 10−5 | 2.2 × 10−4 |
| Signaling.receptor kinases.wheat LRK10 like | 30.2.20 | 1.2 × 10−2 | 3.2 × 10−4 | Not significant | Not significant |
| Development.late embryogenesis abundant | 33.2 | Not significant | 1.7 × 10−2 | 3.9 10−2 | 1.8 × 10−2 |
| Not assigned.no ontology. pentatricopeptide (PPR) repeat-containing protein | 35.1.5 | 1.3 × 10−2 | 7.3 × 10−20 | 7.2 × 10−6 | 1.7 × 10−7 |
Abbreviations: CHO, carbohydrate; 125AA, Kober 125AA (Vitis berlandieri × Vitis riparia); 110R, Richter 110 (Vitis berlandieri × Vitis rupestris).
Table 2. Significantly enriched ontology categories identified by Wilcoxon test at E-L 36 in 2009 and 2010 for Log2 110R-S/110R-C, and Log2 125AA-S/125AA-C respectively.
| Category name | Bin | 110R |
125AA |
||
|---|---|---|---|---|---|
| P-value | P-value | P-value | P-value | ||
| E-L 36 2009 | E-L 36 2010 | E-L 36 2009 | E-L 36 2010 | ||
| PS.lightreaction | 1.1 | 1.3 × 10−2 | 0.0 | Not significant | Not significant |
| Cell wall.cellulose synthesis | 10.4 | Not significant | Not significant | 2.8 × 10−2 | 1.1 × 10−2 |
| Cell wall.degradation | 10.6 | 3.7 × 10−2 | 7.6 × 10−6 | 1.6 × 10−2 | 3.1 × 10−2 |
| Lipid metabolism.FA synthesis and FA elongation | 11.1 | 2.8 × 10−2 | 1.9 × 10−3 | Not significant | Not significant |
| Lipid metabolism.lipid degradation | 11.9 | 2.6 × 10−2 | 1.5 × 10−3 | 7.5 × 10−3 | 7.8 × 10−3 |
| Secondary metabolism.isoprenoids | 16.1 | 3.0 × 10−2 | 3.4 × 10−2 | 3.6 × 10−2 | Not significant |
| Secondary metabolism.phenylpropanoids.lignin biosynthesis | 16.2.1 | 3.2 × 10−4 | 3.0 × 10−4 | Not significant | 1.4 × 10−9 |
| Secondary metabolism.sulfur-containing. glucosinolates.degradation | 16.5.1.3 | 2.8 × 10−4 | 7.2 × 10−6 | 1.4 × 10−4 | 8.0 × 10−3 |
| Secondary metabolism.flavonoids.chalcones. naringenin-chalcone synthase | 16.8.2.1 | 5.6 × 10−4 | 1.3 × 10−18 | 5.4 × 10−14 | 1.9 × 10−10 |
| Secondary metabolism.flavonoids.anthocyanins | 16.8.1 | 4.4 × 10−4 | 9.9 × 10−3 | Not significant | 3.8 × 10−2 |
| Secondary metabolism.flavonoids. dihydroflavonols.flavonoid 3-monooxygenase | 16.8.3.3 | 2.3 × 10−3 | 1.0 × 10−2 | 1.9 × 10−5 | 2.0 × 10−2 |
| Secondary metabolism.flavonoids.flavonols | 16.8.4 | 6.2 × 10−5 | 3.6 × 10−4 | Not significant | Not significant |
| Secondary metabolism.simple phenols | 16.10 | 1.2 × 10−7 | 3.8 × 10−5 | Not significant | 1.4 × 10−3 |
| Hormone metabolism.jasmonate.synthesis-degradation | 17.7.1 | 1.1 × 10−2 | 4.9 × 10−2 | 1.3 × 10−2 | Not significant |
| Stress.abiotic | 20.2 | 1.0 × 10−3 | 5.5 × 10−6 | 6.9 × 10−5 | 2.9 × 10−2 |
| Stress.biotic | 20.1 | 6.1 × 10−3 | 2.1 × 10−4 | 1.5 × 10−3 | 1.2 × 10−7 |
| Redox | 21 | 8.4 × 10−3 | 5.5 × 10−3 | 2.2 × 10−2 | Not significant |
| Misc.glutathione S transferases | 26.9 | 8.2 × 10−3 | 4.9 × 10−2 | Not significant | Not significant |
| RNA.regulation of transcription | 27.3 | 2.7 × 10−2 | 4.8 × 10−2 | 2.4 × 10−3 | 1.6 × 10−2 |
| Protein.synthesis.ribosomal protein | 29.2.1 | 2.9 × 10−13 | 1.9 × 10−2 | Not significant | Not significant |
| Signaling.receptor kinases.leucine rich repeat XI | 30.2.11 | Not significant | Not significant | 3.6 × 10−2 | 2.9 × 10−2 |
| Signaling.receptor kinases.thaumatin like | 30.2.15 | 1.7 × 10−3 | 1.0 × 10−11 | 3.2 × 10−8 | 1.0 × 10−5 |
| Signaling.receptor kinases.wheat LRK10 like | 30.2.20 | 1.7 × 10−3 | 3.4 × 10−7 | 1.3 × 10−15 | 3.1 × 10−8 |
| Development.late embryogenesis abundant | 33.2 | 1.4 × 10−4 | 4.6 × 10−2 | Not significant | 3.0 × 10−4 |
| Transport | 35.1.5 | Not significant | 3.9 × 10−3 | 2.5 × 10−3 | 9.6 × 10−3 |
Abbreviations: 125AA, Kober 125AA (Vitis berlandieri × Vitis riparia); 110R, Richter 110 (Vitis berlandieri × Vitis rupestris).
Some of the genes differentially expressed under water deficit for both years are detailed in supplemental Tables S2 (110R at E-L 35), S3 (125AA at E-L 35), S4 (110R at E-L 36), and S5 (125AA at E-L 36).
Differential gene expression triggered by water stress between the berries grafted on the two rootstocks
In order to highlight the effect of rootstock under drought stress conditions on the transcriptional response of the berries, the ratios 110R-stressed (S)/110R-control (C) vs. 125AA-S/125AA-C were compared by using the relation log2(110R-S/110R-C) – log2(125AA-S/125AA-C). A positive ratio (log2(110R-S/110R-C) – log2(125AA-S/125AA-C)) means that genes are more induced or less repressed by drought stress in berries grafted on 110R (abbreviated: ‘110R’ condition) in comparison to berry grafted on 125AA (abbreviated: ‘125AA’ condition). Conversely, a negative ratio (log2(110R-S/110R-C) – log2(125AA-S/125AA-C)) means that genes are more induced or less repressed by drought stress in 125AA berries than in 110R berries.
Using Wilcoxon rank sum test, we identified functional categories differentially affected among the genes differentially expressed between 110R and 125AA under drought stress (Table 3). Interestingly, genes involved in secondary metabolism (phenylpropanoid and flavonoids), hormone metabolism (JA), and biotic stress response (pathogenesis-related proteins (PR proteins)) were among the highest differentially expressed genes (P-value < 0.05) for both years (2009 and 2010) and both developmental stages (E-L 35 and E-L 36; Table 3).
Table 3. Significantly enriched ontology categories identified by Wilcoxon test for the log2(110R-S/110R-C) – log2(125AA-S/125AA-C) condition that are common for the two developmental stages in 2009 and 2010.
| Category name | Bin | P-value | P-value | P-value | P-value |
|---|---|---|---|---|---|
| E-L 35 2009 | E-L 36 2009 | E-L 35 2010 | E-L 36 2010 | ||
| Secondary metabolism | 16 | 2.9 × 10−3 | 4.8 × 10−10 | 9.6 × 10−4 | 5.7 × 10−8 |
| Phenylpropanoids | 16.2 | 4.7 × 10−3 | 5.4 × 10−4 | 7.9 × 10−3 | 4.6 × 10−7 |
| Flavonoids | 16.8 | 2.0 × 10−2 | 6.9 × 10−5 | 3.3 × 10−3 | 1.0 × 10−5 |
|
Hormones metabolism. Jasmonate | |||||
| Synthesis degradation | 17.7.1 | 3.5 × 10−2 | 1.5 × 10−2 | 2.1 × 10−3 | 1.7 × 10−2 |
| Biotic stress | 20.1 | 5.5 × 10−3 | 2.3 × 10−6 | 2.26 × 10−6 | 4.3 × 10−6 |
| PR proteins | 20.1.7 | 5.8 × 10−4 | 7.4 × 10−3 | 9.3 × 10−14 | 4.1 × 10−12 |
Abbreviation: PR proteins, pathogenesis-related proteins.
The next section will describe and discuss the functional categories cited above. Given that some differences in amino acid composition were described earlier with these samples,23 we will also detail the genes related to amino acid metabolism. Some of the genes differentially expressed in the microarray data were confirmed by qPCR in the same samples (from 2009 to 2010) and results are presented in Table S6. The direction of gene expression changes could generally be repeated by qPCR; however, the magnitude of change was frequently increased in the qPCR data.
Effects of rootstock genotype on phenolic compounds metabolism under water deficit
The phenolic compounds derived from the shikimic acids pathway are precursors of simple phenols, lignin, flavonoids, isoflavonoids, coumarin, and stilbenes. Polyphenolic compounds play an important role in the quality of grapes and wines.38 Flavonoids are important determinants of quality in red grapes and wines. Color and taste of red wines are strongly related to the amount of anthocyanins, flavonols, and proanthocyanidins. Many of these phenolics may be beneficial for human health.39
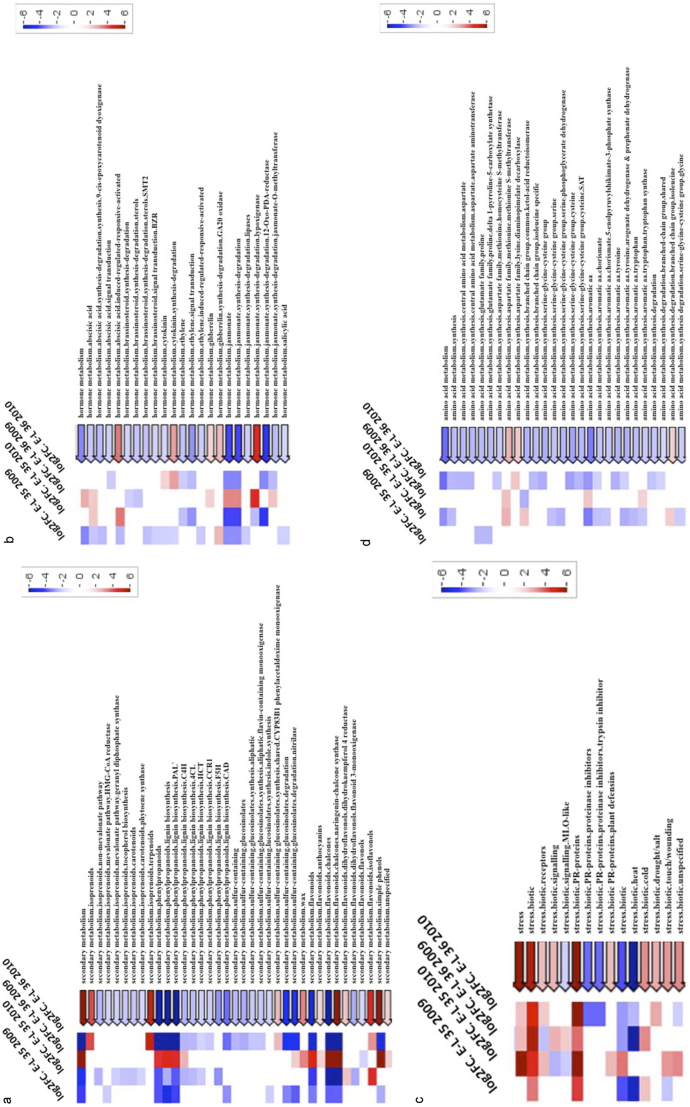
Our transcriptomic data agree with previous observations according to which water deficit affects phenylpropanoid metabolism and increases the berry concentration in anthocyanins and other phenolics.10,11,40 Furthermore, we show that the effects of water deficit on the expression of genes of this family in the berries depend on the rootstock. Indeed, the Pageman representation shows that, overall, genes from the functional category ‘secondary metabolism’ (more particularly phenylpropanoids and flavonoids) are more induced or less repressed by drought stress in berries grown on the drought-sensitive rootstock 125AA compared to the drought-tolerant rootstock 110R, except for E-L 36 developmental stage in 2009 (Figure 2A).
Figure 2.
Pageman visualization of MapMan functional categories: secondary metabolism (A), hormone metabolism (B), stress (C), and amino acid metabolism (D), enriched in the genes differentially expressed between berries grafted on 110R and berries grafted on 125AA, under drought stress. The degree of enrichment of functional categories in up- and downregulated genes for the log2(110R-S/110R-C) – log2(125AA-S/125AA-C) condition is given by shades of red and blue, respectively.
Both water deficit in the field and postharvest dehydration increase the accumulation of phenylalanine ammonia lyase (PAL) transcripts8,9,41 which may indicate a general activation of the phenylpropanoid pathway. No evidence for any significant differential regulation of PAL genes between 125AA and 110R was found. However, other genes involved in the first steps of the phenylpropanoid pathway are differentially expressed between 110R and 125AA under drought stress: a 4-coumarate-CoA ligase (4CL, Vv18s0001g00290) and a trans-cinnamate 4-monooxygenase (C4H, Vv11s0078g00290) in E-L 35 2010 (Table 5) and E-L 36 2009 (Table 6), respectively. The C4H gene is the most differentially affected by water deficit between the two rootstocks due to both its upregulation (1.6X) in 110R stressed berries coupled with a downregulation (2X) in 125AA stressed berries (Table 6; Figure 2A).
Table 5. Selected genes affected in condition E-L 35 (2010) associated with JA biosynthesis, phenylpropanoid metabolism, PR proteins, and amino acid metabolism and transport ontology categories.
| Name | ID | Fold change Log2(110R-S/110R-C) – log2(125AA-S/125AA-C) | P-value | Fold change Log2(110R-S/110R-C) | Fold change Log2(125AA-S/125AA-C) |
|---|---|---|---|---|---|
|
Jasmonate synthesis-related genes | |||||
| Jasmonate-O-methyltransferase | Vv18s0001g12900 | −1.18 | 0.01 | −0.40 | 0.78 |
|
Phenylpropanoids | |||||
| 4CL | Vv18s0001g00290 | −0.63 | 0.00 | −0.28 | 0.35 |
| Simple phenols | Vv08s0007g01910 | 1.77 | 0.01 | 1.25 | −0.52 |
| Laccase 1a | Vv09s0018g00950 | 0.94 | 0.04 | 1.11 | 0.17 |
| Laccase | Vv11s0037g00440 | −1.09 | 0.04 | −0.36 | 0.66 |
| Lignins | Vv18s0001g14910 | −1.02 | 0.00 | −0.20 | 0.89 |
| HCT | Vv15s0046g01960 | −0.78 | 0.04 | −0.31 | 0.47 |
| CAD | Vv00s0218g00190 | −0.65 | 0.00 | −0.06 | 0.59 |
| Flavonoids | Vv04s0044g01530 | 0.90 | 0.04 | 0.24 | −0.66 |
| Anthocyanin 3-glucoside rhamnosyltransferase | |||||
| Anthocyanin 3-glucoside rhamnosyltransferase | Vv00s0361g00030 | 0.94 | 0.01 | 0.69 | −0.25 |
| Cyanidin-3-O-glucoside 2-O-glucuronosyltransferase | |||||
| ANR | Vv03s0088g00060 | 2.30 | 0.03 | 2.34 | 0.03 |
| Isoflavone reductase 2 | |||||
| Isoflavone reductase 2 | Vv03s0088g00250 | 1.35 | 0.01 | 1.37 | 0.01 |
| Isoflavone reductase 2 | Vv03s0088g00140 | 0.90 | 0.04 | 1.00 | 0.10 |
| Isoflavone reductase 6 | Vv03s0038g04620 | 0.60 | 0.03 | 0.52 | −0.08 |
|
PR proteins | |||||
| Endochitinase PR4-like | Vv05s0094g00270 | −0.90 | 0.02 | −0.35 | 0.55 |
| Basic form of pathogenesis-related protein 1 | Vv03s0088g00940 | −0.95 | 0.00 | −0.15 | 0.79 |
|
Amino acid metabolism | |||||
| AAAs | |||||
| Arogenate dehydrogenase | Vv09s0002g08060 | 0.80 | 0.01 | 0.65 | −0.15 |
| Chorismate mutase | Vv01s0010g00480 | 0.99 | 0.03 | 1.42 | 0.44 |
| Methionine | |||||
| MMT | Vv18s0089g01290 | 0.66 | 0.03 | 0.20 | −0.46 |
|
Amino acid transport | |||||
| Amino acid perméase | Vv04s0008g00230 | 1.13 | 0.03 | 1.70 | 0.57 |
| LHT | Vv00s0302g00030 | 0.94 | 0.02 | 0.52 | −0.42 |
| LHT | Vv01s0010g01490 | 0.74 | 0.02 | 0.59 | −0.15 |
Abbreviations: AAAs, aromatic amino acids; ANR, anthocyanidin reductase; CAD, cinnamyl alcohol dehydrogenase; 4CL, 4-coumarate-CoA ligase; HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; JA, jasmonate; 125AA, Kober 125AA (Vitis berlandieri × Vitis riparia); LHT, lysine/histidine transporter; MMT, methionine S-methyltransferase; PAT, glutamate/aspartate-prephenate aminotransferase; PR proteins, pathogenesis-related proteins; 110R, Richter 110 (Vitis berlandieri × Vitis rupestris).
Table 6. Selected genes affected in condition E-L 36 (2009) associated with JA biosynthesis, phenylpropanoid metabolism, PR proteins, amino acid metabolism, and polyamines ontology categories.
| Name | ID | Fold change Log2(110R-S/110R-C) – log2(125AA-S/125AA-C) | P-value | Fold change Log2(110R-S/110R-C) | Fold change Log2(125AA-S/125AA-C) |
|---|---|---|---|---|---|
|
Jasmonate synthesis-related genes | |||||
| Jasmonate-O-methyltransferase | Vv04s0023g02290 | 1.06 | 0.01 | 0.68 | −0.38 |
|
Phenylpropanoids | |||||
| C4H | Vv11s0078g00290 | 1.55 | 0.03 | 0.67 | −0.88 |
| Simple phenols | |||||
| Laccase 15 | Vv18s0075g01090 | −1.20 | 0.02 | −0.12 | 1.08 |
| Flavonoids | |||||
| Flavonoid 3-hydroxylase | Vv07s0031g01380 | 0.71 | 0.02 | −0.33 | −1.04 |
| LDOX | Vv08s0105g00380 | 0.63 | 0.02 | 0.05 | −0.58 |
| Anthocyanin 3-O-glucoside-6″-O-malonyltransferase | Vv12s0134g00590 | −0.85 | 0.02 | −0.14 | 0.71 |
| Flavanone 3-dioxygenase | Vv16s0098g00860 | 0.84 | 0.02 | −0.19 | −1.03 |
| Stilbenes | |||||
| STS | Vv16s0100g00770 | 1.22 | 0.00 | −0.25 | −1.46 |
| STS | Vv16s0100g01140 | 1.07 | 0.03 | 0.05 | −1.02 |
|
PR proteins | |||||
| Endochitinase PR4-like | Vv05s0094g00200 | 0.87 | 0.03 | 0.22 | −0.65 |
|
Amino acid metabolism | |||||
| Aminotransferase ALD1 | Vv12s0055g00920 | 1.24 | 0.02 | −0.46 | −1.70 |
| Polyamines | |||||
| Ornithine decarboxylase | Vv18s0001g00740 | 1.35 | 0.01 | −0.24 | −1.59 |
Abbreviations: JA, jasmonate; 125AA, Kober 125AA (Vitis berlandieri × Vitis riparia); LDOX, leucoanthocyanidin dioxygenase; PR proteins, pathogenesis-related proteins; 110R, Richter 110 (Vitis berlandieri × Vitis rupestris); STS, stilbene synthase; C4H, trans-cinnamate 4-monooxygenase.
In addition to the upstream steps, downstream steps of the phenylpropanoid pathway were also affected. Water deficit accelerates ripening and alters flavonoid biosynthesis, especially the anthocyanin pathway.11 General biosynthetic genes such as DFR (Vv15s0048g00980) and F3H (Vv07s0031g01380 and Vv16s0098g00860) were differentially expressed between 110R and 125AA in 2009 (Tables 4 and 6): DFR was downregulated by drought stress in 110R in comparison to 125AA for E-L 35 (Table 4). At E-L 36, F3H genes were more downregulated by drought stress in 125AA than in 110R (Table 6). Besides these general biosynthetic genes, genes related to anthocyanin modification (including flavonoid 3′5′-hydroxylase and O-methyltransferase) can also be upregulated in water stressed berries.13 Our results further show that the rootstocks also participated in the control of genes related to anthocyanins modification, more particularly anthocyanin 3-O-glucoside-6′-O-malonyltransferase (Vv12s0134g00590), UDP rhamnose-anthocyanidin-3-glucoside-rhamnosyltransferase (Vv19s0014g02020, Vv15s0046g01960, Vv00s0218g00190) and cyanidin-3-O-glucoside-2-O-glucuronosyltransferase (Vv04s0044g01530; Tables 4, 5, 6).
Table 4. Selected genes affected in condition E-L 35 (2009) associated with JA biosynthesis, phenylpropanoid metabolism, PR proteins, amino acid metabolism, and transport and polyamines ontology categories.
| Name | ID | Fold change Log2(110R-S/110R-C) – log2(125AA-S/125AA-C) | P-value | Fold change Log2(110R-S/110R-C) | Fold change Log2(125AA-S/125AA-C) |
|---|---|---|---|---|---|
|
Jasmonate synthesis-related genes | |||||
| Phospholipase A1 | Vv15s0021g01510 | −1.13 | 0.04 | −0.78 | 0.35 |
| Linoleate 13S-lipoxygenase | Vv06s0004g01470 | −1.13 | 0.01 | −0.40 | 0.56 |
|
Phenylpropanoids | |||||
| Simple phenols | |||||
| Laccase 15 | Vv18s0075g00950 | 0.95 | 0.00 | 0.40 | −0.56 |
| Multicopper oxidase | Vv18s0164g00110 | −0.65 | 0.02 | −0.46 | 0.18 |
| Laccase 1a | Vv08s0007g01910 | −0.75 | 0.03 | −0.69 | 0.06 |
| Laccase 15 | Vv13s0019g02160 | −0.80 | 0.01 | −0.16 | 0.64 |
| Laccase 110b | Vv08s0007g00600 | −0.82 | 0.02 | −0.21 | 0.61 |
| Laccase 11 | Vv04s0069g00950 | −0.96 | 0.02 | −0.52 | 0.44 |
| Laccase 1 | Vv17s0000g02590 | −1.06 | 0.00 | −0.45 | 0.61 |
| Laccase | Vv13s0019g0194 | −1.11 | 0.02 | −0.77 | 0.34 |
| Lignins | |||||
| HCT | Vv11s0037g00590 | −0.75 | 0.03 | −0.47 | 0.29 |
| Sinapyl alcohol dehydrogenase | Vv00s0346g00100 | −0.94 | 0.05 | −0.58 | 0.36 |
| CAD | Vv13s0019g05260 | −1.02 | 0.02 | −0.96 | 0.06 |
| CCoAOMT | Vv01s0010g03460 | −0.67 | 0.04 | −0.41 | 0.27 |
| COMT | Vv18s0001g02610 | −0.99 | 0.02 | −0.34 | 0.65 |
| Flavonoids | |||||
| 8-hydroxyquercetin 8-O-methyltransferase | Vv12s0028g02890 | 0.68 | 0.01 | 0.33 | −0.35 |
| Quercetin 3-O-methyltransferase | Vv15s0048g02490 | 1.01 | 0.01 | 0.53 | −0.48 |
| Anthocyanin 3-O-glucoside-6″-O-malonyltransferase | Vv12s0134g00600 | 0.79 | 0.00 | 0.33 | −0.46 |
| UDP rhamnose-anthocyanidin-3-glucoside rhamnosyltransferase | Vv19s0014g02020 | −0.89 | 0.03 | −1.07 | −0.18 |
| ANR | Vv00s0361g00030 | 1.07 | 0.04 | 0.48 | −0.59 |
| DFR | Vv15s0048g00980 | −1.08 | 0.02 | −0.67 | 0.41 |
| Isoflavone reductase 3 | Vv07s0031g03070 | −1.76 | 0.01 | −0.89 | 0.86 |
|
PR proteins | |||||
| Endochitinase PR4-like | Vv05s0094g00220 | −1.05 | 0.05 | −0.35 | 0.70 |
|
Amino acid metabolism | |||||
| AAAs | |||||
| Arogenate dehydratase | Vv10s0116g01670 | −1.30 | 0.00 | −0.99 | 0.31 |
| Arogenate dehydrogenase | Vv09s0002g08030 | 0.74 | 0.04 | 0.60 | −0.15 |
| PAT | Vv14s0083g00900 | −1.13 | 0.02 | −0.72 | 0.41 |
| Methionine | |||||
| MET6 | Vv10s0042g00970 | −1.02 | 0.03 | −0.72 | 0.30 |
| Lysine | |||||
| Aminotransferase ALD1 | Vv18s0001g04630 | 0.75 | 0.03 | 0.57 | −0.19 |
| Valine. leucine. isoleucine | |||||
| Acetolactate synthase | Vv16s0022g01100 | −1.23 | 0.02 | −0.86 | 0.37 |
| Amino acid transport | |||||
| AAP | Vv00s0188g00130 | −1.04 | 0.01 | −0.59 | 0.44 |
| AAP | Vv04s0008g00230 | −1.41 | 0.00 | −0.71 | 0.70 |
| Polyamines | |||||
| Ornithine decarboxylase | Vv18s0001g00740 | 0.81 | 0.01 | 1.08 | 0.27 |
| S-adenosylmethionine decarboxylase | Vv14s0083g00580 | 0.60 | 0.01 | 0.62 | 0.02 |
Abbreviations: AAAs, aromatic amino acids; AAP, amino acid permease; ANR, anthocyanidin reductase; CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA O-methyltransferase; COMT, caffeic acid 3-O-methyltransferase; DFR, dihydroflavonol-4-reductase; HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; JA, jasmonate; 125AA, Kober 125AA (Vitis berlandieri × Vitis riparia); PAT, glutamate/aspartate-prephenate aminotransferase; PR proteins, pathogenesis-related proteins; 110R, Richter 110 (Vitis berlandieri × Vitis rupestris).
Flavonols, which are also products of the flavonoid biosynthetic pathway, contribute to the bitter taste and color of red wine by stabilizing anthocyanin pigments.38 Deficit irrigation moderately affects flavonol synthesis in red grapevines42 and Chardonnay.8,9 Like anthocyanins, changes induced by water deficit affect more the range of flavonols than their global concentration.43 In our study, two genes encoding flavonol modifying enzymes were differentially expressed between 110R and 125AA under drought stress: 8-hydroxyquercetin-8-O-methyltransferase (Vv12s0028g02890) and quercetin-3-O-methyltransferase (Vv15s0048g02490) at E-L 35 2009 (Table 4).
Isoflavonoids are defense compounds mostly found in legumes44 that may have beneficial effects in humans and animals.45 Little information is available related to the involvement of isoflavonoids in grape ripening. IFR6 (isoflavone reductase) which catalyzes the reduction of isoflavones to isoflavonones decreases from veraison to the end of ripening.46 Accumulation of isoflavones is promoted by well-watered conditions and long-term progressive drought significantly decreases isoflavone content,47 but the molecular basis of this response remains elusive. Proteomic studies showed that IFR6 protein is more abundant in the pulp of berries under water-deficit stress,48 which is in agreement with its transcript profile.8 Our study also suggests a role for IFR in drought stress response since IFR3 (Vv07s0031g03070) in 2009 (Table 4), IFR2 (Vv03s0088g00060), and IFR6 (Vv03s0038g04620) in 2010 (Table 5) are strongly differentially expressed between 110R and 125AA under water deficit at E-L 35.
Proanthocyanidins or condensed tannins are flavan-3-ol oligomers responsible for wine bitterness and astringency. Changes occurring in proanthocyanidins during grape development are complex, involving increases in the degree of polymerization, in the proportion of (-)-epigallocatechin extension units, and in polymer-associated anthocyanins.49 In grapevine, both anthocyanidin reductase (ANR) and leucoanthocyanidin reductase contribute to proanthocyanidins synthesis in leaves, flowers, and in the skin and seeds of the developing fruit.50 In seeds, water deficit does not affect ANR transcript levels.8 The slight increases in skin tannin that accompany water deficit51 would result more from differential growth sensitivity of the inner mesocarp and the exocarp than from direct effects on phenolic biosynthesis.10 Our study differs somewhat from these earlier data since we found a differential expression of ANR (Vv00s0361g00030, for E-L 35 2009 and 2010 (Tables 4 and 5)), (Vv00s0361g00040 for E-L 36 2010 (Table 7)), and LDOX (Vv08s0105g00380, for E-L 36 2009 (Table 6)) between 110R and 125AA under water deficit.
Table 7. Selected genes affected in condition E-L 36 (2010) associated with JA biosynthesis, phenylpropanoid metabolism, PR proteins, and amino acid transport ontology categories.
| Name | ID | Fold change Log2(110R-S/110R-C) – log2(125AA-S/125AA-C) | P-value | Fold change Log2(110R-S/110R-C) | Fold change Log2(125AA-S/125AA-C) |
|---|---|---|---|---|---|
|
Jasmonate synthesis-related genes | |||||
| Jasmonate-O-methyltransferase | Vv04s0023g02240 | −0.75 | 0.01 | −0.22 | 0.53 |
|
Phenylpropanoids | |||||
| Simple phenols | |||||
| Laccase 15 | Vv18s0075g01090 | −1.19 | 0.04 | −1.03 | 0.16 |
| Laccase | Vv18s0117g00480 | −1.76 | 0.02 | −1.37 | 0.39 |
| Laccase | Vv00s0444g00010 | −1.95 | 0.03 | −1.51 | 0.44 |
| Laccase | Vv18s0117g00450 | −1.99 | 0.03 | −1.49 | 0.50 |
| Laccase | Vv18s0075g00610 | −2.01 | 0.04 | −1.52 | 0.49 |
| Laccase | Vv18s0075g00780 | −2.43 | 0.04 | −1.84 | 0.59 |
| Laccase | Vv00s1212g00020 | −2.43 | 0.02 | −2.08 | 0.36 |
| Laccase | Vv18s0075g00670 | −2.48 | 0.03 | −1.87 | 0.61 |
| Laccase | Vv00s0731g00010 | −2.83 | 0.02 | −2.41 | 0.42 |
| Flavonoids | |||||
| ANR | Vv00s0361g00040 | 0.51 | 0.04 | 0.53 | 0.02 |
| Stilbenes | |||||
| STS | Vv16s0100g01110 | −0.61 | 0.01 | −0.45 | 0.16 |
| STS | Vv16s0100g01190 | −2.16 | 0.03 | −1.60 | 0.56 |
|
PR proteins | |||||
| PR-1 protein | Vv03s0088g00780 | −1.05 | 0.04 | −0.51 | 0.54 |
| Basic form of pathogenesis-related protein 1 | Vv03s0088g00710 | −1.95 | 0.02 | −0.98 | 0.97 |
|
Amino acid transport | |||||
| LHT | Vv01s0010g02500 | −0.63 | 0.04 | −0.68 | −0.05 |
Abbreviations: ANR, anthocyanidin reductase; JA, jasmonate; 125AA, Kober 125AA (Vitis berlandieri × Vitis riparia); LHT, lysine/histidine transporter; PR proteins, pathogenesis-related proteins; 110R, Richter 110 (Vitis berlandieri × Vitis rupestris); STS, stilbene synthase.
Lignins are cell wall phenolic heteropolymers mainly synthesized by the oxidative coupling of p-hydroxycinnamyl alcohol monomers. Lignification and biosynthesis of lignin precursors is upregulated by drought.52 The general phenylpropanoid pathway is branched to the biosynthesis of monolignol by two enzymatic steps catalyzed by cinnamoyl-CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD). CAD enzyme may also be involved in the synthesis of cinnamyl alcohol derivatives responsible for fruit flavor and aroma.53 In the present study, CAD genes are less repressed (Vv13s0019g05260) or more induced (Vv18s0001g14910) by drought stress in 125AA than in 110R berries at E-L 35 in 2009 and 2010, respectively (Tables 4 and 5). Another alcohol dehydrogenase (SAD, Vv00s0346g00100) was also more repressed by drought stress in 110R than in 125AA berries at E-L 35 in 2009 (Table 4). A similar expression pattern was observed for other transcripts involved the biosynthesis of monolignols, such as caffeoyl-CoA O-methyltransferase (CCoAOMT, Vv01s0010g03460) and caffeic acid 3-O-methyltransferase (COMT, Vv18s0001g02610; Table 4). Both CCoAOMT and CAD transcrits were upregulated during water stress in rice54 and Citrullus lanatus sp.55 Another key lignin biosynthesis-related gene, HCT (Vv11s0037g00590, Vv11s0037g00590), encoding hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, is more positively regulated by drought stress in 125AA than in 110R berries at E-L 35 for both years (Tables 4 and 5). HCT leads to the biosynthesis of two major monolignols, namely, the guaiacyl and syringyl units.56 In line with our results, drought treatment caused an early and biphasic expression of HcHCT in Hibiscus cannabinus.57
Stilbenes are other nonflavonoid compounds present in small quantities in wine.58 Their synthesis increases upon pathogen infection and in response to abiotic stress. Stilbene synthase (STS) is the key enzyme leading to the biosynthesis of resveratrol, which may be beneficial to human health. Water deficit increases the amounts of STS transcripts, which suggests an increase in resveratrol accumulation.8 However, little effect of drought was observed on resveratrol concentrations in skins.59 Water deficit increased the accumulation of trans-piceid (the glycosylated form of resveratrol) in Cabernet Sauvignon but not in Chardonnay berries.60 In the present study, four genes encoding STSs were differentially expressed under water stress depending on the rootstock genotype. Two genes (Vv16s0100g00770, Vv16s0100g01140) were downregulated in 125AA berries in 2009 at E-L 36 (Table 6) and two others (Vv16s0100g01110, Vv16s0100g01190) were strongly downregulated in 110R berries at E-L 36 in 2010 (Table 7).
Finally, several laccase-encoding genes were strongly differentially expressed between 110R and 125AA at both developmental stages for both years (Tables 4, 5, 6, 7). Laccases are widely present in eukaryotes and in prokaryotes.61 In addition to their role in the oxidative polymerization of monolignols, they are involved in the oxidative polymerization of flavonoids.62 Although valued in tea and wine processing, browning reactions are particularly detrimental for fresh fruits.63 Laccases play a role in defending the plant against various biotic and abiotic stresses64 more particularly during acclimation to salinity stress.65 The present study identified several laccases such as LAC 15, 1a, 1, 110B, and 11 that were differentially expressed in berries grown on 110R and 125AA stressed rootstocks (Tables 4, 5, 6, 7). Overall, they were found to be more induced or less repressed in 125AA than in 110R berries for both years (Tables 4, 6, and 7).
Effects of rootstock genotype on hormone metabolism under water deficit
The rootstock genotype may affect JAs metabolism under water stress condition. For both years and both developmental stages, transcripts related to JAs metabolism were significantly more induced or less repressed in 125AA than in 110R berries (except for E-L 36 2009; Figure 2B). JAs is well known to regulate responses to wounding and defense against pathogens but has also been reported to play a role in drought response.66 For instance, JAs were found to cause stomatal closure52 and JA-insensitive mutants (jar1 and coi1) 67,68 show impaired stomatal response to exogenous JAs. There is a cross-talk between JAs and ABA because they use a similar cascade of events to promote stomatal closure.68 Furthermore, a role of JAs in plant response to water deficit has been suggested because this stress induces the expression of several genes that also respond to JAs.69 Microarray analysis in wild-type and coi1 Arabidopsis plants that were wounded or exposed to water stress reveals a surprisingly large overlap of COI1-dependent genes. Moreover, like ABA, JAs levels were increased by drought stress in soybean70 and Pinus pinaster.71 The same was observed with rice, for which drought and high salinity increased leaf and root JAs, resulting in the induction of stress-related PR and JAs biosynthetic genes.72 However, these abiotic stress-induced increases in JAs levels were observed only in vegetative tissues. Whether drought conditions increase JAs levels in reproductive organs remains to be determined. Our data emphasize that JAs may play a role in grape berry acclimation during drought stress. Indeed, key JAs biosynthesis genes such as phospholipase A1 (Vv15s0021g01510), linoleate 13S-lipoxygenase (Vv06s0004g01470), and jasmonate-O-methyltransferase (Vv18s0001g12900, Vv04s0023g02290, Vv04s0023g02240) were differentially regulated under water deficit between the drought-tolerant rootstock 110R and the drought-sensitive rootstock 125AA (Tables 4, 5, 6, 7).
JA compounds regulate the biosynthesis of many different types of secondary metabolites in different plant species including alkaloids, terpenoids, glucosinolates, and phenylpropanoids.73 For instance, exposure of Hypericum perforatum L. suspension culture to JA upregulates the expression of PAL and chalcone isomerase (CHI) genes. This observation was correlated with an increased production of phenylpropanoids such as phenolic, flavanol and flavonol in JA-treated cells.74 In Arabidopsis, JAs induce anthocyanin accumulation through the upregulation of the ‘late’ anthocyanin biosynthetic genes DFR, LDOX, and UFGT.75 In grapes, JAs promote the synthesis and accumulation of stilbenes in berry cell cultures76 and during berry development.59 The present work pinpoints a possible link between JAs and phenylpropanoid pathway in grape berry under water-deficit condition.59 Indeed, genes related to JA metabolism and those related to phenylpropanoid/flavonoid metabolism follow a similar expression pattern for both developmental stages and for both years (Figure 2A and 2B). At E-L 35 (2009 and 2010) and E-L 36 (2010), the genes related to these two functional categories are more induced or less repressed in berries grafted on 125AA than on those grafted on 110R (Figure 2A and 2B). At E-L 36 in 2009, those genes were more induced or less repressed in 110R than in 125AA berries (Figure 2A and 2B).
Effects of rootstock genotype on pathogen-related proteins under water stress
JA is a well-known inducer of PR protein during plant defense responses.77 In grapevine, the most frequent and best characterized defense reactions to fungal infection are the accumulation of phytoalexins and pathogen-related PR proteins.78 The latter accumulate in berries and leaves upon pathogen attack and may contribute to grapevine resistance.79 Besides, due to their resistance to proteolysis and stability at acidic pH, PR proteins are the most abundant class of protein present in wines and adversely affect their clarity and stability.80,81 PR proteins include 17 categories of structurally and functionally unrelated proteins,82 some on which have been detected in grapevine under water stress conditions.83 PR proteins designated as PR 1, 2, 3, 4, 5, 6, 10, and 14 are differentially expressed in several berry tissues.8 The expression pattern of genes related to stress in the present study is shown in Figure 2C. Among them, the transcript abundance of some specific PR genes differed between 110R and 125AA under drought stress, e.g. PR-4 and PR1 (Tables 4, 5, 6, 7). Interestingly, PR4 has been reported to be one of the markers for the JA pathway84 and the subfamily PR-1 may be involved in the response to environmental stresses82 such as drought stress.85
Effects of rootstock genotype on amino acid metabolism under water stress
The discussion above highlighted the functional categories differentially expressed between 110R and 125AA under drought stress for both developmental stages and both years. Our previous study, describing the effects of rootstock genotype under water stress on berry composition, revealed that specific amino acids accumulated more in 125AA than in 110R berries.23 Even though the ‘amino acid metabolism’ functional category as a whole is not statistically affected (Table 3), some genes belonging to this category display differential expression pattern between 110R and 125AA under water deficit (Tables 4, 5, 6). Figure 2D shows that transcripts related to ‘amino acids synthesis’ were more induced or less repressed in 125AA than in 110R berries in drought stress conditions. This observation fits well with our biochemical analysis showing that berries from 125AA accumulated more amino acids than 110R berries.23 Furthermore, genes related to aromatic amino acids (AAAs) (Figure 2D) present the same expression pattern as the ontology categories ‘secondary metabolism’ (Figure 2A) and ‘hormone metabolism jasmonate’ (Figure 2B).
First of all, our data suggest that the shikimate pathway is differentially affected between 110R and 125AA under drought stress (Figure 2D). It is well known that exposure of plants to various stresses generally induces the expression of genes encoding enzymes of the shikimate pathway and AAAs pathway. AAAs include phenylalanine (PHE), tyrosine (TYR), and tryptophan (TRP). This pathway also provides precursors for a variety of plant hormones, such as auxin and salicylate, as well as for a very wide range of secondary metabolites (e.g. alkaloids, flavonoids, and lignin)86 with multiple biological functions and environmental responses.87 TRP is a precursor of alkaloids, phytoalexins, and indole glucosinolates as well as the plant hormone auxin, whereas TYR is a precursor of isoquinoline alkaloids, pigment betalains, and quinones.88 PHE is a common precursor of numerous phenolic compounds, including flavonoids, condensed tannins, lignans, lignin, and phenylpropanoid/benzenoid volatiles.89 TYR and PHE are synthesized from arogenate by the key enzymes arogenate dehydrogenase and arogenate dehydratase, respectively, with the relative flux to each being tightly controlled. The partitioning between TYR and PHE is controlled by the sensitivity of arogenate dehydrogenase to feedback inhibition by TYR.90 The present study shows a differential expression between 110R and 125AA at EL-35 concerning arogenate dehydrogenase (Vv09s0002g08030 and Vv13s0067g02120 for 2009, Vv09s0002g08060 for 2010) and arogenate dehydratase (Vv10s0116g01670 for 2009; Tables 4 and 5). Interestingly, arogenate dehydrogenase and arogenate dehydratase transcripts were respectively more induced and strongly more repressed in 110R berries than in 125AA berries under drought stress (Tables 4 and 5). This observation may partially explain that the phenylpropanoid pathway, which starts from PHE, is more active in 125AA than in 110R berries at E-L 35 under stress (Figure 2A). Arogenate dehydrogenase genes are also differentially regulated in drought-challenged root tissues of two parental genotypes of chickpea.91
Others genes involved in amino acids biosynthesis via the arogenate route are also differentially expressed between 110R and 125AA under drought stress at E-L 35, such as glutamate/aspartate-prephenate aminotransferase (PAT, Vv14s0083g00900 for 2009) and chorismate mutase (Vv01s0010g00480 for 2010; Tables 4 and 5). The same is true for genes related to methionine (MET) metabolism (MET6, Vv10s0042g00970 and MMT, Vv18s0089g01290; Tables 4 and 5). In the literature, there are few and ambiguous data on MET accumulation under drought92 and osmotic stress conditions.93 Our data also reveal a differential expression for amino acid transferase (ALD1: Vv18s0001g04630 and Vv12s0055g00920; Tables 4 and 6). In Arabidopsis, ALD1 generates an amino acid derivative important for the activation of defense signaling, and recombinant ALD1 exhibits aminotransferase activity with a strong substrate preference for lysine.94
No gene directly related to PRO metabolism, well known to play a role in adaptation to drought stress, was differentially expressed between 110R and 125AA. In contrast, our data reveal a differential expression of genes related to polyamines biosynthesis (ornithine decarboxylase, Vv18s0001g00740 and S-adenosylmethionine decarboxylase, Vv14s0083g00580; Tables 4 and 6). Polyamines (PAs) are small aliphatic amines implicated in a wide range of developmental processes and environmental stresses.95 Remarkably, drought stress induces changes in PAs amounts that broadly correlate with drought resistance traits.96 PAs have also been reported to play a role in grape ripening.97 Interestingly, genes related to PAs biosynthesis (and more particularly ornithine decarboxylase) may also regulate the entire subset of pathways for glutamic acid (GLU) to arginine (ARG) and to PRO, a common osmolyte.98 Therefore, we can hypothesize that the differences in ARG, PRO, and GLU content between 125AA and 110R under drought stress we observed in our previous study23 could be partially due to a differential transcriptional regulation of ornithine decarboxylase (Vv18s0001g00740; Tables 4 and 6).
Finally, under drought stress, the rootstock genotypes also impact genes related to amino acid transport (Tables 4, 5, and 7). Plant amino acid transporters (ATFs) are classified into two superfamilies; the amino acid, polyamine, and choline transport superfamily and the ATF family superfamily.99,100 The ATF superfamily contains five sub-classes of transporters including the amino acid permeases (AAPs), lysine/histidine transporters (LHTs), and the PRO transporters. At E-L 35 in 2009, two AAPs (Vv00s0188g00130 and Vv04s0008g00230) were upregulated by drought stress in 125AA and downregulated in 110R (Table 4). In contrast, two LHTs (Vv00s0302g00030 and Vv01s0010g01490) were upregulated in 110R and downregulated in 125AA by drought stress at E-L 35 in 2010 (Table 5).
Conclusions
Although systems biology is increasingly used to study the effects of abiotic stress in plants,101,102 no report has yet investigated the effects of rootstock genotype on the transcriptional response of grape berries to water deficit. The present data provide an overall view of the dynamic changes of the grape berry (cv. Pinot noir) transcriptome during drought stress for berries grafted on a drought-tolerant and a drought-sensitive rootstock. Although the variable environmental conditions occurring in 2009 and 2010 resulted in different levels of water stress, the comparative transcriptome analysis highlighted significant rootstock-dependent differences in the response of genes related to secondary metabolism, JA metabolism, and PR proteins for both years and both developmental stages. Among these, the changes occurring in secondary metabolism are of particular interest for selecting the rootstocks and as a consequence influencing grape berry and wine quality. Overall, genes related to secondary metabolism are more induced and/or less repressed by drought stress in the berries grafted on the drought-sensitive rootstock 125AA. Furthermore, our data also suggest that JA may play a role during a weak to moderate water stress, and may be responsible for the subsequent modulation of the secondary metabolism (e.g. stilbenes) of grape berries.
There are many ways in which the genotype of the rootstock may ultimately affect gene expression in the berries. These include water and nutritional signals depending on root extension and activities, vigor, hormone signaling, physical signaling, and macromolecular signals. This comprehensive transcriptomic analysis will be very useful not only to investigate the molecular network controlling secondary metabolism during drought stress but also to explore further the molecular links between JA and secondary metabolism.
Acknowledgments
We would like to dedicate this paper in deepest recognition to the memory of Dr. Anthony Peccoux, assistant professor of viticulture at the College of Agriculture, Food and Natural Resources, University of Missouri, who passed away in November 2012. Anthony enthusiastically designed the field trial and collected data during season 2009 in the course of his PhD work. We thank Pr. Hans Schultz and Pr. Manfred Stoll (Hochschule Geisenheim University) for helpful discussions. The PhD work of M. Berdeja was supported by an Erasmus Mundus grant. Part of this work was supported by the Conseil Régional d′Aquitaine (France) and the land of Hesse (Germany).
The authors declare no conflict of interest.
References
- Vivier MA, Pretorius IS. Genetically tailored grapevines for the wine industry. Trends Biotechnol 2002; 20: 472–478. [DOI] [PubMed] [Google Scholar]
- Jones GV, Davis RE. Using a synoptic climatological approach to understand climate-viticulture relationships. Int J Climatol 2000; 20: 813–837. [Google Scholar]
- Webb LB, Whetton PH, Barlow EW. Modelled impact of future climate change on the phenology of winegrapes in Australia. Austral J Grape Wine Res 2007; 1: 165–175. [Google Scholar]
- Hannah L, Roehrdanz PR, Ikegami M, Shepard AV, Shaw MR et al. Climate change, wine, and conservation. Proc Natl Acad Sci USA 2013; 110: 6907–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR, Ergül A, Grimplet J, Tillett RL, Tattersall EA et al. Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 2007; 7: 111–134. [DOI] [PubMed] [Google Scholar]
- Chaves MM, Zarrouk O, Francisco R, Costa JM, Santos T et al. Grapevine under deficit irrigation: hints from physiological and molecular data. Ann Bot 2010; 105: 661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 2007; 10: 296–302. [DOI] [PubMed] [Google Scholar]
- Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA et al. Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 2007; 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Quilici DR, Decendit A, Grimplet J, Wheatley MD et al. Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genomics 2009; 10: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby G, Harbertson JF, Adams DA, Matthews MA. Berry size and vine water deficits as factors in winegrape composition: anthocyanins and tannins. Austral J Grape Wine Res 2004; 10: 100–107. [Google Scholar]
- Castellarin SD, Matthews MA, Di Gaspero G, Gambetta GA. Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 2007; 227: 101–112. [DOI] [PubMed] [Google Scholar]
- Koundouras S, Hatzidimitriou E, Karamolegkou M, Dimopoulou E, Kallithraka S et al. Irrigation and rootstock effects on the phenolic concentration and aroma potential of Vitis vinifera L. cv. Cabernet Sauvignon grapes. J Agric Food Chem 2009; 57: 7805–7813. [DOI] [PubMed] [Google Scholar]
- Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E et al. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ 2007; 30: 1381–1399. [DOI] [PubMed] [Google Scholar]
- Gambetta GA, Matthews MA, Shaghasi TH, McElrone AJ, Castellarin SD. Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 2010; 232: 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn N, Guan L, Dai ZW, Wu BH, Lauvergeat V et al. Berry ripening: recently heard through the grapevine. J Exp Bot 2014; 65: 4543–4559, 2014. [DOI] [PubMed] [Google Scholar]
- Lecourieux F, Kappel C, Lecourieux D, Serrano A, Torres E et al. An update on sugar transport and signalling in grapevine. J Exp Bot 2014; 65: 821–832. [DOI] [PubMed] [Google Scholar]
- Stevens RM, Pech JM, Gibberd MR, Walker RR, Nicholas PR. Reduced irrigation and rootstock effects on vegetative growth, yield and its components, and leaf physiological responses of Shiraz. Austral J Grape Wine Res 2010; 16: 413–425. [Google Scholar]
- Tandonnet JP, Cookson SJ, Vivin P, Ollat N. Scion genotype controls biomass allocation and root development in grafted grapevine. Austral J Grape Wine Res 2010; 16: 290–300. [Google Scholar]
- Ollat N, Tandonnet JP, Bordenave L, Decrocq S, Geny L et al. Vigour conferred by rootstock: hypotheses and direction for research. Bull OIV 2003; 76: 581–595. [Google Scholar]
- Cortell JM, Halbleib M, Gallagher AV, Righetti TL, Kennedy JA. Influence of vine vigor on grape (Vitis vinifera L. cv. Pinot Noir) anthocyanins. 1. Anthocyanin concentration and composition in fruit. J Agric Food Chem 2007; 55: 6575–6584. [DOI] [PubMed] [Google Scholar]
- Gambetta GA, Manuck CM, Drucker ST, Shaghasi T, Fort K et al. The relationship between root hydraulics and scion vigour across Vitis rootstocks: what role do root aquaporins play? J Exp Bot 2012; 63: 6445–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Ollat N. Grafting with rootstocks induces extensive transcriptional re-programming in the shoot apical meristem of grapevine. BMC Plant Biol 2013; 13: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdeja M, Hilbert G, Dai ZW, Lafontaine M, Stoll M et al. Effects of water stress and rootstock genotype on Pinot Noir berry composition. Austral J Grape Wine Res 2014; 20: 409–421. [Google Scholar]
- Coombe BG. Adoption of a system for identifying grapevine growth stages. Austral J Grape Wine Res 1995; 1: 104–110. [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 2006; 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 18: 7: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev Genet 2002; 3: 579–588. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2013. ISBN 3-900051-07-0. http://www.R-project.org/. [Google Scholar]
- Gentleman R, Carey V, Bates D et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. ArrayQualityMetrics a bioconductor package for quality assessment of microarray data. Bioinformatics 2009; 25: 415–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Boistad BM, Collin F, Cope LM, Hobbs B et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31: E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: 26. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 2004; 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Blaäsing OE et al. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 2006; 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiol 2005; 138: 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR et al. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 2007; 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati S, Perazzolli M, Malossini A, Cestaro A, Dematte L et al. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and occurrence of an oxidative burst at veraison. BMC Genomics 2007; 8: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheynier V, Duenas-Paton M, Salas E, Maury C, Souquet J-M et al. Structure and properties of wine pigments and tannins. Am J Enol Vitic 2006; 57: 298–305. [Google Scholar]
- Pezzuto JM. Grapes and human health. A perspective. J Agric Food Chem 2008; 56: 6777–6784. [DOI] [PubMed] [Google Scholar]
- Tuccio L, Remorini D, Pinelli P, Fierini E, Tonutti P et al. Rapid and non-destructive method to assess in the vineyard grape berry anthocyanins under different seasonal water condition. Austral J Grape Wine Res 2011; 17: 181–189. [Google Scholar]
- Zamboni A, Di Carli M, Guzzo F, Stocchero M, Zenoni S et al. Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol 2010; 154: 1439–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lüscher J, Sanchez-Diaz M, Delrot S, Aguirreolea J, Pascual I et al. Ultraviolet-B radiation and water deficit interact to alter flavonol and anthocyanin profiles in grapevine berries through transcriptomic regulation. Plant Cell Physiol 2014; 55: 1925–1936. [DOI] [PubMed] [Google Scholar]
- Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R. Metabolite profiling of grape: flavonols and anthocyanins. J Agric Food Chem 2006; 54: 7692–7702. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Steele CL. Flavonoids and isoflavonoids – a gold mine for metabolic engineering. Trends Plant Sci 1999; 4: 394–400. [DOI] [PubMed] [Google Scholar]
- Sirtori CR, Lovati MR, Manzoni C, Castiglioni S, Duranti M et al. Proteins of white lupin seed, a naturally isoflavone-poor legume, reduce cholesterolemia in rats and increase LDL receptor activity in HepG2 cells. J Nutr 2004; 134: 18–23. [DOI] [PubMed] [Google Scholar]
- Giribaldi M, Perugini I, Sauvage FX, Schubert A. Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics 2007; 7: 3154–3170. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Gonzalez JJ, Guttikonda SK, Tran LSP, Aldrich DL, Zhong R et al. Differential expression expression of isoflavone biosynthetic genes in soybean during water deficits. Plant Cell Physiol 2010; 51: 936–948. [DOI] [PubMed] [Google Scholar]
- Grimplet J, Wheatley MD, Jouira HB, Deluc LG, Cramer GR et al. Proteomic and selected metabolite analysis of grape berry tissues under well-watered and water-deficit stress conditions. Proteomics 2009; 9: 2503–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy JA, Matthews MA, Waterhouse AL. Effect of maturity and vine water status on grape skin and wine flavonoids. Am J Enol Vitic 2002; 53: 268–274. [Google Scholar]
- Schijlen EGWM, De Vos CH, Van Tunen AJ, Bovy AG. Modification of flavonoid biosynthesis in crop plants. Phytochemistry 2004; 65: 2631–2648. [DOI] [PubMed] [Google Scholar]
- Downey MO, Dokoozlian NK, Krstic MP. Cultural practice and environmental impacts on the flavonoid composition of grapes and wine: a review of recent research. Am J Enol Vitic 2006; 57: 257–268. [Google Scholar]
- Moura JCMS, Bonine CAV, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 2010; 52: 360–376. [DOI] [PubMed] [Google Scholar]
- Mitchell WC, Jelenkovic G. Characterizing NAD- and NADP- dependent alcohol dehydrogenase enzymes of strawberries. J Am Soc Hortic Sci 1995; 120: 798–801. [Google Scholar]
- Yang L, Wang CC, Guo WD, Li XB, Lu M et al. Differential expression of cell wall related genes in the elongation zone of rice roots under water deficit. Russian J Plant Physiol 2006; 53: 390–395. [Google Scholar]
- Yoshimura K, Masuda A, Kuwano M, Yokota A, Akashi K. Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol 2008; 49: 226–241. [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D et al. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyl- transferase affects phenylpropanoid biosynthesis. Plant Cell 2004; 16: 1446–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury AM, Choi BS, Park SU, Lim HS, Bae H. Transcriptional analysis of hydroxycinnamoyl transferase (HCT) in various tissues of Hibiscus cannabinus in response to abiotic stress conditions. Plant Omics J 2012; 5: 305–313. [Google Scholar]
- Conde C, Silva P, Fontes N, Dias ACP, Tavares RM et al. Biochemical changes throughout grape berry development and fruit and wine quality. Food 2007; 1: 1–22. [Google Scholar]
- Vezzulli S, Civardi SFF, Bavaresco L. Methyl jasmonate treatment as a trigger of resveratrol synthesis in cultivated grapevine. Am J Enol Vitic 2007; 4: 2–5. [Google Scholar]
- Deluc LG, Decendit A, Papastamoulis Y, Mérillon JM, Cushman JC et al. Water deficit increases stilbene metabolism in Cabernet Sauvignon berries. J Agric Food Chem 2011; 59: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus H. Laccases: structure, reactions, distribution. Micron 2004; 35: 93–96. [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I. Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 2006; 12: 1. [DOI] [PubMed] [Google Scholar]
- Fraignier MP, Marques L, Fleuriet A, Macheix JJ. Biochemical and immunochemical characteristics of polyphenol oxidases from different fruits of Prunus. J Agric Food Chem 1995; 43: 2375–2380. [Google Scholar]
- Treutter D. Significance of flavonoids in plant resistance: a review. Environ Chem Lett 2006; 4: 147–157. [Google Scholar]
- Liang M, Haroldsen V, Cai X, Wu Y. Expression of a putative laccase gene, ZmLac1, in maize primary roots under stress. Plant Cell Environ 2006; 29: 746–753. [DOI] [PubMed] [Google Scholar]
- Zhang H, Memelink J. Regulation of secondary metabolism by jasmonate hormones. In: Osbourn A E, Lanzotti V, editors. Plant-derived Natural Products: Synthesis, Function and Application. : Springer-Verlag; 2009: pp181–194. [Google Scholar]
- Munemasa S, Oda K, Watanabe-Sugimoto M, Nakamura Y, Shimoishi Y et al. The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells: specific impairment of ion channel activation and second messenger production. Plant Physiol 2007; 143: 1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate and abscisic acid induced stomatal closure. Plant Physiol 2004; 134: 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, Mullet JE. Lipoxygenase gene expression is modulated in plants by water deficit, wounding and methyl jasmonate. Mol Gen Genet 1991; 230: 456–462. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 1997; 48: 355–381. [DOI] [PubMed] [Google Scholar]
- Pedranzani H, Sierra-de-Grado R, Vigliocco A, Miersch O, Abdala G. Cold and water stresses produce changes in endogenous jasmonates in two populations of Pinus pinaster Ait. Plant Growth Regul 2007; 52: 111–116. [Google Scholar]
- Kiribuchi K, Jikumaru Y, Kaku H, Minami E, Hasegawa M et al. Involvement of the basic helix-loop-helix transcription factor RERJ1 in wounding and drought stress responses in rice plants. Biosci Biotechnol Biochem 2005; 69: 1042–1044. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Inze D, Goossens A. Jasmonate-inducible gene: what does it mean? Trends Plant Sci 2009; 14: 87–91. [DOI] [PubMed] [Google Scholar]
- Gadzovska S, Maury S, Delaunay A, Spasenoski M, Joseph C et al. Jasmonic acid elicitation of Hypericum perforatum L. cell suspensions and effects on the production of phenylpropanoids and naphtodianthrones. Plant Cell Tissue Organ Cult 2007; 89: 1–13. [Google Scholar]
- Shang X, Zhang Y, Peng W, Wang Z, Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot 2009; 60: 3849–3860. [DOI] [PubMed] [Google Scholar]
- Tassoni A, Fornale S, Franceschetti M, Musiani F, Michael AJ et al. Jasmonates, Na-orthovanadate promote resveratrol production in Vitis vinifera cv. Barbera cell cultures. New Phytol 2005; 166: 895–905. [DOI] [PubMed] [Google Scholar]
- Seo S, Sano H, Ohashi Y. Jasmonic acid in wound signal transduction pathways. Physiol Plant 1997; 101: 740–745. [Google Scholar]
- Derckel JP, Baillieul F, Manteau S, Audran JC, Haye B et al. Differential induction of grapevine defenses by two strains of Botrytis cinerea. Phytopathology 1999; 89: 197–203. [DOI] [PubMed] [Google Scholar]
- Giannakis C, Bucheli CS, Skene KGM, Robinson SP, Scott NS. Chitinase and b-1, 3-glucanase in grapevine leaves: a possible defense against powdery mildew infection. Austral J Grape Wine Res 1998; 4: 14–22. [Google Scholar]
- Ferreira RB, Monteiro SS, Piçarra-Pereira MA, Teixeira AR. Engineering grapevine for increased resistance to fungal pathogens without compromising wine stability. Trends Biotechnol 2004; 22: 168–173. [DOI] [PubMed] [Google Scholar]
- Waters EJ, Hayasaka Y, Tattersall DB, Adams KS, Williams PJ. Sequence analysis of grape (Vitis vinifera) berry chitinases that cause haze formation in wines. J Agric Food Chem 1998; 46: 4950–4957. [Google Scholar]
- Van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 2006; 44: 135–162. [DOI] [PubMed] [Google Scholar]
- Park CJ, Kim KJ, Shin R, Park JM, Shin YC et al. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J 2004; 37: 186–198. [DOI] [PubMed] [Google Scholar]
- Naidoo R, Myburg A, Berger D, Naidoo S. Expression profiling of putative Eucalyptus grandis defence marker genes in response to treatment with methyl jasmonate and salicylic acid. BMC Proc 2011; 5: 96. [Google Scholar]
- Liu WX, Zhang FC, Zhang WZ, Song LF, Wu WH et al. Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol Plant 2013; 6: 1487–1502. [DOI] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W. Lignin engineering. Curr Opin Plant Biol 2008; 11: 1–8. [DOI] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol Plant 2010; 3: 2–20. [DOI] [PubMed] [Google Scholar]
- Kutchan TM. Alkaloid biosynthesis: the basis for metabolic engineering of medicinal plants. Plant Cell 1995; 7: 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaga Y, Schnepp J, Peel G, Kish CM, Ben-Nissan G et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J Biol Chem 2006; 281: 23357–23366. [DOI] [PubMed] [Google Scholar]
- Rippert P, Matringe M. Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly similar and active protein domains. Plant Mol Biol 2002; 48: 361–368. [DOI] [PubMed] [Google Scholar]
- Hiremath PJ, Farmer A, Canon SB, Woodward J, Kudapa H et al. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnol J 2011; 9: 922–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Zaplachinski ST. The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiol Plantarum 1994; 90: 9–14. [Google Scholar]
- Gzik A. Accumulation of proline and pattern of alpha-amino acids in sugar beet plants in response to osmotic, water and salt stress. Environ Exp Bot 1996; 36: 29–38. [Google Scholar]
- Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J 2004; 40: 200–212. [DOI] [PubMed] [Google Scholar]
- Alcázar R, Altabella T, Marco F, Bortolotti C, Reymond M et al. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 2010; 231: 1237–1249. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Polyamines and abiotic stress tolerance in plants. Plant Signal Behav 2010; 5: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudelo-Romero P, Erban A, Sousa L, Pais MS, Kopka J et al. Search for transcriptional and metabolic markers of grape pre-ripening and ripening and insights into specific aroma development in three Portuguese cultivars. PLoS One 2013; 8: e60422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R, Shao L, Minocha R, Long S, Minocha SC. Ornithine: the overlooked molecule in the regulation of polyamine metabolism. Plant Cell Physiol 2013; 54: 990–1004. [DOI] [PubMed] [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB. Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chemistry 1995; 270: 16315–16320. [DOI] [PubMed] [Google Scholar]
- Waditee R, Hibino T, Tanaka Y, Nakamura T, Incharoensakdi A et al. Functional characterization of betaine/proline transporters in betaine-accumulating mangrove. J Biol Chem 2002; 277: 18373–18382. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 2011; 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Vannozzi A, Wang G, Liang YH, Tornielli GB et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hort Res 2014; 1: 14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.