Abstract
The apple dwarfing rootstock ‘Malling9’ (‘M9’) has been used worldwide both to reduce scion vigour and as a genetic source for breeding new rootstocks. Progeny of ‘M9’ segregate for rootstock-induced dwarfing of the scion, indicating that this trait is controlled by one or more genetic factors. A quantitative trait locus (QTL) analysis of a rootstock population derived from the cross between ‘M9’ × ‘Robusta5’ (non-dwarfing) and grafted with ‘Braeburn’ scions identified a major QTL (Dw1) on linkage group (LG) 5, which exhibits a significant influence on dwarfing of the scion. A smaller-effect QTL affecting dwarfing (Dw2) was identified on LG11, and four minor-effect QTLs were found on LG6, LG9, LG10 and LG12. Phenotypic analysis indicates that the combination of Dw1 and Dw2 has the strongest influence on rootstock-induced dwarfing, and that Dw1 has a stronger effect than Dw2. Genetic markers linked to Dw1 and Dw2 were screened over 41 rootstock accessions that confer a range of effects on scion growth. The majority of the dwarfing and semi-dwarfing rootstock accessions screened carried marker alleles linked to Dw1 and Dw2. This suggests that most apple dwarfing rootstocks have been derived from the same genetic source.
Introduction
Dwarfing rootstocks have revolutionized the production of some tree and vine crops by permitting high-density plantings that increase fruit yield in the early years of orchard establishment.1–3 The widespread use of dwarfing rootstocks has led to a steady increase in the efficiency of apple production over the past century.4,5 ‘Malling9’ (‘M9’) is the most frequently used apple dwarfing rootstock in both commercial and home orchards.4
‘M9’, originally called ‘Jaune de Metz’, was discovered as a single seedling in the 1800s and was clonally propagated as a rootstock because of its effects on both precocity and vigour control of the grafted scion.6 At the beginning of the twentieth century, all the apple rootstocks grown in Western Europe were collected at the East Malling Research Station (UK) and classified according to their effect on the grafted scion.7 Many of the apple rootstock varieties bred worldwide have parentage derived from this ‘Malling’ series, particularly ‘M9’.1,8 Progeny of ‘M9’ segregate for rootstock-induced dwarfing, indicating that this trait is determined by one or more genetic factors.
Architectural analyses of scion development demonstrate that the earliest measurable effects of the ‘M9’ rootstock are to reduce both the number and length of sylleptic branches that develop in the first year after grafting and to increase the proportion of buds that become floral along the primary axis.9–11 The increase in floral buds results in fewer new extension growth units in the successive annual cycles and a concomitant reduction in the average internode length.12 These early differences in growth are compounded over successive growth cycles and, after several years, trees grafted onto ‘M9’ are 50%–60% the size of genetically identical scions on vigorous rootstocks. Despite their being so widely used and the subjects of numerous studies, the underlying mechanism by which dwarfing rootstocks control both scion vigour and flowering remains unresolved.
Rootstock-induced dwarfing is a complex trait that is affected by environmental conditions, scion genotype, and growth parameters. One of the most powerful methods for dissecting variable and pleiotropic effects is quantitative trait locus (QTL) analysis. Genomic regions that influence apple tree growth, juvenility and fruiting have been identified by several earlier QTL analyses of scion populations segregating for distinct growth and fruiting habits.13–15 These studies have highlighted the importance of using one large family derived from the same parents and making repeated measurements over several years to distinguish traits that are under genetic control, from those that vary with environmental conditions, tree ontogeny or from year to year.
A previous study in a ‘M9’ × ‘R5’ rootstock population, enabled us to identify a major dwarfing locus (Dw1) derived from ‘M9’ and located at the top of linkage group (LG) 5.16 Some of the vigorous individuals in this population carried Dw1, suggesting that there are one or more additional rootstock loci that influence dwarfing of the scion. Using an enlarged population from the same cross, a genetic map was constructed17 which enabled a QTL analysis of rootstock-induced dwarfing. In this paper, we present a multi-trait QTL analysis that demonstrates that Dw1 has a major influence on the overall dwarfing phenotype. Our findings confirm the presence of a second genomic region (Dw2) at the upper end of LG11 of ‘M9’ that has a moderate influence on the majority of dwarfing-related traits studied.18 Several other minor-effect QTLs were also identified, derived from either ‘M9’ or ‘R5’.
Our results demonstrate that the combination of Dw1 and Dw2 has the strongest influence on rootstock-induced dwarfing, that Dw1 has a stronger effect than Dw2, and that Dw2 alone cannot induce dwarfing.
Given that many apple rootstock breeding programmes worldwide have used the East Malling selections as parents, we speculated that other dwarfing rootstocks might also carry Dw1 and Dw2. To test this hypothesis, we genotyped 41 rootstock accessions that confer a range of effects on the scion with simple sequence repeat (SSR) markers linked closely to the dwarfing loci Dw1 and Dw2. Most dwarfing and semi-dwarfing rootstocks carry alleles of markers linked to Dw1 and Dw2, suggesting that the majority of apple dwarfing rootstocks have been derived from the same genetic source.
Materials and methods
Plant material
A segregating rootstock family derived from crosses between Malus × domestica ‘Malling9’ (‘M9’) and M. robusta 5 (‘R5’) was used for QTL analysis. For the first population, 135 seedlings were planted in 1998 and grown as stoolbeds to produce multiple rooted stocks of each genotype. The rootstocks were cleft grafted with ‘Braeburn’ scions, grown in the nursery for 2 years, then transplanted into the Plant & Food Research orchard (Havelock North, New Zealand) as described by Pilcher et al.16 Replicates of the original 135 rootstocks were propagated in 2000 and planted in the orchard as 1-year-old grafted trees. Some of the original 135 rooted stocks died, and some only had enough rootstock for one replicate. Of the replicated trees, 112 individuals from replicate two and 57 individuals from replicate three were phenotyped for QTL analysis. The second population consisted of 350 seedlings, which were generated and grafted as described above and planted in the orchard as 1-year-old trees in 2004. From the second population, between 73 and 81 individuals were evaluated for the QTL analysis and 314 survived until final phenotypic assessment in year 7. Trees were grown with in-row spacing of 1.5 m, 2.5m between rows, and a double wire trellis as support, in a complete randomized block design. ‘Braeburn’ scions grafted onto ‘M9’ and ‘R5’ were planted throughout as controls. Trees were not pruned, to allow full expression of the rootstock effects on scion growth. Once trees began fruiting, chemical thinning sprays were applied to avoid over-cropping and limb breakage.
Forty-one apple rootstock accessions (Malus spp.) representing rootstock varieties used in major apple-growing regions in the world were used for pedigree analysis of Dw1 and Dw2.
Phenotypic analysis
Rootstock effects on the development of ‘Braeburn’ scions were assessed using multiple methods, over 7 years, within the two populations. Table 1 presents the specific traits that were assessed for the QTL analysis in each population/replicate and the sample size phenotyped. Height, internode number and average internode length of the scion were recorded at the end of the first year of growth after grafting (year 1). Flowering was scored by estimating the total number of flower clusters on each tree in the spring of year 2, and placing them into quartiles relative to the most highly floral trees, i.e., 1%–25% had the fewest flowers; 75%–100% had the most flowers. Trees without any flowers in year 2 were recorded as ‘0’. Trunk cross-sectional area (TCA) was measured 20 cm above the graft junction at the end of each year when the trees were dormant. The overall vigour of each tree was assessed annually by comparing trunk size, crown height and spread, branch density and vigour from year 2 to year 7. For the QTL analysis, an overall dwarfing phenotype (DW%) was assigned in year 7, with 100%=very vigorous, 80%=vigorous, 60%=intermediate, 40%=semi-dwarfed and 20%=dwarfed. ANOVA analysis was performed in Origin 8.5®, and graphs were generated in Microsoft excel®.
Table 1. Summary of the phenotypic data collected for QTL analysis of the ‘M9’ × ‘R5’ rootstock population. The first population included three replicates (Rep1–3). Year refers to the year of growth after grafting. Shoot height and node number were measured at the end of year 1, flowering was assessed in spring of year 2 and TCA was measured in June (winter) of each year.
| Number of individuals phenotyped |
||||
|---|---|---|---|---|
| Population 1 |
||||
| Scion trait phenotyped for QTL analysis | Rep1 | Rep2 | Rep3 | Population 2 |
| Flowering in year 2 | 109 | |||
| TCA in year 2 | 103 | 112 | 57 | 81 |
| TCA increase years 2–3 | 103 | 112 | 57 | |
| TCA increase years 3–4 | 103 | 112 | 57 | |
| TCA increase years 4–5 | 103 | 112 | 57 | |
| TCA increase years 5–6 | 103 | 112 | 57 | |
| TCA increase years 6–7 | 103 | 112 | 57 | |
| TCA increase years 2–7 | 103 | 112 | 57 | |
| Primary shoot height in year 1 | 73 | |||
| Node number in year 1 | 73 | |||
| Average internode length in year 1 | 73 | |||
| Overall dwarfing phenotype year 7 (DW%) | 135 | |||
The 41 rootstocks accessions used for the pedigree analysis were classified according to their dwarfing effect in accordance with the literature and in-house Plant & Food Research professional expertise.
DNA isolation and genotyping in the ‘M9’ × ‘R5’ rootstock population and rootstock accessions
Total genomic DNA was extracted from leaves and quantified according to Gardiner et al.19 Leaf material was collected from 135 seedlings from the first ‘M9’ × ‘R5’ population and 350 from the second population. Leaves of the rootstock accessions were collected from the Plant & Food Research germplasm collection in Havelock North, New Zealand, or from the USDA-ARS collection in Geneva, NY, USA.
For Dw1 and Dw2 genotyping of the entire population of ‘M9’ × ‘R5’ rootstocks, polymerase chain reaction products containing single nucleotide polymorphisms were amplified on a LightCycler480 instrument (Roche Diagnostics, Basel, Switzerland) and screened using the High Resolution Melting technique as described by Chagné et al.20 Supplementary Table S1 lists the position of markers on the ‘Golden Delicious’ genome21 and primer sequence.
Markers detecting SSRs located on LG5 and LG11 were employed to genotype the 41 rootstock accessions. Hi01c04, Hi04a08, CH03a09 (LG5) and CH02d08 (LG11) were developed by Silfverberg-Dilworth et al.22 and Liebhard et al.23 Two new SSR markers (MDP0000365711 and MDP00024703) located at the top of LG11 were developed using the Plant & Food Research Malus genome database24, with the programmes Sputnik and Primer3. The M13 sequence TGT AAA ACG ACG GCC AGT was added to the 5′ end of the forward primer to enable the use of Schuelke’s25 approach to fluorescent labeling. polymerase chain reaction reactions were performed and analysed on an ABI 3500 Genetic Analyzer (Applied Biosystems, Waltham Massachusetts, USA) as described by Hayden et al.26
QTL analysis
The parental genetic maps for ‘M9’ and ‘R5’ were constructed using a total of 316 loci amplified from 296 primer pairs as described in Celton et al.17 The maps span a total of 1175.7 and 1086.7 cM for ‘M9’ and ‘R5’, respectively.17 The linkage phase of the markers was determined using JoinMap® 3.027. QTL analysis was performed for all growth traits using MapQTL® 5.28 Traits evaluated over multiple years and in replicates of the first population were analysed separately. Interval mapping, followed by multiple QTL model analysis, using the best markers obtained by interval mapping as co-factors, was used for normally distributed quantitative traits. Only additive models were considered for the QTL analysis. The threshold for QTL genome-wide significance was calculated after 1000 permutations. Kruskal–Wallis analysis was used for ordinal traits such as the estimated number of flower clusters and expert assessment of dwarfing.
Results
Phenotypic variability for scion flowering and architectural traits in the ‘M9’ × ‘R5’ rootstock population
There was a wide variation in overall dwarfing phenotype (DW%), increase in scion TCA, primary axis height, node number and the number of flowers in the spring of year 2 (Supplementary Figure S1). Based on phenotypic assessment from year 2 to year 7, individuals were placed into one of five classes: dwarfed, semi-dwarfed, intermediate, vigorous and very vigorous. Some individuals were more dwarfed than ‘M9’, and some were more vigorous than ‘R5’ (very vigorous). While the differences in scion vigour were apparent by year 3 (Figure 1), the distinction between classes became more pronounced over successive growth cycles.
Figure 1.

Three-year-old compound trees with ‘Braeburn’ scions grafted to sibling rootstocks from a segregating population of ‘M9’ × ‘R5’. The tree on the left is grafted to a rootstock with Dw1 and Dw2, the one on the right has a rootstock with neither. Red arrowheads indicate graft junction, 2-m measure for scale.
Over 5 years, dwarfed, semi-dwarfed and intermediate groups showed a relatively constant rate of TCA gain each year, whereas the vigorous and very vigorous classes exhibited a steady increase in the rate of annual TCA gain (Supplementary Figure S2). When the average TCA gain was compared from year 2 to 7, there was considerable overlap between the dwarfed, semi-dwarfed and intermediate classes; however, there was a clear distinction between these first three classes and the two vigorous classes (Supplementary Figure S3).
There was wide variability in flowering behaviour within the population; some trees did not flower until year 3, whereas others flowered heavily from year 2 onwards. On average, scions grafted onto dwarfing and semi-dwarfing rootstocks had more flowers in year 2 than those on vigorous rootstocks. This observation was consistent within the three replicates from the first population and the second population. Significant positive correlations were found between the increases in TCA over 5 years, the number of flower clusters the spring of year 2 and overall dwarfing phenotype (Supplementary Table S2).
QTL analysis
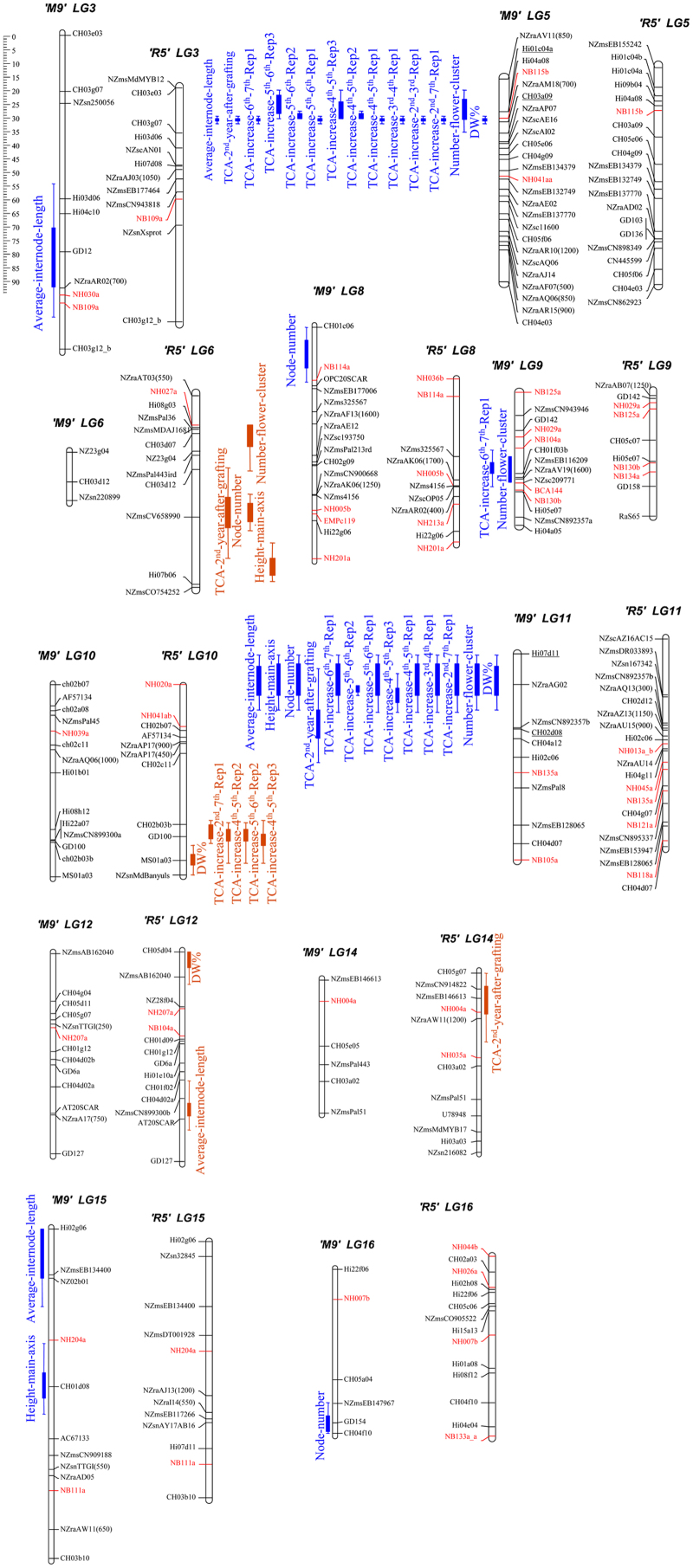
The chromosome locations of QTLs identified in this study are shown in Figure 2 and Table 2. From the analysis of the first population, two QTLs from ‘M9’ were identified for control of overall dwarfing phenotype (DW%). The first one on the top of LG5 (Dw1) explained 57.2% of the phenotypic variation, and the second QTL on the top of LG11 (Dw2) explained 11.4% of the variation. From ‘R5’, QTLs affecting DW% were identified on LG10 and LG12, explaining 7.6% and 5.4% respectively of the phenotypic variation.
Figure 2.
Representation of rootstock QTLs influencing dwarfing and flowering traits on the LGs of ‘M9’ and ‘R5’. The solid part of the bars indicates the most likely position of the QTL and the lines represent the confidence interval. Traits phenotyped are listed in Table 1. The QTLs identified from ‘M9’ are in blue and located on the left side of the LGs, and the QTLs identified from ‘R5’ are in orange and located on the right side of the LGs. The markers flanking Dw1 and Dw2 are underlined and Pyrus SSR markers are indicated in red. Scale bar indicates genetic distance in cM. Details on the markers used to construct the ‘M9’ and ‘R5’ genetic maps are given in Celton et al.17 cM, centiMorgans.
Table 2. Rootstock QTLs identified from the ‘M9’ × ‘R5’ population that influence the growth and flowering of the grafted scion. QTLs are derived from three replicates (Rep1–3) of the first population and a subset of the second population. For each QTL detected with the MQM the LOD score is given by the number before ‘/’. For QTLs detected with the Kruskal–Wallis test, the significance level (indicated by asterisks: *P<0.01; ***P<0.005, ****P<0.0001) is given. The percentage of variance explained by each QTL is indicated by the number after ‘/’.
| Trait | Map | LG3 | LG5 | LG6 | LG8 | LG9 | LG10 | LG11 | LG12 | LG14 | LG15 | LG16 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population 1 | Rep1 | DW% | ‘M9’ | ****/50.2 | ****/11.4 | |||||||||
| ‘R5’ | ***/7.6 | */5.4 | ||||||||||||
| Flowering | ‘M9’ | ****/21.1 | */6.8 | ****/16.5 | ||||||||||
| ‘R5’ | ***/9.0 | |||||||||||||
| TCA year 2 | ‘M9’ | 3.1/7.1 | 2.4/5.4 | |||||||||||
| ‘R5’ | 1.8/5.3 | 2.0/7.4 | ||||||||||||
| TCA increase years 2–3 | ‘M9’ | 11.3/36.3 | ||||||||||||
| ‘R5’ | ||||||||||||||
| TCA increase years 3–4 | ‘M9’ | 11.5/34.4 | 2.7/6.6 | |||||||||||
| ‘R5’ | ||||||||||||||
| TCA increase years 4–5 | ‘M9’ | 13.3/39.9 | 2.5/5.7 | |||||||||||
| ‘R5’ | ||||||||||||||
| TCA increase years 5–6 | ‘M9’ | 12.8/38.9 | 1.6/4.2 | |||||||||||
| ‘R5’ | ||||||||||||||
| TCA increase years 6–7 | ‘M9’ | 8.2/23.9 | 2.4/6.2 | 2.0/5.2 | ||||||||||
| ‘R5’ | ||||||||||||||
| TCA increase years 2–7 | ‘M9’ | 14.2/40.7 | 2.9/6.3 | |||||||||||
| ‘R5’ | 1.9/8.0 | |||||||||||||
| Rep2 | TCA increase years 4–5 | ‘M9’ | 21.6/57.2 | |||||||||||
| ‘R5’ | 1.8/7.1 | |||||||||||||
| TCA increase years 5–6 | ‘M9’ | 12.4/39.8 | 2.2/11.7 | |||||||||||
| ‘R5’ | 2.2/8.7 | |||||||||||||
| Rep3 | TCA increase years 4–5 | ‘M9’ | 9.8/56.5 | 1.6/10.1 | ||||||||||
| ‘R5’ | 1.7/14.9 | |||||||||||||
| TCA increase years 5–6 | ‘M9’ | 9.8/57.1 | ||||||||||||
| ‘R5’ | ||||||||||||||
| Population 2 | Height year 1 | ‘M9’ | 3.8/17.8 | 3.1/17.3 | ||||||||||
| ‘R5’ | 1.6/8.8 | |||||||||||||
| Node number year 1 | ‘M9’ | 2.2/15.8 | 2.4/11.3 | 2.6/12.5 | ||||||||||
| ‘R5’ | 2.6/13.7 | |||||||||||||
| Average internode length | ‘M9’ | 3.3/14 | 3.1/13 | 1.8/7.5 | 2.0/8.3 | |||||||||
| ‘R5’ | 1.6/11.4 | |||||||||||||
Abbreviation: MQM, multiple QTL model.
Four QTLs were detected for control of the number of flower clusters in the spring of year 2. Of the three QTLs from ‘M9’, Dw1 explained 21.1% of the variation, Dw2 explained 16.5% of the variation and a third QTL identified on LG9 explained 6.8% of the variation. The single QTL detected from ‘R5’ was on LG6 and explained 9.0% of the variation.
From the second population, three QTLs controlling the height of the primary axis in year 1 were identified. Two of these QTLs were Dw1 and Dw2, which explained 17.8% and 17.3% of the phenotypic variation, respectively. The third QTL was detected on ‘R5’ LG6 and explained 8.8% of the phenotypic variation.
Four QTLs controlling the number of nodes initiated on the primary axis in year 1 were identified. Three of these QTLs were derived from ‘M9’, on LG8, LG11 and LG16, explaining 15.8%, 11.3% and 12.5% of the variation, respectively. One QTL was located on ‘R5’ LG6, explaining13.7% of the variation.
QTLs controlling all traits analysed co-located on LG5 (Dw1) and LG11 (Dw2). Of the nine minor-effect QTLs identified, only four stable QTLs were detected for more than one trait. Three of these QTLs were on LG 6, 10 and 12 on the ‘R5’ map, and one was located on ‘M9’ LG9.
Phenotypic analysis of individuals genotyped for Dw1 and Dw2
To elucidate the relative contributions of Dw1 and Dw2 to dwarfing of the scion, we examined three of the most robust phenotypes associated with dwarfing, i.e., early flowering (spring of year 2), final TCA (year 7) and overall visual assessment (year 7) of scions grafted to rootstocks carrying various combinations of Dw1 and Dw2. Five markers linked to Dw1 and six markers linked to Dw2 were used to genotype individuals from the ‘M9’ × ‘R5’ rootstock population by High Resolution Melting analysis of single nucleotide polymorphisms (Supplementary Table S1).
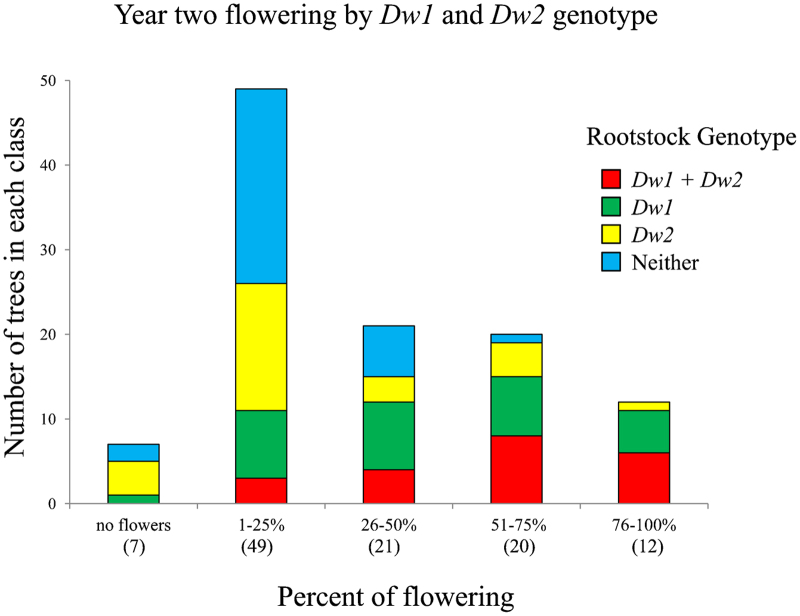
Early flowering was assessed in the spring of year 2 by estimating the number of floral clusters on 109 trees from the first population. The majority of the trees with the highest degree of flowering had been grafted onto rootstocks carrying both Dw1 and Dw2 (50%), or Dw1 alone (41.7%) (Figure 3). Conversely, the trees with no flowers or the fewest flowers were predominantly grafted onto rootstocks carrying Dw2 alone (33.9%), or neither dwarfing locus (44.6%).
Figure 3.
Number of trees in each flowering class and composition of classes by Dw1 and Dw2 genotype. Flowering was assessed by estimating the total number of flower clusters on each tree in the spring of year 2, and placing them into quartiles relative to the most highly floral trees, i.e., 1%–25%, 26%–50%, 51%–75% and 76%–100%. Trees with no flowers were also recorded. Data are from 109 trees from the first population, replicate 1.
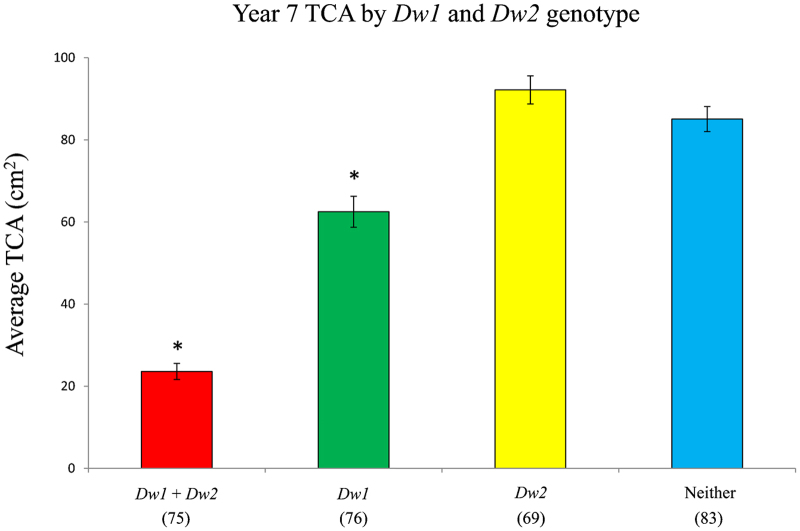
After 7 years of growth, the TCA of 303 trees from the second population were measured. Trees grafted onto rootstocks carrying both Dw1 and Dw2 exhibited the lowest average TCA, only 23% of that of scions on rootstocks with neither loci. Rootstocks with Dw1 alone reduced scion TCA to 73% of those with neither rootstock loci. Surprisingly, trees grafted onto rootstocks with Dw2 alone had the highest TCA of all (Figure 4).
Figure 4.
Average year 7 TCA of trees in each genotypic class. The number of individuals in each class is given in parentheses; error bars indicate standard error. Average TCAs were compared to the group with neither Dw1 nor Dw2 by ANOVA; asterisks indicate the means are significantly different with a P value of <0.001. Data are from 303 trees from the second population.
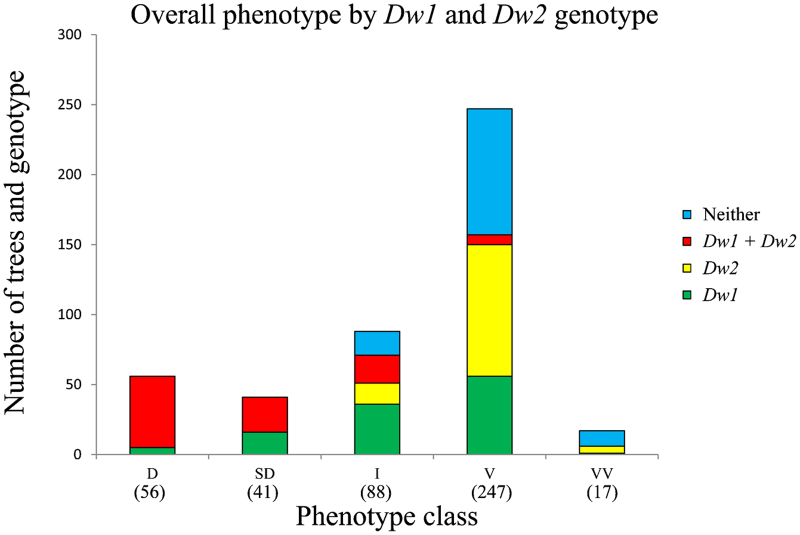
As rootstock-induced dwarfing becomes more pronounced over successive growth cycles, an expert visual assessment of the whole tree phenotype after 7 years provided an overall measure of scion vigour. When 449 grafted trees from both populations were compared, a clear trend relating rootstock genotype to phenotypic class was observed. All the dwarfed and semi-dwarfed trees were grafted onto rootstocks with Dw1 and Dw2 or Dw1 alone, whereas the vigorous and very vigorous trees had rootstocks carrying Dw2 alone, Dw1 alone, or neither locus (Figure 5). Nearly 40% of the vigorous trees were on rootstocks carrying Dw2, indicating that this locus alone is not sufficient to dwarf the scion.
Figure 5.
Composition of each phenotypic class by Dw1 and Dw2 genotype. Trees from both populations (449 trees in total) were visually assessed after 7 years of growth and placed into one of five phenotypic classes, D=dwarf, SD=semi-dwarf, I=intermediate, V=vigorous and VV=very vigorous.
Genotyping rootstock accessions for Dw1 and Dw2
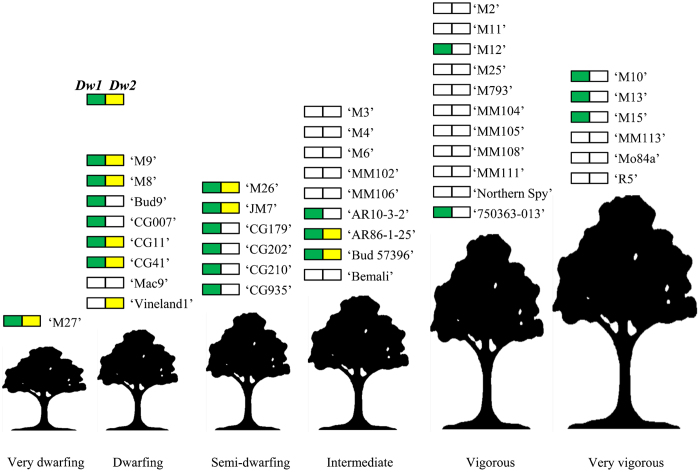
To determine whether Dw1 and Dw2 are present in other known dwarfing rootstocks, we employed multi-allelic SSR markers linked to either Dw1 or Dw2 to genotype 41 rootstock accessions that confer a range of phenotypes on the scion. The use of SSR genotyping provided more detailed information about the allelic status of each locus, which cannot be determined from High Resolution Melting-based single nucleotide polymorphism markers. Three markers, Hi01c04, Hi04a08, and CH03a09, linked to Dw1 on LG5, and three markers spanning from 3.51 to 8.98 Mb on LG11 of ‘Golden Delicious’ were employed to test for the presence of Dw2 (Supplementary Table S1). Figure 6 summarizes the results of SSR genotyping for Dw1 and Dw2.
Figure 6.
Summary of Dw1 and Dw2 genotyping of rootstock accessions. A green square indicates the presence of a single allele of Dw1; yellow represents Dw2. Details on the markers employed to genotype Dw1 and Dw2 and the sizes of products amplified by each accession are specified in Supplementary Tables S1 and S3, respectively.
For each marker, the allele associated with Dw1 or Dw2 was identified from dwarfing individuals in the ‘M9’ × ‘R5’ population. Accessions were scored as having Dw1 if they amplified the same allele as ‘M9’ for all three markers linked to Dw1. To account for the relatively large Dw2 interval, accessions that amplified the same allele as ‘M9’ for two of the three LG11 markers were scored as carrying Dw2. The sizes of amplicons generated for each marker and which alleles are associated with Dw1 and Dw2 are specified in Supplementary Table S3.
Thirteen of the 15 very dwarfing to semi-dwarfing rootstocks genotyped carried marker alleles linked to Dw1 (Supplementary Table S3). Eight of these 13 also carried marker alleles associated with Dw2. Although ‘Mac9’ and ‘Vineland1’ are both derived from ‘M9’ and considered dwarfing rootstocks, neither carried Dw1. Of the nine intermediate rootstocks analysed, three carried Dw1 and two accessions, ‘Bud 57396’ and ‘AR 86-1-25’, carried both Dw1 and Dw2. Of the 17 vigorous to very vigorous rootstocks, only five carried Dw1 and none had Dw2.
Discussion
In a previous study, we identified the major dwarfing locus (Dw1) at the top of LG5 by using a segregating ‘M9’ × ‘R5’ rootstock family.16 However, some of the vigorous individuals in this population carried Dw1, suggesting that are other loci may influence dwarfing. The aim of the present study was to identify additional genetic loci that confer rootstock-induced dwarfing of the scion and to determine their relative contributions to the dwarfing phenotype of apple rootstocks.
Rootstock-induced dwarfing is a complex trait that is influenced by genotype of both rootstock and scion, as well as by growth conditions and environmental effects. To minimize fluctuations in phenotype due to any of these variables, we conducted a QTL analysis based on a large ‘M9’ × ‘R5’ rootstock population, grafted with ‘Braeburn’ scions and grown in a single orchard under identical management. Thus, the wide variation that was observed in scion traits was primarily due to the genotype of the rootstock, rather than to environmental or management effects.
The earliest observed manifestation of rootstock-induced dwarfing in this study was increased flowering in the spring of the second year after grafting. An increased proportion of axillary floral buds along the primary axis can have a profound impact on the subsequent growth of the scion. In a floral bud, the sympodial ‘bourse’ shoot that develops from an axillary meristem is much less vigorous than the monopodial shoot that continues growth from the apex of a vegetative bud.9,11 Bourse shoots do not begin extension until anthesis of the flowers and are developmentally delayed relative to monopodial shoots, which begin growth immediately after budbreak. The effects of increased flowering and reduced sylleptic shoot number and length in year 1 became amplified in successive growth seasons, and within 3 years, scions grafted on dwarf or semi-dwarf rootstocks exhibited a distinctly reduced canopy size and branching density (Figure 1).
This study confirms that scion TCA is a reliable indicator of the degree of dwarfing exerted by a rootstock. Reduced scion TCA is also associated with vigour-controlling rootstocks in peach and kiwifruit.29,30 We found that the differences in TCA were not apparent until at least 2 years after grafting, but became increasingly pronounced over time, suggesting that this trait may be a consequence of the reduced leaf area for light interception in trees with a smaller canopy size. Another factor might be the differential allocation of carbon from different shoot types. It has been shown that sylleptic shoots translocate more carbon to the stem and root than proleptic shoots, which undergo winter dormancy prior to growth.31 Vigorous rootstocks increase sylleptic branching, especially in the early development of the compound tree.11,32
Two major QTLs, Dw1 and Dw2, strongly influence rootstock-induced dwarfing
Our QTL analysis confirmed that Dw1 located on LG5 has a major influence on early flowering and scion architecture, and a second QTL (Dw2) affecting the same traits was validated at the top of LG11, but at a slightly different location from that reported by Fazio et al.18 Four out of nine minor-effect QTLs identified (located on LGs 6, 9, 10 and 12) were stable and controlled more than one trait. Two of the QTLs affecting the overall dwarfing phenotype (DW%) were from the ‘R5’ parent, which was unexpected as ‘R5’ is considered a vigorous rootstock. Two QTLs affecting TCA and early flowering co-located near the centre of LG9 of ‘M9’. A QTL affecting axillary shoot number located in the same region of LG9 was previously identified from a scion population segregating for diverse architectural traits.15 Given the negative relationship between axillary shoots and axillary flowers, these QTLs could be influencing the same trait.
Fazio and co-workers18 have recently reported the results of a QTL analysis involving multiple rootstock families, either grafted to one of several scion genotypes or self-rooted, and grown under multiple conditions. Their study confirmed the presence of Dw1 at the top of LG5 and identified a second QTL in the middle of LG11 (Dw2). However, the position of the Dw2 QTL in the Fazio et al.'s18 study differs significantly from the one that we mapped. The LG11 QTL that we identified is located distally to marker CH02d08, while theirs is proximal. Using a scion population, Kenis and Keulemans14 identified two QTLs in the middle of LG11 that affect sylleptic branch number and length. The markers flanking these QTLs would place them in the same genomic interval as the QTL identified by Fazio et al.18 Other studies have identified two QTLs at the top of LG11 co-locating with ours, one influencing stem diameter13 and a second affecting branching behaviour.15 Further research is required to fine map Dw2 and to develop markers that are more closely linked to this locus and hence more useful for marker assisted selection.
Other reasons for the discrepancies between these studies could include differences in population makeup and size, the genotype of scion(s) grafted to rootstock populations (or left ungrafted), the growth conditions, the traits analysed and the duration of phenotyping. Hatton33 demonstrated that the dwarfing phenotype can take much longer to express in some scion genotypes, and the only reliable measure of rootstock effects is based on phenotyping after at least 3 years of growth. Scion genotype, environment and growth conditions are also known to have a significant effect on the expression of a dwarfing rootstock. For example, root restriction has been demonstrated to mimic the effect of a dwarfing rootstock on a scion, which would complicate identification of genetic factors influencing dwarfing.32 This could explain why Dw1 exhibited a smaller effect on dwarfing in the Fazio study than in the current study, as well as their observation that the Dw2 QTL disappeared entirely in some scion genotypes when trees were grown in small (14.5-L) pots.
Dw1 has a stronger effect than Dw2 on rootstock-induced dwarfing
Analysis of all traits associated with dwarfing indicates that rootstocks carrying both Dw1 and Dw2 had the most significant effects on the scion phenotype. Although Dw1 alone was able to influence growth and flowering of the scion, Dw2 alone was not. Of the trees with the highest degree of flowering, half had rootstocks with both Dw1 and Dw2, and 42% carried Dw1 alone. Rootstocks with Dw1 alone had a statistically significant effect on scion TCA, whereas rootstocks carrying Dw2 only did not. Of the dwarfed and semi-dwarfed trees, 18% had rootstocks with just Dw1, while none had rootstocks with Dw2 alone. Based on these observations, we suggest that Dw1 has a stronger effect on rootstock-induced dwarfing than Dw2, and that Dw2 may act as an enhancer of Dw1. Identification of the genes that underlie Dw1 and Dw2 QTLs will help to elucidate the interaction between these two loci.
Fazio and co-workers have also reported that the strongest degree of dwarfing was conferred by rootstocks with both Dw1 and Dw2.18 However, our results disagree with their conclusion that Dw1 has no effect in the absence of Dw2. If the two QTLs known as Dw2 are in fact the same, the discrepancy in the effect of Dw2 could be due to differences in genotypes of rootstock and scion, growth conditions, sample size and duration of phenotyping between the two studies.
Most dwarfing rootstocks carry Dw1
Our genotypic analysis of rootstock accessions demonstrates that most dwarfing and semi-dwarfing rootstocks carry Dw1 and about half of them also carry Dw2. Four of the six semi-dwarfing rootstocks tested carried Dw1, but not Dw2. This finding supports our conclusion that the strongest degree of dwarfing is conferred by Dw1 and Dw2 in combination, and that Dw1 alone can reduce scion vigour, whereas Dw2 alone cannot.
It is well known that rootstock-induced dwarfing segregates in progeny of ‘M9’; in fact many of the rootstocks classified as dwarfing have ‘M9’ in their lineage.34 The parentage of the dwarf accessions ‘M8’ and ‘Bud9’ are not known, but the fact that both carry marker alleles associated with Dw1 and Dw2 suggests they are also related to ‘M9’. The results of our pedigree analysis further support the role of Dw1 and Dw2 in conferring rootstock-induced dwarfing and suggest that there might only be one allele of Dw1 and Dw2 that enables dwarfing. Unexpectedly, ‘Mac9’ and ‘Vineland1’, both thought to have ‘M9’ as a parent, did not appear to carry Dw1. It is possible these rootstocks are recombinant between the linked SSR markers and the Dw1 locus itself.
It is unclear if all dwarfing rootstocks are derived directly from ‘M9’ itself, or derived from a more distant genetic source. The discovery that approximately one third of the ‘M9’ genome is from the wild European crabapple Malus sylvestris35 raises the possibility that Dw1 and Dw2 originated from M. sylvestris and are present only in some cultivated apple varieties. This could be tested by screening a large number of M. sylvestris accessions with markers linked to Dw1 and Dw2.
Three of the nine rootstock accessions classified as intermediate amplified alleles linked to Dw1. This proportion is consistent with our previous analysis of a ‘M9’ × ‘R5’ population and this study, which found that 40% of intermediate rootstocks carried Dw1.16 In our mapping population, the intermediate class was more similar to the dwarf and semi-dwarf than the vigorous classes in terms of TCA and overall tree phenotype (Figure 2). Furthermore, the phenotypic classifications given in the literature are based on evaluations carried out by different researchers in multiple sites and using different scion genotypes, all of which make it difficult to make direct comparisons between their effects on scion vigour.
Five of the 17 vigorous and very vigorous rootstock accessions carried Dw1, and none carried Dw2. Vigorous rootstocks with Dw1 may have modifiers in their genetic backgrounds that suppress or reduce the influence of Dw1. It is not surprising to have identified some vigorous individuals with the Dw1 locus. The accession ‘750363-013’ is derived from a cross with ‘Ottawa 3’, which itself has ‘M9’ as a parent. The accessions ‘M10’, ‘M12’, ‘M13’, ‘M15’ and ‘M16’ all originate from the initial East Malling selection, as do the dwarfing rootstocks ‘M9’, ‘M8’ and ‘M20’. This result indicates that many of the rootstocks collected together at East Malling could be related. Most rootstocks were selected from seedling populations; growers and nurserymen would have exchanged the best rootstocks, resulting in a narrow genetic base.
Conclusion
The results of this investigation confirm that Dw1 and Dw2 both play a crucial role in rootstock-induced dwarfing of the apple scion. All but two of the dwarf and semi-dwarf rootstock accessions screened amplified marker alleles linked to Dw1. These results also indicate that there may be a unique genetic source of dwarfing in the apple rootstocks used commercially throughout the world. The outcomes of this study will have direct applications in apple rootstock breeding.
Acknowledgments
The authors would like to thank Gennaro Fazio for providing Geneva rootstock accessions, and colleagues John Palmer, Ben van Hooijdonk, Gail Timmerman-Vaughan, and Bruce Veit for helpful comments on the manuscript. This research was funded by the New Zealand Ministry of Business, Innovation, and Employment (contract #30467).
The authors declare no conflict of interest.
References
- Webster AD, Wertheim SJ. Apple rootstocks. In: Ferree D C, Warrington I J (ed.) Apples: Botany, Production and Uses. Wallingford: CABI Publishing, CAB International, 2003: 91–124. [Google Scholar]
- Ferree DC, Carlson RF. Apple rootstocks. In: Rom R C, Carlson R F (ed.) Rootstocks for Fruit Crops. New York: John Wiley & Sons, 1987: 107–143. [Google Scholar]
- Gregory PJ, George TS. Feeding nine billion: the challenge to sustainable crop production. J Exp Botany 2011; 62: 5233–5239. [DOI] [PubMed] [Google Scholar]
- Webster AD. Rootstock and interstock effects on deciduous fruit tree vigour, precocoity and yield productivity. NZ J Crop Hort Sci 1995; 23: 373–382. [Google Scholar]
- Hirst PM, Ferree DC. Rootstock effects on the flowering of ‘delicious’ apple. I. Bud development. J Am Soc Hort Sci 1995; 120: 1010–1017. [Google Scholar]
- Carrière EA. Un nouveau sujet pour greffer les pommes. Rev Hort 1897:436–437.
- Hatton RG. ‘Paradise’ apple socks. J R Hort Soc 1917; 42: 361–399. [Google Scholar]
- Manhart W. Apples for the Twenty-First Century. Portland, OR: North American Tree, 1995. [Google Scholar]
- Seleznyova A, Tustin DS, Thorp TG. Apple dwarfing rootstocks and interstocks affect the type of growth units produced during the annual growth cycle: precocious transition to flowering affects the composition and vigour of annual shoots. Ann Botany 2008; 101: 679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooijdonk BM, Woolley DJ, Warrington IJ, Tustin DS. Initial alteration of scion architecture by dwarfing apple rootstocks may involve shoot–root–shoot signalling by auxin, gibberellin, and cytokinin. J Hort Sci Biotechnol 2010; 85: 59–65. [Google Scholar]
- Foster T, Watson A, van Hooijdonk B, Schaffer R. Key flowering genes including FT-like genes are upregulated in the vasculature of apple dwarfing rootstocks. Tree Genet Genomes 2014; 10: 189–202. [Google Scholar]
- Seleznyova AN, Thorp TG, White M, Tustin S, Costes E. Application of architectural analysis and AMAPmod methodology to study dwarfing phenomenon: the branch structure of ‘Royal Gala’ apple grafted on dwarfing and non-dwarfing rootstock/interstock combinations. Ann Botany 2003; 91: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhard R, Kellerhals M, Pfammatter W, Jertmini M, Gessler C. Mapping quantitative physiological traits in apple (Malus × domestica Borkh.). Plant Mol Biol 2003; 52: 511–526. [DOI] [PubMed] [Google Scholar]
- Kenis K, Keulemans J. Study of tree architecture of apple (Malus × domestica Borkh.) by QTL analysis of growth traits. Mol Breeding 2007; 19: 193–208. [Google Scholar]
- Segura V, Durel CE, Costes E. Dissecting apple tree architecture into genetic, ontogenetic and environmental effects: QTL mapping. Tree Genet Genomes 2009; 5: 165–179. [Google Scholar]
- Pilcher RLR, Celton JM, Gardiner SE, Tustin DS. Genetic markers linked to the dwarfing trait of apple rootstock ‘Malling 9’. J Am Soc Hort Sci 2008; 133: 100–106. [Google Scholar]
- Celton JM, Tustin DS, Chagné D, Gardiner SE. Construction of a dense genetic linkage map for apple rootstocks using SSRs developed from Malus ESTs and Pyrus genomic sequences. Tree Genet Genomes 2009; 5: 93–107. [Google Scholar]
- Fazio G, Wan Y, Kviklys D et al. Dw2, a new dwarfing locus in apple rootstocks and its relationship to induction of early bearing in apple scions. J Am Soc Hort Sci 2014; 139: 87–98. [Google Scholar]
- Gardiner S, Bassett H, Noiton D et al. A detailed linkage map around an apple scab resistance gene demonstrates that two disease resistance classes both carry the Vf gene. Theor Appl Genet 1996; 93: 485–493. [DOI] [PubMed] [Google Scholar]
- Chagné D, Gasic K, Crowhurst RN et al. Development of a set of SNP markers present in expressed genes of the apple. Genomics 2008; 92: 353–358. [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 2010; 42: 833–839. [DOI] [PubMed] [Google Scholar]
- Silfverberg-Dilworth E, Matasci CL, van de Weg WE et al. Microsatellite markers spanning the apple (Malus × domestica Borkh.) genome. Tree Genet Genomes 2006; 2: 202–224. [Google Scholar]
- Liebhard R, Gianfranceschi L, Koller B et al. Development and characterisation of 140 new microsatellites in apple (Malus × domestica Borkh.). Mol Breeding 2002; 10: 217–241. [Google Scholar]
- Newcomb RD, Crowhurst RN, Gleave AP et al. Analyses of expressed sequence tags from apple. Plant Physiol 2006; 141: 147–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 2000; 18: 233–234. [DOI] [PubMed] [Google Scholar]
- Hayden M, Nguyen T, Waterman A, Chalmers K. Multiplex-ready PCR: a new method for multiplexed SSR and SNP genotyping. BMC Genomics 2008; 9: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van OoiejnJ, Voorrips R. JoinMap 3.0®, software for the calculation of genetic linkage maps. Wageningen: Plant Research International 2001.
- Van Ooiejn J. MapQTL5®, software for the mapping of quantitative trait loci in experimental populations. Wageningen 2004.
- Clearwater MJ, Seleznyova AN, Thorp TG et al. Rootstock effects on shoot growth and leaf area development of kiwifruit. Tree Physiol 2006; 26: 505–515. [DOI] [PubMed] [Google Scholar]
- Tombesi S, Johnson RS, Day KR, DeJong TM. Interactions between rootstock, inter-stem and scion xylem vessel characteristics of peach trees growing on rootstocks with contrasting size-controlling characteristics. AoB Plants 2010; 2010: plq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarascia-Mugnozza GE, Hinckley TM, Stettler RF, Heilman PE, Isebrands JG. Production physiology and morphology of Populus species and their hybrids grown under short rotation. III. Seasonal carbon allocation patterns from branches. Can J Forest Res 1999; 29: 1419–1432. [Google Scholar]
- van Hooijdonk B, Woolley D, Warrington I, Tustin S. Rootstocks modify scion architecture, endogenous hormones, and root growth of newly grafted ‘Royal Gala’ apple trees. J Am Soc Hort Sci 2011; 136: 93–102. [Google Scholar]
- Hatton RG. The influence of different rootstocks upon vigour and productivity of the variety budded or grafted thereon. J Pomol Hort Sci 1927; 6: 1–28. [Google Scholar]
- Menendez RA, Larsen FE, Fritss R. Identification of apple roostock cultivars by isozyme analysis. J Am Soc Hort Sci 1986; 111: 933–937. [Google Scholar]
- Cornille A, Gladieux P, Smulders MJM et al. New insight into the history of domesticated apple: secondary contribution of the european wild apple to the genome of cultivated varieties. PLoS Genet 2012; 8: e1002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.