Abstract
Citrus canker, caused by the bacterial pathogen Xanthomonas citri ssp. citri (Xcc), has been attributed to millions of dollars in loss or damage to commercial citrus crops in subtropical production areas of the world. Since identification of resistant plants is one of the most effective methods of disease management, the ability to screen for resistant seedlings plays a key role in the production of a long-term solution to canker. Here, an inverse correlation between reactive oxygen species (ROS) production by the plant and the ability of Xcc to grow and form lesions on infected plants is reported. Based on this information, a novel screening method that can rapidly identify citrus seedlings that are less susceptible to early infection by Xcc was devised by measuring ROS accumulation triggered by a 22-amino acid sequence of the conserved N-terminal part of flagellin (flg22) from X. citri ssp. citri (Xcc-flg22). In addition to limiting disease symptoms, ROS production was also correlated with the expression of basal defense-related genes such as the pattern recognition receptors LRR8 and FLS2, the leucine-rich repeat receptor-like protein RLP12, and the defense-related gene PR1, indicating an important role for pathogen-associated molecular pattern-triggered immunity (PTI) in determining resistance to citrus canker. Moreover, the differential expression patterns observed amongst the citrus seedlings demonstrated the existence of genetic variations in the PTI response among citrus species/varieties.
Introduction
Asiatic citrus canker (ACC) is caused by the gram-negative bacterial pathogen Xanthomonas citri ssp. citri (Xcc) (reclassified from ‘A’ pathotype Xanthomonas axonopodis pv. citri), which is not only the most virulent bacteria that cause citrus canker but also the one with the widest host range.1,2 For most citrus species, ACC is a devastating disease, resulting in extensive damage to commercial citrus cultivars in subtropical production areas of the world.3 It is one of Florida’s most notorious plant diseases because of the impact of citrus on the state’s economy.4 Although several eradication efforts have been attempted in Florida, the disease is still present today.5 Even with an estimated US$12 million per year spent on control methods, the disease continues to spread.6
Citrus canker symptoms are initially seen as small lesions on the leaf that expand over time, eventually becoming erumpent. As the disease progresses, necrotic lesions surrounded by oily, water-soaked margins, and yellow chlorotic rings appear on the fruits, leaves, and young stems. The bacteria remain alive in the margins of the lesions on the leaves and fruits until they fall4 with older lesions taking on a crater-like appearance.7 The bacterium multiplies to a high level in the lesions, which then disperses to new growth and other plants when free moisture is present.8–10 Rain and wind increase its dispersal,11 while extreme weather conditions such as hurricanes and tropical storms are associated with long-distance dissemination and a local increase the level of disease.12 Insect feeding can also lead to significant field infections. It has been noted that the Asiatic citrus leafminer (Phyllocnists citrella) damages citrus not only by feeding on new leaf growth but also by greatly exacerbating the severity of citrus canker.13,14
The canker bacterium enters the host through stomata or wounds15 where it colonizes the mesophyll parenchyma and multiplies in the apoplast. Virulence factors encoded by the bacterium aid in its survival within a host. For example, flagellar-dependent motility in Xcc has been deemed necessary for proper development of a mature biofilm structure, which directly correlates with X. citri pv. citri pathogenicity.16,17 The Xcc pathogen has evolved virulence factors in addition to biofilm formation, which can modulate the plant’s basal defense response and promote infection.18–20 These include secretion systems (including: type III secretion system genes), cell–wall degrading enzymes, effectors, toxins, and bacterial adhesins.21
Survival of the plant, on the other hand, relies upon the development of an immune response to counter the attack of the microbial pathogens through a combination of diverse molecular mechanisms.22 Upon attack, the plant can trigger several categories of defense mechanisms involved in disease resistance.23 The first pathway, pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), is a basal defense response prompted by the recognition of PAMPs, like flagellin and elongation factor Tu (EF-Tu), which through a set of specialized plant extracellular receptor kinase proteins can stop and prevent further colonization.24–26 The leucine-rich repeat receptor kinase (LRR-RK) FLS2 acts as the pattern recognition receptor (PRR) for flagellin,25 while the LRR-RK known as receptor kinase essential for EF-Tu perception (EFR) is the PRR for EF-Tu.27 Both the flagellin protein and synthetic peptides derived from the flagellin N-terminus are recognized by most plants and can induce a multitude of defense responses including: accumulation of reactive oxygen species (ROS), induction of defense genes, and cell–wall reinforcement by callose deposition.28,29
Identification and utilization of host resistance are probably the most effective way to manage various crop and fruit diseases, including ACC, but, unfortunately, no true resistance to citrus canker has yet been identified. Since most of the commercially grown citrus types are susceptible to canker,7,30,31 current control measures for ACC worldwide are aimed at reducing inoculum levels, which consist of copper-based bactericidal sprays to protect the expanding foliar and fruit surfaces from infection or eradication of infected trees upon detection.7 However, newer approaches ranging from conventional breeding methods to production of transgenic plants are being utilized in an effort to produce plants resistant to ACC.32–37 Plants suspected to provide resistance must then be screened for their susceptibility to ACC. Various methods have been described for susceptibility testing including traditional inoculation studies, leaf-mesophyll susceptibility,31 a leaf enrichment test,38 and pathogen detection via qPCR.39 Several pitfalls are associated with the use of these methods including the fact that the success of whole plant sprays can be dependent upon leaf age at the time of inoculation, the need to propagate the plants in order to produce replicates for testing, and the growth/utilization of a regulated pathogen for such testing methods. This paper reports a rapid screening method with the ability to identify citrus seedlings that are less susceptible to early infection by Xcc based on early ROS accumulation triggered by a conserved 22-amino acid sequence of the flagellin (flg22) from X. citri pv. citri (Xcc-flg22). Here, ROS production is correlated to the expression of defense-related genes and the limitation of disease symptoms amongst seedling, indicating a role for PTI in resistance to canker and the existence of genetic variations within that response among citrus species/varieties.
Materials and methods
Citrus seedlings
The seedlings of grapefruit (C. paradise ‘Duncan’, mandarin (C. reticulate ‘Cleo’), sweet orange (C. sinensis ‘Valencia’), sour orange (C. aurantium ‘Karun Jamir’), and pomelo (C. maxima ‘Thong Dee’) were grown in the screened US Horticultural Research Laboratory greenhouse in Fort Pierce, FL, USA from the seeds with the exception of the Citrus × Poncirus trifoliate US-802, which was obtained from Phillip Rucks Citrus Nursery in Frostproof, FL, USA. All the seeds were collected from the citrus collection located at Pico farm, USDA, Fort Pierce, FL, USA, using a routine horticultural practice.
Oxidative burst measurements
A peptide corresponding to the flg22 of Xcc was ordered from GenScript (Piscataway, NJ, USA) with >90% purity using the amino sequence QRLSSGLRINSAKDDAAGLAIS. The peptide was resuspended in sterile water and used to measure induction of ROS. Six leaf disks from three newly expanded young leaves were punched out with a 4-mm-diameter cork borer. Leaf discs were floated abaxial side up in an individual well of a black 96-well plate with 200 µl of water per well at room temperature overnight and covered with plastic bags. The next day the water was removed, and ROS production was triggered with 200 nM flg22 applied together with 20 μM luminol and 1 μg per 100 μL of horseradish peroxidase. Luminescence was measured by the LUMIstar microplate luminometer (BMG Labtech, Cary, NC, USA). Each plate was measured over a period of 40 min. A total of 32 one-year-old seedlings from each variety were used. The assay was repeated twice. Controls lacking flg22 were included for each species with the control lane displayed in Figure 1 consisting of a combination of five data points from each of the species tested (30 total data points). Data were analyzed using the Optima microplate reader software (BMG Labtech).
Figure 1.
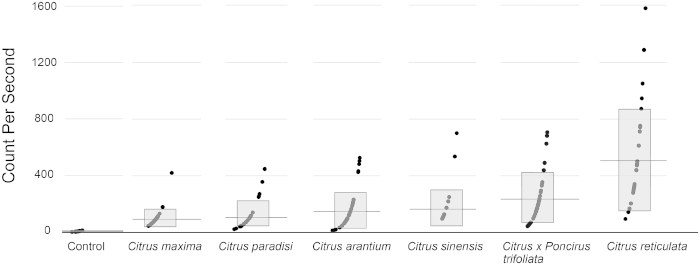
Xcc-flg22 induced varying levels of ROS production in Citrus seedlings. Flg22-triggered ROS production was measured using luminol as a chemiluminescence probe in the following Citrus seedlings: C. maxima, C. paradisi, C. arantium, C. sinensis, Citrus × Poncirus trifoliata, and C. reticulata. Each reaction was monitored for 40 min with the ROS production peak from within the time course shown in CPS. Error bars represent the standard deviation of two independent experiments. The level of ROS was also measured for non-Xcc-flg22-induced samples as a negative control, which is a combination of five seedlings for each species used in the assay. Gray boxes represent one standard deviation of the mean while the black horizontal lines represent the mean.
Attached leaf-infiltration and leaf dip inoculations
Xanthomonas citri pv. citri (A strain 3213)40 cells were pelleted from a 3-mL overnight culture grown in LB liquid medium at 28 °C with a rotational speed of 220 rpm. The pellet was resuspended in sterile tap water and adjusted to an appropriate concentration (3 × 104 cells/mL for macroscopic symptoms examination and 3 × 105 cells/mL for gene expression analysis). The bacteria suspension was infiltrated into three whole, attached leaves for each seedling by pressing the opening of a needless syringe onto the abaxial leaf surface supported by one finger. As a control, the same number of leaves were mock-inoculated by infiltrating sterile tap water. Plants were maintained in the greenhouse at 28–30 °C. Tests for disease symptom severity were repeated three times with photographs taken 20 days post inoculation. Bacterial populations and gene expression profiles were determined using infiltrated leaf samples, as described below, using qPCR and RT-qPCR.
Leaf dip inoculations were performed by growing 5 mL of Xcc in LB overnight as stated above. Cells from the culture were pelleted and resuspended in 1 L of a sterile 0.05% Silwet-L77 solution made with autoclaved tap water. Attached leaves were then dipped into the bacterial suspension. Bags were placed around dipped leaves to keep the humidity level high. Plants were grown under greenhouse conditions for 20 days before being analyzed for the presence of canker lesions. Undipped leaves, acting as controls, did not contain any canker lesions.
DNA sampling and processing
Samples were collected 4, 8, 12, 16, and 20 days after inoculation from three leaves of each seedling. A 1-cm diameter disc, centered on the infiltrated area of each leaf, was removed using a cork borer. Each leaf disc was transferred into an autoclaved 2-mL Fast Prep tube (MP Bio., Solon, OH, USA) containing two 4-mm silicone-carbide sharp particles and three 2.3 mm chrome-steel beads in 800 µL of extraction buffer (100 mM Tris-Base, 50 mM EDTA, 500 mM NaCl, 2.5% polyvinylpyrrolidone, and 10 mM β-mercaptoethanol). They were homogenized by a Fast Prep®-24 homogenizer (MP Bio.) at the speed of 6.0 for 60 sec and incubated at 65 °C for 30 min after the addition of 20% SDS. After adding one-third volume of 5 M potassium acetate, the tubes were incubated on ice for 5 min and centrifuged at maximum speed for 5 min to remove plant debris. The supernatants were centrifuged for an additional 10 min at the same speed, and 800 µL of supernatant was transferred to a new 1.5-mL tube containing two-third volume of cold isopropanol. The sample was then placed in a Genesee column (Genesee Scientific, San Diego, CA, USA), centrifuged 1 min at 8000 rpm, and washed twice with 70% ethanol. Samples were eluted with 100 µl nuclease-free water, analyzed by Nanodrop, and then adjusted to the concentration of 50 ng/µl DNA prior to qPCR analyses.
Estimation of bacterial populations of Xcc by qPCR
Three inoculated leaves were sampled in triplicate at 4, 8, 12, 16, and 20 days after inoculation. Fresh tissues were homogenized for genomic DNA extractions and used for PCR analyses. The presence of the pthA gene was determined using two pairs of gene-specific primers designed for Xcc: VM3 (GCATTTGATGACGCCATGAC) and VM4 (TCCCTGATGCCTGGAGGATA),41 which were added at 0.5 µM in a 15 µL reaction with 7.5 µL of 2× FAST SYBR Green Master Mix (Life Technologies, Carlsbad, CA, USA) reagent and 2 µL of DNA template. The following standard thermal profile was used for all amplifications: 95 °C for 20 sec followed by 40 cycles of 95 °C for 3 sec and 60 °C for 30 sec. To analyze dissociation curve profiles, the following program was run after the 40 cycles of PCR: 95 °C for 15 sec followed by a constant increase in temperature between 60 °C and 95 °C. Primers targeting the pthA gene were used for quantification of Xcc.39
Total RNA extraction from sour orange leaves and preparation of cDNA
At 12 hours and 24 hours post Xcc infiltration, five leaf discs from three leaves per seedling were cut using a circular 5-mm-diameter cork borer. Samples were quickly frozen in liquid nitrogen and grounded to a powder in autoclaved 2-mL Fast Prep tubes containing glass beads using a Fast Prep®-24 homogenizer (MP Bio.) at speed 6.0 for 30 sec. Total RNA was extracted using the TRIzol Reagent (Invitrogen, Grand Island, NY, USA). Total RNA concentration and purity were determined from the ratio of absorbance readings at 260 and 280 nm, using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The synthesis of cDNA was performed with poly-T primers using the M-MLV reverse transcriptase system (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Real-time quantitative reverse transcriptase (RT)-PCR
The SYBR® Green (Applied Biosystems, Foster City, CA, USA) qPCR assay was used to determine the genes expression in sour orange seedlings after Xcc inoculation. SYBR® real-time PCR amplifications were performed in a Mastercycler ep realplex (Eppendorf, Hauppauge, NY, USA). The reaction mixture contained 7.5 µL 2× FAST SYBR Green Master Mix reagent (Applied Biosystems), 2 µL of cDNA, 150 nM of each gene-specific primer pair in a final volume of 15 µL.
The following standard thermal profile was used for all amplifications: 95 °C for 20 sec followed by 40 cycles of 95 °C for 3 sec and then 60 °C for 30 sec. All assays were performed using three technical replicates and a non-template control. To analyze dissociation curve profiles, the following program was run after the 40 cycles of PCR: 95 °C for 15 sec followed by a constant increase in temperature between 60 °C and 95 °C. The following five genes that were identified by RNA-seq42 were selected for the analysis: PRf (ACAAACACACATCTCCGAAATGA), PRr (TTGAAATGAGCAGCAGCAAAA); LRR8f (TGCACCAGAGCTAGCCTACA), LRR8r (GATGGATATGGAAGCCGAGA); RLP12f (ACGGATTCCTCAACAACTGG), RLP12r (TTGTTGCAGCTTCACCACTC); FLS2f (AGCCCTCCAGGTTCTTGATT), FLS2r (GAGACCAAGCGCTAACAAGG), GSLf (CGAAGAGGCTTTGGCTACAC), and GSLr (TCCTCGACCAGTAGCTCGAT. The UPL7 (ubiquitin protein ligase 7) identified as superior reference gene was selected UPL7f (CAAAGAAGTGCAGCGAGAGA) and UPL7r (TCAGGAACA GCAAAAGCAAG).43
For the real-time qRT-PCR experiments, the expression levels of the target genes were normalized using the following model. The relative expression ratio (R) of a target gene is calculated through the E and crossing points (CP) deviation of a sample versus a control and expressed in comparison to the reference gene UPL7.44
Results
Production of ROS triggered by Xcc-flg22 varies among seedlings from various citrus species
One of the earliest cellular responses following successful pathogen recognition, known as the oxidative burst, is characterized by a rapid and transient accumulation of ROS.45,46 This response can be induced during pathogen invasion or via elicitor treatment such as that triggered by the perception of the flagellin peptide flg22.47 To trigger ROS production in our seedlings collection, we used the flagellin peptide from X. citri (Xcc-flg22). When leaf discs were incubated with 100 nM of Xcc-flg22, variations in ROS production were observed both among the different species and between the individual seedlings within a particular species (Figure 1). Typically, the photon counts per second (CPS) for seedlings within a species oscillated around an average specific for that particular species, which ranged from 101 to 509 depending upon the species.
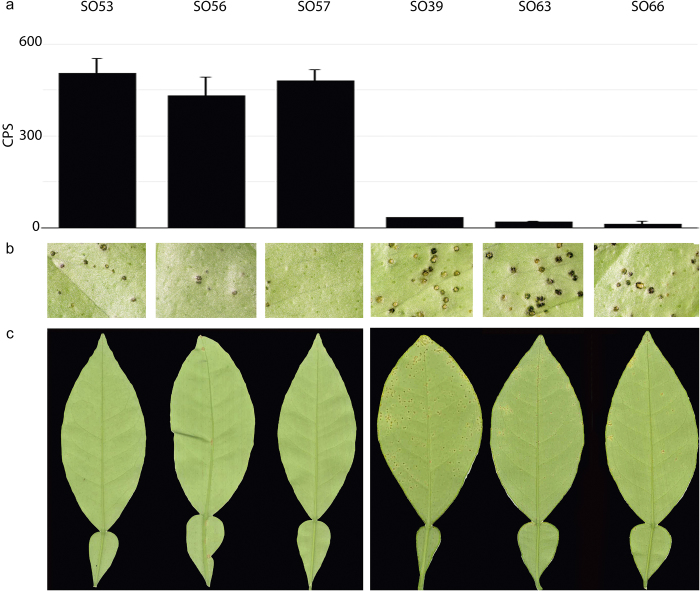
Seedling with high ROS production affects Xcc growth and canker lesions
Typically, the level of ROS produced in response to Xcc-flg22 measured via CPS in the sour orange seedlings ranged between 50 and 300; however, a small group of seedlings showed a CPS above 400 whereas another showed a CPS below 50. To determine if the results of the ROS assay correlated with disease suppression, we used the standard deviation of the mean to identify several plants as having high ROS production (SO53, SO56, and SO57) and low ROS production (SO39, SO63, and SO66; Figure 2a) and inoculated their leaves with the Xcc pathogen using both a leaf-infiltration method (Figure 2b) and a leaf dip inoculation (Figure 2c). Autoclaved tap water was used as a control in these experiments. A close correlation between the magnitude of the lesions and the number of lesions was observed. By 20 days post Xcc inoculation, lesions on the leaf surface were both more numerous, larger in size, and more severe in seedlings SO39, SO63, and SO66 compared to SO53, SO56, and SO57 (Figure 2). Moreover, a close correlation between the magnitude of the lesions and the number of Xcc cells was seen despite the leaves being inoculated with the same cell concentration. Differences in bacterial Xcc cell concentrations could be seen in the leaves of the sour orange seedlings by 4 days post Xcc infiltration (Figure 3). Although none of the seedlings were completely resistant to ACC, it was possible to separate the seedlings into two groups, which corresponded with high ROS production or low ROS production, based on the number of cells present 20 days after infiltration (Figure 3). Leaves averaged 104–105 cells for seedlings that showed high ROS production post Xcc-flg22 treatment and 106–107 cells for the seedling with low ROS production post Xcc-flg22 treatment.
Figure 2.
Severity of canker symptoms in sour orange leaves correlate with ROS production. (a) ROS production in sour orange seedlings corresponding to both high and low level producers and the resulting symptom severity on leaves, (b) 20 days post-infiltration with Xcc, and (c) 20 days post leaf-dip inoculation with Xcc. Note that the water-soaked margins and yellow halos typically associated with sever canker infection and the substantial increase in the number of lesions on the seedling that produce a low level of ROS. A representative of three separate experiments is shown.
Figure 3.
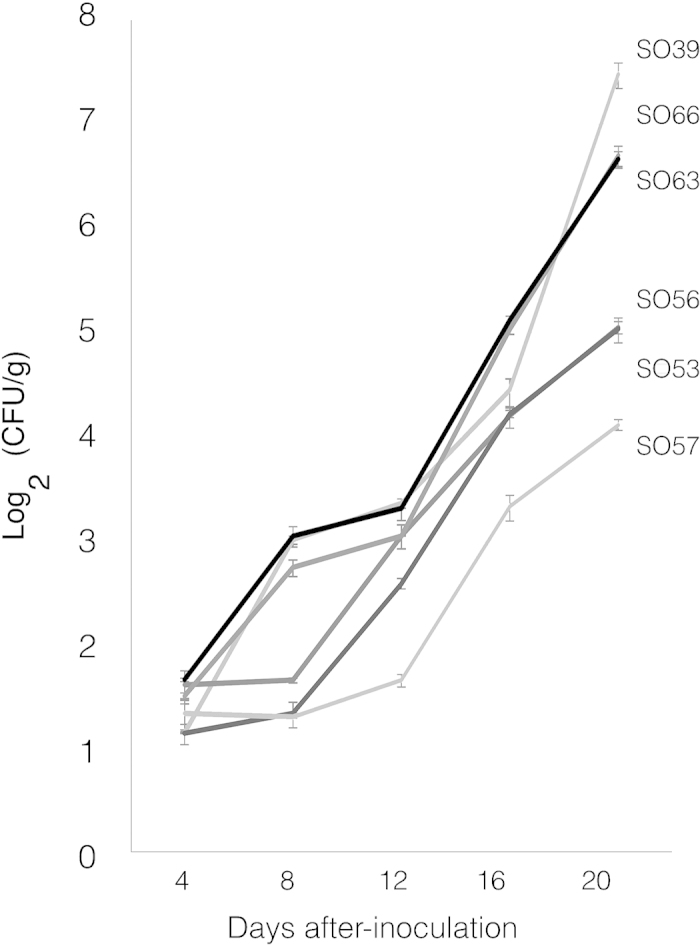
Seedlings with high ROS accumulation show reduced growth of Xcc. Growth of the Xcc bacterium in seedlings with high levels of ROS (SO53, SO56, and SO57) compared to seedlings with a low production of ROS (SO39, SO63, and SO66) was determined by qPCR for the pthA gene using two pairs of gene-specific primers designed for Xcc. Growth is represented as CFUs per gram of tissue tested. Error bars represent the standard deviation of two independent experiments.
Xcc induces the expression of defense genes in seedlings with high ROS levels
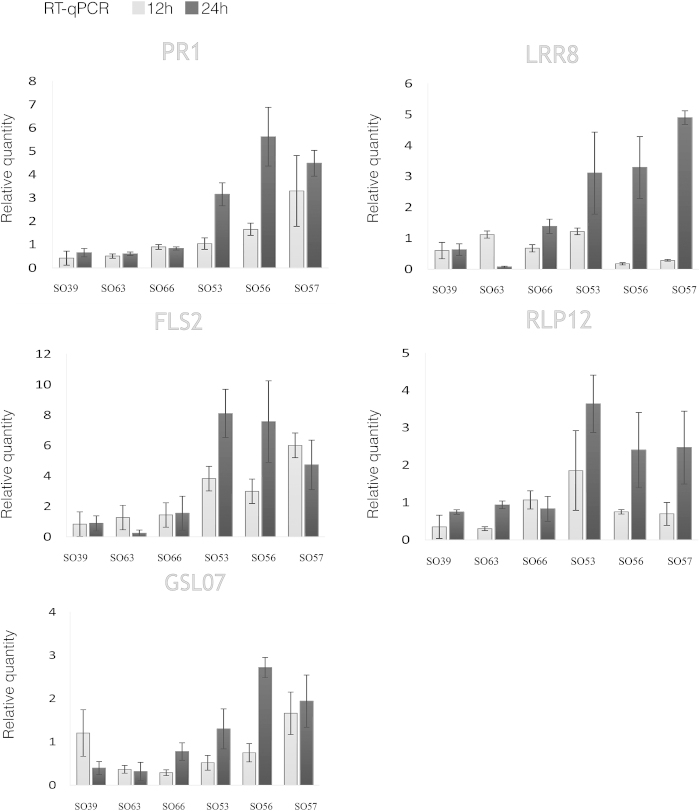
To examine whether PTI was activated in those seedlings that displayed an increased level of ROS and a decreased level of pathogen, we analyzed the transcript levels of a set of known PTI marker genes. To do so, RT-PCR was performed on RNA isolated from sour orange leaves that had been syringe-infiltrated with either the Xcc pathogen or autoclaved tap water. Overall, the group of seedlings with high ROS production (SO53, SO56, and SO57) showed an increase in the level of transcript of the PTI-related genes in response to the bacterial pathogen Xcc (Figure 4). In particular, the genes that showed increased expression were as follows: the PRR LRR8, which is similar to a LRR receptor-like protein kinase in Arabidopsis; the LRR receptor-like protein RLP12, which is associated mainly with disease resistance;48–50 and the defense-related gene PR1. Upregulation of these genes suggests that PAMPs from Xcc might be perceived by PRRs in the sour orange seedlings and could trigger the PTI response. Since callose deposition also plays an important role in a plants defense response, it is worth mentioning that the callose synthase gene GSL07 was also induced in seedlings with high ROS (Figure 4).
Figure 4.
Induction of PTI-related resistance genes in seedlings with high ROS production. The relative quantity of PTI-related genes in SO53, SO56, and SO57 compared to SO39, SO63, and SO66 seedlings at 12 and 24 hours post-infiltration with the Xcc-flg22 peptide. The relative quantity of genes was determined using reverse transcriptase real-time PCR with primers specific for the PR1, LRR8, FLS2, RLP12, and GSL07 genes. Error bars represent the standard deviation of two independent experiments.
Discussion
Although all citrus species are susceptible to citrus canker, previous studies have noted that varying degrees of susceptibility to ACC exist, with grapefruit (C. paradisi) being highly susceptible, mandarin (C. reticulata) being moderately resistant, and many species falling in between, such as sour (C. aurantium) and sweet (C. sinensis) oranges, which are considered less susceptible, and pummelo (C. maxima), which is considered susceptible.7,8,31,51 Results of the ROS assay (Figure 1) paralleled these findings with C. reticulata displaying the highest level of activity (mean = 509 CPS), C. paradisi (mean = 112 CPS) one of the lowest levels, and moderate activity levels for C. sinensis (mean = 155 CPS) and C. aurantium (mean = 158 CPS). ROS production in seedlings not subjected to Xcc-flg22 was near zero (control), indicating a specificity of the response to the peptide.
The secondary cultivated species, C. aurantium, is the result of a cross between the more susceptible species C. maxima and the moderately resistant species C. reticulata.52,53 It was chosen for detailed study here because offspring generated from two parents at opposing ends of the ACC resistance/susceptibility spectrum may be more variable in their response to ACC than offspring generated from two susceptible or two resistant parents. In addition, the parent plant of the C. aurantium seedlings had displayed remarkable resistance/tolerance to disease (including both ACC and huanglongbing) over a 7-year period during an ongoing cultivar study (data not shown). Given the segregation of the plant’s progeny in regards to ACC resistance (Figures 2 and 3), we hypothesize the involvement of a genetic component in this resistance.
In citrus, zygote frequency is affected by factors such as the species/variety, genetic and environmental factors, and even nutritional status.54–57 Since the seedlings screened for their ability to induce ROS production were grown from random seeds (Figure 1), the number of seeds with ROS levels that deviate far from the mean for that species may be reflective of the differences in the number of zygotic seedlings screened for that species; although further experimentation using molecular markers would be needed to establish such a correlation. More importantly, the variation in ROS production also suggests that differences may exist in the perception of flagellin by these seedlings since recognition of PAMPS by the plant initiates some of the earliest basal PTI responses, including an increase in ROS.28 Previous studies have shown that elicitors such as flagellin can activate PAMP recognition through LRR-RK and a MAP kinase cascade, leading to basal resistance.58,59 Induction of defense responses in non-host plants also includes the synthesis and accumulation of ROS. Overall, ROS production plays a dual role in defense since the resulting oxidative burst acts as both an early stress signal and part of the downstream machinery designed to attack invading pathogens.60 We hypothesized that the high ROS levels in the seedlings correlated with an increase in activation of early defense-response genes related to PTI. Therefore, plants with higher ROS levels would be more resistant to Xcc while those with lower levels would correlate with an increased susceptibility to Xcc, allowing a simple ROS production assay to serve as a screening tool for inherited Xcc resistance in seedlings.
Analysis showed a strong correlation between ROS production and the expression of defense-associated genes in the assayed plants (Figure 4). In particular, Xcc inoculation triggered the expression of GSL07, GLUCAN SYNTHASE-LIKE, or CALLOSE SYNTHASE, which functions in callose deposition after injury and pathogen attack.61 Several LRR receptor-like genes were also upregulated in seedlings with high ROS accumulation including RLP, which was previously associated with disease resistance in plants,50 and LRR8, a protein kinase shown to be involved in the perception of pathogens through the detection of PAMPs and the initiation of a well-characterized PTI response that includes ROS production, callose deposition, and the induction of other defense-related genes.26 The expression of the defense-associated gene PR1 was also upregulated in the group of plants with high ROS accumulation post Xcc inoculation. An increase in the expression of PR1 has been reported in Arabidopsis seedlings in response to flg22,62 although it is interesting that in a previous study comparing gene expression in the citrus canker-resistant genotype ‘Nagami’ kumquat to the susceptible genotypes ‘Duncan’ grapefruit, PR1 was downregulated in kumquat even though other defense-related genes and ROS production were shown to increase.63 Since PR1 expression is related to other factors such as salicylic acid production64 and systemic acquired resistance,65,66 it is possible that the molecular mechanism involved in its regulation differs amongst the citrus relatives.
In addition to increased expression of defense-associated genes, seedlings with high ROS production also showed reduced growth of Xcc (Figure 3). A divergence could be detected as early as 4 days post inoculation, with the bacteria growing slower on seedlings with increased ROS (SO56, SO53, and SO57) compared to seedlings with lower ROS production (SO39, SO66, and SO63). By day 20, there was a discernible distinction between the two groups. Moreover, seedlings with higher colony forming units (CFUs) clearly displayed the classical leaf symptomology including yellow-haloed lesions and water-soaked margins, while the other seedlings developed only mild symptoms (Figure 2).
Taken together, seedlings with high ROS production overexpressed defense-associated genes and showed a reduction in bacterial growth after inoculation of Xcc compared to the seedlings with low ROS production. These differences indicate that ROS production induced by Xcc-flg22 is associated with both the induction of defense genes by Xcc and the level of resistance/susceptibility in the citrus genotypes, suggesting that PTI may limit the initial pathogen invasion and multiplication of Xcc. Since susceptible seedlings did not respond to Xcc-flg22 and did not activate the ROS response compared to the more resistant seedlings, differences in PAMP perception by FLS2 may be responsible for the observed variations. Analysis of gene sequences involved in PTI amongst the seedlings could clarify the difference in the PTI response.
Moreover, this analysis reveals that a luminol-based ROS assay can be utilized to rapidly screen citrus seedling for PAMP-elicitation and for the selection of seedling candidates that display resistance to Xcc. This screening technique provides many advantages over other methods used to identify Xcc resistance. For example, the time needed to identify resistant plants is greatly reduced because the need to propagate plants for pathogenicity testing is eliminated and the ROS-based assay can be performed on seedlings, thus eliminating the need to grow the plants to a more mature state. In addition, by substituting the use of live pathogen with a flagellin peptide, testing can be conducted in areas where the pathogen is regulated. Variability may also be minimized since whole plant spray inoculations are dependent upon leaf age at the time of inoculation. Another advantage of the ROS-based assay is that tree weakening resulting from pathogen exposure during testing39 could be curtailed by the elimination of the live pathogen. Overall, this luminol-based ROS assay represents a rapid and novel method for identifying citrus seedlings less susceptible to early infection by Xcc. Because of the similarities in PAMP-elicitation among different plant species, this technique may be applicable to resistance screening for other flagellin-producing pathogens in other crops.
Acknowledgments
We would like to thank C. Latza for her excellent technical assistance. Funding was provided by the U. S. Department of Agriculture. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
All authors certify that they have no affiliations with any organization or entity with a competing financial interest that would influence the outcome of the subject matter or materials discussed in this manuscript.
References
- Cubero J, Graham JH. Genetic relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl Environ Microb 2002; 68: 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova IV et al. Emended classification of xanthomonad pathogens on citrus. Syst Appl Microbiol 2006; 29: 690–695. [DOI] [PubMed] [Google Scholar]
- Das AK. Citrus canker – A review. J Appl Hort 2003; 5: 52–60. [Google Scholar]
- Stall RE, Civerolo EL. Research relating to the recent outbreak of citrus canker in Florida. Annu Rev Phytopathol 1991; 29: 399–420. [DOI] [PubMed] [Google Scholar]
- Leyns F, Decleene M, Swings JG, Deley J. The host range of the genus Xanthomonas. Bot Rev 1984; 50: 308–356. [Google Scholar]
- Schubert TS, Sun X. Bacterial citrus canker. In: Division of Plant Industry, Florida Department of Agriculture and Consumer Services, editor. Plant Pathology Circular. Gainesville, Florida: Division of Plant Industry, 2003: 1–6. [Google Scholar]
- Gottwald T, Graham J, Schubert T. Citrus canker: The pathogen and its impact. Plant Health Progress 2002.
- Brunings AM, Gabriel DW. Xanthomonas citri: Breaking the surface. Mol Plant Pathol 2003; 4: 141–157. [DOI] [PubMed] [Google Scholar]
- Civerolo EL. Bacterial canker disease of citrus. J Rio Grande Valley Hort Assoc 1984; 37: 127–146. [Google Scholar]
- Schubert TS, Rizvi SA, Sun XA, Gottwald TR, Graham JH et al. Meeting the challenge of eradicating citrus canker in Florida - again. Plant Dis 2001; 85: 340–356. [DOI] [PubMed] [Google Scholar]
- Graham JH, Gottwald TR, Cubero J, Achor DS. Xanthomonas axonopodis pv. citri: Factors affecting successful eradication of citrus canker. Mol Plant Pathol 2004; 5: 1–15. [DOI] [PubMed] [Google Scholar]
- Gottwald T, Irey M. Post-hurricane analysis of citrus canker ii: Predictive model estimation of disease spread and area potentially impacted by various eradication protocols following catastrophic weather events. Plant Health Progress 2007.
- Belasque J, Parra-Pedrazzoli AL, Neto JR, Yamamoto PT, Chagas MCM et al. Adult citrus leafminers (Phyllocnistis citrella) are not efficient vectors for Xanthomonas axonopodis pv. citri. Plant Dis 2005; 89: 590–594. [DOI] [PubMed] [Google Scholar]
- Christiano RSC, Dalla Pria M, Jesus WC, Parra JRP, Amorim L et al. Effect of citrus leaf-miner damage, mechanical damage and inoculum concentration on severity of symptoms of Asiatic citrus canker in Tahiti lime. Crop Prot 2007; 26: 59–65. [Google Scholar]
- Graham JH, Gottwald TR, Riley TD, Bruce MA. Susceptibility of citrus-fruit to bacterial spot and citrus canker. Phytopathology 1992; 82: 452–457. [Google Scholar]
- Malamud F, Torres PS, Roeschlin R, Rigano LA, Enrique R et al. The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiol-Sgm 2011; 157: 819–829. [DOI] [PubMed] [Google Scholar]
- Rigano LA, Siciliano F, Enrique R, Sendin L, Filippone P et al. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv citri. Mol Plant Microbe Interact 2007; 20: 1222–1230. [DOI] [PubMed] [Google Scholar]
- Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol 2009; 150: 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laia ML, Moreira LM, Dezajacomo J, Brigati JB, Ferreira CB et al. New genes of Xanthomonas citri subsp citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library. BMC Microbiol 2009; 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Wang N. High-throughput screening and analysis of genes of Xanthomonas citri subsp citri involved in citrus canker symptom development. Mol Plant Microbe Interact 2012; 25: 69–84. [DOI] [PubMed] [Google Scholar]
- da Silva ACR, Ferro JA, Reinach FC, Farah CS, Furlan LR et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002; 417: 459–463. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006; 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Harrison MJ, Lamb CJ. Early events in the activation of plant defense responses. Ann Rev Phytopathol 1994; 32: 479–501. [Google Scholar]
- Espinosa A, Alfano JR. Disabling surveillance: Bacterial type III secretion system effectors that suppress innate immunity. Cell Microbiol 2004; 6: 1027–1040. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 2000; 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature 2006; 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006; 125: 749–760. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Ann Rev Plant Biol 2009; 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S et al. Two Pseudomonas syringae type III effectors inhibit RIM-regulated basal defense in Arabidopsis. Cell 2005; 121(5):749–759. [DOI] [PubMed] [Google Scholar]
- Goto M. Citrus Canker. Englewood Cliff, NJ: Prentice-Hall; 1992. [Google Scholar]
- Gottwald TR, Graham JH, Civerolo EL, Barrett HC, Hearn CJ. Differential host-range reaction of citrus and citrus relatives to citrus canker and citrus bacterial spot determined by leaf mesophyll susceptibility. Plant Dis 1993; 77: 1004–1009. [Google Scholar]
- Amaral AM, Carvalho SA, Silva LFC, Machado MA. Reaction of genotypes of citrus species and varieties to Xanthomonas citri subsp citri under greenhouse conditions. J Plant Pathol 2010; 92: 519–524. [Google Scholar]
- Cardoso SC, Barbosa-Mendes JM, Boscariol-Camargo RL, Christiano RSC, Bergamin A et al. Transgenic sweet orange (Citrus sinensis L. Osbeck) expressing the attacin A gene for resistance to Xanthomonas citri subsp citri. Plant Mol Biol Rep 2010; 28: 185–192. [Google Scholar]
- Mendes BMJ, Cardoso SC, Boscariol-Camargo RL, Cruz RB, Mourao FAA et al. Reduction in susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinensis expressing the rice Xa21 gene. Plant Pathol 2010; 59: 68–75. [Google Scholar]
- Orbovic V, Ananthakrishnan G, Gowda S, Dawson WO, Grosser JW. Genetic engineering of resistance to CTV in grapefruit through Agrobacterium-mediated genetic transformation. In Vitro Cell Dev Biol 2004; 40: 68. [Google Scholar]
- Peltier GL. Susceptibility and resistance to citrus-canker of the wild relative, citrus fruits, and hybrids of the genus Citrus. J Agric Res 1918; 14: 0337–0358. [Google Scholar]
- Prasad MBNV, Singh R, Rekha A, Chand R. Evaluation of lemon cultivars and acid lime X lemon hybrids for resistance to Xanthomonas axonopodis pv. citri. Sci Hortic-Amsterdam 1997; 71: 267–272. [Google Scholar]
- Broadbent P, Fahy PC, Gillings MR, Bradley JK, Barnes D. Asiatic citrus canker detected in a pomelo orchard in Northern Australia. Plant Dis 1992; 76: 824–829. [Google Scholar]
- McCollum G, Stange R, Albrecht U, Bowman K, Niedz R et al. Development of a qPCR technique to screen for resistance to Asiatic citrus canker. Acta Hortic (ISHS) 2011; 892: 173–181. [Google Scholar]
- Al-Saadi A, Reddy JD, Duan YP, Brunings AM, Yuan QP et al. All five host-range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats on citrus, but that determines none determine host-range pathogenicity variation. Mol Plant Microbe Interact 2007; 20: 934–943. [DOI] [PubMed] [Google Scholar]
- Mavrodieva V, Levy L, Gabriel DW. Improved sampling methods for real-time polymerase chain reaction diagnosis of citrus canker from field samples. Phytopathology 2004; 94: 61–68. [DOI] [PubMed] [Google Scholar]
- Rodrigues CM, de Souza AA, Takita MA, Kishi LT, Machado MA. RNA-Seq analysis of Citrus reticulata in the early stages of Xylella fastidiosa infection reveals auxin-related genes as a defense response. BMC Genomics 2013; 14: 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. Plos One 2012; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P. Oxidative burst: An early plant response to pathogen infection. Biochem J 1997; 322: 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomorodipour A, Andersson SGE. Obligate intracellular parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett 1999; 452: 11–15. [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 1999; 18: 265–276. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struc Biol 2001; 11: 725–732. [DOI] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K et al. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 2007; 50: 751–766. [DOI] [PubMed] [Google Scholar]
- Wang GD, Long YC, Thomma BPHJ, de Wit PJGM, Angenent GC et al. Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity. Plant Physiol 2010; 152: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover EW, McCollum TG. Levels of Candidatus Liberibacter asiaticus and Xanthomonas citri in diverse citrus genotypes and relevance to potential transmission from pollinations. Hortscience 2011; 46: 854–857. [Google Scholar]
- Garcia-Lor A, Luro F, Navarro L, Ollitrault P. Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: A perspective for genetic association studies. Mol Genet Genomics 2012; 287: 77–94. [DOI] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol 2014; 32: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IA, Roose ML. Frequency and characteristics of nucellar and zygotic seedlings in 3 cultivars on trifoliate orange. J Am Soc Hortic Sci 1988; 113: 105–110. [Google Scholar]
- Koltunow AM, Hidaka T, Robinson SP. Polyembryony in citrus - Accumulation of seed storage proteins in seeds and in embryos cultured in vitro. Plant Physiol 1996; 110: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MN, Soneji JR, Chen CX, Huang S, Gmitter FG. Characterization of zygotic and nucellar seedlings from sour orange-like citrus rootstock candidates using RAPD and EST-SSR markers. Tree Genet Genomes 2008; 4: 113–124. [Google Scholar]
- Xiang C, Roose ML. Frequency and characteristics of nucellar and zygotic seedlings in 12 citrus rootstocks. Sci Hortic-Amsterdam 1988; 37: 47–59. [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002; 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. Flagellin perception: A paradigm for innate immunity. Trends Plant Sci 2002; 7: 251–256. [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiol 2006; 141: 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Liu L, Lee E, Han X, Rim Y et al. The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol 2009; 150: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D et al. Activation of defense response pathways by OGs and flg22 elicitors in Arabidopsis seedlings. Mol Plant 2008; 1: 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Febres VJ, Jones JB, Moore GA. Responsiveness of different citrus genotypes to the Xanthomonas citri ssp. citri-derived pathogen-associated molecular pattern (PAMP) flg22 correlates with resistance to citrus canker. Mol Plant Pathol 2014; 16: 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol 2004; 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Alexander D, Goodman RM, Gutrella M, Glascock C, Weymann K et al. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein-1a. P Natl Acad Sci USA 1993; 90: 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Winter AM, Delaney T, Vernooij B, Morse A et al. Biological induction of systemic acquired-resistance in Arabidopsis. Mol Plant Microbe Interact 1993; 6: 692–698. [Google Scholar]