Abstract
We identified a novel, evolutionarily conserved receptor encoded within the human Leukocyte Receptor Complex (LRC) and syntenic region of mouse chromosome 7, named T cell-interacting, activating receptor on myeloid cells-1 (TARM1). The transmembrane region of TARM1 contained a conserved arginine residue, consistent with association with a signaling adaptor. TARM1 associated with the ITAM adaptor Fc receptor common γ chain but not with DAP10 or DAP12. In healthy mice, TARM1 is constitutively expressed on the cell-surface of mature and immature CD11b+ Gr-1+ neutrophils within the bone marrow. Following intraperitoneal lipopolysaccharide (LPS) treatment or systemic bacterial challenge TARM1 expression was upregulated by neutrophils and inflammatory monocytes and TARM1+ cells were rapidly recruited to sites of inflammation. TARM1 expression was also upregulated by bone marrow-derived macrophages and dendritic cells following stimulation with TLR agonists in vitro. Ligation of TARM1 receptor in the presence of TLR ligands, such as LPS, enhanced the secretion of pro-inflammatory cytokines by macrophages and primary mouse neutrophils, whereas TARM1 stimulation alone had no effect. Finally, an immobilized TARM1-Fc fusion protein suppressed CD4+ T cell activation and proliferation in vitro. These results suggest that a putative T cell ligand can interact with TARM1 receptor resulting in bi-directional signaling, raising the T cell activation threshold whilst co-stimulating the release of pro-inflammatory cytokines by macrophages and neutrophils.
INTRODUCTION
The initiation and resolution of immune responses are orchestrated through a complex interplay between activating and inhibitory signals transmitted by an array of cell surface receptors. Many immunoreceptors are found as clusters of closely related genes encoding “paired” cell-surface receptors with activating and inhibitory functions and, in some instances, share the same ligands. Families of such “paired” immunoglobulin-like receptors are encoded within the leukocyte receptor complex (LRC) on chromosome 19q13.4 (1). These include the killer immunoglobulin-like receptors (KIR), expressed mainly on NK cells (2), and the leukocyte immunoglobulin-like receptors (LILR), expressed on lymphoid and myeloid cells (3).
The inhibitory members of the LRC gene family contain long cytoplasmic domains with one or more Immunoreceptor Tyrosine-based Inhibition Motif (ITIM) (4); whereas the activating counterparts have a short cytoplasmic tail and encode a charged amino acid in their transmembrane (TM) domain. This charged amino acid, usually an arginine or a lysine, facilitates the association with an Immunoreceptor Tyrosine-based Activation Motif (ITAM)-bearing signaling adaptor such as the Fc Receptor common γ chain (FcRγ) (5-8). Examples of such paired receptors include some with antagonistic signaling properties, such as KIR3DL1/KIR3DS1 and LILRB3/LILRA6, which bind MHC I ligands. The mechanism of signal integration from such receptors and their ability to modulate cellular immune responses is not fully understood.
Although some KIRs and LILRs are known to bind MHC I ligands, non-MHC ligands have also been identified as targets for several LRC-encoded receptors. For example, the activating LRC receptor OSCAR interacts with collagen and is involved in bone metabolism (9). The inhibitory LRC receptor LAIR-1 also binds to collagen, an interaction thought to be involved in tumor immune evasion (10).
The concept that multiple activating receptors encoded within the LRC have closely related inhibitory counterparts encoded in close proximity (1), prompted us to investigate whether OSCAR has an inhibitory paralog. We examined the human genomic sequence adjacent to OSCAR and identified a novel gene encoding an orphan receptor related to OSCAR named T cell interacting, activating receptor on myeloid cells (TARM1). Another related receptor, SIRL-1, encoded by the VSTM1 gene (Genbank NM_198481) is located close to TARM1 and has recently been shown to negatively regulate oxidative burst in human phagocytes (11, 12). The amino acid sequences of SIRL-1 and TARM1 are closely related and they may represent another example of “paired” receptors that duplicated from a common ancestor and acquired antithetical functions in terms of cellular activation.
Neutrophils have traditionally been viewed as short-lived, terminally differentiated effectors of the innate immune response. However, this view has recently been challenged by emerging evidence that circulating neutrophils may live longer than previously appreciated, can undergo reverse transmigration, display plasticity and functional and phenotypic heterogeneity (13) (14). There is compelling evidence that neutrophils engage in bi-directional interactions with a variety of immune cells to modulate adaptive immune responses (15, 16). For instance, ex vivo culture of human and murine neutrophils in the presence of IFN-γ, GM-CSF and IL-3 induces a DC-like phenotype, whereby neutrophils become less susceptible to apoptosis whilst acquiring the ability to prime antigen-specific T cell responses (13, 14, 17-19). Similarly, in the absence of exogenous cytokines, antigen-pulsed neutrophils can present in an MHC II-dependent manner to antigen specific T cells and induce their polarization towards a proinflammatory Th1 or Th17 phenotype (20, 21). In addition, in vivo, neutrophils were found to deliver antigens or live bacilli to the lymph nodes (22, 23) and cross-present to prime naive CD8+ T cells (24, 25).
Neutrophils can also inhibit T cell responses through release of soluble factors such as reactive oxygen species (26, 27), arginase-I (28-30), thromboxanes (31) and, in mice, production of the anti-inflammatory cytokine IL-10 (32, 33). Cell contact-dependent suppression has also been demonstrated. Activated neutrophils were shown to inhibit lymphocyte proliferation through the PD-L1/PD1 pathway in vitro (34) and in HIV-1 infected patients (35). CD11b on human neutrophils from patients with acute systemic inflammation was shown to facilitate the formation of immunological synapses with T cells and enabled a local release of H2O2 (36).
Since identifying the expression profiles and immunoregulatory properties of polymorphic immunoreceptors, such as those encoded in the LRC, contributes to understanding the delicate balance of immune responses, we investigated the role of TARM1 in relation to these findings. We demonstrate that TARM1 is expressed by human and murine neutrophils and associates with the ITAM-bearing adaptor Fc receptor common γ (FcRγ) chain. We also show that cell-surface expression of TARM1 is upregulated by certain TLR agonists in neutrophils, macrophages and dendritic cells and that the concomitant stimulation of TARM1 and TLRs enhances the secretion of pro-inflammatory cytokines by macrophages and neutrophils. In addition, an immobilized TARM1-Fc fusion protein inhibits CD4+ T cell activation and proliferation in vitro.
MATERIALS AND METHODS
Ethics statement
Female NOD mice were bred and maintained under barrier conditions in the Biological Services facility of the Department of Pathology, University of Cambridge (Cambridge, U.K.). Female C57BL/6 mice were purchased at 6 wk of age from Harlan Laboratories (Loughborough, U.K.) and maintained under barrier conditions at the Department of Veterinary Medicine, University of Cambridge. Animals received standard laboratory food and water ad libitum. All animal experiments were approved by the Ethical Review Committee of the University of Cambridge. All human studies were approved by the local Research Ethics Committee and complied with the Declaration of Helsinki. Informed written consent was obtained from healthy adult volunteers (HV) to purify circulating PMNs (ethical approval UK06/Q0108/281).
Sequence information
cDNAs encoding the full coding sequence of human TARM1 (Genbank, DQ479398) and murine TARM1 (DQ973493) were amplified by RT-PCR from total RNA of bone marrow and spleen, respectively, using the following primers: human TARM1 forward primer 5′-actctgggagggctaaggag-3′ was specific to exon1 5’ UTR and reverse primer 5′-gaatgcagtccagcaggttg-3′ was specific to exon 5 3’ UTR. Murine TARM1 forward primer 5′-agacctgctgaagacctttg-3′ was specific to exon1 5’ UTR and reverse primer 5′-agggtttatttggagacagc-3′ was specific to exon 5’ 3’ UTR.
RT-PCR
Total RNA was extracted from tissues of 8-10 week old C57BL/6 female mice with TRIzol reagent (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized from 2 μg total RNA using oligo dT primer and Superscript III (Invitrogen). PCR screening was performed using the following primers: forward primer 5’-agacctgctgaagacctttg-3’ was specific to 5’ UTR region of TARM1 and reverse primer 5’-ttcaaccaggaagcctcccactatta-3’ was specific to exon 6. Mouse Gapdh was used as a reference gene with the following primers: forward 5’-gcagtgccagcctcgtcc-3’ and reverse 5’-tgaggtcaatgaaggggtcgt-3’. Human total RNA Master Panel II was purchased from Clontech (cat. 636643). cDNA was synthesized from 2 μg total RNA using oligo-dT primer and Superscript III (Invitrogen). forward primer 5’-cacaaggggagatgggtcac-3’ was specific to the junction of exons 2 and 3; reverse primer 5’-agccccggttcaagatggag-3’ was specific to exon 5. Human GAPDH was used as a reference gene with the following primers: forward 5’-gaaggtgaaggtcggagtc-3’ and reverse 5’-catcacgccacagtttccc-3’
Quantitative PCR
Mouse tissues were harvested at indicated time points following Salmonella infection and stored in RNAlater (Qiagen) at −20 °C until further processing. Total RNA was extracted using RNeasy kit (Qiagen) and cDNA was synthesized from 2.5 μg total RNA using oligo dT primer and Superscript III (Invitrogen). qPCR was performed using GoTaq qPCR Master Mix (Promega), according to the manufacturer’s instructions on an ABI 7500 Fast Real-Time PCR System. Forward primer sequence 5’-tctgtgatagacaaccatctgcctc-3’ was designed to span the junction of exons 4 and 5. Reverse primer sequence 5’-acaccgacccggatgagatt-3’ was specific to exon 6.
Gapdh was used as a reference gene using Gapdh QuantiTect Primer mix (Qiagen, cat QT01658692). Primer amplification efficiencies (E=1.9 for both TARM1 and Gapdh) were calculated from the slope of a standard curve prepared with a 2-fold serial dilution of splenic cDNA. TARM1 gene expression levels were calculated as fold change over the levels detected in control animals using Fold change=2−ΔΔCT method (37). Primer specificity was verified by melt curve analysis and amplicon sequencing.
Generation of Fc fusion proteins
DNA sequences encoding the extracellular portions of murine (aa 16-255) and human (aa 16-233) TARM1 were cloned into the mammalian expression vector SigpIg Plus consisting of the CD33 signal sequence and a human IgG1 Fc tail. The plasmids were transfected into HEK 293 cells and stable cell lines were established. Cells were maintained in DMEM with 2% low IgG FCS (Invitrogen). Culture medium containing the secreted Fc fusion proteins was collected and TARM1-Fc was purified on columns packed with protein A agarose FastFlow beads (Sigma). Proteins were secreted predominantly as dimers as assessed by Western blot.
Monoclonal antibody production
Monoclonal mouse anti-human TARM1 and rat anti-mouse TARM1 antibodies were raised against the ectodomain of mouse and human TARM1 proteins using TARM1-Fc as immunogens. The performance and specificity of several antibody clones were validated by flow cytometry and Western blot against transfected and primary cells.
Analysis of glycosylation
Hek293T cells were transfected with full-length Flag-tagged TARM1 using FugeneHD (Promega). Total cell lysates were prepared with Nonidet P-40 (NP-40) lysis buffer (1% NP-40, 0.15M NaCl, 0.05M Tris-HCl pH8) containing 1mM AEBSF (Sigma) for 25 min on ice, and then centrifuged at 14,000 × g and 4 °C for 15 min. Supernatants were collected and N-linked carbohydrates were removed by incubation with peptide-N-glycosidase F (PNGaseF) or endoglycosidase H (Endo H) (New England Biolabs) for 4 h at 37 °C, according to the manufacturer’s instructions. Following the treatment, protein mobility shift was examined by SDS-PAGE and Western blotting.
Co-immunoprecipitation
Hek293 cells stably expressing HA-tagged Dap10, Dap12 and FcRγ adaptors were transiently transfected with full-length Flag-tagged human TARM1 and cell lysates were prepared with Tris-buffered saline (TBS) lysis buffer (1% Triton X-100, 0.15M NaCl, 0.02M Tris-HCl pH8) containing 0.5 mM AEBSF (Sigma) and ProteoBlock (Fermentas) for 25 min on ice. Lysates were centrifuged at 10,000 × g and 4 °C for 5 min. Supernatants were collected and incubated with monoclonal anti-TARM1 antibody for 1h at 4 °C on a rotor. Protein A agarose FastFlow beads (Sigma) were added and incubated on a rotor for 1h at 4 °C. The agarose beads were then washed three times with TBS lysis buffer and two times in TBS. Proteins were eluted from the beads by heating at 70 °C for 10 min in LDS sample buffer and separated by SDS-PAGE. Co-immunoprecipitated proteins were analyzed by Western blotting.
SDS PAGE and Western blotting
Cell lysates were denatured in lithium dodecyl sulfate sample buffer (Invitrogen), separated on 10% or 12% Tris-Glycine gels and transferred to Immobilon-P PVDF membrane (Millipore). Blocking and immunoblotting steps were carried out in PBS with 0.05% Tween-20 and 5% milk. Detection was performed with mouse anti-human TARM1 and goat anti-mouse HRP Ab (Invitrogen), or rat anti–HA High affinity Ab (Roche) and anti-rat HRP mouse-adsorbed Ab (Serotech). All blots were developed with ECL Western blotting reagent (GE Healthcare).
Sterile inflammation and S. enterica serovar Typhimurium infection
To induce sterile inflammation C57BL/6 mice were injected with ultra pure LPS (3 μg/mouse) (InvivoGen) intraperitoneally, and tissues were harvested for immunophenotyping 24 h later. Systemic infection was induced with an aroA attenuated strain of S. enterica serovar Typhimurium, SL3261. Bacteria were grown overnight in Luria-Bertani broth until stationary-phase and C57BL/6 mice were injected i.v. with 1 × 106 CFU diluted in PBS. At indicated times following infection, mice were euthanized and TARM1 expression in cells and tissues was analyzed by flow cytometry, qPCR and confocal microscopy.
Confocal microscopy
Spleen fragments from control or Salmonella infected mice were harvested at 2 wpi and fixed in 4% paraformaldehyde, paraffin-embedded and stored until analysis. Tissue sections were deparaffinized, and heat-induced epitope retrieval was performed in citrate buffer pH 5.5. Sections were sequentially stained with anti-TARM1 and goat F(ab’)2 anti-rat FITC (AbD Serotec) secondary Ab. Nuclei were visualized with DAPI. Multiple images of each section were taken to ensure accurate representation.
Murine leukocyte isolation
A single-cell suspension was prepared from spleen, lymph nodes, liver, bone marrow and peritoneal exudate cells (PECs) of mice using standard procedures. Spleens and lymph nodes were dissociated by gently passing the tissues through a 70-μm cell strainer (BD Falcon) followed by hypotonic red blood cell (RBC) lysis. Bone marrow was extracted from femurs and tibias by flushing with 5 ml ice-cold PBS. To prepare total liver cells, livers were treated with 1 mg/ml Collagenase D (Roche) for 30 min at 37 °C and then passed through a 70-μm cell strainer. Total liver cell suspension was washed with PBS-2.5% FCS and the pellet was resuspended in 33% Percoll (GE Healthcare) solution. The suspension was centrifuged for 20 min at 2100 rpm at room temperature and the pellet was washed in PBS-2.5% FCS followed by hypotonic RBC lysis. PECs were isolated by three successive washes of the peritoneal cavity with 4 ml ice-cold PBS.
Isolation of peripheral blood human neutrophils
For flow cytometric analysis of whole blood, 10 ml was collected from healthy volunteers, and leukocyte-rich plasma was prepared by sedimentation of red blood cells using HetaSep (StemCell Technologies) according to manufacturer’s instructions. Circulating neutrophils were purified from healthy volunteers using Dextran sedimentation and discontinuous plasma-Percoll gradients as previously described (38). Briefly, 40 ml of blood was collected into 50 ml tubes (containing 4 mL 3.8% (w/v) sodium citrate) using a 19 gauge needle, and centrifuged for 20 minutes at 300 × g. Platelet-rich plasma upper layer was removed for later use. RBC were removed from the leukocyte/erythrocyte pellet by dextran sedimentation. The upper leukocyte-rich layer was removed and centrifuged at 275×g for 5 minutes. The cell pellet was resuspended in platelet-poor plasma and neutrophils were isolated by discontinuous plasma-Percoll gradient centrifugation. Neutrophil purity as determined by cytospins was routinely greater than 95% using this method.
Generation of bone marrow-derived macrophages and DC
Bone marrow (BM) was flushed from femurs and tibias of 8-12 week old female C57BL/6 mice, dispersed by passage through a 70 μm cell strainer and seeded into 100 mm cell culture dishes at 1×106 cells/ml in complete IMDM (Gibco) containing 10% FBS (Gibco), Penicillin 100 U/ml, Streptomycin 100 μg/ml, L-Glutamine 2 mM (all from Life Technologies) and 50 μM 2-ME (Sigma-Aldrich). For BM macrophage (BMM) differentiation, the medium was supplemented with 10% L-cell conditioned medium as a source of M-CSF. For BMDC differentiation the cells were allowed to adhere for 30 min at 37 °C and the non-adherent cells were reseeded into new plates in complete IMDM supplemented with 20 ng/ml GM-CSF and 10 ng/ml IL-4 (both from PeproTech). Cells were harvested on day 7 and the cellular phenotype was confirmed by flow cytometry using CD11b and F4/80 markers for BMM and CD11c and CD80 for BMDC.
Isolation of murine bone marrow granulocytes
BM was flushed from femurs and tibias of C57BL/6 mice and the total granulocyte population was sorted on a MoFlo cell sorter (Dako Cytomation) using side and forward scatter characteristics. Subsequent staining with anti-CD11b and Gr1 markers showed that over 94% of sorted cells were neutrophils.
Cellular stimulation and measurement of cytokine secretion
For the analysis of TARM1 cell-surface expression, BMM and BMDC were stimulated for 24h with the following TLR agonists (all from Apotech): 100 ng/ml Pam3CSK4 (TLR1/2 agonist), 50 μg/ml Poly(I:C) (TLR3 agonist), 100ng/ml LPS from E. coli R515 (TLR4 agonist), 50 ng/ml Flagellin from S. Typhimurium (TLR5 agonist), 80 ng/ml MALP-2 (TLR2/6 agonist), 1 μg/ml Imiquimod (TLR7 agonist), 2 μg/ml CpG ODN 1585 (TLR9 agonist), and 250 ng/ml Profilin from Toxoplasma gondii (TLR11 agonist). For analysis of cytokine secretion, 1 × 105 of BMM or 1.3 × 105 sorted neutrophils were cultured in 96-well tissue culture plates coated with goat anti-rat capture Ab and rat anti-TARM1 crosslinking Ab or a rat IgG2a isotype control Ab (all at 10 μg/ml). Neutrophils were stimulated in the presence or absence of 10 ng/ml Ultrapure LPS from E. coli K12 (InvivoGen) for 8 h at 37 °C. BMM were stimulated in the presence or absence of either 0.5 μg/ml Pam3CSK4, 0.5 μg/ml PolyI:C, 1 ng/ml LPS, or 10 μM Imiquimod for 16 h at 37 °C. Tissue culture supernatants were collected and cytokine production determined using the Cytometric Bead Array mouse inflammation kit (Becton Dickinson). DuoSet sandwich ELISA (R&D) was used to determine IL-2 secretion by stimulated mouse T cells.
Flow cytometric cell-surface phenotyping
For flow cytometric cell phenotyping, 0.5 - 1 × 106 cells were stained per well. The following antibodies were used: goat anti-mouse IgG Alexa Fluor 647 secondary Ab (Molecular Probes), goat anti-rat IgG (H+L) PE mouse adsorbed secondary Ab (SouthernBiotech). Anti-CD3-FITC (2C11), CD19-APC (1D3), CD11b-APC (M1/70), CD11c (N418), Gr-1 (RB6-8C5), CD25 (PC61), CD69 (H1.2F3) were from eBioscience; Ly6G FITC (1A8), Ly6C APC (HK1.4) were from BioLegend; AnnexinV FITC (BD Pharmingen). Flow cytometry was performed on BD FACScan with CyTek DxP three laser setup and data analysis was carried out using FlowJo 10 software (Tree Star).
T cell isolation and in vitro proliferation assays
For CFSE dilution assay CD4+ T cells were purified from naive NOD female mice using EasySep positive selection magnetic beads (StemCell Technologies) to 93–98% purity as assessed by flow cytometric analysis. Cells were loaded with 1 μM CFSE (Molecular Probes) in PBS for 15 min at 37 °C, washed and seeded into a 96-well flat-bottomed plates coated, in two layers, with first 10 μg/ml anti-human IgG Fc-specific antibody (ImmunoReagents) and 1 μg/ml anti-CD3 (clone 145-2C11, eBioscience) together with 0.25 μg/ml anti-CD28 (clone 37.51, Miltenyi) over night at 4 °C, washed and then coated with 10 μg/ml mouse TARM1-Fc or human IgG1 control. A total of 1.5 × 105 T cells were added to each well in a volume of 200 μl RPMI 1640 with 10% FCS, penicillin/streptomycin and 50 μM 2-ME. CFSE dilution was assessed after 90 h of culture. For metabolic MTT assay, CD4+ T cells were purified using EasySep negative selection magnetic beads (StemCell Technologies) and activated as described above, but excluding anti-CD28 antibody. Cells were cultured for 20 h, 40 h and 80 h. Two hours prior to each time point, MTT (Sigma) stock solution was added directly to the wells to give 500 μg/ml final concentration and following 2 h incubation at 37 °C, DMSO was used to dissolve formazan crystals and absorbance (570 nm) was measured on a Synergy HT plate reader. Human T cells were isolated from peripheral blood of healthy donors (n=2) using CD4+ positive selection magnetic beads (Miltenyi) to 95-98% purity and activated with plate-bound anti-CD3 (1.2 μg/ml, clone OKT3) in the presence of plate-bound human TARM1-Fc (10 μg/ml) or hIgG1 (10 μg/ml). Recombinant human IL-2 (50 U/ml) was added to some wells as indicated. T cells were cultured for 3 days and the expression of CD25 and CD69 was analyzed by flow cytometry.
RESULTS
TARM1 is a novel LRC-encoded immunoreceptor
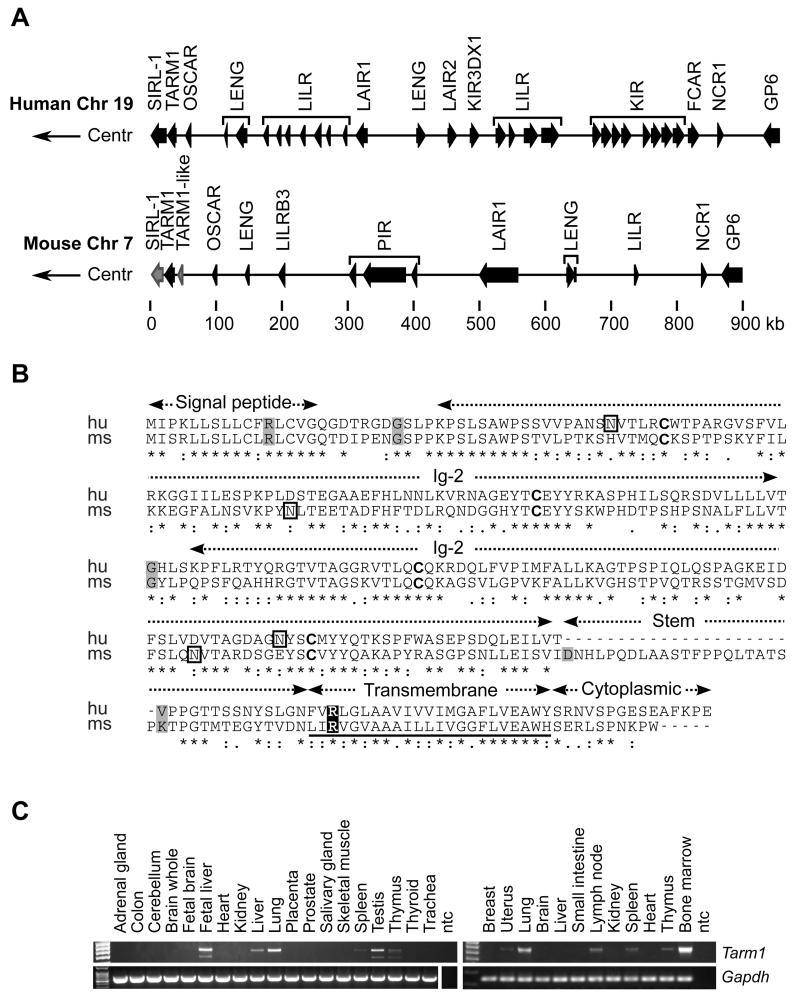
We performed a bioinformatics analysis of open reading frames encoded at the centromeric end of the human LRC on chromosome 19q13.4 and identified a putative gene TARM1 (Genbank NM_001135686) closely related to VSTM1 and OSCAR (Fig. 1A). We also identified the analogous murine TARM1 gene, which resembles closely its human ortholog both in sequence and exon structure, and is encoded within the syntenic region of the murine chromosome 7A1 (Fig. 1B). The TARM1 gene spans <11.4 kb of human genomic sequence encoding a polypeptide of 271 amino-acids (aa), and <13.4 kb of mouse sequence encoding a polypeptide of 288 aa. Protein alignment of human and mouse orthologs indicated <47% sequence identity and 62% sequence similarity. Both are type Ia membrane proteins composed of a 16-aa hydrophobic signal peptide followed by two extracellular Ig-like domains, a transmembrane (TM) region with a conserved arginine and a short cytoplasmic tail devoid of signaling motifs.
FIGURE 1.
Genomic location of human and mouse TARM1 genes and comparison of predicted amino-acid sequences. (A) Genomic organization of the human LRC (top) on chromosome 19q13.4 and the syntenic mouse region (bottom) on chromosome 7A1. Gene orientation is indicated by arrows; pseudogenes are in grey; Centr, centromeric. (B) Amino-acid sequence alignment of human and mouse TARM1 orthologs. Structural protein domains are labeled above the alignment. Grey-shaded amino-acids are encoded at exonic splice junctions. Conserved cysteines potentially involved in the formation of the IgSF-fold intra-chain disulfide bridge are in bold. The transmembrane region is underscored and contains a conserved arginine (highlighted) predicted to mediate association with FcRγ chain. Asparagine residues predicted to be glycosylated are enclosed in black rectangles. (C) Expression of full-length TARM1 mRNA in human and mouse tissues. PCR was performed on cDNA synthesized from Human Total RNA Master Panel II, ClonTech (left panel). Mouse tissue total RNA panel was produced in-house (right panel). TARM1 specific primers amplified full-length coding region of the transcript. Gapdh was used as a reference gene. ntc, no template control.
A BLAST search of the NCBI human and mouse sequence databases using nucleotide and protein sequences of TARM1 revealed that it is most closely related to members of the immunoglobulin superfamily (IgSF) encoded within the LRC, in particular SIRL-1 and OSCAR. The single Ig-V domain of SIRL-1 is <48% identical and <58% similar to the first Ig-like domain of TARM1. The ectodomains of OSCAR and TARM1 proteins are <36% identical and <46% similar in their amino acid sequences.
We amplified a full-length transcript of TARM1 and examined its expression in normal human and mouse tissues by RT-PCR. TARM1 mRNA was expressed in lymphoid organs thymus, spleen, mesenteric lymph nodes, with most transcript detected in the bone marrow. TARM1 expression was also detected in non-lymphoid tissues lung and uterus (Fig. 1C).
TARM1 protein is glycosylated and associates with Fc receptor common γ chain
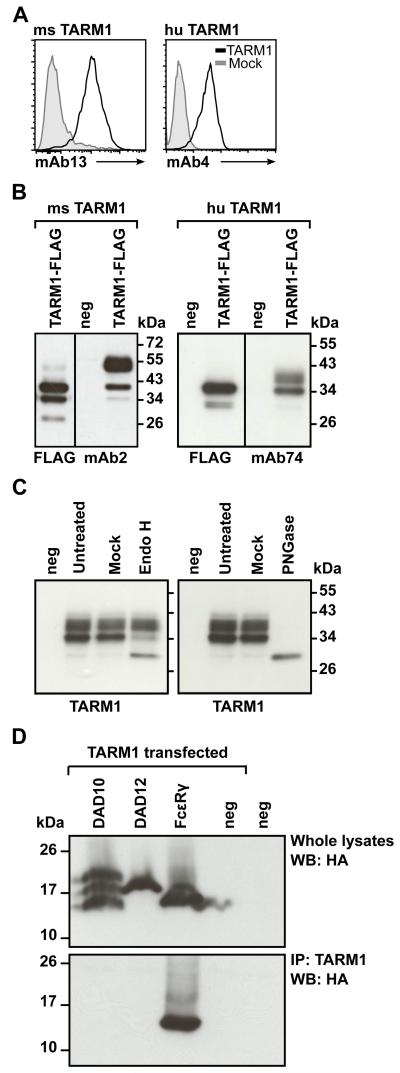
To characterize TARM1 further we raised specific monoclonal antibodies to the human and mouse TARM1 receptors for use in flow cytometry and Western blotting. Antibody clones mAb13 (rat anti-mouse TARM1) and mAb4 (mouse anti-human TARM1) stained TARM1-transfected Hek293T cells but not mock-transfected cells, as determined by flow cytometry (Fig. 2A). Clones mAb2 (rat anti-mouse TARM1) and mAb74 (mouse anti-human TARM1) detected Flag-tagged TARM1 in transfected cell lysates by Western blot (Fig. 2B). Surprisingly, the anti-Flag antibody failed to detect the higher molecular mass (Mr) band detected by the TARM1 specific antibodies in both human and mouse blots. We speculated that this discrepancy may be due to the steric occlusion of the Flag epitope caused by TARM1 glycosylation.
FIGURE 2.
TARM1 is N-glycosylated and associates with the ITAM-containing adaptor FcRγ. (A) Anti-TARM1 monoclonal Abs bind specifically to TARM1. Hek293T cells transiently transfected with TARM1 (open histograms) or mock-transfected (filled histograms) were stained with monoclonal anti-mouse TARM1 mAb13 (left panel) or anti-human TARM1 mAb4 (right panel) and fluorescently labeled secondary Abs and analyzed by flow cytometry. (B) Western blot analysis of the total lysate from TARM1-transfected cells. Hek293T cells were transfected with full-length Flag-tagged TARM1 and the specificity of anti-TARM1 antibodies determined by Western blot. Anti-Flag and anti-TARM1 monoclonal antibodies detected bands of <40 kDa for mouse TARM1 (left panel, mAb2) and <34 kDa for human TARM1 (right panel, mAb74). Anti-Flag antibody failed to detect the higher molecular mass band of both mouse and human TARM1 proteins. No bands were detected in untransfected (neg) cell lysates. (C) TARM1 is N-glycosylated. Total cell lysates of TARM1-transfected cells were either left untreated (Untreated), mock treated (Mock) or treated with EndoH (left panel) or PNGase (right panel) and analyzed by Western blot. neg, untransfected cells. (D) TARM1 co-immunoprecipitated with the ITAM-containing adaptor FcRγ chain. Hek293 cells stably expressing either the HA-tagged adaptors DAP10, DAP12 or FcRγ or parental Hek293 cells (neg) were transiently transfected with full-length TARM1. Whole cell lysates were analyzed by Western blot with anti-HA antibody to confirm the presence of adaptor proteins (top panel). TARM1 was then immunoprecipitated using monoclonal anti-TARM1 antibody and the complex analyzed by Western blot (bottom panel) with anti-HA antibody to detect the co-immunoprecipitated ITAM adaptor.
Two N-glycosylation sites were predicted for both human and mouse TARM1 polypeptides. To determine the glycosylation state, Hek293T cells were transiently transfected with Flag-tagged full-length human TARM1 and the total cell lysates were treated with either PNGase F or Endo H. The protein mobility shift was examined by SDS-PAGE (Fig. 2C). The predicted Mr of the unglycosylated TARM1 protein is <29.5 kDa. Immunoblotting of untreated or mock-treated protein with anti-TARM1 mAb74 showed two bands of <40 and <34 kDa indicating the presence of post-translational protein modification. Treatment with Endo H resulted in a conversion of only the 34 kDa band to <30 kDa (Fig. 2C, left panel). This suggests that the immature, Endo H sensitive form of TARM1 protein is recognized by both the anti-Flag and the anti-TARM1 Abs, however, the fully glycosylated higher Mr TARM1 species may not be recognized by anti-Flag Ab. Treatment with PNGase F resulted in a single TARM1 that migrated at <30 kDa, which represented the fully deglycosylated form (Fig. 2C, right panel).
An arginine residue is embedded within the TM region of mouse and human TARM1. In the OSCAR TM this residue mediates association with FcRγ, resulting in increased cell surface expression and transduction of activating ITAM signals (6, 8, 39, 40). To examine whether TARM1, like OSCAR, associates with an ITAM-containing adaptor, we generated Hek293 cell-lines stably expressing hemagglutinin (HA) epitope-tagged FcRγ (HA-FcRγ), DAP12 (HA-DAP12) and DAP10 (HA-DAP10) adaptors, respectively. The cell lines were transiently transfected with a full-length Flag-tagged human TARM1 and used in co-immunoprecipitation experiments. Only FcRγ, but not DAP10 or DAP12, co-immunoprecipitated with TARM1 (Fig. 2D). These data demonstrate that TARM1 can specifically associate with the ITAM-containing adaptor FcRγ, consistent with an activating function (6, 8, 39, 40).
TARM1 is expressed constitutively by bone marrow granulocytes, monocytes and neutrophils that home to sites of inflammation
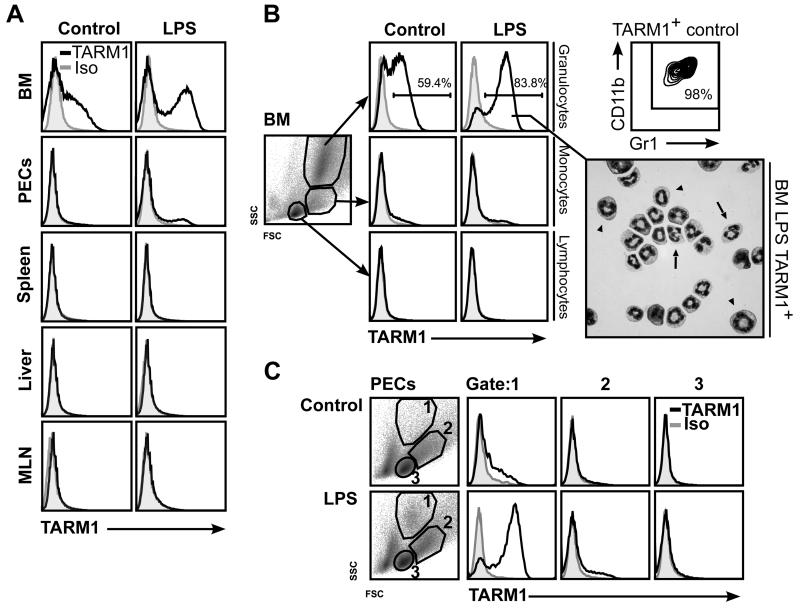
High expression of mouse TARM1 mRNA was detected in the bone marrow (BM) of healthy C57BL/6 mice by RT-PCR analysis. Constitutive cell-surface expression of TARM1 protein was confirmed by flow cytometry and was only observed within the BM and not at peripheral sites (Fig. 3A). Closer inspection revealed that TARM1 expression was restricted to granulocytes with a CD11b+Gr1+ cell-surface phenotype (Fig. 3B).
FIGURE 3.
In healthy mice, TARM1 expression is restricted to bone marrow neutrophils and granulocyte precursors. C57BL/6 mice were injected i.p. with either PBS or ultra-pure LPS (3 μg/mouse) and sacrificed 24 h later. (A) Flow cytometric analysis of cell-surface expression of TARM1 in the bone marrow (BM), peritoneal exudate cells (PECs), liver and mesenteric lymph nodes (MLN) following LPS treatment. (B) BM granulocytes expressed TARM1. Inset: cytospin preparation of FACS-sorted TARM1+ cells from the BM of LPS treated animals (stained with DiffQuick, 100 × magnification) is a mixture of immature (arrowheads) neutrophils distinguished by their large ring-shaped nuclei, and mature neutrophils (arrows) with characteristic segmented nuclei. (C) Flow cytometric analysis of the peritoneal lavage from LPS treated or control animals showed an influx of TARM1+ cells following LPS treatment. Individual gates were set according to the Fsc:Ssc characteristics of granulocytes (gate 1), monocytes (gate 2) and lymphocytes (gate 3). Anti-TARM1 mAb (open histograms) or isotype control (filled histograms). Analysis is based on live cells. The plots are representative of three independent experiments with three mice in each.
TARM1 mRNA expression could be further enhanced by intraperitoneal (i.p.) injection of low dose (150 μg/kg) LPS (data not shown). To assess whether this increase in TARM1 transcript correlated with the increase in cell-surface protein expression, we repeated the low dose LPS treatment and examined TARM1 expression in several organs 24 h following i.p. injection. LPS treatment caused a strong upregulation of TARM1 cell-surface expression within the CD11b+Gr1+ population (Fig. 3B). Microscopy of TARM1+ cells isolated from the BM of LPS treated animals revealed a heterogenous population composed of mature and immature neutrophils, as determined by cell morphology (Fig. 3B inset).
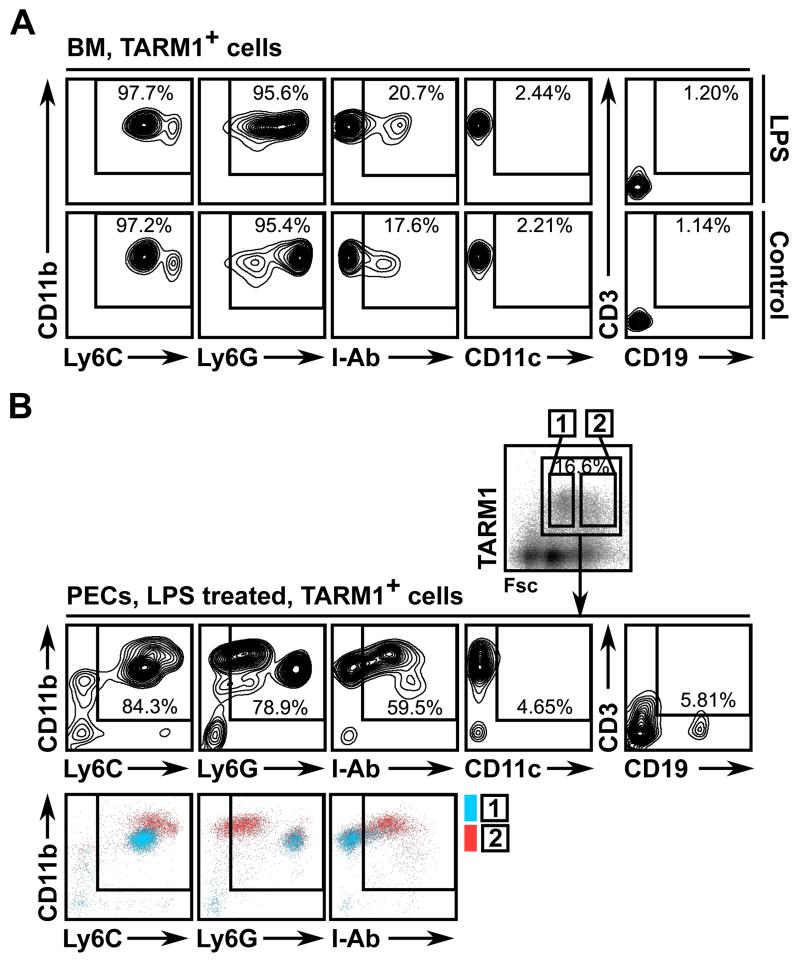
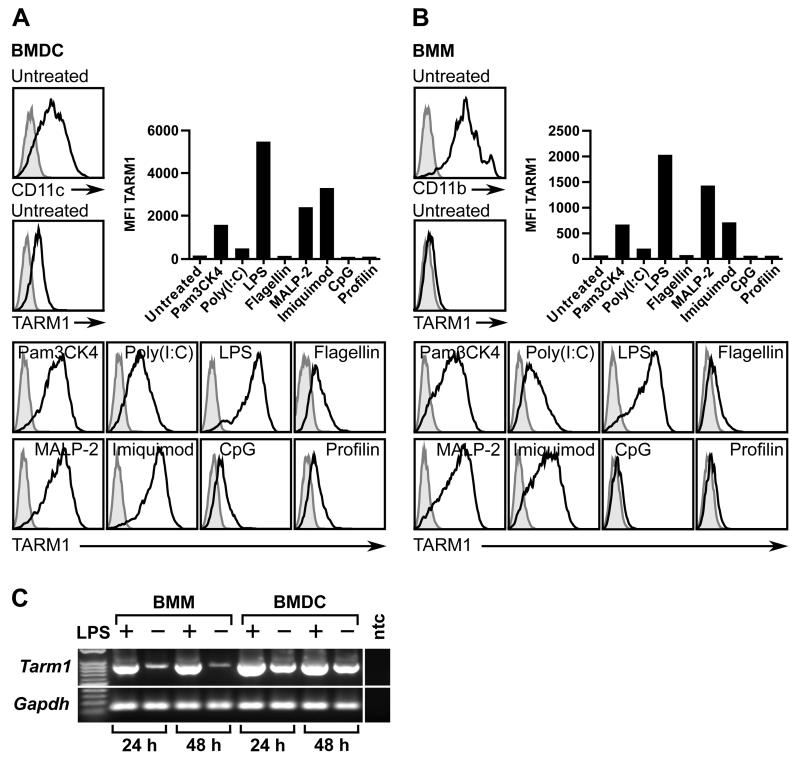
Interestingly, following i.p. LPS administration, peritoneal exudate cells (PECs) contained a population with forward scatterint side scatterhi (FscintSschi) phenotype comprised predominantly of TARM1+ cells (Fig. 3C). This population was not present in PECs from control animals. The anti-Gr1 (RB6-8C5) antibody recognizes epitopes present in both Ly6C and Ly6G. We therefore used Ly6C and Ly6G-specific antibodies for further phenotypic characterization of TARM1+ cells. We found that TARM1+ cells in the BM co-expressed classical markers of the granulo-monocytic lineage, such as CD11b, Ly6G and Ly6C, but were negative for markers of B cells (CD19), T cells (CD3) and DC (CD11c) (Fig. 4A). Twenty-four hours following LPS injection, TARM1+ cells infiltrating the peritoneum formed two distinct populations based on their Fsc characteristics (Fig. 4B). The predominant population in gate 1 (Fig. 4B) was CD11b+Ly6CintLy6GhiMHCII−. The population in gate 2 was CD11b+Ly6ChiLy6GloMHCII+. These phenotypic characteristics are consistent with granulocytic origin of the population in gate 1, and monocytic origin of the population in gate 2. Since TARM1+ cells did not express DC/macrophage marker CD11c in either the bone marrow or the peritoneum, the MHCII+ cells in gate 2 are most likely inflammatory monocytes. These cells were shown to be rapidly recruited to the sites of inflammation where they can give rise to pro-inflammatory DC and macrophages (41). To investigate whether DC and macrophages express TARM1 under inflammatory conditions, we generated bone marrow-derived DC (BMDC) and macrophages (BMM) and stimulated the cells with a panel of TLR agonists (Fig. 5). Unstimulated BMDC (Fig. 5A), but not BMM (Fig. 5B) expressed low levels of cell surface TARM1. The cell surface expression of TARM1 was upregulated to varying degrees following stimulation of BMDC and BMM with agonists for TLR-1/2, -3, -4, -2/6, and -7, respectively. In contrast, TARM1 expression remained unchanged following stimulation with agonists for TLR-5, -9, or -11, respectively (Fig. 5A and B). Upregulation of Tarm1 transcript by BMM and BMDC following 24 h LPS stimulation was confirmed by semi-quantitative RT-PCR (Fig. 5C). These results show that DC and macrophages upregulate TARM1 at the transcriptional and translational level under specific pro-inflammatory conditions.
FIGURE 4.
TARM1 is expressed by CD11b+Ly6G+Ly6C+ neutrophils and infiltrating CD11b+Ly6GloLy6ChiMHCII+ inflammatory monocytes. C57BL/6 mice were injected i.p. with either PBS or ultra-pure LPS (3 μg/kg) and sacrificed 24 h later. (A) BM and (B) PECs were isolated and stained with anti-TARM1 mAb and cell lineage markers. Plots show TARM1-gated cells.
FIGURE 5.
TARM1 expression by BMDC and BMM is upregulated by specific TLR agonists. (A) BMDC and (B) BMM were stimulated with a panel of TLR agonists (see Materials and Methods) and the median fluorescence intensity (MFI) of immunostaining with either TARM1 mAb (open histograms) or rat IgG2a isotype control (filled histograms) was determined by flow cytometry. (C) Expression of Tarm1 mRNA by LPS stimulated and control BMM and BMDC as determined by semi-quantitative RT-PCR 24 h or 48 h following stimulation. Gapdh was used as a reference ‘house-keeping gene’ (ntc, no template control).
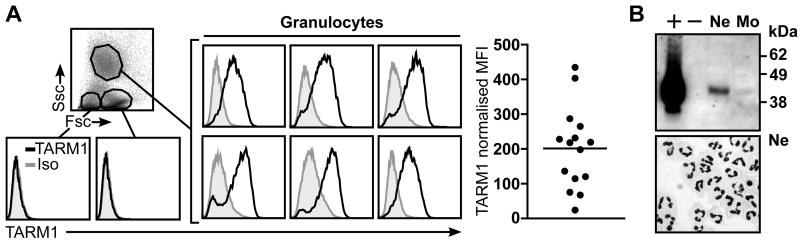
We also examined TARM1 protein expression in human peripheral blood (Fig. 6). As in mice, human TARM1 was detected on circulating granulocytes, but the expression varied greatly among donors (n=15) (Fig. 6A). Western blot analysis of purified peripheral blood neutrophils confirmed the presence of TARM1 protein within these cells but not in neutrophil-depleted mononuclear cell fraction (Fig. 6B).
FIGURE 6.
TARM1 is expressed on the surface of circulating human neutrophils. (A) TARM1 expression on circulating human leukocytes from healthy donors (n=15) was analyzed by flow cytometry. Only granulocytes were positive for TARM1. Representative histograms are shown. (B) Western blot analysis of purified neutrophils (Ne) and neutrophil-depleted mononuclear cell fraction (Mo) (top panel). Neutrophil purity was assessed by cytospin stained with DiffQuick and 100 × magnification (bottom panel) and was routinely >95%. +, TARM1-transfected cells; −, mock-transfected cells.
These results indicate that in healthy mice TARM1 expression is restricted to the BM CD11b+Ly6C+Ly6G+ mature and immature neutrophils. The inflammatory response was strongly associated with increased TARM1 expression on BM neutrophils and with homing of TARM1+ cells to peripheral sites. Monocytes infiltrating the site of inflammation also expressed cell-surface TARM1. As in mice, human TARM1 was also expressed on granulocytes suggesting evolutionary conservation of TARM1 function.
Systemic Salmonella infection causes upregulation of TARM1 expression by granulocytes and their accumulation in the spleen
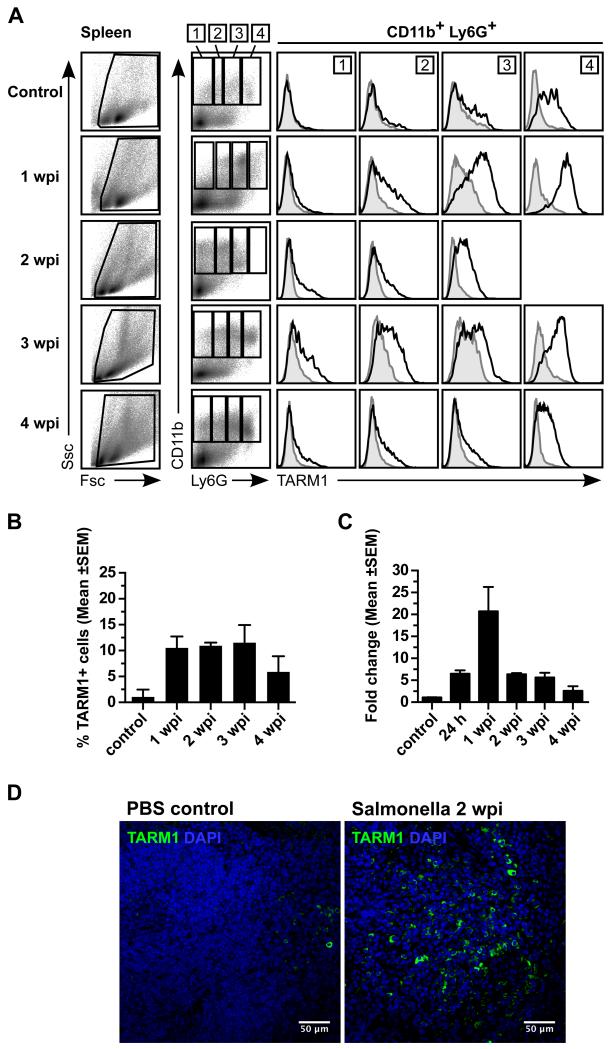
To study the effects of a systemic immune challenge on TARM1 expression, we administered an aroA attenuated strain of Salmonella enterica serovar Typhimurium (S. Typhimurium) SL3261 i.v. to C57BL/6 mice. Flow cytometric analysis of spleens at 1, 2, 3 and 4 weeks post infection (wpi) showed an influx of TARM1+ granulocytes as evidenced by the accumulation of Fscint/hiSschi population (Fig. 7A). TARM1 receptor density correlated positively with Ly6G expression. TARM1+ cells reached ~10% of total splenocytes by 1 wpi and declined between 3 and 4 wpi (Fig. 7B). Quantitative PCR (qPCR) analysis of Tarm1 mRNA expression in total splenocytes of infected mice indicated a ~6-fold upregulation over control animals at 24 h following infection, with a peak expression of ~22-fold at 1 wpi (Fig. 7C). The presence of infiltrating TARM1+ cells was also confirmed by confocal microscopy 2 wpi in the spleens of infected but not control animals (Fig. 7D). A strong upregulation of TARM1 expression on the cell-surface of CD11b+Ly6C+ and CD11b+Ly6G+ cells was observed at the same time point p.i. in the BM (Supplemental Fig. 1). Thus systemic infection with Salmonella resulted in upregulation of TARM1 cell-surface expression on granulocytes and their migration to the spleen.
FIGURE 7.
Systemic infection with S. Typhimurium causes upregulation of TARM1 expression by CD11b+Ly6G+ neutrophils and their accumulation in the spleen. C57BL/6 mice (n=3) were injected i.v. with either PBS or aroA attenuated S. Typhimurium strain SL3261 (1 × 106 CFU) and sacrificed at the indicated time points. TARM1 expression was determined by flow cytometry, qPCR and confocal microscopy. Representative plots from one of three independent experiments are shown. (A) Flow cytometric analysis of the expression of TARM1, CD11b and Ly6G on total splenocytes at the indicated time points following infection. CD11b+ cells were gated according to their Ly6G expression (gates 1-4). Histogram overlays show TARM1 expression in corresponding Ly6G gates. (B) Percentage of TARM1+ cells in total splenocytes at indicated time points following infection. (C) qPCR analysis of TARM1 mRNA expression in total splenocytes over the course of infection. (D) Confocal microscopy of the spleen at 2 wpi and uninfected control. Green, TARM1; Blue, DAPI.
TARM1 co-stimulates pro-inflammatory cytokine secretion in BMM and primary neutrophils
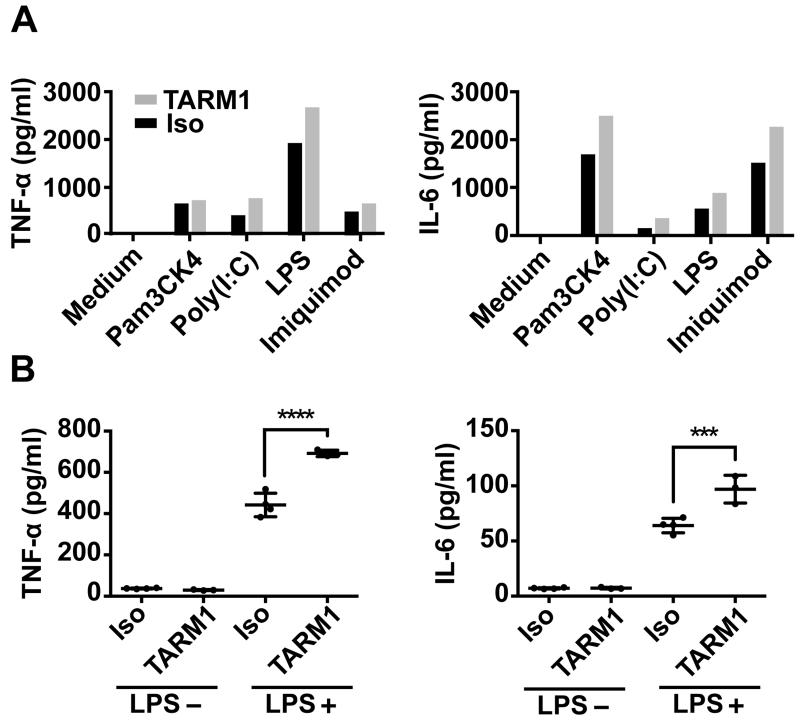
ITAM receptors such as OSCAR and TREM-1 have been shown to regulate cytokine secretion (42) and pro-inflammatory responses (43, 44) of myeloid cells. Since TARM1 expression was regulated by TLR ligands we examined whether TARM1 ligation in vitro might influence the secretion of pro-inflammatory cytokines by BMM and neutrophils. BMM or primary mouse BM neutrophils were cultured in tissue culture plates coated with either TARM1-specific or isotype control mAbs in the presence or absence of TLR agonists (Fig. 8). Cross-linking of TARM1 in the absence of TLR agonists did not stimulate cytokine secretion by either BMM or neutrophils (Fig. 8A,B). However, the concomitant stimulation of TARM1 and TLR-1/2, -3, -4 and -7, respectively, enhanced secretion of the pro-inflammatory cytokines TNF-α and IL-6 by BMM (Fig. 8A). Similarly, concomitant stimulation of TARM1 and TLR4 enhanced the secretion of TNF-α and IL-6 by neutrophils (Fig. 8B). These results show that TARM1 cooperates with TLR stimulation to enhance the secretion of pro-inflammatory cytokines by macrophages and neutrophils.
FIGURE 8.
TARM1 co-stimulates the secretion of pro-inflammatory cytokines by macrophages and neutrophils in a TLR-dependent manner. Concentrations of TNF-α or IL-6 in supernatants derived from (A) BMM or (B) primary neutrophil cultures stimulated with either immobilized anti-TARM1 (TARM1) or rat IgG2a isotype control mAb (Iso), with or without agonists (see Materials and Methods) for TLR-1/2, -3, -4 and -7, respectively (***, P ≤ 0.001; ****, P ≤ 0.0001).
TARM1 inhibits T cell activation and proliferation in vitro
Numerous studies have demonstrated a role for neutrophils in the regulation of T cell responses. We therefore examined the immunomodulatory effects of TARM1 on anti-CD3/-CD28 induced CD4+ T cell activation and proliferation. For this purpose, we generated soluble mouse TARM1-Fc fusion protein comprised of the ectodomain of TARM1 fused to the Fc portion of human IgG1.
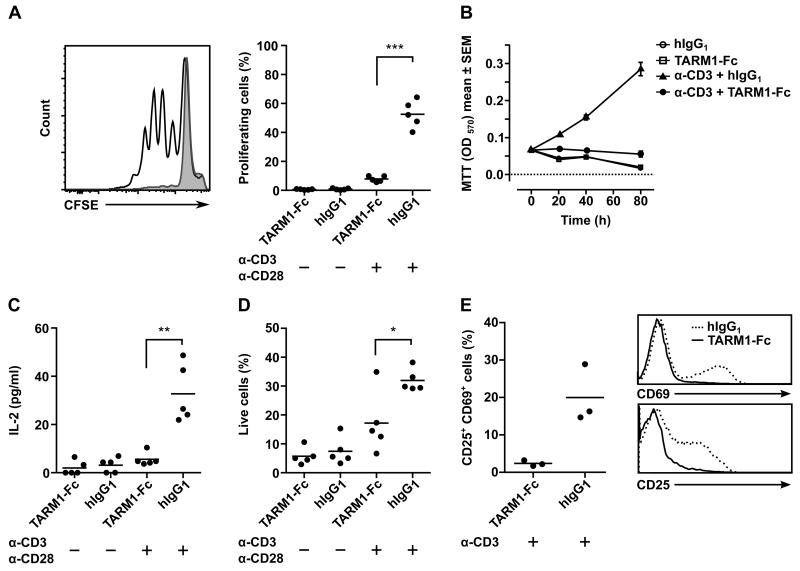
When purified primary mouse CD4+ T cells were activated with plate-bound anti-CD3 and anti-CD28 in the presence of plate-bound TARM1-Fc fusion protein, T cell proliferation was inhibited as measured by CFSE dilution assay (Fig. 9A). Cells proliferated normally when control IgG1 was used instead of TARM1-Fc. Since cellular activation and proliferation is accompanied by an increase in cellular metabolism, we used a tetrazolium salt MTT as a redox indicator in a metabolic proliferation assay. This assay allows multiple measurements to be carried out throughout the course of cell stimulation and enables the study of T cell activation dynamics. We used MTT to measure TARM1-Fc mediated inhibition of CD4+ T cell activation over the course of 4 days and observed a significant inhibition as early as 20 h (Fig. 9B). The inhibition was accompanied by a significantly reduced IL-2 secretion (Fig.9C). The proliferation and IL-2 secretion were not inhibited when human IgG1 replaced TARM1-Fc. The inhibition could not be attributed to cytotoxicity as TARM1-Fc had no impact on T cell viability (Fig. 9D). The decreased live cell count seen in the presence of TARM1-Fc is most likely due to the T cell apoptosis as a consequence of the absence of stimulation. In support of this, the T cell activation markers CD25 and CD69 were lower on T cells stimulated in the presence of TARM1-Fc (Fig. 9E). Immobilized human TARM1-Fc also inhibited the activation of human CD4+ T cells compared to control hIgG1 (Supplemental Fig. 2). Collagen was identified as the ligand for the closest TARM1 homologue OSCAR (9). However, we did not detect specific binding of human TARM1-Fc to collagens I-V (data not shown). These results show that the TARM1 receptor can interact with an as yet unidentified ligand on T cells to inhibit T cell activation and proliferation.
FIGURE 9.
TARM1 inhibits CD4+ T cell activation and proliferation in vitro. (A) CFSE dilution assay. CD4+ T cells isolated by positive selection from spleens and mesenteric lymph nodes of NOD mice (n=5) were labeled with CFSE and activated with plate-bound anti-CD3 (1 μg/ml) and anti-CD28 (0.25 μg/ml) Abs in the presence of either plate-bound TARM1-Fc (10 μg/ml) or control hIgG1 (10 μg/ml). Proliferation was determined by CFSE dilution after 90 h of culture. (B) Metabolic MTT assay. CD4+ T cells were isolated by negative selection from spleens of NOD mice (n=4) and activated with plate-bound anti-CD3 antibody (1 μg/ml) in the presence of either plate-bound TARM1-Fc (10 μg/ml) or control hIgG1 (10 μg/ml). T cell activation and proliferation was determined at 0 h, 20 h, 40 h and 80 h of culture. (C) TARM1-Fc inhibits IL-2 secretion by stimulated T cells. T cells were isolated and stimulated as described for CFSE dilution assay and IL-2 concentration in the culture supernatant was measured at 24 h of stimulation. (D) TARM1-Fc does not affect T cell viability. T cells were isolated and stimulated as described for CFSE dilution assay and the viability was determined by staining with AnnexinV and PI after 90 h of culture. AnnexinV−PI− cells were considered viable. (E) T cells stimulated with anti-CD3 (1 μg/ml) in the presence of plate-bound TARM1-Fc (10 μg/ml), but not hIgG1 (10 μg/ml), fail to upregulate activation markers CD25 and CD69. The plot shows percentage of CD25+CD69+ double positive T cells on day 3 of stimulation. Representative histograms of CD69 (top) and CD25 (bottom) expression in the presence of TARM1 or hIgG1.
DISCUSSION
We report the identification and characterization of a novel LRC-encoded receptor, TARM1. Like many other LRC members, TARM1 has two Ig-like folds and thus belongs to the broader IgSF. Like the activating LRC receptors, TARM1 lacks signaling motifs in its short cytoplasmic tail and, instead, associates with FcRγ via a conserved arginine in its TM suggesting an activating function (4). Phylogenetic analysis shows that human TARM1 is most closely related to an inhibitory receptor SIRL-1 (11) encoded by the neighboring VSTM-1 gene. SIRL-1 expression is restricted to phagocytes such as neutrophils and monocytes, where it plays a role in the negative regulation of the oxidative burst (12) and can prevent the pathogenic release of neutrophil extracellular traps in cells from SLE patients (45).
Our flow cytometric analysis of human peripheral blood showed that, similarly to SIRL-1, human TARM1 is also expressed on neutrophils but not monocytes from healthy individuals, although the expression level varied greatly amongst donors. The expression of murine TARM1 is restricted to the bone marrow CD11b+Ly6G+Ly6C+ granulocyte precursors and mature neutrophils under steady-state conditions. LPS challenge, or systemic infection with S. Typhimurium, induced a strong upregulation of cell-surface TARM1 expression by bone marrow CD11b+Ly6G+Ly6C+ cells and their migration to the site of inflammation. Following LPS administration i.p., TARM1+ cells homed rapidly to the peritoneum, whereas systemic Salmonella infection induced an accumulation of TARM1+ cells in the spleen. Since the pro-inflammatory environment has been shown to increase neutrophil lifespan (13, 14), and inflammation upregulates TARM1 expression by neutrophils, increased TARM1 expression (TARMHi) may be a marker for neutrophils with enhanced in vivo lifespans.
Over 50% of TARM1+ cells that migrated to the peritoneum upregulated MHC II expression and had a CD11b+Ly6ChiLy6Glo phenotype but lacked the DC/macrophage marker CD11c. Given the rapid recruitment to the peritoneum following LPS injection, the CD11b+Ly6ChiLy6GloMHCII+TARM1+ cells likely represent inflammatory monocytes. These cells were shown to rapidly migrate to the sites of inflammation where they can give rise to pro-inflammatory DC and macrophages (41). Indeed, TARM1 expression could be induced on BMM and BMDC following stimulation with ligands for TLR-1/2, -3, -4, -2/6, and -7, respectively, but not TLR-5, -9 or -11 agonists. Further studies are required to determine whether human TARM1 can be expressed by additional immune cell types other than neutrophils (e.g. monocytes, macrophages or DC), and whether human TARM1 cell-surface expression is modulated by TLR agonists and other pro-inflammatory stimuli.
Interestingly, the amino acid sequence of the single IgV domain of SIRL-1 is <48% identical to the first Ig-like domain of TARM1. However, owing to its two ITIMs, SIRL-1 exerts inhibitory effects on neutrophil function (12, 45), whereas TARM1 associates with ITAM-bearing adaptor FcR© suggesting an activating function (4). Indeed, TARM1 engagement enhances the secretion of pro-inflammatory cytokines by murine BMM and BM neutrophils stimulated with TLR ligands, such as LPS. Therefore, TARM1 and the related SIRL-1 may be examples of “paired” receptors. The LRC contains several sets of closely related, paired receptors with activating and inhibitory functions such as the LAIRs, KIRs and LILRs. The integration of opposing signals delivered by these receptors may balance the modulation of the innate and adaptive immune cell responses (1).
A large body of work suggests that both human and mouse neutrophils can be activated by, and may exert immunoregulatory effects on, T cells (17, 20, 46-48). Notably, T cells from the synovial fluid of rheumatoid arthritis patients were shown to activate neutrophils at the site of inflammation (47). In patients with renal cell carcinoma, neutrophils were shown to suppress T cell responses through arginase I release (30). The secretion of IL-10 (32, 33, 49) and PD-L1 expression (50) have been proposed as the major components of the immunosuppressive mechanisms employed by neutrophils during bacterial infections. We demonstrate that the TARM1 receptor ectodomain is capable, at least in in vitro assay, of potently inhibiting anti-CD3/-CD28 induced CD4+ T cell activation and proliferation. Bearing in mind that TARM1 cell-surface expression is rapidly upregulated by BMM and BMDC and by neutrophils and inflammatory monocytes in vivo during immune challenge, and TARM1+ cells home to the sites of inflammation, TARM1 may constitute a previously uncharacterized negative regulator of T cell activation expressed by neutrophils and inflammatory monocytes, macrophages and DC to modulate the early stages of T cell responses.
Our data indicate the TARM1 receptor ectodomain interacts with an as yet unidentified ligand on T cells that can mediate the inhibition of T cell activation. In contrast, TARM1 receptor stimulation on macrophages and neutrophils co-stimulated the secretion of pro-inflammatory cytokines induced by TLR ligands, such as LPS. These data suggest that TARM1 receptor interaction with an unidentified molecule on T cells may result in a bi-directional signaling between macrophages and neutrophils expressing TARM1 and T cells expressing an unidentified inhibitory molecule, respectively (Supplemental Fig. 3). Bi-directional signaling has been previously observed between myeloid cells and innate lymphocytes, such as NK cells, as exemplified by the interaction between AICL and NKp80 (51). Similarly to TARM1, the cell surface expression of AICL is also regulated by TLR ligands. The AICL:NKp80 bi-directional signaling interaction resulted in the activation of both NK cells and monocytes, respectively. The identification of the inhibitory TARM1 ligand expressed by T cells would be an interesting subject for further research.
In conclusion, our data shows that TARM1 expression is a hallmark of neutrophils and their precursors in the steady-state and that TARM1 is upregulated by mature, activated neutrophils, monocytes, macrophages and DC under specific inflammatory conditions. TLR-specific pro-inflammatory cytokine release by BMM and primary bone marrow neutrophils was co-stimulated by TARM1 engagement and the ability of TARM1-Fc to suppress T cell activation and proliferation in vitro, likely points to a role for TARM1-mediated bi-directional signaling in the regulation of immune cell function.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Professor Jim Kaufman for critically evaluating this manuscript.
This work was supported by grants from the Cancer Research UK, the Wellcome Trust, Medical Research Council, UK, and a Marie Curie International Outgoing Fellowship awarded to A.D.B. with additional support from the Wellcome Trust and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre.
Abbreviations used in this article
- BMDC
bone marrow-derived DC
- BMM
bone marrow-derived macrophage
- ECM
extracellular matrix
- FcRγ
Fc Receptor common γ chain
- Fsc
Forward Scatter
- Hi
High
- h
human
- ILT
Immunoglobulin-like Transcript
- ITAM
Immunoreceptor Tyrosine-based Activation Motif
- KIR
killer immunoglobulin-like receptors
- LAIR
Leukocyte-Associated Ig-like Receptor
- LILR
leukocyte immunoglobulin-like receptors
- Lo
Low
- OSCAR
Osteoclast-associated receptor
- SIRL-1
Signal Inhibitory Receptor on Leukocytes 1
- TARM1
T-cell interacting, activating receptor on myeloid cells 1
- Ssc
Side Scatter
- TM
transmembrane
- TREM
Triggering Receptor Expressed on Myeloid cells
REFERENCES
- 1.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunological reviews. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunological reviews. 2006;214:143–154. doi: 10.1111/j.1600-065X.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 3.Colonna M, Navarro F, Bellón T, Llano M, García P, Samaridis J, Angman L, Cella M, López-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. The Journal of experimental medicine. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daëron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunological reviews. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 5.Tsuji M, Ezumi Y, Arai M, Takayama H. A novel association of Fc receptor γ-chain with glycoprotein VI and their co-expression as a collagen receptor in human platelets. Journal of Biological Chemistry. 1997;272:23528–23531. doi: 10.1074/jbc.272.38.23528. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima H, Samaridis J, Angman L, Colonna M. Cutting edge: human myeloid cells express an activating ILT receptor (ILT1) that associates with Fc receptor γ-chain. The Journal of Immunology. 1999;162:5–8. [PubMed] [Google Scholar]
- 7.Morton HC, van den Herik-Oudijk IE, Vossebeld P, Snijders A, Verhoeven AJ, Capel PJ, van de Winkel JG. Functional Association between the Human Myeloid Immunoglobulin A Fc Receptor (CD89) and FcR γ-Chain Molecular Basis for CD89/FcR γ-Chain Association. Journal of Biological Chemistry. 1995;270:29781–29787. doi: 10.1074/jbc.270.50.29781. [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 9.Barrow AD, Raynal N, Andersen TL, Slatter DA, Bihan D, Pugh N, Cella M, Kim T, Rho J, Negishi-Koga T. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. The Journal of clinical investigation. 2011;121:3505. doi: 10.1172/JCI45913. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyaard L. The inhibitory collagen receptor LAIR-1 (CD305) Journal of leukocyte biology. 2008;83:799–803. doi: 10.1189/jlb.0907609. [DOI] [PubMed] [Google Scholar]
- 11.Steevels TA, Lebbink RJ, Westerlaken GH, Coffer PJ, Meyaard L. Signal inhibitory receptor on leukocytes-1 is a novel functional inhibitory immune receptor expressed on human phagocytes. The Journal of Immunology. 2010;184:4741–4748. doi: 10.4049/jimmunol.0902039. [DOI] [PubMed] [Google Scholar]
- 12.Steevels TA, Avondt K, Westerlaken GH, Stalpers F, Walk J, Bont L, Coffer PJ, Meyaard L. Signal inhibitory receptor on leukocytes-1 (SIRL-1) negatively regulates the oxidative burst in human phagocytes. European journal of immunology. 2013;43:1297–1308. doi: 10.1002/eji.201242916. [DOI] [PubMed] [Google Scholar]
- 13.Brach MA, Gruss HJ, Herrmann F. Prolongation of survival of human polymorphonuclear neutrophils by granulocyte-macrophage colony-stimulating factor is caused by inhibition of programmed cell death. Blood. 1992;80:2920–2924. others. [PubMed] [Google Scholar]
- 14.Colotta F, Re F, Polentarutti N, Sozzani S, Mantovani A. Modulation of granulocyte survival and programmed cell death by cytokines and bacterial products. Blood. 1992;80:2012–2020. [PubMed] [Google Scholar]
- 15.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nature Reviews Immunology. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 16.Pelletier M, Maggi L, Micheletti A, Lazzeri E, Tamassia N, Costantini C, Cosmi L, Lunardi C, Annunziato F, Romagnani S. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115:335–343. doi: 10.1182/blood-2009-04-216085. others. [DOI] [PubMed] [Google Scholar]
- 17.Gosselin EJ, Wardwell K, Rigby WF, Guyre PM. Induction of MHC class II on human polymorphonuclear neutrophils by granulocyte/macrophage colony-stimulating factor, IFN-gamma, and IL-3. The Journal of Immunology. 1993;151:1482–1490. [PubMed] [Google Scholar]
- 18.Smith WB, Guida L, Sun Q, Korpelainen EI, Van Den Heuvel C, Gillis D, Hawrylowicz CM, Vadas MA, Lopez AF. Neutrophils activated by granulocyte-macrophage colony-stimulating factor express receptors for interleukin-3 which mediate class II expression. Blood. 1995;86:3938–3944. [PubMed] [Google Scholar]
- 19.Oehler L, Majdic O, Pickl WF, Stöckl J, Riedl E, Drach J, Rappersberger K, Geissler K, Knapp W. Neutrophil Granulocyte--committed Cells Can Be Driven to Acquire Dendritic Cell Characteristics. The Journal of experimental medicine. 1998;187:1019–1028. doi: 10.1084/jem.187.7.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdallah DSA, Egan CE, Butcher BA, Denkers EY. Mouse neutrophils are professional antigen-presenting cells programmed to instruct Th1 and Th17 T-cell differentiation. International immunology. 2011;23:317–326. doi: 10.1093/intimm/dxr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radsak M, Iking-Konert C, Stegmaier S, Andrassy K, Hänsch GM. Polymorphonuclear neutrophils as accessory cells for T-cell activation: major histocompatibility complex class II restricted antigen-dependent induction of T-cell proliferation. Immunology. 2000;101:521–530. doi: 10.1046/j.1365-2567.2000.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abadie V, Badell E, Douillard P, Ensergueix D, Leenen PJ, Tanguy M, Fiette L, Saeland S, Gicquel B, Winter N. Neutrophils rapidly migrate via lymphatics after Mycobacterium bovis BCG intradermal vaccination and shuttle live bacilli to the draining lymph nodes. Blood. 2005;106:1843–1850. doi: 10.1182/blood-2005-03-1281. [DOI] [PubMed] [Google Scholar]
- 23.Chtanova T, Schaeffer M, Han S-J, van Dooren GG, Nollmann M, Herzmark P, Chan SW, Satija H, Camfield K, Aaron H. Dynamics of neutrophil migration in lymph nodes during infection. Immunity. 2008;29:487–496. doi: 10.1016/j.immuni.2008.07.012. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tvinnereim AR, Hamilton SE, Harty JT. Neutrophil involvement in cross-priming CD8+ T cell responses to bacterial antigens. The Journal of Immunology. 2004;173:1994–2002. doi: 10.4049/jimmunol.173.3.1994. [DOI] [PubMed] [Google Scholar]
- 25.Beauvillain C, Delneste Y, Scotet M, Peres A, Gascan H, Guermonprez P, Barnaba V, Jeannin P. Neutrophils efficiently cross-prime naive T cells in vivo. Blood. 2007;110:2965–2973. doi: 10.1182/blood-2006-12-063826. [DOI] [PubMed] [Google Scholar]
- 26.Schmielau J, Finn OJ. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of T-cell function in advanced cancer patients. Cancer research. 2001;61:4756–4760. [PubMed] [Google Scholar]
- 27.Sabbione F, Gabelloni ML, Ernst G, Gori MS, Salamone G, Oleastro M, Trevani A, Geffner J, Jancic CC. Neutrophils suppress γδ T-cell function. European journal of immunology. 2013 doi: 10.1002/eji.201343664. [DOI] [PubMed] [Google Scholar]
- 28.Munder M, Schneider H, Luckner C, Giese T, Langhans C-D, Fuentes JM, Kropf P, Mueller I, Kolb A, Modolell M. Suppression of T-cell functions by human granulocyte arginase. Blood. 2006;108:1627–1634. doi: 10.1182/blood-2006-11-010389. others. [DOI] [PubMed] [Google Scholar]
- 29.Kropf P, Baud D, Marshall SE, Munder M, Mosley A, Fuentes JM, Bangham CR, Taylor GP, Herath S, Choi B-S. Arginase activity mediates reversible T cell hyporesponsiveness in human pregnancy. European journal of immunology. 2007;37:935–945. doi: 10.1002/eji.200636542. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, Ochoa AC. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer research. 2009;69:1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang CW, Unanue ER. Neutrophils control the magnitude and spread of the immune response in a thromboxane A2-mediated process. The Journal of experimental medicine. 2013;210:375–387. doi: 10.1084/jem.20122183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Majlessi L, Deriaud E, Leclerc C, Lo-Man R. Coactivation of Syk kinase and MyD88 adaptor protein pathways by bacteria promotes regulatory properties of neutrophils. Immunity. 2009;31:761–771. doi: 10.1016/j.immuni.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Boari JT, Vesely MCA, Bermejo DA, Ramello MC, Montes CL, Cejas H, Gruppi A, Rodríguez EVA. IL-17RA signaling reduces inflammation and mortality during Trypanosoma cruzi infection by recruiting suppressive IL-10-producing neutrophils. PLoS pathogens. 2012;8:e1002658. doi: 10.1371/journal.ppat.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Kleijn S, Langereis JD, Leentjens J, Kox M, Netea MG, Koenderman L, Ferwerda G, Pickkers P, Hermans PW. IFN-γ-Stimulated Neutrophils Suppress Lymphocyte Proliferation through Expression of PD-L1. PloS one. 2013;8:e72249. doi: 10.1371/journal.pone.0072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowers NL, Helton ES, Huijbregts RP, Goepfert PA, Heath SL, Hel Z. Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway. PLoS pathogens. 2014;10:e1003993. doi: 10.1371/journal.ppat.1003993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillay J, Kamp VM, van Hoffen E, Visser T, Tak T, Lammers J-W, Ulfman LH, Leenen LP, Pickkers P, Koenderman L. A subset of neutrophils in human systemic inflammation inhibits T cell responses through Mac-1. The Journal of clinical investigation. 2012;122:327. doi: 10.1172/JCI57990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Haslett C, Guthrie LA, Kopaniak MM, Johnston RB, Jr, Henson PM. Modulation of multiple neutrophil functions by preparative methods or trace concentrations of bacterial lipopolysaccharide. The American journal of pathology. 1985;119:101. [PMC free article] [PubMed] [Google Scholar]
- 39.Merck E, Gaillard C, Gorman DM, Montero-Julian F, Durand I, Zurawski SM, Menetrier-Caux C, Carra G, Lebecque S, Trinchieri G. OSCAR is an FcRgamma-associated receptor that is expressed by myeloid cells and is involved in antigen presentation and activation of human dendritic cells. Blood. 2004;104:1386–1395. doi: 10.1182/blood-2004-03-0850. others. [DOI] [PubMed] [Google Scholar]
- 40.Tomasello E, Olcese L, Vély F, Geourgeon C, Bléry M, Moqrich A, Gautheret D, Djabali M, Mattei M-G, Vivier E. Gene structure, expression pattern, and biological activity of mouse killer cell activating receptor-associated protein (KARAP)/DAP-12. Journal of Biological Chemistry. 1998;273:34115–34119. doi: 10.1074/jbc.273.51.34115. [DOI] [PubMed] [Google Scholar]
- 41.Henderson RB, Hobbs JA, Mathies M, Hogg N. Rapid recruitment of inflammatory monocytes is independent of neutrophil migration. Blood. 2003;102:328–335. doi: 10.1182/blood-2002-10-3228. [DOI] [PubMed] [Google Scholar]
- 42.Barrow AD, Palarasah Y, Bugatti M, Holehouse AS, Byers DE, Holtzman MJ, Vermi W, Skjødt K, Crouch E, Colonna M. OSCAR Is a Receptor for Surfactant Protein D That Activates TNF-α Release from Human CCR2+ Inflammatory Monocytes. The Journal of Immunology. 2015;194:3317–3326. doi: 10.4049/jimmunol.1402289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merck E, Gaillard C, Scuiller M, Scapini P, Cassatella MA, Trinchieri G, Bates EE. Ligation of the FcRγ chain-associated human osteoclast-associated receptor enhances the proinflammatory responses of human monocytes and neutrophils. The Journal of Immunology. 2006;176:3149–3156. doi: 10.4049/jimmunol.176.5.3149. [DOI] [PubMed] [Google Scholar]
- 44.Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 45.Van Avondt K, Fritsch-Stork R, Derksen RH, Meyaard L. Ligation of Signal Inhibitory Receptor on Leukocytes-1 Suppresses the Release of Neutrophil Extracellular Traps in Systemic Lupus Erythematosus. PloS one. 2013;8:e78459. doi: 10.1371/journal.pone.0078459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alemán M, Silvia S, Schierloh PL, Alves L, Yokobori N, Baldini M, Abbate E, Sasiain MC. In Tuberculous Pleural Effusions, Activated Neutrophils Undergo Apoptosis and Acquire a Dendritic Cell-Like Phenotype. Journal of Infectious Diseases. 2005;192:399–409. doi: 10.1086/431680. [DOI] [PubMed] [Google Scholar]
- 47.Iking-Konert C, Ostendorf B, Sander O, Jost M, Wagner C, Joosten L, Schneider M, Hänsch GM. Transdifferentiation of polymorphonuclear neutrophils to dendritic-like cells at the site of inflammation in rheumatoid arthritis: evidence for activation by T cells. Annals of the rheumatic diseases. 2005;64:1436–1442. doi: 10.1136/ard.2004.034132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostanin DV, Kurmaeva E, Furr K, Bao R, Hoffman J, Berney S, Grisham MB. Acquisition of antigen-presenting functions by neutrophils isolated from mice with chronic colitis. The Journal of Immunology. 2012;188:1491–1502. doi: 10.4049/jimmunol.1102296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doz E, Lombard R, Carreras F, Buzoni-Gatel D, Winter N. Mycobacteria-Infected Dendritic Cells Attract Neutrophils That Produce IL-10 and Specifically Shut Down Th17 CD4 T Cells through Their IL-10 Receptor. The Journal of Immunology. 2013;191:3818–3826. doi: 10.4049/jimmunol.1300527. [DOI] [PubMed] [Google Scholar]
- 50.McNab FW, Berry MP, Graham CM, Bloch SA, Oni T, Wilkinson KA, Wilkinson RJ, Kon OM, Banchereau J, Chaussabel D. Programmed death ligand 1 is over-expressed by neutrophils in the blood of patients with active tuberculosis. European journal of immunology. 2011;41:1941–1947. doi: 10.1002/eji.201141421. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welte S, Kuttruff S, Waldhauer I, Steinle A. Mutual activation of natural killer cells and monocytes mediated by NKp80-AICL interaction. Nature immunology. 2006;7:1334–1342. doi: 10.1038/ni1402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.