Abstract
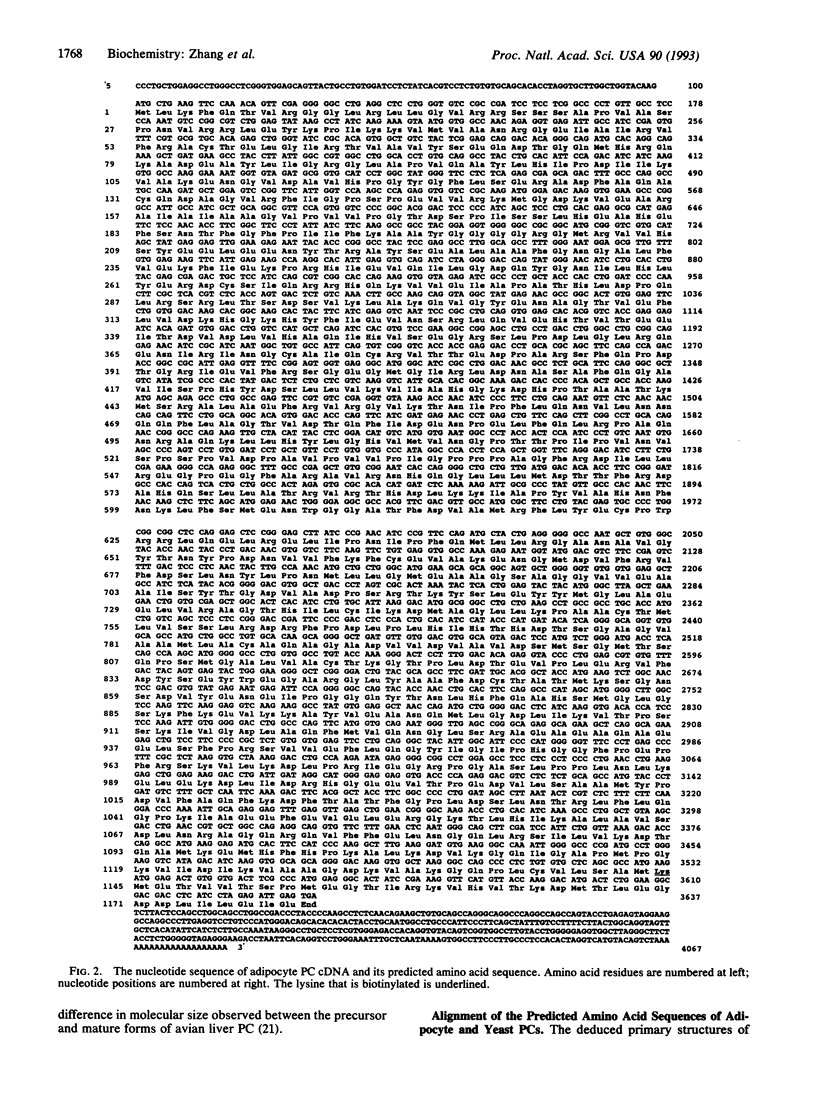
The complete amino acid sequence of 3T3-L1 adipocyte pyruvate carboxylase (PC) [pyruvate:carbon-dioxide ligase (ADP-forming), EC 6.4.1.1] has been deduced from sequencing overlapping cDNA clones obtained from an adipocyte cDNA library constructed in the lambda Zap vector. The encoding mRNA for PC promoter contains 4067 nt, including a 3534-nt coding sequence and noncoding regions of 100 and 433 nt at the 5' and 3' ends, respectively. The biotinylated lysine of the encoded PC promoter (1178 amino acids with a calculated M(r) of apocarboxylase = 129,784) is located 35 residues from the COOH-terminal end and, as in most other biotin enzymes, is in the consensus sequence AMKM. The adipocyte PC is closely similar (53% identity) to the yeast enzyme and contains different segments that are homologous with regions from the biotin carboxylase component of Escherichia coli acetyl-CoA carboxylase, the keto acid-binding subunits of Propionibacterium shermanii oxaloacetate transcarboxylase and Klebsiella pneumoniae oxaloacetate decarboxylase, and to the biotin carboxyl-carrier protein of the bacterial biotin enzymes. In addition to the putative mitochondrial targeting signal, functional domains are readily identifiable in the sequence and are in the following order: biotin carboxylase-carboxyltransferase-biotin carboxyl-carrier protein, as proposed for yeast PC.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad F., Ahmad P. M., Mendez A. Rat liver pyruvate carboxylase. Purification, detection and quantification of apo and holo forms by immuno-blotting and by an enzyme-linked immunosorbent assay. Biochem J. 1986 Jun 1;236(2):527–533. doi: 10.1042/bj2360527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Feel W., Chirala S. S., Wakil S. J. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4534–4538. doi: 10.1073/pnas.89.10.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus C. W., Lane M. D., Rosenfeld P. J., Kelly T. J. Increase in translatable mRNA for mitochondrial pyruvate carboxylase during differentiation of 3T3 preadipocytes. Biochem Biophys Res Commun. 1981 Dec 31;103(4):1216–1222. doi: 10.1016/0006-291x(81)90252-7. [DOI] [PubMed] [Google Scholar]
- Attwood P. V., Keech D. B. Pyruvate carboxylase. Curr Top Cell Regul. 1984;23:1–55. doi: 10.1016/b978-0-12-152823-2.50005-2. [DOI] [PubMed] [Google Scholar]
- Browner M. F., Taroni F., Sztul E., Rosenberg L. E. Sequence analysis, biogenesis, and mitochondrial import of the alpha-subunit of rat liver propionyl-CoA carboxylase. J Biol Chem. 1989 Jul 25;264(21):12680–12685. [PubMed] [Google Scholar]
- Freytag S. O., Collier K. J. Molecular cloning of a cDNA for human pyruvate carboxylase. Structural relationship to other biotin-containing carboxylases and regulation of mRNA content in differentiating preadipocytes. J Biol Chem. 1984 Oct 25;259(20):12831–12837. [PubMed] [Google Scholar]
- Freytag S. O., Utter M. F. Induction of pyruvate carboxylase apoenzyme and holoenzyme in 3T3-L1 cells during differentiation. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1321–1325. doi: 10.1073/pnas.77.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbauffe F. S., Cooper T. G. The urea amidolyase (DUR1,2) gene of Saccharomyces cerevisiae. DNA Seq. 1991;2(1):19–32. doi: 10.3109/10425179109008435. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hodges P. E., Rosenberg L. E. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N. E., Pichersky E., Cashmore A. R. A tomato cDNA encoding a biotin-binding protein. Nucleic Acids Res. 1987 May 11;15(9):3928–3928. doi: 10.1093/nar/15.9.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Shiratsuchi K., Yoshimoto T., Masuda T., Kitazono A., Tsuru D., Anai M., Sekiguchi M., Tanabe T. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G. K., Beegen H., Wood H. G. Involvement of tryptophans at the catalytic and subunit-binding domains of transcarboxylase. Biochemistry. 1988 Aug 9;27(16):5972–5978. doi: 10.1021/bi00416a021. [DOI] [PubMed] [Google Scholar]
- Lamhonwah A. M., Quan F., Gravel R. A. Sequence homology around the biotin-binding site of human propionyl-CoA carboxylase and pyruvate carboxylase. Arch Biochem Biophys. 1987 May 1;254(2):631–636. doi: 10.1016/0003-9861(87)90146-9. [DOI] [PubMed] [Google Scholar]
- Lane M. D., Moss J., Polakis S. E. Acetyl coenzyme A carboxylase. Curr Top Cell Regul. 1974;8(0):139–195. [PubMed] [Google Scholar]
- Li S. J., Cronan J. E., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992 Jan 15;267(2):855–863. [PubMed] [Google Scholar]
- Libor S., Sundaram T. K., Warwick R., Chapman J. A., Grundy S. M. Pyruvate carboxylase from a thermophilic Bacillus: some molecular characteristics. Biochemistry. 1979 Aug 21;18(17):3647–3653. doi: 10.1021/bi00584a001. [DOI] [PubMed] [Google Scholar]
- Lim F., Morris C. P., Occhiodoro F., Wallace J. C. Sequence and domain structure of yeast pyruvate carboxylase. J Biol Chem. 1988 Aug 15;263(23):11493–11497. [PubMed] [Google Scholar]
- López-Casillas F., Bai D. H., Luo X. C., Kong I. S., Hermodson M. A., Kim K. H. Structure of the coding sequence and primary amino acid sequence of acetyl-coenzyme A carboxylase. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5784–5788. doi: 10.1073/pnas.85.16.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackall J. C., Student A. K., Polakis S. E., Lane M. D. Induction of lipogenesis during differentiation in a "preadipocyte" cell line. J Biol Chem. 1976 Oct 25;251(20):6462–6464. [PubMed] [Google Scholar]
- Möller W., Amons R. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 1985 Jul 1;186(1):1–7. doi: 10.1016/0014-5793(85)81326-0. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Neupert W. The mitochondrial protein import apparatus. Annu Rev Biochem. 1990;59:331–353. doi: 10.1146/annurev.bi.59.070190.001555. [DOI] [PubMed] [Google Scholar]
- Robinson B. H., Oei J., Sherwood W. G., Applegarth D., Wong L., Haworth J., Goodyer P., Casey R., Zaleski L. A. The molecular basis for the two different clinical presentations of classical pyruvate carboxylase deficiency. Am J Hum Genet. 1984 Mar;36(2):283–294. [PMC free article] [PubMed] [Google Scholar]
- Rohde M., Lim F., Wallace J. C. Electron microscopic localization of pyruvate carboxylase in rat liver and Saccharomyces cerevisiae by immunogold procedures. Arch Biochem Biophys. 1991 Oct;290(1):197–201. doi: 10.1016/0003-9861(91)90608-l. [DOI] [PubMed] [Google Scholar]
- Samols D., Thornton C. G., Murtif V. L., Kumar G. K., Haase F. C., Wood H. G. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988 May 15;263(14):6461–6464. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E., Oesterhelt D., Reinke H., Beyreuther K., Dimroth P. The sodium ion translocating oxalacetate decarboxylase of Klebsiella pneumoniae. Sequence of the biotin-containing alpha-subunit and relationship to other biotin-containing enzymes. J Biol Chem. 1988 Jul 15;263(20):9640–9645. [PubMed] [Google Scholar]
- Srivastava G., Borthwick I. A., Brooker J. D., Wallace J. C., May B. K., Elliott W. H. Hemin inhibits transfer of pre-delta-aminolevulinate synthase into chick embryo liver mitochondria. Biochem Biophys Res Commun. 1983 Nov 30;117(1):344–349. doi: 10.1016/0006-291x(83)91582-6. [DOI] [PubMed] [Google Scholar]
- Sternberg M. J., Taylor W. R. Modelling the ATP-binding site of oncogene products, the epidermal growth factor receptor and related proteins. FEBS Lett. 1984 Oct 1;175(2):387–392. doi: 10.1016/0014-5793(84)80774-7. [DOI] [PubMed] [Google Scholar]
- Stucka R., Dequin S., Salmon J. M., Gancedo C. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol Gen Genet. 1991 Oct;229(2):307–315. doi: 10.1007/BF00272171. [DOI] [PubMed] [Google Scholar]
- Sutton M. R., Fall R. R., Nervi A. M., Alberts A. W., Vagelos P. R., Bradshaw R. A. Amino acid sequence of Escherichia coli biotin carboxyl carrier protein (9100). J Biol Chem. 1977 Jun 10;252(11):3934–3940. [PubMed] [Google Scholar]
- Takai T., Yokoyama C., Wada K., Tanabe T. Primary structure of chicken liver acetyl-CoA carboxylase deduced from cDNA sequence. J Biol Chem. 1988 Feb 25;263(6):2651–2657. [PubMed] [Google Scholar]
- Thampy K. G., Huang W. Y., Wakil S. J. A rapid purification method for rat liver pyruvate carboxylase and amino acid sequence analyses of NH2-terminal and biotin peptide. Arch Biochem Biophys. 1988 Oct;266(1):270–276. doi: 10.1016/0003-9861(88)90258-5. [DOI] [PubMed] [Google Scholar]
- Utter M. F., Barden R. E., Taylor B. L. Pyruvate carboxylase: an evaluation of the relationships between structure and mechanism and between structure and catalytic activity. Adv Enzymol Relat Areas Mol Biol. 1975;42:1–72. doi: 10.1002/9780470122877.ch1. [DOI] [PubMed] [Google Scholar]
- Wood H. G., Barden R. E. Biotin enzymes. Annu Rev Biochem. 1977;46:385–413. doi: 10.1146/annurev.bi.46.070177.002125. [DOI] [PubMed] [Google Scholar]



