To the Editor
The International Mouse Phenotyping Consortium (IMPC) aims1 to create a comprehensive functional catalog of the mammalian genome. It currently provides access to over 22 million data points concerning the knockout effects of 1,633 mouse genes on 44 classes of well-defined phenotypic traits. These data were collected2 at ten phenotyping centers worldwide. Because data are only as good as the tools available to analyze them, the IMPC has developed specialized tools3 to reliably extract useful information4 from these data.
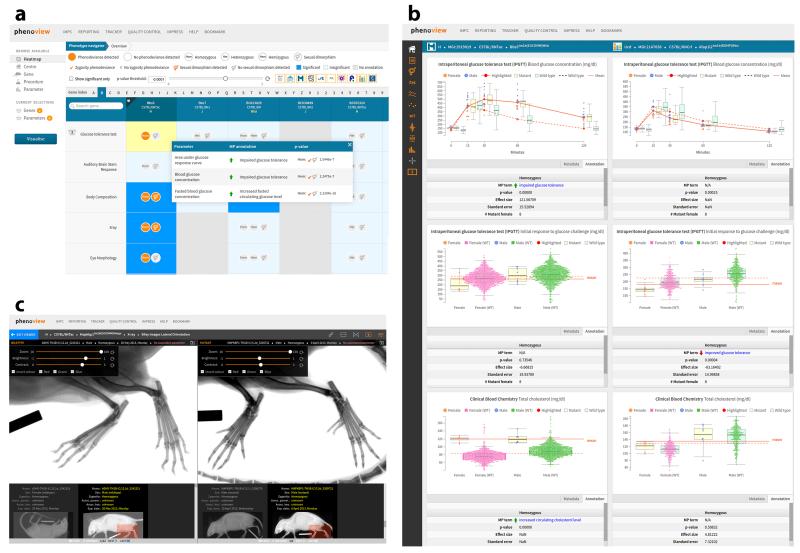
Phenoview is a publicly accessible web-based tool (https://www.mousephenotype.org/phenoview) developed by the IMPC for visualizing genotype-phenotype relationships. It provides access to the IMPC phenotype data through a grid-like interface. This phenotype heat map is organized into columns for genes and rows for phenotypes (Fig. 1a). Each cell in this grid displays statistical analysis5 results, highlighting sexual dimorphism and ‘phenodeviance’, in which the mutant phenotype deviates significantly from that of the wild type. This provides an overview of what data are available and which are of potential interest. Clicking on a cell displays the data for the genotype-phenotype combination.
Figure 1.
Defining features of Phenoview for comparative visualization of genotype-phenotype relationships. (a) The genotype-phenotype heat map displays data availability and a summary of statistical analysis results, with phenodeviance and sexual dimorphism highlighted. (b) Comparison of three metabolic phenotypes in two mutant mouse strains. A toolbar on the left allows interaction with the graphs. (c) Integrated media viewer showing side-by-side comparison of X-ray images for wild-type controls and a mutant mouse strain, with slider settings for adjusting brightness, contrast and zoom.
A defining feature of Phenoview is its ability to comparatively visualize data for multiple genotype-phenotype combinations simultaneously. This mix-and-match option is suited to investigating the knockout effects of a genotype on multiple traits, and also to the investigation of multiple genotypes that affect the same trait. Selecting the comparative visualization mode displays multiple graphs that reflect selections made by the user (Fig. 1b). These geno-type-phenotype combinations are selectable on the basis of user-defined P-value thresholds configurable on the heat-map interface.
Phenoview enables real-time interaction with the displayed data, allowing users to interactively filter out data points by gender and zygosity and to dynamically configure the statistics displayed. It also allows the generated graphs to be saved as image files. Phenoview is context sensitive and automatically chooses the appropriate display type to match the data. It can also display digital media (e.g., X-ray or LacZ expression), with the flexibility to adjust brightness, contrast, zooming and panning (Fig. 1c). To permit further analysis of the data using alternative tools, Phenoview also allows downloading of the raw data (Supplementary Note).
By collating data from multiple centers, Phenoview provides a single, convenient data access point for researchers, increasing accessibility to the phenotype data. Through these tools, the IMPC aims to advance our understanding of genotype-phenotype relationships6 by enabling systematic analysis of large-scale phenotype data.
Supplementary Material
ACKNOWLEDGMENTS
The US National Institutes of Health (NIH) funded Phenoview (U54 HG006370). We thank the phenotyping centers for collecting the data, and EMBL-EBI for hosting the data archive.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper (doi:10.1038/nmeth.3477).
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Brown SDM, Moore MW. Dis. Model. Mech. 2012;5:289–292. doi: 10.1242/dmm.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koscielny G, et al. Nucleic Acids Res. 2014;42:D802–D809. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle J. Nature. 2013;499:7. doi: 10.1038/499007a. [DOI] [PubMed] [Google Scholar]
- 4.Frankel F, Reid R. Nature. 2008;455:30. [Google Scholar]
- 5.Karp NA, Melvin D, Sanger Mouse Genetics Project. Mott RF. PLoS ONE. 2012;7:e52410. doi: 10.1371/journal.pone.0052410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowell RD, et al. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.