Abstract
Context:
We previously showed that a long noncoding RNA gene, PTCSC3, located close to the variant rs944289 that predisposes to papillary thyroid carcinoma (PTC) might target the S100A4 gene.
Objective:
The aim was to investigate the impact of PTCSC3 on S100A4 expression and its role in cancer development.
Design:
S100A4 abundance was analyzed by quantitative PCR (qPCR) in unaffected and tumor tissue from n = 73 PTC patients. The expression of PTCSC3 and S100A4 was studied in BCPAP and TPC-1 cell lines with forced expression of PTCSC3 by qPCR. Expression of S100A4 target genes (VEGF and MMP-9) was studied in the BCPAP cell line with forced expression of PTCSC3 by qPCR, reverse transcriptase PCR, and Western blot. The impact of PTCSC3 on BCPAP motility and invasiveness was analyzed by the Transwell and Matrigel assays, respectively.
Setting:
This was a laboratory-based study using cells from clinical samples and thyroid cancer cell lines.
Main Outcome and Measure:
We aimed to find evidence for a link between the expression of PTCSC3 and thyroid carcinogenesis.
Results:
Expression data from PTC cell lines pinpointed S100A4 as the most significantly downregulated gene in the presence of PTCSC3. S100A4 was upregulated in tumor tissue (P = 9.33 × 10−7) while PTCSC3 was strongly downregulated (P = 2.2 × 10−16). S100A4 transcription was moderately correlated with PTCSC3 expression in unaffected thyroid tissue (r = 0.429, P = .0001), and strongly in unaffected tissue of patients with the risk allele of rs944289 (r = 0.685, P = 7.88 × 10−5). S100A4, VEGF, and MMP-9 were suppressed in the presence of PTCSC3 (P = .0051, P = .0090, and P =.0037, respectively). PTC cells expressing PTCSC3 showed reduction in motility and invasiveness (P = 4.52 × 10−5 and P = 1.0 × 10−4, respectively).
Conclusions:
PTCSC3 downregulates S100A4, leading to a reduction in cell motility and invasiveness. We propose that PTCSC3 impacts PTC predisposition and carcinogenesis through the S100A4 pathway.
Among all cancers the incidence of thyroid carcinoma has shown one of the most rapid increases over the past two decades (1, 2). The dominant histological type of thyroid carcinoma is papillary thyroid carcinoma (PTC) that accounts for 80–85% of all cases (3). PTC has a highly favorable overall 5-year survival rate reaching approximately 98%. However, in the presence of distant metastases, the survival rate decreases to approximately 51% (www.cancer.org, accessed January 2015). The spread of tumors is a complex process involving the proliferation of cancer cells, breaking of the extracellular matrix, angiogenesis, hematogenous and lymphatic dissemination, homing in distant tissues, and proliferation at novel sites that eventually results in metastatic tumors (4, 5). It is important to identify individuals with a genetic predisposition to PTC and patients with potentially more aggressive types of PTC who might need novel therapeutic approaches or at least more rigorous postoperative follow-up.
By genome-wide association analysis, a single nucleotide polymorphism (SNP) rs944289 was originally associated with PTC susceptibility and this finding has been replicated in different populations albeit with relatively low odds ratios (1.21–1.33) (6–10). We previously cloned a novel long noncoding RNA gene PTCSC3 located in close vicinity of SNP rs944289. The PTCSC3 gene is exclusively expressed in thyroid tissue and significantly suppressed in PTC tumor tissue. The risk allele of SNP rs944289 suppresses PTCSC3 by destroying a transcription factor–binding site in the promoter of PTCSC3. Microarray expression assay data suggested that PTCSC3 alters the expression of genes of major importance for PTC development and progression such as S100A4, RHOB, MOAP1, AKT, CTSA, MECP2, FLNA, C19orf33, GDF15, and others (11).
We have studied the mechanistic aspects of PTCSC3 regulation in PTC. We show that stable restoration of PTCSC3 expression in PTC cell lines is associated with suppression of S100A4 expression. This in turn leads to diminished abundance of the downstream genes VEGF and MMP-9 causing decreased PTC cancer cell motility and invasiveness. This links the risk allele of rs944289 directly to a role in the development and progression of PTC.
Materials and Methods
Thyroid tissue samples
The paired samples used in this study (n = 73) consisted of tumor tissue and adjacent unaffected thyroid tissue from PTC patients collected at The Ohio State University as part of ongoing studies beforehand approved by the Institutional Review Board. Written informed consent was obtained from all patients included in the study. The samples were snap-frozen in liquid nitrogen. Subsequently, total RNA was extracted by using TRIzol reagent according to the manufacturer's protocol and stored at −80°C. Genomic DNA was extracted from blood and PTC tissue by using the phenol-chloroform method and stored at 4°C for further analysis.
PTC cell lines
TPC-1 cells were cultured at 37°C in 5% carbon dioxide (CO2) in DMEM media (Gibco) supplemented with 10% fetal calf serum (Gibco) and with 1× MEM nonessential amino acids (Gibco). BCPAP cells were cultured in RPMI media (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and incubated at 37°C in 5% CO2.
Microarray RNA hybridization
To assess the gene transcription patterns downstream of PTCSC3, complementary DNA from TPC-1 cells with and without transient expression of PTCSC3 was hybridized to the Agilent Sure Print G3 Human Gene Expression 8 × 60K arrays. The method has been described in detail previously (11).
Establishment of PTC cell line expressing PTCSC3
BCPAP and TPC-1 cells were seeded onto 24 well culture plates (Corning) and transfected at 80% cell confluence with 100 ng of pcDNA3.1 vector (control) or the pcDNA3.1-PTCSC3 construct by using 2 μl of Lipofectamine2000 reagent (Invitrogen) according to the manufacturer's protocol. Stable expressing empty pcDNA3.1 (controls) or pcDNA3.1-PTCSC3 clones of BCPAP and TPC-1 were selected by culturing for 4 weeks in media containing 400 μg/ml and 1.2 mg/ml geneticin (G418), respectively. The restoration of PTCSC3 expression in BCPAP and TPC-1 stable clones was confirmed by reverse transcriptase PCR (RT-PCR) and quantitative PCR (qPCR).
qPCR measurement of gene expression
One microgram of RNA was reverse transcribed by using the High Capacity Reverse Transcriptase kit (Applied Biosystems). The expression of PTCSC3 and S100A4 in clones of BCPAP and TPC-1 stably expressing PTCSC3, and the expression of MMP-9 as well as VEGF in BCPAP clones stably expressing PTCSC3 were assessed by qPCR in the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Fast Taqman assays were used for PTCSC3 and S100A4. The conditions were as follows: 95°C for 5 seconds followed by 40 cycles of 95°C, 5 seconds of denaturation at 60°C, and 5 seconds of annealing/elongation. SybrGreen assays were used for MMP-9 and VEGF. The conditions were as follows: 95°C for 10 minutes followed by 40 cycles of denaturation (95°C, 15 seconds) and annealing/elongation (60°C, 1 minute). A dissociation curve was obtained for SybrGreen plates to monitor any unspecific double stranded DNA. GAPDH served as a control gene for calculation of the relative gene abundance based on the 2−ΔCt equation, where ΔCt = CtGENE − CtGAPDH.
Transwell motility assay
Stable control or PTCSC3 expressing BCPAP cells were seeded (3 × 105 cells suspended in 1 ml of no FBS media) in duplicates into six-well Transwell chambers with an 8-μm pore size polycarbonate membrane. As a chemotaxis factor, FBS (10%) was added to the lower compartment of the chambers. After 24 hours of incubation at 37°C in 5% CO2, the cells that had migrated through the pores were stained by JorVet DipQuick Stain assay and counted under a microscope (12 slides per well, each experiment repeated 6 times).
Matrigel invasion assay
Stable control or PTCSC3 expressing BCPAP cells were seeded (7.5 × 104 cells suspended in 0.5 ml of no FBS media) in duplicate into 12-well Corning BioCoat Matrigel Invasion Chambers with an 8 μm pore size polyethylene terephthalate membrane. As a chemotaxis factor, FBS (10%) was added to the lower compartment of the Matrigel Invasion Chambers. After 24 hours of incubation at 37°C in 5% CO2 cells that had migrated through the pores were stained by JorVet DipQuick Stain assay and counted under a microscope (six slides per well; each experiment was repeated six times).
Western blot analysis
For protein extracts, cells were collected, washed with 1× PBS, resuspended with 2× SDS loading buffer (125 mM Tris pH 6.8, 4% SDS, 20% glycerol, 200 mM dithiothreitol, 200 mM β-mercaptoethanol, 0.2% w/v bromophenol blue), and boiled for 10 minutes. Protein lysates were loaded onto 4–20% Criterion Tris-HCl precast gels (Bio-Rad), proteins were separated by electrophoresis (300V for 30 minutes), and subsequently transferred to 0.45 μm polyvinylidene fluoride membranes. Membranes were incubated in 5% milk solution of Tris-buffered saline buffer containing 0.1% Tween-20 (TBST) and incubated with primary antibodies diluted in 5% bovine serum albumin TBST solution containing 0.1% sodium azide overnight at 4°C. Membranes were washed in TBST and probed with secondary antibodies. Following the final wash in TBST, membranes were incubated in ECL Western blotting detection reagents (GE Healthcare) and exposed to film (Denville). Antibodies used were anti-Mst-1 (Santa Cruz, sc-19948), anti-hsp90 (Santa Cruz, sc-69703), antirabbit horseradish peroxidase–linked (GE Healthcare), antimouse horseradish peroxidase–linked (GE Healthcare).
Genotyping
Genomic DNA from blood of PTC patients was used for genotyping SNP rs944289 by a SNaPshot assay according to the manufacturer's protocol (Applied Biosystems). The PCR primers and extension primer sequences are provided in Supplemental Table 1. The PCR reaction was performed in GeneAmp PCR System 9700 as follows: 2 minutes at 94°C; followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 58°C, and 30 seconds at 72°C; followed by a final extension of 10 minutes at 72°C. An ABI 3730 DNA Analyzer was used for allele analysis. The results were analyzed using GeneMapper v3.7 software (Applied Biosystems).
Statistical analysis
The impact of PTCSC3 on gene expression in TPC-1 cells was assessed by applying random variance model t test using BRB-Array Tools software (11). ANOVA was used to compare expression levels or average cell counts between the control cells and cells with stable expression of PTCSC3, controlling for day effects when necessary. Repeated measure ANOVA analysis was used to compare tumor and adjacent unaffected thyroid samples from PTC patients. Pearson correlation coefficients between PTCSC3 and S100A4 abundance in adjacent unaffected samples and P values were obtained. The results presented in the figures are with mean ± SEM. A two-sided comparison with a P value less than .05 was considered statistically significant.
Results
Target genes of PTCSC3
In step 1, TPC-1 cells were transfected with the pcDNA3-PTCSC3 construct and empty vector (pcDNA3) as a control, grown for 24 hours and harvested followed by testing in an Agilent expression array as described in Materials and Methods. Transient expression of PTCSC3 in TPC-1 cells significantly (P < .05) affected the transcription of n = 467 genes. The Agilent data are shown in Supplemental Table 2.
We previously analyzed gene expression changes between unaffected thyroid tissue and tumor tissue of nine PTC patients by using Affymetrix HG-U133 (12). The data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the deposition number GSE3467. In step 2, we used these two microarray data sets to identify genes with both altered expression caused by PTCSC3 in TPC-1 cells (fold change >1.5) and significant alterations in expression between unaffected tissue and tumor tissue in PTC patients (P < .05). This narrowed the number of potentially important genes to 77.
In step 3, we searched the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed, accessed in 2012) for genes that were associated with solid cancers, preferably with nonmedullary thyroid cancer. This further narrowed the potential candidate list to seven genes (Supplemental Table 3).
In step 4, we analyzed by qPCR the abundance of the selected transcripts (n = 7) in the PTCSC3 stable expression BCPAP clones. We uncovered transcript S100A4 as the most significant downregulated gene in the presence of PTCSC3 and selected it for further analysis (Supplemental Table 3, Supplemental Figure 1).
S100A4 expression correlates with PTCSC3 abundance in unaffected thyroid tissue
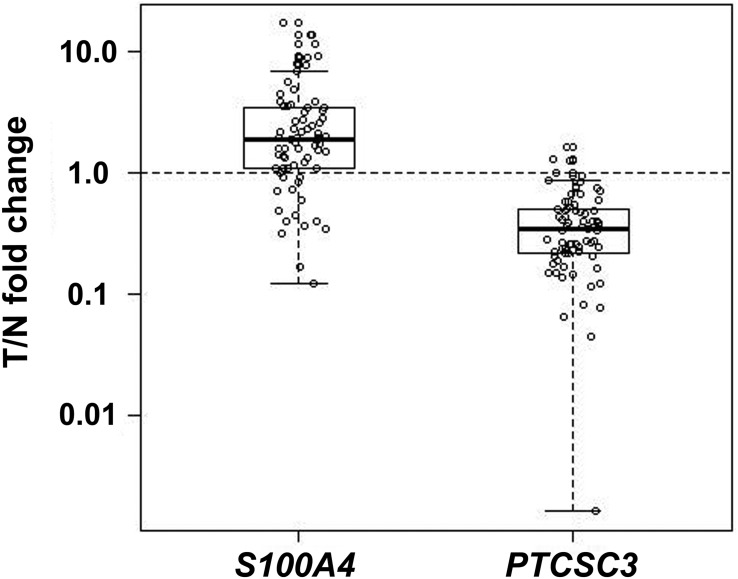
We analyzed S100A4 expression in paired unaffected thyroid tissue and tumor tissue from 73 patients with PTC. The S100A4 abundance was significantly greater in tumor tissue compared to unaffected thyroid tissue (P = 9.33 × 10−7), whereas the expression of PTCSC3 was significantly suppressed in tumor tissue (P = 2.2 × 10−16) (Figure 1). The S100A4 abundance level correlated moderately with PTCSC3 expression in unaffected thyroid tissue (r = 0.429, P = .0001). The correlation between SNP rs944289 genotype and the expression of PTCSC3 and S100A4 was strong in patients homozygous for the [TT] risk allele (n = 27, r = 0.685, P = 7.88 × 10−5, Table 1). The correlation was weak in tumor tissue (n = 73, r = 0.254, P = .0297; Table 1). These data tie S100A4 to the carcinogenesis in PTC and reconfirm the role of the risk allele of the SNP in PTC predisposition.
Figure 1.
PTCSC3 and S100A4 expressions assessed by qPCR in unaffected thyroid tissue and PTC tumor tissue in 73 patients. The results are expressed as relative expression in tumor tissue normalized to paired unaffected tissue (T/N fold change). S100A4 showed overexpression (relative expression in tumor > 1.0) (P = 9.33 × 10−7) and PTCSC3 showed underexpression (relative expression in tumor < 1.0) (P = 2.2 × 10−16). x-axis: genes; y-axis: relative expression in tumor.
Table 1.
Correlation Between the Expression Levels of PTCSC3 and S100A4 in Unaffected Thyroid Tissue and PTC Tumors Using All the Samples and Within rs944289 Genotype Groups
| Genotypes | n | Adjacent Unaffected Tissue |
Tumor Tissue |
||
|---|---|---|---|---|---|
| Pearson Correlation | P Value | Pearson Correlation | P Value | ||
| All genotypesa | 73 | 0.429 | .0001 | 0.254 | .0297 |
| rs944289[CC] | 11 | 0.533 | .0913 | 0.410 | .2108 |
| rs944289[CT] | 33 | 0.237 | .1834 | 0.245 | .1688 |
| rs944289[TT] | 27 | 0.685 | 7.88 × 10−5 | 0.066 | .7440 |
Including two cases that could not be genotyped.
Rs944289[T] is the risk allele.
Forced PTCSC3 expression inhibits the motility and invasion potential of BCPAP cells
We used Transwell and Matrigel assays to assess whether forced expression of PTCSC3 in BCPAP cells impacts their motility and invasion, respectively. We analyzed the difference between the average number of control cells (BCPAP-C) and of cells from stable clones (BCPAP-P) that migrated through the pores. The results for both assays are presented as a ratio of the average cell number for BCPAP-P divided by the average cell number for BCPAP-C. BCPAP cells with restored PTCSC3 expression showed significant reduction in motility (P = 4.52 × 10−5) and invasiveness (P = 1.0 × 10−4) (Figure 2). These data suggest that PTCSC3 has tumor suppressor properties.
Figure 2.
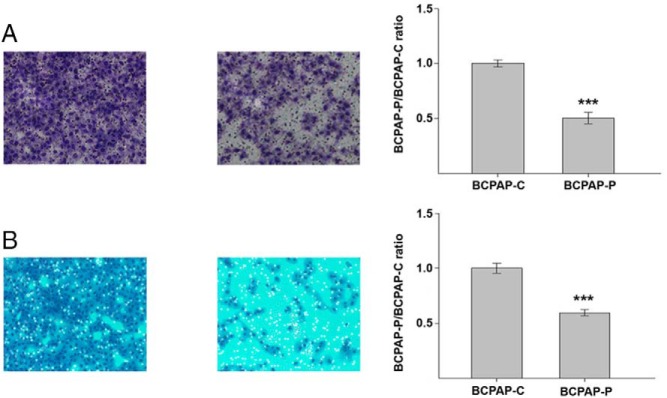
The results of motility and invasion assays. A, Representative images (10× magnification) of slides from the Transwell assay of control cells (BCPAP-C) and BCPAP-P cells expressing PTCSC3 showing suppression of motility in BCPAP-P when compared to BCPAP-C (left). The effect on motility potential in BCPAP cells generated by the restoration of PTCSC3 expression calculated as a ratio of the average cell number in BCPAP-P divided by the average cell number in BCPAP-C. The thyroid cancer cells with restored PTCSC3 expression showed significant reduction in cell motility (P = 4.52 × 10−5) (right). B, Representative images (10× magnification) of slides from the Matrigel assay of control cells (BCPAP-C) and BCPAP-P cells with stable expression of PTCSC3 showing suppression of BCPAP-P invasiveness when compared to BCPAP-C (left). The BCPAP-P/BCPAP-C ratio calculated in the same way as for the motility assay showed significant reduction in invasiveness in BCPAP cells with restored expression of PTCSC3 (P = 1.0 × 10−4, respectively) (right). ***, P < .0001.
S100A4 expression is suppressed in PTC cells with forced expression of PTCSC3
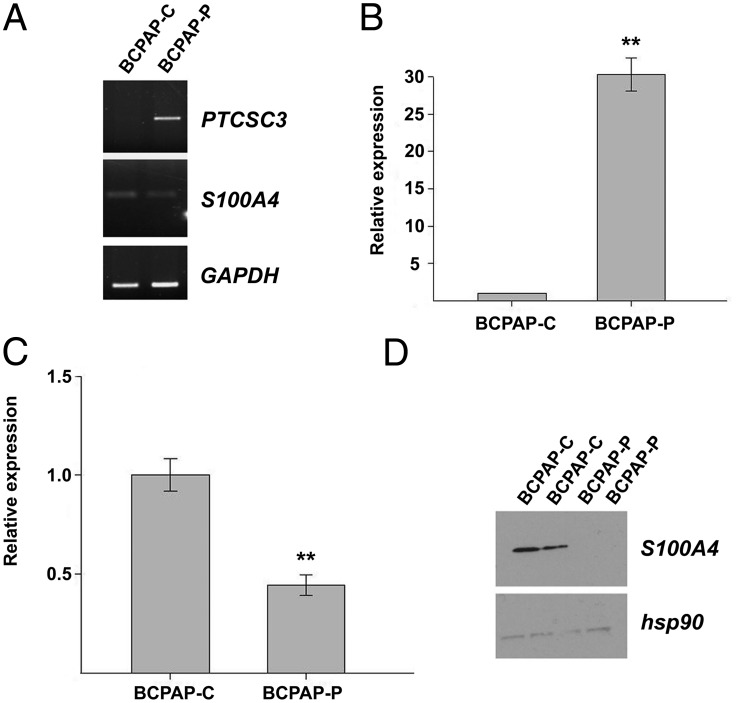
We analyzed S100A4 expression in BCPAP-C and BCPAP with stable expression of PTCSC3 cells (BCPAP-P) by RT-PCR, qPCR, and Western blot. As evidenced by RT-PCR, PTCSC3 was not expressed in BCPAP-C and its expression was restored in BCPAP-P, whereas S100A4 transcription was suppressed in BCPAP-P cells (Figure 3). Next, we analyzed S100A4 transcription in control and stable clones by qPCR. As shown in Figure 3, S100A4 expression was significantly suppressed in BCPAP cells with restored PTCSC3 expression (P = .0051). Western blot data disclosed that the strong inhibition of S100A4 expression occurred also at the protein level (Figure 3). These data show that PTCSC3 suppresses the abundance of S100A4.
Figure 3.
PTCSC3 and S100A4 expression analyzed in control cells (BCPAP-C) and cells with stable expression of PTCSC3 (BCPAP-P). A, PTCSC3 and S100A4 expression analyzed by RT-PCR showed restoration of PTCSC3 expression and suppression of S100A4 in BCPAP-P. GAPDH served as loading control. B, PTCSC3 shows strong abundance in stable clones (fold change 30, P = 2.32 × 10−6). The data are presented as relative expression (BCPAP-P/BCPAP-C ratio). C, S100A4 expression analyzed by qPCR showed significant suppression in BCPAP cells with restored PTCSC3 expression (P = .0051). The data are presented as relative expression (BCPAP-P/BCPAP-C ratio). D, S100A4 expression analyzed by Western blot showed suppression at the protein level in BCPAP-P. hsp90 served as loading control. **, P = .01–.001.
VEGF and MMP-9 transcription is suppressed in BCPAP cells with stable expression of PTCSC3
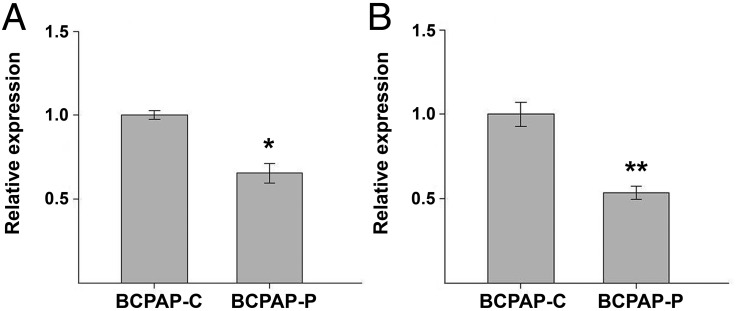
It has been shown that S100A4 suppresses invasion of thyroid cancer cells through downregulation of the VEGF and MMP-9 genes (13). To analyze whether downstream events in the S100A4 pathway are affected by PTCSC3, we analyzed the expression of VEGF and MMP-9 in controls and stable BCPAP clones by qPCR. The abundance of VEGF and MMP-9 was calculated as BCPAP-P/BCPAP-C relative ratio. As expected, both genes were significantly suppressed in BCPAP-P cells (fold change BCPAP-P/BCPAP-C 0.66, P = .0090 and .54, P = .0037, for VEGF and MMP-9, respectively) (Figure 4). This result was expected in view of previous studies and suggests that these downstream genes have an impact on thyroid carcinogenesis.
Figure 4.
Expression of VEGF and MMP-9 in control cells (BCPAP-C) and BCPAP-P cells expressing PTCSC3 assessed by qPCR and expressed as relative BCPAP-P/BCPAP-C expression ratios. A, Expression of VEGF at messenger RNA level shows significant suppression in BCPAP-P cells (P = .0090). B, Expression of MMP-9 at messenger RNA level shows significant suppression in BCPAP-P cells (P = .0037). *, P < .05; **, P = .01–.001.
PTCSC3 impacts S100A4 expression in the TPC-1 cell line
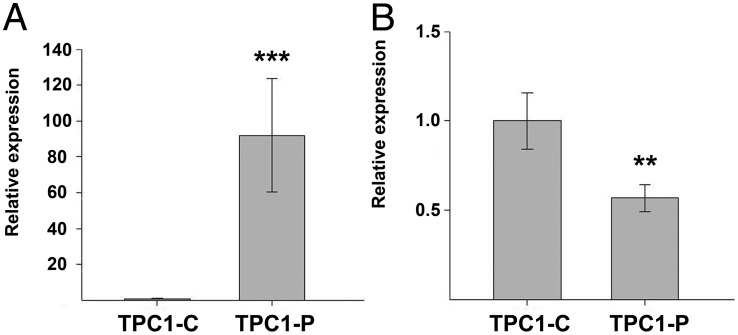
We showed that ectopic expression of PTCSC3 in the TPC-1 cell line impacted S100A4 expression (11). To confirm this effect, we created TPC-1 clones with stable expression of PTCSC3. We showed by qPCR that forced expression of PTCSC3 in the TPC-1 cell line led to suppression of S100A4 expression (P = .0137) (Figure 5). Hence, we confirmed that the effect of PTCSC3 on S100A4 transcription occurs in different PTC cell models.
Figure 5.
PTCSC3 and S100A4 expression analyzed in control cells (TPC1-C) and cells with stable expression of PTCSC3 (TPC1-P). A, PTCSC3 shows strong abundance in stable clones (fold change 91, P = 1.23 × 10−8). The data are presented as relative expression (TPC1-P/TPC1-C ratio). B, S100A4 expression analyzed by qPCR showed significant suppression in TPC-1 cells with restored PTCSC3 expression (fold change 0.53, P = .0137). The data are presented as relative expression (TPC1-P/TPC1-C ratio). *, P = .05–.01; ***, P < .001.
Discussion
S100A4 belongs to a large family of EF-hand domain calcium-binding proteins (14). S100 proteins are involved in many processes important for cancer growth and progression, such as proliferation, apoptosis, motility, and invasiveness (15). Among S100 transcripts, S100A4 has emerged as a well-recognized marker for cancer metastasis. S100A4 overexpression has been demonstrated in breast, pancreas, lung, bladder, and gastric tumors associating with a more aggressive phenotype and poor outcome (16–19). Conversely, suppression of S100A4 transcription in osteosarcoma cells results in reduced metastasis potential (20). Transgenic mice overexpressing S100A4 developed aggressive metastatic mammary cancer, whereas mice lacking S100A4 displayed suppression of tumor development and metastasis formation (21, 22). S100A4 has also been proven to be associated with metastasis of nonmedullary thyroid carcinoma (23, 24). It was strongly expressed in an anaplastic thyroid carcinoma cell line with high metastatic potential (23). In PTC, the overexpression of S100A4 was associated with increased tumor invasiveness and metastasis (24). Our data showed that restoration of PTCSC3 expression in the BCPAP and TPC-1 cell lines of PTC origin was associated with a significant reduction of S100A4 expression (P = .0051 and P = .0137, respectively). We also tested the expression of other putative target genes for PTCSC3. Differences between BCPAP control and stable clones were seen, but were not as significant as for S100A4 (Supplemental Table 3). Nonetheless, we assume that S100A4 is not the only target gene for PTCSC3; other genes and their pathways might be affected by PTCSC3 as well. According to a recent report, the expression of PTCSC3 correlates with the expression of the NRG1 gene (25). Interestingly, a SNP rs2439302, within the NRG1 gene predisposes to PTC (26). In our Agilent Ingenuity Pathway analysis, neuregulin signaling was the third most significant canonical pathway (P = 9.54 × 10−4) (11). However, NRG1 itself did not show up as a candidate gene in the Agilent dataset analysis shown here (Supplemental Table 2). Obviously, more work needs to be done to clarify the possible interactions between the neuregulin and S100A4 pathways.
To assess whether S100A4 suppression in PTC cells might result in a less aggressive phenotype, we compared BCPAP cells with and without PTCSC3 expression. BCPAP cells with restored PTCSC3 transcription showed lower motility and invasiveness (P = 4.52 × 10−5 and P = 1.0 × 10−4, respectively). These data suggest that suppression of S100A4 associated with PTCSC3 overexpression reduces PTC cell motility and invasiveness.
Our data showed that PTCSC3 was strongly downregulated in PTC tumor tissue (n = 73, P = 2.2 × 10−16). We hypothesize that the strong suppression of the tumor suppressor PTCSC3 seen in PTC tumors allows cancer cells to increase their invasiveness and motility ability through overexpression of S100A4. The precise mechanism by which PTCSC3 acts on S100A4 remains unidentified and needs to be further investigated.
Several genes have been reported as downstream targets of S100A4. Among them, VGEF and MMP-9 are prominent (27). It has been shown that S100A4 silencing in thyroid cancer cells suppresses their invasion potential through downregulation of MMP-9 and VEGF (13). Here we analyzed VEGF and MMP-9 expression in BCPAP cells by qPCR and showed that they were significantly downregulated in the presence of PTCSC3. Thus, the likely mechanism is that PTCSC3 suppresses S100A4, causing the downstream suppression of VEGF and MMP-9.
S100A4 has been shown to be overexpressed in many solid tumors. Our data add PTC to the list and demonstrate a strong upregulation of S100A4 in PTC tumor tissue when compared to unaffected thyroid tissue (P = 9.33 × 10−7). In tumor tissue, PTCSC3 shows strong downregulation, whereas S100A4 is upregulated; however, there was no correlation of their expressions (r = 0.254, P = .0297). In unaffected tissue, the correlation is present but at moderate levels (r = 0.429, P = .0001). Interestingly, the correlation was in the opposite direction (upregulation of both PTCSC3 and S100A4), seemingly contradicting the findings reported here. We previously showed that PTCSC3 expression was strongly suppressed in PTC tumors but upregulated in unaffected thyroid tissue in patients homozygous for the rs944289 risk allele when compared to individuals with other genotypes (11). Hence, we propose that the action of PTCSC3 is complex, with dual roles as a tumor suppressor (in tumor) and oncogene (in unaffected tissue). Our findings indicating involvement of PTCSC3 in PTC invasiveness are in agreement with observations showing an association between many long noncoding RNAs such as HOTAIR, MALAT1, PCAT-1, or H19 and solid tumor progression and metastasis potential (28). Our analysis of genotypes of rs944289 has indicated that the abundances of PTCSC3 and S100A4 correlate strongly in patients homozygous for the risk allele of rs944289 (r = 0.685, P = 7.88 × 10−5). We hypothesize that overexpression of PTCSC3 in unaffected thyroid tissue stimulates S100A4 expression, whereas in thyroid tissue that has already undergone malignant transformation, further upregulation of S100A4 is prevented. The concept of a dual nature of genes being both tumor suppressors and oncogenes is well-recognized in modern genetics. Several genes such as AKT1, WT1, MYC, MDM2, and others have been shown to possess oncogenic and antioncogenic activity depending on the constellation of other genes at a given point in carcinogenesis stage (29–32). Of note, the 14q13.3 region harboring PTCSC3 also contains another thyroid-expressed gene NKX2-1, also known as TTF-1, located 336 kb upstream of PTCSC3. TTF-1 was first reported as an oncogene based on its overexpression in lung cancer (33, 34). Surprisingly, follow-up studies focusing on its molecular function revealed that it also might act as a tumor suppressor by preventing primary tumors from metastasizing (35, 36).
In summary, we have shown that S100A4 expression is suppressed in cells with forced stable abundance of PTCSC3. Downregulation of S100A4 in PTCSC3-expressing clones results in reduction of cell motility and invasiveness. Cells with stable expression of PTCSC3 display suppression of downstream target genes of S100A4, such as VEGF and MMP-9. S100A4 is upregulated in PTC tumor tissue and its transcription correlates with PTCSC3 expression in unaffected thyroid tissue from PTC patients. Further, we have confirmed the role of SNP rs944289 as a variant predisposing to PTC via the long noncoding RNA gene PTCSC3.
Acknowledgments
This work was supported by National Cancer Institute Grants P30CA16058 and P50CA168505, and an American Thyroid Association Thy-Ca Research Grant 2013.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BCPAP-C
- BCPAP control cells
- BCPAP-P
- BCPAP stable clone cells
- CO2
- carbon dioxide
- FBS
- fetal bovine serum
- PTC
- papillary thyroid carcinoma
- q
- quantitative
- RT
- reverse transcriptase
- SNP
- single nucleotide polymorphism
- TBST
- Tris-buffered saline buffer containing 0.1% Tween-20.
References
- 1. American Cancer Society. Cancer Facts & Figures 2012. Atlanta: American Cancer Society; 2012. http://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2012/index. [Google Scholar]
- 2. Eheman C, Henley SJ, Ballard-Barbash R, et al. Annual report to the nation on the status of cancer, 1975–2008, featuring cancers associated with excess weight and lack of sufficient physical activity. Cancer. 2012;118:2338–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. [DOI] [PubMed] [Google Scholar]
- 4. Woodhouse EC, Chuaqui RF, Liotta LA. General mechanisms of metastasis. Cancer. 1997;80:1529–1537. [DOI] [PubMed] [Google Scholar]
- 5. Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. [DOI] [PubMed] [Google Scholar]
- 6. Jones AM, Howarth KM, Martin L, et al. Thyroid cancer susceptibility polymorphisms: confirmation of loci on chromosomes 9q22 and 14q13, validation of a recessive 8q24 locus and failure to replicate a locus on 5q24. J Med Genet. 2012;49:158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matsuse M, Takahashi M, Mitsutake N, et al. The FOXE1 and NKX2-1 loci are associated with susceptibility to papillary thyroid carcinoma in the Japanese population. J Med Genet. 2011;48:645–648. [DOI] [PubMed] [Google Scholar]
- 9. Wang YL, Feng SH, Guo SC, et al. Confirmation of papillary thyroid cancer susceptibility loci identified by genome-wide association studies of chromosomes 14q13, 9q22, 2q35 and 8p12 in a Chinese population. J Med Genet. 2013;50:689–695. [DOI] [PubMed] [Google Scholar]
- 10. Rogounovitch T, Bychkov A, Takahashi M, et al. The common genetic variant rs944289 at chromosome 14q13.3 associates with risk of both malignant and benign thyroid tumors in Japanese population. Thyroid. 2015;25:333–340. [DOI] [PubMed] [Google Scholar]
- 11. Jendrzejewski J, He H, Radomska HS, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A. 2012;109:8646–8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jia W, Gao XJ, Zhang ZD, Yang ZX, Zhang G. S100A4 silencing suppresses proliferation, angiogenesis and invasion of thyroid cancer cells through downregulation of MMP-9 and VEGF. Eur Rev Med Pharmacol Sci. 2013;17:1495–1508. [PubMed] [Google Scholar]
- 14. Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. [DOI] [PubMed] [Google Scholar]
- 15. Chen H, Xu C, Jin Q, Liu Z. S100 protein family in human cancer. Am J Cancer Res. 2014;4:89–115. [PMC free article] [PubMed] [Google Scholar]
- 16. Rosty C, Ueki T, Argani P, et al. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. 2002;160:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yonemura Y, Endou Y, Kimura K, et al. Inverse expression of S100A4 and E-cadherin is associated with metastatic potential in gastric cancer. Clin Cancer Res. 2000;6:4234–4242. [PubMed] [Google Scholar]
- 18. Davies BR, O'Donnell M, Durkan GC, et al. Expression of S100A4 protein is associated with metastasis and reduced survival in human bladder cancer. J Pathol. 2002;196:292–299. [DOI] [PubMed] [Google Scholar]
- 19. Lee WY, Su WC, Lin PW, Guo HR, Chang TW, Chen HH. Expression of S100A4 and Met: potential predictors for metastasis and survival in early-stage breast cancer. Oncology. 2004;66:429–438. [DOI] [PubMed] [Google Scholar]
- 20. Maelandsmo GM, Hovig E, Skrede M, et al. Reversal of the in vivo metastatic phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme. Cancer Res. 1996;56:5490–5498. [PubMed] [Google Scholar]
- 21. Ambartsumian NS, Grigorian MS, Larsen IF, et al. Metastasis of mammary carcinomas in GRS/A hybrid mice transgenic for the mts1 gene. Oncogene. 1996;13:1621–1630. [PubMed] [Google Scholar]
- 22. Grum-Schwensen B, Klingelhofer J, Berg CH, et al. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 2005;65:3772–3780. [DOI] [PubMed] [Google Scholar]
- 23. Zou M, Famulski KS, Parhar RS, et al. Microarray analysis of metastasis-associated gene expression profiling in a murine model of thyroid carcinoma pulmonary metastasis: identification of S100A4 (Mts1) gene overexpression as a poor prognostic marker for thyroid carcinoma. J Clin Endocrinol Metab. 2004;89:6146–6154. [DOI] [PubMed] [Google Scholar]
- 24. Zou M, Al-Baradie RS, Al-Hindi H, Farid NR, Shi Y. S100A4 (Mts1) gene overexpression is associated with invasion and metastasis of papillary thyroid carcinoma. Br J Cancer. 2005;93:1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rogounovitch TI, Bychkov A, Takahashi M, et al. The common genetic variant rs944289 on chromosome 14q13.3 associates with risk of both malignant and benign thyroid tumors in the Japanese population. Thyroid. 2015;25:333–340. [DOI] [PubMed] [Google Scholar]
- 26. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet. 2012;44:319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Donato R, Cannon BR, Sorci G, et al. Functions of S100 proteins. Curr Mol Med. 2013;13:24–57. [PMC free article] [PubMed] [Google Scholar]
- 28. Shen XH, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol. 2015;28:4–13. [DOI] [PubMed] [Google Scholar]
- 29. Liu H, Radisky DC, Nelson CM, et al. Mechanism of Akt1 inhibition of breast cancer cell invasion reveals a protumorigenic role for TSC2. Proc Natl Acad Sci USA. 2006;103:4134–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. [DOI] [PubMed] [Google Scholar]
- 31. Liu H, Radisky DC, Yang D, et al. MYC suppresses cancer metastasis by direct transcriptional silencing of alphav and beta3 integrin subunits. Nat Cell Biol. 2012;14:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ordonez NG. Thyroid transcription factor-1 is a marker of lung and thyroid carcinomas. Adv Anat Pathol. 2000;7:123–127. [DOI] [PubMed] [Google Scholar]
- 34. Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winslow MM, Dayton TL, Verhaak RG, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maeda Y, Tsuchiya T, Hao H, et al. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest. 2012;122:4388–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]