Abstract
Background and Purpose
Multiple sclerosis (MS) is a demyelinating and inflammatory disease of the central nervous system. The aim of this study was to identify more genes associated with MS.
Methods
Based on the publicly available data of the single-nucleotide polymorphism-based genome-wide association study (GWAS) from the database of Genotypes and Phenotypes, we conducted a powerful gene-based GWAS in an initial sample with 931 family trios, and a replication study sample with 978 cases and 883 controls. For interesting genes, gene expression in MS-related cells between MS cases and controls was examined by using publicly available datasets.
Results
A total of 58 genes was identified, including 20 "novel" genes significantly associated with MS (p<1.40×10-4). In the replication study, 44 of the 58 identified genes had been genotyped and 35 replicated the association. In the gene-expression study, 21 of the 58 identified genes exhibited differential expressions in MS-related cells. Thus, 15 novel genes were supported by replicated association and/or differential expression. In particular, four of the novel genes, those encoding myelin oligodendrocyte glycoprotein (MOG), coiled-coil alpha-helical rod protein 1 (CCHCR1), human leukocyte antigen complex group 22 (HCG22), and major histocompatibility complex, class II, DM alpha (HLA-DMA), were supported by the evidence of both.
Conclusions
The results of this study emphasize the high power of gene-based GWAS in detecting the susceptibility genes of MS. The novel genes identified herein may provide new insights into the molecular genetic mechanisms underlying MS.
Keywords: multiple sclerosis, gene-based GWAS, gene expression, human leukocyte antigen
INTRODUCTION
Multiple sclerosis (MS) is a chronic demyelinating and inflammatory disease of the the central nervous system (CNS).1,2 Genetic factors contribute markedly to the susceptibility to MS, since the children of affected parents have a tenfold higher risk of developing the condition than the general population.3
It has been demonstrated that the human leukocyte antigen (HLA) gene is closely related to MS susceptibility. The HLA gene contains a great many genes (HLA class I, II, and III) residing on chromosome 6, which is related to immune system function in humans. The proteins encoded by the HLA class I and II regions are involved in antigen processing and presentation, and play a major role in autoimmune events. MS is believed to be an immune-mediated disorder that leads to recurrent immune attacks on the CNS.4
Recent genome-wide association studies (GWASs) have identified many loci with modest effects.5,6,7 However, previous GWASs used single-nucleotide polymorphisms (SNPs) as a basic analysis unit,5,6,8,9 and adopted stringent thresholds of significance to control for the false-positive rate. This approach resulted in a large number of SNPs with potential effects being filtered out and ignored.
The gene-based GWAS study strategy, involving analyzing all variants within a putative gene, has proved to be more powerful than regular single-SNP-based GWASs for detecting disease susceptibility genes.10,11,12 To detect "novel" genes associated with MS, we performed a gene-based GWAS using the Knowledge-based mining system for Genome-wide Genetic studies (KGG; http://statgenpro.psychiatry.hku.hk/limx/kgg/index.html)10 in an initial study sample containing 931 family trios.8 We also performed other functional analyses to supplement the evidence regarding the relevance of the novel genes to MS.
METHODS
Samples
The initial GWAS sample included a total of 931 family trios, each of which consisted of an affected MS child and both parents. The replication sample contained 978 cases and 883 controls. The MS patient was diagnosed according to the McDonald criteria.13 Both the initial and replication study samples included all clinical subtypes and partial clinically isolated syndromes. However, the replication study had the priority to include patients with the relapsing onset form of MS. Institutional Review Board was exempted because this study used public available database of Genotypes and Phenotypes (dbGaP) which dose not involve any personal information. The study samples, genotyping, quality control, and SNP exclusion criteria have been described previously.8,9
Gene-based GWAS
The gene-based GWAS and replication analysis were based on the probability values generated in previous genome-wide SNP-based association studies and downloaded from the dbGaP (http://www.ncbi.nlm.nih.gov/gap/?term=multiple+sclerosis, accession number: phs000139 and phs000171). We used the Gene-Based Association Test Using Extended Simes Procedure analysis method modeled in KGG 2.5 (http://statgenpro.psychiatry.hku.hk/limx/kgg/index.html).10 The SNPs, ranging from the upstream 5 kb at the 5' end to the downstream 5 kb at the 3' end, were assigned to one gene. In total, 48% of SNPs across the whole genome were assigned to genes.
Differential expression analysis for MS-associated genes
We downloaded four gene-expression data sets from the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo; GSE21942, GSE27694, GSE16461, and GSE52139). A case-control study design was used to investigate all four data sets, and multiple cell types were investigated, including peripheral blood mononuclear cells (PBMCs), CD34+ hematopoietic progenitor cells (HPCs), CD8+ T lymphocytes, and spinal cord. The study design data analysis are described in detail elsewhere.14,15,16,17 Differentially expressed genes between MS cases and controls were identified by comparing mean gene-expression signals in MS cases versus controls and analyzing the findings using t-tests.
Protein-protein interaction network
MS-associated gene interactions and associations were detected by protein-protein interaction analysis, conducted by searching the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/). The STRING database integrates known and predicted associations derived from a genomic context, high-throughput experiments, coexpression, and previous knowledge (text mining).18
Functional annotation clustering analysis
The probability of the identified MS-associated genes clustering in a Gene Ontology (GO) term or a particular biological pathway as defined by the GO project and Kyoto Encyclopedia of Genes and Genomes database was tested by performing a functional annotation clustering analysis using the Database for Annotation, Visualization and Integrated Discovery integrated database query tools (http://david.abcc.ncifcrf.gov/).19,20 The enrichment was measured quantitatively using Fisher's exact test, and the Bonferroni method21 was adopted to correct for multiple testing.
RESULTS
Quantile-quantile plots of the association results for the original genome scan and the present gene-based GWAS are shown in Fig. 1, in which probability values (shown as -log10 values) for all 20,761 genes and all SNPs are plotted against the expected null distribution. The tail of the distribution of gene-based probability values deviated more significantly than those of SNPs inside or outside of the gene. The distribution of the probability values for the present gene-based GWAS is shown in Fig. 2.
Fig. 1. Quantile-quantile plots of the association results for the initial genome scan. The tail of the distribution of gene-based probability values deviated more significantly than those of SNPs inside or outside of the gene, which suggests that the power was higher for a gene-based association analysis than a single SNP-based GWAS in detecting associations. GWAS: genome-wide association study, SNP: single-nucleotide polymorphism.
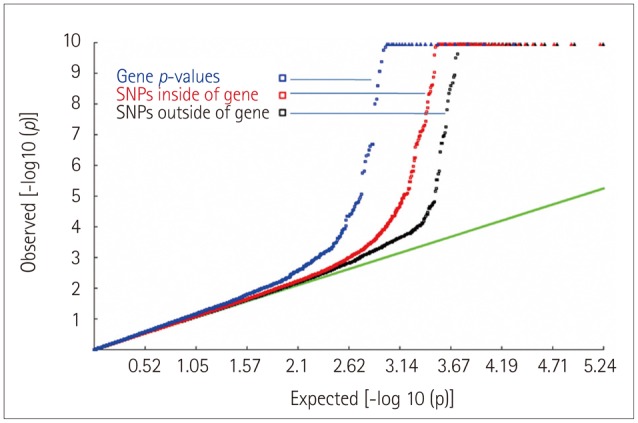
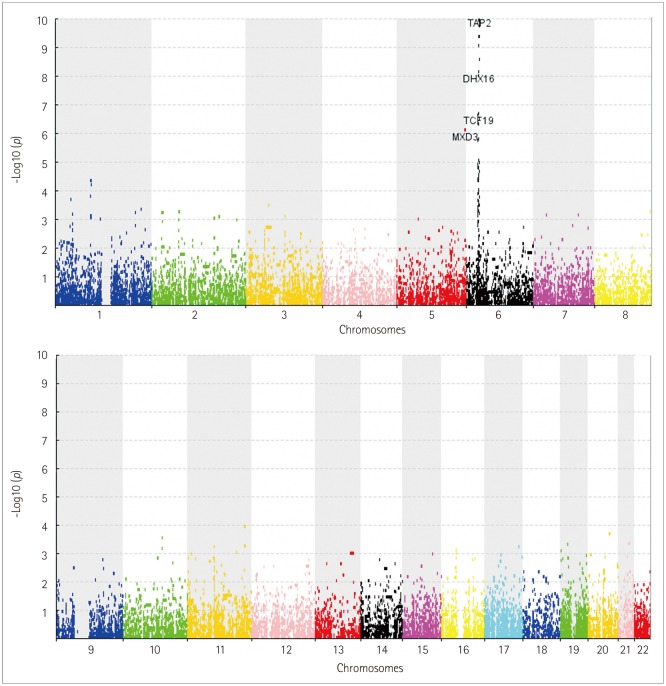
Fig. 2. Manhattan plot of gene probability values on chromosomes. Most of the genes significantly associated with MS are mapped to the HLA region (chromosome 6). HLA: human leukocyte antigen, MS: multiple sclerosis.
Based on a false discovery rate threshold of 1.40×10-4, 58 genes were found to be significantly associated with MS in the initial gene-based GWAS (Supplementary Table 1 in the online-only Data Supplement). Among those 58 genes, 53 were located in the HLA region on chromosome 6. In contrast, according to the raw SNP-based probability values, 75 SNPs in 31 genes were found to be significantly associated with MS (threshold of p<1.49×10-7). Therefore, the present gene-based GWAS detected 27 novel genes that had not previously been detected in the original SNP-based GWAS. After searching the Phenotype-Genotype Integrator (www.ncbi.nlm.nih.gov/gap/phegeni/), a database that archives previous association results, we found that 7 of the 27 genes had already been reported for significant associations (threshold of p<5.0×10-5). Therefore, among the 58 MS-associated genes that we identified, 20 were considered as novel MS candidate genes. Of note, five of these novel genes were located outside the HLA region.
Among the total 58 identified MS-associated genes, 44 genes with genotype data in the replication sample were subjected to further association tests, which revealed that 35 of them (80%) were still significantly associated with MS (p<0.05). Furthermore, 21 of these 58 genes, including 8 novel genes, exhibited differential expression in the differential expression analysis (Table 1). Most interestingly, the genes encoding histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EHMT2), major histocompatibility complex, Class I, A (HLA-C), and negative elongation factor E (RDBP) exhibited significantly differential expressions between cases and controls in both PBMCs and CD34+ HPCs, and the gene encoding myelin oligodendrocyte glycoprotein (MOG) in PBMCs, CD8+ T lymphocytes, and spinal cord, simultaneously (Table 1).
Table 1. MS-associated genes with significantly differential expressions in MS-related cells/tissue.
| Sample | S1 | S2 | S3 | S4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Target cells/tissue | PBMC | CD34+ HPC | CD8+ T lymphocytes | Spinal cord | ||||||||
| Sample size | 12:15 | 8:5 | 4:4 | 8:8 | ||||||||
| Platform | [HG-U133_Plus_2] Affymetrix Human | Agilent-014850 Whole Human | [HG-U133_Plus_2] Affymetrix Human | [HG-U133_Plus_2] Affymetrix Human | ||||||||
| Genome U133 Plus 2.0 Array | Genome Microarray 4×44K G4112F | Genome U133 Plus 2.0 Array | Genome U133 Plus 2.0 Array | |||||||||
| PMID | 22021740 | 22252466 | 21216829 | 24910450 | ||||||||
| GSE No. | GSE21942 | GSE27694 | GSE16461 | GSE52139 | ||||||||
The ratio listed in the line of "Sample size" is the number of multiple sclerosis (MS) cases compared with that of controls. The genes listed here that are not defined in the main text are major histocompatibility complex, class II, DO beta (HLA-DOB), major histocompatibility complex, class II, DR alha (HLA-DRA), POU class 5 homeobox 1 (POU5F1), uncharacterized LOC100294145 (LOC100294145), tenascin XB (TNXB), psoriasis susceptibility 1 candidate 1 (PSORS1C1), major histocompatibility complex, class II, DM beta (HLA-DMB), transporter 2, ATP-binding cassette, sub-family B (TAP2), allograft inflammatory factor 1 (AIF1), major histocompatibility complex, class I, F (HLA-F), major histocompatibility complex, class II, DQ alpha 1 (HLA-DQA1), respectively.
*Only the most significant probe is listed, even if more than one probe was detected for a gene.
GSE No.: Gene Expression Omnibus number, www.ncbi.nlm.nih.gov/geo/, PMID: PubMed unique identifier.
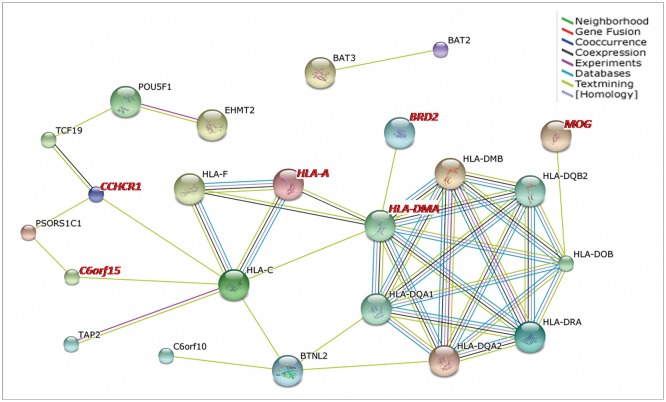
The 58 identified MS-associated genes were retrieved from the STRING database. Only 36 genes, including 10 novel genes, were annotated in this database. The genes at the HLA regions were clearly enriched into two clusters: HLA class I and II clusters (Fig. 3). Two novel genes, those encoding major histocompatibility complex, class, I, A (HLA-A) and major histocompatibility complex, class II, DM alpha (HLA-DMA), were involved in HLA-class I and II clusters, respectively. Another four novel genes, MOG, and those encoding coiled-coil alpha-helical rod protein 1 (CCHCR1), bromodomain-containing protein 2 (BRD2), and chromosome 6 open reading frame 15 (C6orf15), were directly connected with the HLA clusters (Fig. 3).
Fig. 3. Protein-protein interactions between MS-associated genes. Only the connected genes are shown. The novel genes are labeled in red. Different line colors represent the types of evidence for the association. HLA class I cluster: major histocompatibility complex, class I, A (HLA-A), Major histocompatibility complex, class I, C (HLA-C), and major histocompatibility complex, class I, F (HLA-F); HLA class II cluster: major histocompatibility complex, class II, DQ alpha 1 (HLA-DQA1), major histocompatibility complex, class II, DQ alpha 2 (HLA-DQA2), major histocompatibility complex, class II, DQ beta 2 (HLA-DQB2), major histocompatibility complex, class II, DR alha (HLA-DRA), major histocompatibility complex, class II, DO beta (HLA-DOB), major histocompatibility complex, class II, DM alpha (HLA-DMA), and major histocompatibility complex, class II, DM beta (HLA-DMB). HLA: human leukocyte antigen, MS: Multiple sclerosis.
Taking both the replication data and gene-expression data together, we found that 15 novel genes were supported by replicated association and/or differential expression (Table 2). Most interestingly, the significance of four genes—HLA-DMA, CCHCR1, MOG, and the gene encoding HLA complex group 22 (HCG22)—in MS was supported by both the replication and gene-expression studies. In addition, the significance of two non-HLA genes—those encoding family with sequence similarity 69, member A (FAM69A) and POU domain class 2 transcription factor 3 (POU2F3)—in MS was supported by evidence of differential expressions in PBMCs and CD8+ T lymphocytes, respectively.
Table 2. Fifteen novel MS-associated genes, supported by replication studies and/or expression studies.
| Gene | Chromosome | Start position | Length | #SNP* | Initial p | Replication p | Differential expression p, sample† | |
|---|---|---|---|---|---|---|---|---|
| MOG | 6 | 29619758 | 25391 | 2 | 1.21×10-5 | 5.09×10-4 | 2.12×10-2 | S1 |
| 3.65×10-2 | S3 | |||||||
| 2.92×10-2 | S4 | |||||||
| HLA-DMA | 6 | 32911391 | 14508 | 2 | 3.15×10-7 | 4.08×10-2 | 8.62×10-4 | S1 |
| CCHCR1 | 6 | 31105216 | 25799 | 2 | 4.03×10-5 | 1.08×10-3 | 1.05×10-2 | S1 |
| HCG22 | 6 | 31016984 | 15669 | 6 | 1.21×10-4 | 2.29×10-2 | 3.85×10-2 | S1 |
| BRD2 | 6 | 32931437 | 22845 | 1 | 1.07×10-5 | NS | 5.65×10-6 | S1 |
| FAM69A | 1 | 93302721 | 129358 | 11 | 4.47×10-5 | NA | 1.63×10-2 | S1 |
| LOC100294145 | 6 | 32856953 | 19582 | 3 | 2.27×10-5 | NA | 4.21×10-2 | S2 |
| POU2F3 | 11 | 120000000 | 89702 | 3 | 1.13×10-4 | NS | 8.41×10-3 | S3 |
| TRIM26P | 6 | 30201078 | 13978 | 2 | 3.72×10-5 | 7.80×10-6 | NA | NA |
| HCP5P14 | 6 | 29733324 | 12339 | 1 | 1.33×10-4 | 9.94×10-5 | NA | NA |
| 3.8-1.4 (HLA complex group 26) | 6 | 29828692 | 11172 | 7 | 1.58×10-5 | 1.08×10-3 | NA | NA |
| C6orf15 | 6 | 31074000 | 11332 | 3 | 9.26×10-5 | 2.22×10-3 | NS | NS |
| MUC21 | 6 | 30946485 | 16190 | 4 | 4.40×10-5 | 3.42×10-3 | NA | NA |
| HLA-A | 6 | 29905309 | 13352 | 4 | 1.61×10-5 | 8.38×10-3 | NS | NS |
| HLA-F-AS1 | 6 | 29689378 | 32448 | 7 | 1.83×10-6 | 2.52×10-2 | NA | NA |
The genes listed here that are not defined in the main text are tripartite motif containing 26 B (TRIM26P), HLA complex P5 pseudogene 14 (HCP5P14), HLA complex group 26 pseudogene (3.8-1.4), mucin 21, cell surface associated (MUC21), HLA-F antisense RNA 1 (HLA-F-AS1), respectively.
*Number of SNPs included in the gene with p<0.05, †Multiple sclerosis (MS)-related cells/tissue sample used for the differential expression analysis; S1, PBMC; S2, CD34+ HPC; S3, CD8+ T lymphocytes; S4, spinal cord.
NA: not available, NS: not significant.
The identified 58 MS-associated genes were found enriched in 54 GO terms, even after Bonferroni correction (p<0.05) (Supplementary Table 2 in the online-only Data Supplement). Most significantly, 12 genes were enriched in "antigen processing and presentation" (GO: 0019882; p=271.09×10-16), and 11 genes were enriched in "MHC protein complex" (GO: 0012611; p=1.06×10-16).
DISCUSSION
Gene-based association analysis is an efficient method for detecting associations between candidate genes and complex diseases, as it combines signals for all variants within a putative gene. By using this method, several studies have identified new associations between genes and diseases.22,23 The present study again highlights the superior power of gene-based association analysis for detecting associations for MS. Specifically, we detected 58 genes significantly associated with MS, including 20 novel genes that were undetected in previous single SNP-based GWASs. Furthermore, associations for 80% of the identified MS-associated genes were replicated.
Most previous association studies have identified only the statistical relevance of genes to MS (at the DNA level), without dissecting the functional mechanisms underlying those associations. In contrast, in the present study we not only established statistical associations between genetic markers and MS at the DNA level, but also performed follow-up differential gene expression analyses and functional annotation clustering analyses as important supplementary methods to analyze the function of causal variants. This supplementary evidence strengthens the likelihood that the eight novel genes identified in this study with significantly differential expressions are directly involved in the pathogenesis of MS.
Recent GWASs have identified a great number of MS-associated genetic loci,5,9,24,25,26 most of which have been mapped to the HLA region. Most of the loci identified in the present study were also located in this region. As we know, the major HLAs are essential elements for immune function. Therefore, our findings also highlight the importance of the autoimmune system in the etiology of MS.
Most interestingly, evidence from association, replication, and differential expression studies strongly supports the significance of the following four genes to MS: MOG, HLA-DMA, CCHCR1, and HCG22. Up to now, the biological function of HCG22 in MS or immunity is unknown. HLA-DMA and CCHCR1 exhibit suggestive associations with rheumatoid arthritis (RA) and psoriasis,27,28,29 respectively, suggesting that both genes are involved in the immune response. The protein encoded by MOG, myelin oligodendrocyte glycoprotein (MOG) is a membrane protein expressed on the surface of oligodendrocyte cells and myelin sheaths.30 MOG is an important candidate target antigen in MS.31,32,33 Monoclonal antibodies against MOG were used to develop an animal model of MS.34 The functions of these genes in MS need further investigation.
In addition, three genes (POU2F3, BRD2, and FAM69A) were expressed differentially in MS-related cells. POU2F3, a non-HLA gene (chromosome 11), is associated with melanoma and cervical cancer.35,36 BRD2 is associated with cancer, obesity, type 2 diabetes, RA, and inflammation,37,38,39 and was differentially expressed in the PBMCs of patients with RA (p=2.31×10-3, data sets from GEO; www.ncbi.nlm.nih.gov/geo, GSE#: GSE15573). The associations with other autoimmune diseases suggested the possible relevance of BRD2 and POU2F3 to MS. The specific functions of FAM69A in MS are unknown. A previous study found that 21 SNPs located at the GFI-EVI5-RPL5-FAM69A locus were positively associated with MS.40 In-depth studies are needed to disclose the functional mechanism of these genes in MS.
It should be noted that since there is strong linkage disequilibrium (LD) at the HLA region, it is reasonable to infer that the significant signals for some of the genetic markers are partially due to their strong LD with true functional variants within the HLA region. Although the supplementary analysis could play a part in analyzing the function of causal variants, it is also necessary to research the functional mechanisms underlying the associations further, especially those between the HLAs and MS.
In conclusion, this gene-based GWAS has identified 20 novel MS-associated genes. The results highlight the advantages of gene-based association analysis over single SNP-based GWASs for detecting susceptibility genes for MS. The new findings may provide novel insights into the molecular mechanisms underlying MS.
Acknowledgements
The study was supported by Natural Science Foundation of China (81473046, 81401343, 31401079, 31271336, 31071097, and 81373010), the Natural Science Foundation of Jiangsu Province (BK20130300), the Startup Fund from Soochow University (Q413900112, Q413900712), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, and a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Conflicts of Interest: The authors have no financial conflicts of interest.
Supplementary Materials
The online-only Data Supplement is available with this article at http://dx.doi.org/10.3988/jcn.2015.11.4.311.
Supplementary Table 1. Results of gene-based and differential expression analyses of 58 MS-associated genes.
| Gene | Chromosome | Start position | Length | #SNP* | Initial p | Genes significant in initial study based on SNP-based p | Genes in PheGenI | Replication p | Differential expression p | Sample† |
|---|---|---|---|---|---|---|---|---|---|---|
| C6orf10 | 6 | 32255475 | 89181 | 21 | 3.97×10-39 | C6orf10 | C6orf10 | 3.16×10-3 | NS | NS |
| NOTCH4 | 6 | 32157620 | 39224 | 7 | 2.51×10-31 | NOTCH4 | NOTCH4 | NS | NS | NS |
| TNXB | 6 | 32003932 | 78221 | 2 | 6.61×10-28 | TNXB | TNXB | 3.37×10-3 | 4.45×10-2 | S2 |
| HLA-DRA | 6 | 32402619 | 15204 | 9 | 3.73×10-24 | HLA-DRA | HLA-DRA | 4.08×10-3 | 5.82×10-5 | S1 |
| AIF1 | 6 | 31577994 | 11804 | 1 | 3.15×10-23 | AIF1 | AIF1 | NA | 2.04×10-2 | S1 |
| SNORA38 | 6 | 31585856 | 10131 | 1 | 3.15×10-23 | SNORA38 | NA | NA | NA | NA |
| BAT2 | 6 | 31583450 | 27104 | 3 | 1.18×10-22 | BAT2 | BAT2 | 3.42×10-2 | NS | NS |
| BAT3 | 6 | 31601805 | 23672 | 3 | 4.41×10-19 | BAT3 | BAT3 | 9.52×10-3 | NS | NS |
| PPIAP9 | 6 | 31481654 | 11525 | 1 | 1.46×10-15 | PPIAP9 | PPIAP9 | 4.67×10-2 | NA | NA |
| HLA-DQB2 | 6 | 32718875 | 17455 | 6 | 9.02×10-15 | HLA-DQB2 | HLA-DQB2 | 1.01×10-2 | NS | NS |
| HLA-DQA2 | 6 | 32704163 | 15501 | 6 | 1.25×10-14 | HLA-DQA2 | HLA-DQA2 | 3.93×10-3 | NS | NS |
| EHMT2 | 6 | 31842536 | 27928 | 1 | 8.91×10-14 | EHMT2 | EHMT2 | NS | 4.30×10-3 | S1 |
| 4.50×10-2 | S2 | |||||||||
| LOC100287272 | 6 | 31238352 | 13179 | 5 | 1.79×10-13 | LOC100287272 | NA | 4.76×10-3 | NA | NA |
| MCCD1 | 6 | 31491739 | 11269 | 2 | 4.39×10-13 | MCCD1 | MCCD1 | NA | NS | NS |
| LOC100462812 | 6 | 31340494 | 10311 | 5 | 4.71×10-13 | LOC100462812 | NA | 8.07×10-5 | NA | NA |
| HCG27 | 6 | 31160537 | 16208 | 3 | 6.34×10-13 | HCG27 | HCG27 | 3.84×10-2 | NS | NS |
| HLA-DRB9 | 6 | 32422597 | 10269 | 6 | 9.42×10-13 | HLA-DRB9 | HLA-DRB9 | 1.60×10-3 | NA | NA |
| HLA-S | 6 | 31344346 | 10918 | 6 | 4.89×10-12 | HLA-S | HLA-S | 3.36×10-2 | NA | NA |
| HLA-DQA1 | 6 | 32600183 | 16246 | 1 | 1.68×10-11 | HLA-DQA1 | HLA-DQA1 | 5.96×10-4 | 3.51×10-2 | S1 |
| TAP2 | 6 | 32784610 | 26937 | 2 | 8.02×10-11 | TAP2 | TAP2 | NS | 3.35×10-3 | S1 |
| BTNL2 | 6 | 32357513 | 22387 | 7 | 1.43×10-10 | BTNL2 | BTNL2 | 8.76×10-5 | NS | NS |
| HLA-DOB | 6 | 32775540 | 14285 | 4 | 1.78×10-10 | HLA-DOB | HLA-DOB | NA | 1.63×10-5 | S1 |
| RDBP | 6 | 31914864 | 17000 | 2 | 4.31×10-10 | RDBP | RDBP | 4.16×10-2 | 2.01×10-2 | S1 |
| 3.70×10-2 | S2 | |||||||||
| PSORS1C1 | 6 | 31077608 | 35261 | 10 | 8.59×10-10 | PSORS1C1 | PSORS1C1 | NS | 4.85×10-2 | S2 |
| HLA-DMB | 6 | 32897406 | 16441 | 1 | 2.63×10-9 | HLA-DMB | HLA-DMB | NS | 4.45×10-2 | S3 |
| DHX16 | 6 | 30615896 | 29934 | 1 | 7.13×10-9 | DHX16 | DHX16 | 2.73×10-3 | NS | NS |
| POU5F1 | 6 | 31127114 | 16337 | 4 | 9.96×10-9 | POU5F1 | POU5F1 | 4.68×10-3 | 8.28×10-3 | S2 |
| TCF19 | 6 | 31121303 | 15689 | 2 | 2.02×10-7 | TCF19 | TCF19 | 1.78×10-2 | NS | NS |
| 3.8-1.5 (HLA complex group 26) | 6 | 29727893 | 11180 | 4 | 2.40×10-7 | 3.8-1.5 | NA | 1.27×10-3 | NA | NA |
| HLA-F | 6 | 29686117 | 13956 | 7 | 4.67×10-7 | HLA-F | NA | 1.17×10-2 | 2.64×10-2 | S1 |
| LOC100507436 | 6 | 31362561 | 76025 | 8 | 4.98×10-7 | LOC100507436 | NA | NS | NA | NA |
| HLA-X | 6 | 31424623 | 10644 | 1 | 1.55×10-6 | NA | HLA-X | NA | NA | NA |
| HCG4P4 | 6 | 29917982 | 10428 | 1 | 8.60×10-6 | NA | HCG4P4 | 6.98×10-4 | NA | NA |
| RPL3P2 | 6 | 31243108 | 11240 | 2 | 1.66×10-5 | NA | RPL3P2 | NA | NA | NA |
| HLA-C | 6 | 31231529 | 13326 | 2 | 1.72×10-5 | NA | HLA-C | 4.29×10-2 | 6.15×10-3 | S1 |
| 2.74×10-2 | S2 | |||||||||
| DHFRP2 | 6 | 31326244 | 13498 | 2 | 2.89×10-5 | NA | DHFRP2 | NA | NA | NA |
| MICE | 6 | 29704234 | 17646 | 1 | 4.63×10-5 | NA | MICE | NA | NA | NA |
| TRIM31 | 6 | 30065674 | 20193 | 1 | 1.18×10-4 | NA | TRIM31 | 1.31×10-4 | NS | NS |
| MOG | 6 | 29619758 | 25391 | 2 | 1.21×10-5 | NA | NA | 5.09×10-4 | 2.12×10-2 | S1 |
| 3.65×10-2 | S3 | |||||||||
| 2.92×10-2 | S4 | |||||||||
| HLA-DMA | 6 | 32911391 | 14508 | 2 | 3.15×10-7 | NA | NA | 4.08×10-2 | 8.62×10-4 | S1 |
| CCHCR1 | 6 | 31105216 | 25799 | 2 | 4.03×10-5 | NA | NA | 1.08×10-3 | 1.05×10-2 | S1 |
| HCG22 | 6 | 31016984 | 15669 | 6 | 1.21×10-4 | NA | NA | 2.29×10-2 | 3.85×10-2 | S1 |
| BRD2 | 6 | 32931437 | 22845 | 1 | 1.07×10-5 | NA | NA | NS | 5.65×10-6 | S1 |
| FAM69A | 1 | 93302721 | 129358 | 11 | 4.47×10-5 | NA | NA | NA | 1.63×10-2 | S1 |
| LOC100294145 | 6 | 32856953 | 19582 | 3 | 2.27×10-5 | NA | NA | NA | 4.21×10-2 | S2 |
| POU2F3 | 11 | 120000000 | 89702 | 3 | 1.13×10-4 | NA | NA | NS | 8.41×10-3 | S3 |
| TRIM26P | 6 | 30201078 | 13978 | 2 | 3.72×10-5 | NA | NA | 7.80×10-6 | NA | NA |
| HCP5P14 | 6 | 29733324 | 12339 | 1 | 1.33×10-4 | NA | NA | 9.94×10-5 | NA | NA |
| 3.8-1.4(HLA complex group 26) | 6 | 29828692 | 11172 | 7 | 1.58×10-5 | NA | NA | 1.08×10-3 | NA | NA |
| C6orf15 | 6 | 31074000 | 11332 | 3 | 9.26×10-5 | NA | NA | 2.22×10-3 | NS | NS |
| MUC21 | 6 | 30946485 | 16190 | 4 | 4.40×10-5 | NA | NA | 3.42×10-3 | NA | NA |
| HLA-A | 6 | 29905309 | 13352 | 4 | 1.61×10-5 | NA | NA | 8.38×10-3 | NS | NS |
| HLA-F-AS1 | 6 | 29689378 | 32448 | 7 | 1.83×10-6 | NA | NA | 2.52×10-2 | NA | NA |
| MXD3 | 5 | 177000000 | 16791 | 2 | 7.60×10-7 | NA | NA | NS | NS | NS |
| LOC100287247 | 22 | - | - | 1 | 2.71×10-5 | NA | NA | NA | NA | NA |
| LOC100127934 | 1 | 93392136 | 10509 | 1 | 6.18×10-5 | NA | NA | NA | NA | NA |
| RPL15P4 | 6 | 31490853 | 10645 | 1 | 2.08×10-7 | NA | NA | NA | NA | NA |
| HCG2P8 | 6 | 29767896 | 13543 | 1 | 3.96×10-5 | NA | NA | NA | NA | NA |
PheGenI, Phenotype-Genotype Integrator.
*The number of SNP included in a gene with p-value<0.05, †Ms-related cells sample used to differential expression ananlysis. S1: PBMC, S2: CD34+ HPC, S3: CD8+ T lymphocytes, S4, spinal cord.
The genes listed here that are not defined in the main text are chromosome 6 open reading frame 10 (C6orf10), notch 4 (NOTCH4), small nucleolar RNA, H/ACA box 38 (SNORA38), proline-rich coiled-coil 2A (BAT2), BCL2-associated athanogene 6 (BAT3), peptidylprolyl isomerase A pseudogene 9 (PPIAP9), major histocompatibility complex, class II, DQ beta 2 (HLA-DQB2), major histocompatibility complex, class II, DQ alpha 2 (HLA-DQA2), ubiquitin specific peptidase 8 pseudogene 1 (LOC100287272), mitochondrial coiled-coil domain 1 (MCCD1), fibroblast growth factor receptor 3 pseudogene 1 (LOC100462812), HLA complex group 27 (HCG27), major histocompatibility complex, class II, DR beta 9 (HLA-DRB9), major histocompatibility complex, class I, S (HLA-S), butyrophilin-like 2 (BTNL2), DEAH (Asp-Glu-Ala-His) box polypeptide 16 (DHX16) transcription factor 19 (TCF19), HLA complex group 26 pseudogene (3.8-1.5), MHC class I polypeptide-related sequence A (LOC100507436), major histocompatibility complex, class I, x (HLA-X), HLA complex group 4 pseudogene 4 (HCG4P4), ribosomal protein L3 pseudogene 2 (RPL3P2), dihydrofolate reductase pseudogene 2 (DHFRP2), MHC class I polypeptide-related sequence E (MICE), ripartite motif containing 31 (TRIM31), MAX dimerization protein 3 (MXD3), similar to hCG1987428 (LOC100287247), meiotic nuclear divisions 1 homolog (LOC100127934), ribosomal protein L15 pseudogene 4 (RPL15P4), HLA complex group 2 pseudogene 8 (HCG2P8) respectively.
Supplementary Table 2. Enrichment of GO terms and KEGG pathways of the 58 MS-associated genes.
| Category | Term | Count | % | Genes | List total | Relative enrichment | Bonferroni* |
|---|---|---|---|---|---|---|---|
| GOTERM_BP_FAT | GO:0019882~antigen processing and presentation | 12 | 21.43 | 3112, 285830, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 25 | 78.23 | 1.09×10-16 |
| GOTERM_CC_FAT | GO:0042611~MHC protein complex | 11 | 19.64 | 3112, 285830, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 24 | 102.78 | 1.06×10-16 |
| SP_PIR_KEYWORDS | mhc ii | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3105, 3132, 3122 | 34 | 164.25 | 1.84×10-14 |
| UP_SEQ_FEATURE | Region of interest:Connecting peptide | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 33 | 137.17 | 3.39×10-13 |
| UP_SEQ_FEATURE | Domain:Ig-like C1-type | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 33 | 130.32 | 5.42×10-13 |
| KEGG_PATHWAY | hsa05330:Allograft rejection | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 90.80 | 7.23×10-14 |
| KEGG_PATHWAY | hsa05332:Graft-versus-host disease | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 83.82 | 1.47×10-13 |
| KEGG_PATHWAY | hsa04940:Type I diabetes mellitus | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 77.83 | 2.80×10-13 |
| KEGG_PATHWAY | hsa04612:Antigen processing and presentation | 10 | 17.86 | 3112, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 43.76 | 7.53×10-13 |
| KEGG_PATHWAY | hsa05320:Autoimmune thyroid disease | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 64.10 | 1.50×10-12 |
| INTERPRO | IPR003597:Immunoglobulin C1-set | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 29 | 76.03 | 1.16×10-11 |
| UP_SEQ_FEATURE | Region of interest:Alpha-1 | 7 | 12.50 | 3117, 3118, 3107, 3108, 3134, 3105, 3122 | 33 | 202.71 | 1.56×10-10 |
| UP_SEQ_FEATURE | Region of interest: Alpha-2 | 7 | 12.50 | 3117, 3118, 3107, 3108, 3134, 3105, 3122 | 33 | 202.71 | 1.56×10-10 |
| INTERPRO | IPR014745:MHC class II, alpha/beta chain, N-terminal | 7 | 12.50 | 3112, 3117, 3118, 3109, 3108, 3132, 3122 | 29 | 182.78 | 7.35×10-11 |
| INTERPRO | IPR003006:Immunoglobulin/major histocompatibility complex, conserved site | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 29 | 59.43 | 8.98×10-11 |
| KEGG_PATHWAY | hsa05416:Viral myocarditis | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 46.04 | 2.47×10-11 |
| SMART | SM00407:IGc1 | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 24 | 50.07 | 7.27×10-11 |
| GOTERM_CC_FAT | GO:0042613~MHC class II protein complex | 7 | 12.50 | 3112, 3117, 3118, 3109, 3108, 3132, 3122 | 24 | 128.55 | 5.56×10-10 |
| GOTERM_BP_FAT | GO:0002504~antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 7 | 12.50 | 3112, 3117, 3118, 3109, 3108, 3132, 3122 | 25 | 114.78 | 3.42×10-9 |
| GOTERM_MF_FAT | GO:0032395~MHC class II receptor activity | 6 | 10.71 | 3112, 3117, 3118, 3107, 3108, 3122 | 23 | 178.26 | 7.85×10-9 |
| SP_PIR_KEYWORDS | Immune response | 10 | 17.86 | 3112, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3122 | 34 | 25.26 | 8.42×10-9 |
| KEGG_PATHWAY | hsa04514:Cell adhesion molecules (CAMs) | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 24.76 | 4.04×10-9 |
| GOTERM_CC_FAT | GO:0042825~TAP complex | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 24 | 380.42 | 2.00×10-8 |
| GOTERM_CC_FAT | GO:0042824~MHC class I peptide loading complex | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 24 | 295.88 | 7.18×10-8 |
| GOTERM_BP_FAT | GO:0048002~antigen processing and presentation of peptide antigen | 6 | 10.71 | 6891, 3107, 3108, 3134, 3105, 3122 | 25 | 115.95 | 2.17×10-7 |
| PIR_SUPERFAMILY | PIRSF001991:Class II histocompatibility antigen | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 18 | 94.82 | 2.79×10-8 |
| INTERPRO | IPR007110:Immunoglobulin-like | 11 | 19.64 | 3112, 3117, 3118, 4340, 3107, 3109, 3108, 3134, 56244, 3105, 3122 | 29 | 12.61 | 3.53×10-7 |
| KEGG_PATHWAY | hsa05310:Asthma | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 14 | 75.15 | 1.10×10-7 |
| GOTERM_BP_FAT | GO:0006955~immune response | 12 | 21.43 | 3112, 285830, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 25 | 9.41 | 1.53×10-6 |
| GOTERM_MF_FAT | GO:0042288~MHC class I protein binding | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 23 | 176.40 | 8.88×10-7 |
| INTERPRO | IPR013783:Immunoglobulin-like fold | 11 | 19.64 | 3112, 3117, 3118, 4340, 3107, 3109, 3108, 3134, 56244, 3105, 3122 | 29 | 11.43 | 9.06×10-7 |
| GOTERM_MF_FAT | GO:0042287~MHC protein binding | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 23 | 112.90 | 6.11×10-6 |
| KEGG_PATHWAY | hsa04672:Intestinal immune network for IgA production | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 14 | 44.48 | 1.72×10-6 |
| SP_PIR_KEYWORDS | Heterodimer | 6 | 10.71 | 3117, 3118, 3107, 3134, 3105, 3122 | 34 | 32.96 | 6.98×10-5 |
| SP_PIR_KEYWORDS | Transmembrane protein | 10 | 17.86 | 3112, 3117, 3118, 4340, 6891, 3107, 3108, 3134, 3105, 3122 | 34 | 8.81 | 7.59×10-5 |
| INTERPRO | IPR001003:MHC class II, alpha chain, N-terminal | 4 | 7.14 | 3117, 3118, 3108, 3122 | 29 | 191.48 | 7.32×10-5 |
| KEGG_PATHWAY | hsa05322:Systemic lupus erythematosus | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 14 | 22.01 | 6.04×10-5 |
| GOTERM_BP_FAT | GO:0002474~antigen processing and presentation of peptide antigen via MHC class I | 4 | 7.14 | 6891, 3107, 3134, 3105 | 25 | 127.32 | 6.60×10-4 |
| GOTERM_CC_FAT | GO:0044459~plasma membrane part | 15 | 26.79 | 4855, 3112, 3117, 3118, 6891, 394263, 285830, 199, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 24 | 3.63 | 2.43×10-4 |
| INTERPRO | IPR001039:MHC class I, alpha chain, alpha1, and alpha2 | 4 | 7.14 | 285830, 3107, 3134, 3105 | 29 | 85.10 | 9.56×10-4 |
| INTERPRO | IPR011161:MHC class I-like antigen recognition | 4 | 7.14 | 285830, 3107, 3134, 3105 | 29 | 82.06 | 1.07×10-3 |
| GOTERM_CC_FAT | GO:0042612~MHC class I protein complex | 4 | 7.14 | 285830, 3107, 3134, 3105 | 24 | 76.08 | 1.17×10-3 |
| BIOCARTA | h_mhcPathway:Antigen processing and presentation | 3 | 5.36 | 6891, 3105, 3122 | 3 | 143.70 | 5.67×10-4 |
| UP_SEQ_FEATURE | Sequence variant | 31 | 55.36 | 170679, 3112, 4855, 11074, 54535, 6891, 56244, 7148, 10919, 6941, 199, 6046, 83463, 5460, 3107, 3109, 3108, 3134, 3105, 401250, 3117, 3118, 4340, 29113, 8449, 10665, 7916, 394263, 25833, 3122, 7917 | 33 | 1.50 | 1.94×10-2 |
| SP_PIR_KEYWORDS | Polymorphism | 31 | 55.36 | 170679, 3112, 4855, 11074, 54535, 6891, 56244, 7148, 10919, 6941, 199, 6046, 83463, 5460, 3107, 3109, 3108, 3134, 3105, 401250, 3117, 3118, 4340, 29113, 8449, 10665, 7916, 394263, 25833, 3122, 7917 | 34 | 1.52 | 7.47×10-3 |
| UP_SEQ_FEATURE | Region of interest:Alpha-3 | 3 | 5.36 | 3107, 3134, 3105 | 33 | 193.06 | 2.92×10-2 |
| SP_PIR_KEYWORDS | mhc i | 3 | 5.36 | 3107, 3134, 3105 | 34 | 169.72 | 1.05×10-2 |
| GOTERM_BP_FAT | GO:0002478~antigen processing and presentation of exogenous peptide antigen | 3 | 5.36 | 6891, 3108, 3122 | 25 | 147.58 | 3.25×10-2 |
| PIR_SUPERFAMILY | PIRSF001990:Class I histocompatibility antigen | 3 | 5.36 | 3107, 3134, 3105 | 18 | 136.96 | 2.30×10-3 |
| GOTERM_BP_FAT | GO:0019884~antigen processing and presentation of exogenous antigen | 3 | 5.36 | 6891, 3108, 3122 | 25 | 115.95 | 5.30×10-2 |
| GOTERM_MF_FAT | GO:0042277~peptide binding | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 23 | 13.90 | 2.69×10-2 |
| INTERPRO | IPR000353:MHC class II, beta chain, N-terminal | 3 | 5.36 | 3112, 3109, 3132 | 29 | 101.37 | 2.84×10-2 |
| GOTERM_MF_FAT | GO:0032393~MHC class I receptor activity | 3 | 5.36 | 3107, 3134, 3105 | 23 | 99.61 | 2.89×10-2 |
| GOTERM_CC_FAT | GO:0005886~plasma membrane | 16 | 28.57 | 4855, 3112, 3117, 3118, 4340, 6891, 394263, 285830, 199, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 24 | 2.26 | 2.98×10-2 |
Count, the number of genes enriched in particular GO terms or KEGG pathways; Genes, potential genes enriched in particular GO terms or KEGG pathways.
*p value with Bonferroni correction for multiple tests. Only significant results with p<0.05 are listed.
GO: Gene Ontology, KEGG: Kyoto Encyclopedia of Genes and Genomes, MHC: major histocompatibility complex, MS: mltiple sclerosis.
References
- 1.Selchen D, Bhan V, Blevins G, Devonshire V, Duquette P, Grand'Maison F, et al. MS, MRI, and the 2010 McDonald criteria: a Canadian expert commentary. Neurology. 2012;79(23 Suppl 2):S1–S15. doi: 10.1212/WNL.0b013e318277d144. [DOI] [PubMed] [Google Scholar]
- 2.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 3.Milo R, Kahana E. Multiple sclerosis: geoepidemiology, genetics and the environment. Autoimmun Rev. 2010;9:A387–A394. doi: 10.1016/j.autrev.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Nakahara J, Maeda M, Aiso S, Suzuki N. Current concepts in multiple sclerosis: autoimmunity versus oligodendrogliopathy. Clin Rev Allergy Immunol. 2012;42:26–34. doi: 10.1007/s12016-011-8287-6. [DOI] [PubMed] [Google Scholar]
- 5.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 6.Jakkula E, Leppä V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Netto MJ, Ward H, Morrison KM, Ramagopalan SV, Dyment DA, DeLuca GC, et al. Risk alleles for multiple sclerosis in multiplex families. Neurology. 2009;72:1984–1988. doi: 10.1212/WNL.0b013e3181a92c25. [DOI] [PubMed] [Google Scholar]
- 8.International Multiple Sclerosis Genetics Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 9.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 14.Kemppinen AK, Kaprio J, Palotie A, Saarela J. Systematic review of genome-wide expression studies in multiple sclerosis. BMJ Open. 2011;1:e000053. doi: 10.1136/bmjopen-2011-000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutterotti A, Jelcić I, Schulze C, Schippling S, Breiden P, Mazzanti B, et al. No proinflammatory signature in CD34+ hematopoietic progenitor cells in multiple sclerosis patients. Mult Scler. 2012;18:1188–1192. doi: 10.1177/1352458511434067. [DOI] [PubMed] [Google Scholar]
- 16.Annibali V, Ristori G, Angelini DF, Serafini B, Mechelli R, Cannoni S, et al. CD161(high)CD8+T cells bear pathogenetic potential in multiple sclerosis. Brain. 2011;134(Pt 2):542–554. doi: 10.1093/brain/awq354. [DOI] [PubMed] [Google Scholar]
- 17.Lieury A, Chanal M, Androdias G, Reynolds R, Cavagna S, Giraudon P, et al. Tissue remodeling in periplaque regions of multiple sclerosis spinal cord lesions. Glia. 2014;62:1645–1658. doi: 10.1002/glia.22705. [DOI] [PubMed] [Google Scholar]
- 18.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 21.Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–802. [Google Scholar]
- 22.Hong MG, Reynolds CA, Feldman AL, Kallin M, Lambert JC, Amouyel P, et al. Genome-wide and gene-based association implicates FRMD6 in Alzheimer disease. Hum Mutat. 2012;33:521–529. doi: 10.1002/humu.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buil A, Martinez-Perez A, Perera-Lluna A, Rib L, Caminal P, Soria JM. A new gene-based association test for genome-wide association studies. BMC Proc. 2009;3(Suppl 7):S130. doi: 10.1186/1753-6561-3-s7-s130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patsopoulos NA, et al. Bayer Pharma MS Genetics Working Group; Steering Committees of Studies Evaluating IFNβ-1b and a CCR1-Antagonist; ANZgene Consortium; GeneMSA; International Multiple Sclerosis Genetic Consortium. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol. 2011;70:897–912. doi: 10.1002/ana.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber F, Cepok S, Wolf C, Berthele A, Uhr M, Bettecken T, et al. Single-nucleotide polymorphisms in HLA- and non-HLA genes associated with the development of antibodies to interferon-β therapy in multiple sclerosis patients. Pharmacogenomics J. 2012;12:238–245. doi: 10.1038/tpj.2011.14. [DOI] [PubMed] [Google Scholar]
- 26.International Multiple Sclerosis Genetics Consortium; Wellcome Trust Case Control Consortium 2. Sawcer S, Hellenthal G, Pirinen M, Spencer CC, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song GG, Bae SC, Lee YH. Pathway analysis of genome-wide association studies on rheumatoid arthritis. Clin Exp Rheumatol. 2013;31:566–574. [PubMed] [Google Scholar]
- 28.Tervaniemi MH, Siitonen HA, Söderhäll C, Minhas G, Vuola J, Tiala I, et al. Centrosomal localization of the psoriasis candidate gene product, CCHCR1, supports a role in cytoskeletal organization. PLoS One. 2012;7:e49920. doi: 10.1371/journal.pone.0049920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandhi G, Buttar BS, Albert L, Hasan Q, Aggarwal RK. Psoriasis-associated genetic polymorphism in North Indian population in the CCHCR1 gene and in a genomic segment flanking the HLA-C region. Dis Markers. 2011;31:361–370. doi: 10.3233/DMA-2011-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger T, Reindl M. Multiple sclerosis: disease biomarkers as indicated by pathophysiology. J Neurol Sci. 2007;259:21–26. doi: 10.1016/j.jns.2006.05.070. [DOI] [PubMed] [Google Scholar]
- 31.Wallström E, Khademi M, Andersson M, Weissert R, Linington C, Olsson T. Increased reactivity to myelin oligodendrocyte glycoprotein peptides and epitope mapping in HLA DR2(15)+ multiple sclerosis. Eur J Immunol. 1998;28:3329–3335. doi: 10.1002/(SICI)1521-4141(199810)28:10<3329::AID-IMMU3329>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 32.Raddassi K, Kent SC, Yang J, Bourcier K, Bradshaw EM, Seyfert-Margolis V, et al. Increased frequencies of myelin oligodendrocyte glycoprotein/MHC class II-binding CD4 cells in patients with multiple sclerosis. J Immunol. 2011;187:1039–1046. doi: 10.4049/jimmunol.1001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Aa A, Hellings N, Bernard CC, Raus J, Stinissen P. Functional properties of myelin oligodendrocyte glycoprotein-reactive T cells in multiple sclerosis patients and controls. J Neuroimmunol. 2003;137:164–176. doi: 10.1016/s0165-5728(03)00048-1. [DOI] [PubMed] [Google Scholar]
- 34.Linington C, Bradl M, Lassmann H, Brunner C, Vass K. Augmentation of demyelination in rat acute allergic encephalomyelitis by circulating mouse monoclonal antibodies directed against a myelin/oligodendrocyte glycoprotein. Am J Pathol. 1988;130:443–454. [PMC free article] [PubMed] [Google Scholar]
- 35.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Huettner PC, Nguyen L, Bidder M, Funk MC, Li J, et al. Aberrant promoter methylation and silencing of the POU2F3 gene in cervical cancer. Oncogene. 2006;25:5436–5445. doi: 10.1038/sj.onc.1209530. [DOI] [PubMed] [Google Scholar]
- 37.Denis GV. Bromodomain coactivators in cancer, obesity, type 2 diabetes, and inflammation. Discov Med. 2010;10:489–499. [PMC free article] [PubMed] [Google Scholar]
- 38.Filetici P, P O, Ballario P. The bromodomain: a chromatin browser? Front Biosci. 2001;6:D866–D876. doi: 10.2741/filetici. [DOI] [PubMed] [Google Scholar]
- 39.Mahdi H, Fisher BA, Källberg H, Plant D, Malmström V, Rönnelid J, et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nat Genet. 2009;41:1319–1324. doi: 10.1038/ng.480. [DOI] [PubMed] [Google Scholar]
- 40.Alcina A, Fernández O, Gonzalez JR, Catalá-Rabasa A, Fedetz M, Ndagire D, et al. Tag-SNP analysis of the GFI1-EVI5-RPL5-FAM69 risk locus for multiple sclerosis. Eur J Hum Genet. 2010;18:827–831. doi: 10.1038/ejhg.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Results of gene-based and differential expression analyses of 58 MS-associated genes.
| Gene | Chromosome | Start position | Length | #SNP* | Initial p | Genes significant in initial study based on SNP-based p | Genes in PheGenI | Replication p | Differential expression p | Sample† |
|---|---|---|---|---|---|---|---|---|---|---|
| C6orf10 | 6 | 32255475 | 89181 | 21 | 3.97×10-39 | C6orf10 | C6orf10 | 3.16×10-3 | NS | NS |
| NOTCH4 | 6 | 32157620 | 39224 | 7 | 2.51×10-31 | NOTCH4 | NOTCH4 | NS | NS | NS |
| TNXB | 6 | 32003932 | 78221 | 2 | 6.61×10-28 | TNXB | TNXB | 3.37×10-3 | 4.45×10-2 | S2 |
| HLA-DRA | 6 | 32402619 | 15204 | 9 | 3.73×10-24 | HLA-DRA | HLA-DRA | 4.08×10-3 | 5.82×10-5 | S1 |
| AIF1 | 6 | 31577994 | 11804 | 1 | 3.15×10-23 | AIF1 | AIF1 | NA | 2.04×10-2 | S1 |
| SNORA38 | 6 | 31585856 | 10131 | 1 | 3.15×10-23 | SNORA38 | NA | NA | NA | NA |
| BAT2 | 6 | 31583450 | 27104 | 3 | 1.18×10-22 | BAT2 | BAT2 | 3.42×10-2 | NS | NS |
| BAT3 | 6 | 31601805 | 23672 | 3 | 4.41×10-19 | BAT3 | BAT3 | 9.52×10-3 | NS | NS |
| PPIAP9 | 6 | 31481654 | 11525 | 1 | 1.46×10-15 | PPIAP9 | PPIAP9 | 4.67×10-2 | NA | NA |
| HLA-DQB2 | 6 | 32718875 | 17455 | 6 | 9.02×10-15 | HLA-DQB2 | HLA-DQB2 | 1.01×10-2 | NS | NS |
| HLA-DQA2 | 6 | 32704163 | 15501 | 6 | 1.25×10-14 | HLA-DQA2 | HLA-DQA2 | 3.93×10-3 | NS | NS |
| EHMT2 | 6 | 31842536 | 27928 | 1 | 8.91×10-14 | EHMT2 | EHMT2 | NS | 4.30×10-3 | S1 |
| 4.50×10-2 | S2 | |||||||||
| LOC100287272 | 6 | 31238352 | 13179 | 5 | 1.79×10-13 | LOC100287272 | NA | 4.76×10-3 | NA | NA |
| MCCD1 | 6 | 31491739 | 11269 | 2 | 4.39×10-13 | MCCD1 | MCCD1 | NA | NS | NS |
| LOC100462812 | 6 | 31340494 | 10311 | 5 | 4.71×10-13 | LOC100462812 | NA | 8.07×10-5 | NA | NA |
| HCG27 | 6 | 31160537 | 16208 | 3 | 6.34×10-13 | HCG27 | HCG27 | 3.84×10-2 | NS | NS |
| HLA-DRB9 | 6 | 32422597 | 10269 | 6 | 9.42×10-13 | HLA-DRB9 | HLA-DRB9 | 1.60×10-3 | NA | NA |
| HLA-S | 6 | 31344346 | 10918 | 6 | 4.89×10-12 | HLA-S | HLA-S | 3.36×10-2 | NA | NA |
| HLA-DQA1 | 6 | 32600183 | 16246 | 1 | 1.68×10-11 | HLA-DQA1 | HLA-DQA1 | 5.96×10-4 | 3.51×10-2 | S1 |
| TAP2 | 6 | 32784610 | 26937 | 2 | 8.02×10-11 | TAP2 | TAP2 | NS | 3.35×10-3 | S1 |
| BTNL2 | 6 | 32357513 | 22387 | 7 | 1.43×10-10 | BTNL2 | BTNL2 | 8.76×10-5 | NS | NS |
| HLA-DOB | 6 | 32775540 | 14285 | 4 | 1.78×10-10 | HLA-DOB | HLA-DOB | NA | 1.63×10-5 | S1 |
| RDBP | 6 | 31914864 | 17000 | 2 | 4.31×10-10 | RDBP | RDBP | 4.16×10-2 | 2.01×10-2 | S1 |
| 3.70×10-2 | S2 | |||||||||
| PSORS1C1 | 6 | 31077608 | 35261 | 10 | 8.59×10-10 | PSORS1C1 | PSORS1C1 | NS | 4.85×10-2 | S2 |
| HLA-DMB | 6 | 32897406 | 16441 | 1 | 2.63×10-9 | HLA-DMB | HLA-DMB | NS | 4.45×10-2 | S3 |
| DHX16 | 6 | 30615896 | 29934 | 1 | 7.13×10-9 | DHX16 | DHX16 | 2.73×10-3 | NS | NS |
| POU5F1 | 6 | 31127114 | 16337 | 4 | 9.96×10-9 | POU5F1 | POU5F1 | 4.68×10-3 | 8.28×10-3 | S2 |
| TCF19 | 6 | 31121303 | 15689 | 2 | 2.02×10-7 | TCF19 | TCF19 | 1.78×10-2 | NS | NS |
| 3.8-1.5 (HLA complex group 26) | 6 | 29727893 | 11180 | 4 | 2.40×10-7 | 3.8-1.5 | NA | 1.27×10-3 | NA | NA |
| HLA-F | 6 | 29686117 | 13956 | 7 | 4.67×10-7 | HLA-F | NA | 1.17×10-2 | 2.64×10-2 | S1 |
| LOC100507436 | 6 | 31362561 | 76025 | 8 | 4.98×10-7 | LOC100507436 | NA | NS | NA | NA |
| HLA-X | 6 | 31424623 | 10644 | 1 | 1.55×10-6 | NA | HLA-X | NA | NA | NA |
| HCG4P4 | 6 | 29917982 | 10428 | 1 | 8.60×10-6 | NA | HCG4P4 | 6.98×10-4 | NA | NA |
| RPL3P2 | 6 | 31243108 | 11240 | 2 | 1.66×10-5 | NA | RPL3P2 | NA | NA | NA |
| HLA-C | 6 | 31231529 | 13326 | 2 | 1.72×10-5 | NA | HLA-C | 4.29×10-2 | 6.15×10-3 | S1 |
| 2.74×10-2 | S2 | |||||||||
| DHFRP2 | 6 | 31326244 | 13498 | 2 | 2.89×10-5 | NA | DHFRP2 | NA | NA | NA |
| MICE | 6 | 29704234 | 17646 | 1 | 4.63×10-5 | NA | MICE | NA | NA | NA |
| TRIM31 | 6 | 30065674 | 20193 | 1 | 1.18×10-4 | NA | TRIM31 | 1.31×10-4 | NS | NS |
| MOG | 6 | 29619758 | 25391 | 2 | 1.21×10-5 | NA | NA | 5.09×10-4 | 2.12×10-2 | S1 |
| 3.65×10-2 | S3 | |||||||||
| 2.92×10-2 | S4 | |||||||||
| HLA-DMA | 6 | 32911391 | 14508 | 2 | 3.15×10-7 | NA | NA | 4.08×10-2 | 8.62×10-4 | S1 |
| CCHCR1 | 6 | 31105216 | 25799 | 2 | 4.03×10-5 | NA | NA | 1.08×10-3 | 1.05×10-2 | S1 |
| HCG22 | 6 | 31016984 | 15669 | 6 | 1.21×10-4 | NA | NA | 2.29×10-2 | 3.85×10-2 | S1 |
| BRD2 | 6 | 32931437 | 22845 | 1 | 1.07×10-5 | NA | NA | NS | 5.65×10-6 | S1 |
| FAM69A | 1 | 93302721 | 129358 | 11 | 4.47×10-5 | NA | NA | NA | 1.63×10-2 | S1 |
| LOC100294145 | 6 | 32856953 | 19582 | 3 | 2.27×10-5 | NA | NA | NA | 4.21×10-2 | S2 |
| POU2F3 | 11 | 120000000 | 89702 | 3 | 1.13×10-4 | NA | NA | NS | 8.41×10-3 | S3 |
| TRIM26P | 6 | 30201078 | 13978 | 2 | 3.72×10-5 | NA | NA | 7.80×10-6 | NA | NA |
| HCP5P14 | 6 | 29733324 | 12339 | 1 | 1.33×10-4 | NA | NA | 9.94×10-5 | NA | NA |
| 3.8-1.4(HLA complex group 26) | 6 | 29828692 | 11172 | 7 | 1.58×10-5 | NA | NA | 1.08×10-3 | NA | NA |
| C6orf15 | 6 | 31074000 | 11332 | 3 | 9.26×10-5 | NA | NA | 2.22×10-3 | NS | NS |
| MUC21 | 6 | 30946485 | 16190 | 4 | 4.40×10-5 | NA | NA | 3.42×10-3 | NA | NA |
| HLA-A | 6 | 29905309 | 13352 | 4 | 1.61×10-5 | NA | NA | 8.38×10-3 | NS | NS |
| HLA-F-AS1 | 6 | 29689378 | 32448 | 7 | 1.83×10-6 | NA | NA | 2.52×10-2 | NA | NA |
| MXD3 | 5 | 177000000 | 16791 | 2 | 7.60×10-7 | NA | NA | NS | NS | NS |
| LOC100287247 | 22 | - | - | 1 | 2.71×10-5 | NA | NA | NA | NA | NA |
| LOC100127934 | 1 | 93392136 | 10509 | 1 | 6.18×10-5 | NA | NA | NA | NA | NA |
| RPL15P4 | 6 | 31490853 | 10645 | 1 | 2.08×10-7 | NA | NA | NA | NA | NA |
| HCG2P8 | 6 | 29767896 | 13543 | 1 | 3.96×10-5 | NA | NA | NA | NA | NA |
PheGenI, Phenotype-Genotype Integrator.
*The number of SNP included in a gene with p-value<0.05, †Ms-related cells sample used to differential expression ananlysis. S1: PBMC, S2: CD34+ HPC, S3: CD8+ T lymphocytes, S4, spinal cord.
The genes listed here that are not defined in the main text are chromosome 6 open reading frame 10 (C6orf10), notch 4 (NOTCH4), small nucleolar RNA, H/ACA box 38 (SNORA38), proline-rich coiled-coil 2A (BAT2), BCL2-associated athanogene 6 (BAT3), peptidylprolyl isomerase A pseudogene 9 (PPIAP9), major histocompatibility complex, class II, DQ beta 2 (HLA-DQB2), major histocompatibility complex, class II, DQ alpha 2 (HLA-DQA2), ubiquitin specific peptidase 8 pseudogene 1 (LOC100287272), mitochondrial coiled-coil domain 1 (MCCD1), fibroblast growth factor receptor 3 pseudogene 1 (LOC100462812), HLA complex group 27 (HCG27), major histocompatibility complex, class II, DR beta 9 (HLA-DRB9), major histocompatibility complex, class I, S (HLA-S), butyrophilin-like 2 (BTNL2), DEAH (Asp-Glu-Ala-His) box polypeptide 16 (DHX16) transcription factor 19 (TCF19), HLA complex group 26 pseudogene (3.8-1.5), MHC class I polypeptide-related sequence A (LOC100507436), major histocompatibility complex, class I, x (HLA-X), HLA complex group 4 pseudogene 4 (HCG4P4), ribosomal protein L3 pseudogene 2 (RPL3P2), dihydrofolate reductase pseudogene 2 (DHFRP2), MHC class I polypeptide-related sequence E (MICE), ripartite motif containing 31 (TRIM31), MAX dimerization protein 3 (MXD3), similar to hCG1987428 (LOC100287247), meiotic nuclear divisions 1 homolog (LOC100127934), ribosomal protein L15 pseudogene 4 (RPL15P4), HLA complex group 2 pseudogene 8 (HCG2P8) respectively.
Supplementary Table 2. Enrichment of GO terms and KEGG pathways of the 58 MS-associated genes.
| Category | Term | Count | % | Genes | List total | Relative enrichment | Bonferroni* |
|---|---|---|---|---|---|---|---|
| GOTERM_BP_FAT | GO:0019882~antigen processing and presentation | 12 | 21.43 | 3112, 285830, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 25 | 78.23 | 1.09×10-16 |
| GOTERM_CC_FAT | GO:0042611~MHC protein complex | 11 | 19.64 | 3112, 285830, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 24 | 102.78 | 1.06×10-16 |
| SP_PIR_KEYWORDS | mhc ii | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3105, 3132, 3122 | 34 | 164.25 | 1.84×10-14 |
| UP_SEQ_FEATURE | Region of interest:Connecting peptide | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 33 | 137.17 | 3.39×10-13 |
| UP_SEQ_FEATURE | Domain:Ig-like C1-type | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 33 | 130.32 | 5.42×10-13 |
| KEGG_PATHWAY | hsa05330:Allograft rejection | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 90.80 | 7.23×10-14 |
| KEGG_PATHWAY | hsa05332:Graft-versus-host disease | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 83.82 | 1.47×10-13 |
| KEGG_PATHWAY | hsa04940:Type I diabetes mellitus | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 77.83 | 2.80×10-13 |
| KEGG_PATHWAY | hsa04612:Antigen processing and presentation | 10 | 17.86 | 3112, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 43.76 | 7.53×10-13 |
| KEGG_PATHWAY | hsa05320:Autoimmune thyroid disease | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 64.10 | 1.50×10-12 |
| INTERPRO | IPR003597:Immunoglobulin C1-set | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 29 | 76.03 | 1.16×10-11 |
| UP_SEQ_FEATURE | Region of interest:Alpha-1 | 7 | 12.50 | 3117, 3118, 3107, 3108, 3134, 3105, 3122 | 33 | 202.71 | 1.56×10-10 |
| UP_SEQ_FEATURE | Region of interest: Alpha-2 | 7 | 12.50 | 3117, 3118, 3107, 3108, 3134, 3105, 3122 | 33 | 202.71 | 1.56×10-10 |
| INTERPRO | IPR014745:MHC class II, alpha/beta chain, N-terminal | 7 | 12.50 | 3112, 3117, 3118, 3109, 3108, 3132, 3122 | 29 | 182.78 | 7.35×10-11 |
| INTERPRO | IPR003006:Immunoglobulin/major histocompatibility complex, conserved site | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 29 | 59.43 | 8.98×10-11 |
| KEGG_PATHWAY | hsa05416:Viral myocarditis | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 46.04 | 2.47×10-11 |
| SMART | SM00407:IGc1 | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 24 | 50.07 | 7.27×10-11 |
| GOTERM_CC_FAT | GO:0042613~MHC class II protein complex | 7 | 12.50 | 3112, 3117, 3118, 3109, 3108, 3132, 3122 | 24 | 128.55 | 5.56×10-10 |
| GOTERM_BP_FAT | GO:0002504~antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 7 | 12.50 | 3112, 3117, 3118, 3109, 3108, 3132, 3122 | 25 | 114.78 | 3.42×10-9 |
| GOTERM_MF_FAT | GO:0032395~MHC class II receptor activity | 6 | 10.71 | 3112, 3117, 3118, 3107, 3108, 3122 | 23 | 178.26 | 7.85×10-9 |
| SP_PIR_KEYWORDS | Immune response | 10 | 17.86 | 3112, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3122 | 34 | 25.26 | 8.42×10-9 |
| KEGG_PATHWAY | hsa04514:Cell adhesion molecules (CAMs) | 9 | 16.07 | 3112, 3117, 3118, 3107, 3109, 3108, 3134, 3105, 3122 | 14 | 24.76 | 4.04×10-9 |
| GOTERM_CC_FAT | GO:0042825~TAP complex | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 24 | 380.42 | 2.00×10-8 |
| GOTERM_CC_FAT | GO:0042824~MHC class I peptide loading complex | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 24 | 295.88 | 7.18×10-8 |
| GOTERM_BP_FAT | GO:0048002~antigen processing and presentation of peptide antigen | 6 | 10.71 | 6891, 3107, 3108, 3134, 3105, 3122 | 25 | 115.95 | 2.17×10-7 |
| PIR_SUPERFAMILY | PIRSF001991:Class II histocompatibility antigen | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 18 | 94.82 | 2.79×10-8 |
| INTERPRO | IPR007110:Immunoglobulin-like | 11 | 19.64 | 3112, 3117, 3118, 4340, 3107, 3109, 3108, 3134, 56244, 3105, 3122 | 29 | 12.61 | 3.53×10-7 |
| KEGG_PATHWAY | hsa05310:Asthma | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 14 | 75.15 | 1.10×10-7 |
| GOTERM_BP_FAT | GO:0006955~immune response | 12 | 21.43 | 3112, 285830, 3117, 3118, 6891, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 25 | 9.41 | 1.53×10-6 |
| GOTERM_MF_FAT | GO:0042288~MHC class I protein binding | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 23 | 176.40 | 8.88×10-7 |
| INTERPRO | IPR013783:Immunoglobulin-like fold | 11 | 19.64 | 3112, 3117, 3118, 4340, 3107, 3109, 3108, 3134, 56244, 3105, 3122 | 29 | 11.43 | 9.06×10-7 |
| GOTERM_MF_FAT | GO:0042287~MHC protein binding | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 23 | 112.90 | 6.11×10-6 |
| KEGG_PATHWAY | hsa04672:Intestinal immune network for IgA production | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 14 | 44.48 | 1.72×10-6 |
| SP_PIR_KEYWORDS | Heterodimer | 6 | 10.71 | 3117, 3118, 3107, 3134, 3105, 3122 | 34 | 32.96 | 6.98×10-5 |
| SP_PIR_KEYWORDS | Transmembrane protein | 10 | 17.86 | 3112, 3117, 3118, 4340, 6891, 3107, 3108, 3134, 3105, 3122 | 34 | 8.81 | 7.59×10-5 |
| INTERPRO | IPR001003:MHC class II, alpha chain, N-terminal | 4 | 7.14 | 3117, 3118, 3108, 3122 | 29 | 191.48 | 7.32×10-5 |
| KEGG_PATHWAY | hsa05322:Systemic lupus erythematosus | 6 | 10.71 | 3112, 3117, 3118, 3109, 3108, 3122 | 14 | 22.01 | 6.04×10-5 |
| GOTERM_BP_FAT | GO:0002474~antigen processing and presentation of peptide antigen via MHC class I | 4 | 7.14 | 6891, 3107, 3134, 3105 | 25 | 127.32 | 6.60×10-4 |
| GOTERM_CC_FAT | GO:0044459~plasma membrane part | 15 | 26.79 | 4855, 3112, 3117, 3118, 6891, 394263, 285830, 199, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 24 | 3.63 | 2.43×10-4 |
| INTERPRO | IPR001039:MHC class I, alpha chain, alpha1, and alpha2 | 4 | 7.14 | 285830, 3107, 3134, 3105 | 29 | 85.10 | 9.56×10-4 |
| INTERPRO | IPR011161:MHC class I-like antigen recognition | 4 | 7.14 | 285830, 3107, 3134, 3105 | 29 | 82.06 | 1.07×10-3 |
| GOTERM_CC_FAT | GO:0042612~MHC class I protein complex | 4 | 7.14 | 285830, 3107, 3134, 3105 | 24 | 76.08 | 1.17×10-3 |
| BIOCARTA | h_mhcPathway:Antigen processing and presentation | 3 | 5.36 | 6891, 3105, 3122 | 3 | 143.70 | 5.67×10-4 |
| UP_SEQ_FEATURE | Sequence variant | 31 | 55.36 | 170679, 3112, 4855, 11074, 54535, 6891, 56244, 7148, 10919, 6941, 199, 6046, 83463, 5460, 3107, 3109, 3108, 3134, 3105, 401250, 3117, 3118, 4340, 29113, 8449, 10665, 7916, 394263, 25833, 3122, 7917 | 33 | 1.50 | 1.94×10-2 |
| SP_PIR_KEYWORDS | Polymorphism | 31 | 55.36 | 170679, 3112, 4855, 11074, 54535, 6891, 56244, 7148, 10919, 6941, 199, 6046, 83463, 5460, 3107, 3109, 3108, 3134, 3105, 401250, 3117, 3118, 4340, 29113, 8449, 10665, 7916, 394263, 25833, 3122, 7917 | 34 | 1.52 | 7.47×10-3 |
| UP_SEQ_FEATURE | Region of interest:Alpha-3 | 3 | 5.36 | 3107, 3134, 3105 | 33 | 193.06 | 2.92×10-2 |
| SP_PIR_KEYWORDS | mhc i | 3 | 5.36 | 3107, 3134, 3105 | 34 | 169.72 | 1.05×10-2 |
| GOTERM_BP_FAT | GO:0002478~antigen processing and presentation of exogenous peptide antigen | 3 | 5.36 | 6891, 3108, 3122 | 25 | 147.58 | 3.25×10-2 |
| PIR_SUPERFAMILY | PIRSF001990:Class I histocompatibility antigen | 3 | 5.36 | 3107, 3134, 3105 | 18 | 136.96 | 2.30×10-3 |
| GOTERM_BP_FAT | GO:0019884~antigen processing and presentation of exogenous antigen | 3 | 5.36 | 6891, 3108, 3122 | 25 | 115.95 | 5.30×10-2 |
| GOTERM_MF_FAT | GO:0042277~peptide binding | 5 | 8.93 | 3112, 6891, 3109, 3108, 3122 | 23 | 13.90 | 2.69×10-2 |
| INTERPRO | IPR000353:MHC class II, beta chain, N-terminal | 3 | 5.36 | 3112, 3109, 3132 | 29 | 101.37 | 2.84×10-2 |
| GOTERM_MF_FAT | GO:0032393~MHC class I receptor activity | 3 | 5.36 | 3107, 3134, 3105 | 23 | 99.61 | 2.89×10-2 |
| GOTERM_CC_FAT | GO:0005886~plasma membrane | 16 | 28.57 | 4855, 3112, 3117, 3118, 4340, 6891, 394263, 285830, 199, 3107, 3109, 3108, 3134, 3105, 3132, 3122 | 24 | 2.26 | 2.98×10-2 |
Count, the number of genes enriched in particular GO terms or KEGG pathways; Genes, potential genes enriched in particular GO terms or KEGG pathways.
*p value with Bonferroni correction for multiple tests. Only significant results with p<0.05 are listed.
GO: Gene Ontology, KEGG: Kyoto Encyclopedia of Genes and Genomes, MHC: major histocompatibility complex, MS: mltiple sclerosis.