Abstract
Purpose
To assess the performance of Cepheid® Xpert MTB/RIF® (“Xpert”) and TB Biochip® MDR (“TB-Biochip”).
Methods
Sputum specimens from adults with presumptive tuberculosis (TB) were homogenized and split for (1) direct Xpert and microscopy, and (2) concentration for Xpert, microscopy, culture (Lowenstein-Jensen [LJ] solid media and Mycobacteria Growth Indicator Tube® [MGIT]), indirect drug susceptibility testing (DST) using the absolute concentration method and MGIT, and TB-Biochip.
Results
In total, 109 of 238 (45.8%) specimens were culture-positive for M. tuberculosis complex (MTBC), and of these, 67 isolates were rifampicin resistant (RIF-R) by phenotypic DST, and 64/67 (95.5%) were isoniazid resistant (INH-R). Compared to culture of the same specimen, a single direct Xpert was more sensitive for detecting MTBC (95.3%, 95% confidence interval [CI], 90.0–98.3%) than direct (59.6%, 95% CI, 50.2–68.5%) or concentrated smear (85.3%, 95% CI, 77.7–91.1%) or LJ culture (80.8%, 95% CI, 72.4–87.5%); specificity was 86.0% (95% CI 78.9–91.3%). Compared with MGIT DST, Xpert correctly identified 98.2% (95% CI, 91.5–99.9%) of RIF-R and 95.5% (95% CI, 85.8–99.2%) of RIF-susceptible (RIF-S) specimens. In a subset of 104 specimens, the sensitivity of TB-Biochip for MTBC detection compared to culture was 97.3% (95% CI, 91.0–99.5%); specificity was 78.1% (95% CI, 61.5– 89.9%). TB-Biochip correctly identified 100% (95% CI, 94.2–100%) of RIF-R, 94.7% (95% CI, 76.7–99.7%) of RIF-S, 98.2% (95% CI, 91.4–99.9%) of INH-R, and 78.6% (95% CI, 52.1–94.2%) of INH-S specimens, compared to MGIT DST.
Conclusions
Xpert and Biochip were similar in accuracy for detecting MTBC and RIF resistance compared to conventional culture methods.
Keywords: tuberculosis, drug resistance, Cepheid® Xpert MTB/RIF®, rapid diagnosis, Russia
Introduction
Tuberculosis (TB) is an infectious disease caused by the bacillus Mycobacterium tuberculosis with an estimated 8.8 million new cases and 1.5 million deaths in 2010 [1]. Russia ranks among 22 countries in the world with the largest burden of tuberculosis, including one of the highest rates of drug-resistant TB [1]. According to nationwide surveillance data collected in 2009, 15.5% of not previously treated cases and 33.7% of previously treated TB cases in Russia were identified as multidrug-resistant (MDR) TB and rates continue to rise [2].
Rapid detection of MDR TB would be optimal for patient management but is hampered by the limitations and delays associated with the primary methods available for diagnosis of TB. Significant advances have been made in rapid and accurate diagnosis of TB and drug resistance with the advent of molecular methods. The TB-Biochip MDR (“TB-Biochip”) system (Biochip-IMB Ltd, Moscow, Russia), developed at the Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, is based on a multiplex polymerase chain reaction (PCR) followed by hybridization on a platform of gel-based oligonucleotide microarrays (biochips) that simultaneously detects DNA specific for Mycobacterium tuberculosis complex (MTBC) and mutations that cause rifampicin (RIF) and isoniazid (INH) resistance from either culture isolates or clinical specimens in less than 12 hours [3,4]. It is used primarily in Russia and in neighboring countries that were part of the former Soviet Union. However, because TB-Biochip is labor intensive, technically demanding and requires specialized laboratory infrastructure, its utility is limited to reference laboratories. At the same time, batch-testing increases the turn-around time thereby reducing its clinical utility. More recently, technical progress has resulted in simplified diagnostic tools that overcome several drawbacks of earlier nucleic acid amplification tests (NAAT). The Cepheid® eXpert MTB/RIF® test (“Xpert”, Cepheid Inc., Sunnyvale, CA) is an automated on-demand assay that detects MTBC-specific DNA and mutations associated with RIF resistance directly from sputum within 2 hours. It does not require bio-containment infrastructure or molecular expertise to perform [5,6]. The test utilizes PCR and molecular beacon technology within a closed-system cartridge that integrates sample preparation, DNA extraction, amplification, and detection with minimal risk of contamination.
Our project aimed to (1) validate the Xpert using both unconcentrated and concentrated portions of a single sputum specimen by comparing results with direct and concentrated microscopy, conventional solid or liquid culture and indirect phenotypic drug susceptibility testing (DST) methods, and (2) compare results from a subset of specimens to those obtained by TB-Biochip for detection of MTBC and rifampicin resistance directly from sputum.
Materials and Methods
Project Settings, Patients, and Specimens
Specimens were collected as part of routine medical care from patients at two sites in Russia: (1) the Central TB Research Institute (CTRI) of the Russian Academy of Medical Sciences in Moscow, where the majority of patients are referred with chronic, often drug-resistant TB, and (2) the Regional TB Dispensary in Vladimir Oblast where both hospitalized and ambulatory patients from within the region are treated. Microbiology testing was performed at each site by its respective laboratory. Internal quality control and external quality assessment practices are followed by both laboratories with equally high performance.
Sputum samples of at least 5.0 ml were collected from consecutively evaluated adults (≥18 years old) with presumptive or recently diagnosed pulmonary TB between July and October 2011. Patients having received anti-TB drugs within 60 days prior to specimen collection were excluded. The final diagnosis, based on the clinical and radiographic findings and microbiology results from all specimens obtained from the patient during this illness episode, was assigned by a panel of physicians according to explicit clinical definitions and documented for each specimen. Because the purpose of this project was to evaluate Xpert performance in these two laboratories, results were not reported to the physician and did not influence patient management decisions. Upon review, the U.S. Centers for Disease Control and Prevention determined this evaluation to be a laboratory test validation project and not human subject research. The local sites in Russia relied on the CDC determination.
Laboratory Methods
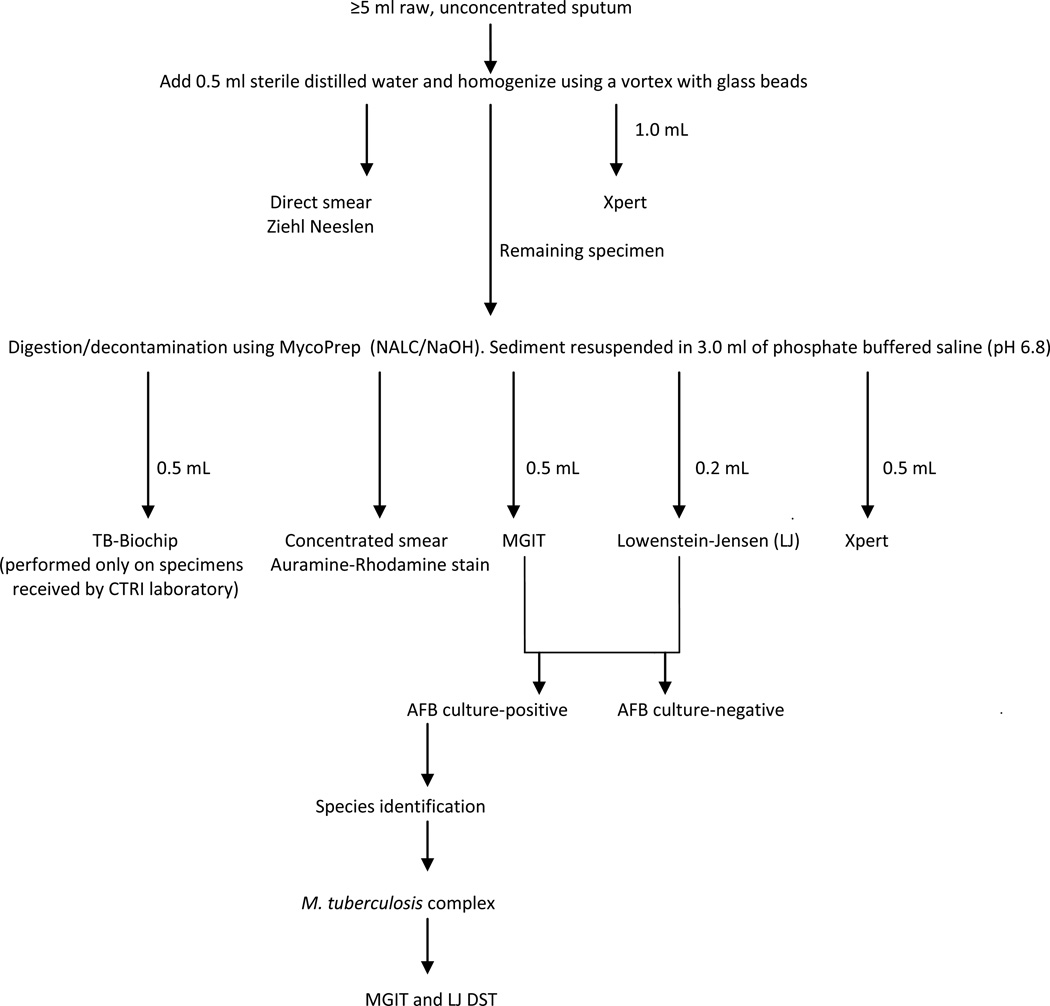
After addition of 0.5 ml sterile distilled water and glass beads, each specimen was homogenized for 60–90 sec with a vortex unit and split into two portions (Figure 1). From one portion 1.0 mL was tested by Xpert and a smear prepared for Ziehl-Neelsen (ZN) microscopy [7]. The remaining portion (≥3 ml) was decontaminated with N-acetyl-L-cysteine/sodium hydroxide (BBL MycoPrep; BD, Sparks, MD, USA) [7] and centrifuged at 3,000 × g for 15 minutes. After centrifugation, the supernatant was decanted and the sediment was resuspended in 3.0 ml of phosphate buffered saline (pH 6.8), from which 0.5 mL was tested with Xpert, 0.2 mL was inoculated onto Lowenstein-Jensen (LJ), 0.5 mL onto BACTEC Mycobacteria Growth Indicator Tube® (MGIT) 960 system (Becton Dickinson [BD], Sparks, MD, USA) culture media, and a smear prepared for auramine-rhodamine fluorescence microscopy [7].
Figure 1.
Validation algorithm.
Smears were scored using the World Health Organization scale [8]. Acid-fast bacillus (AFB) positive cultures were confirmed to be MTBC by the MTBcID test (BD, Sparks, MD) [9], or conventional biochemical tests performed according to standard protocols [7]. For culture confirmation of MTBC, the CTRI laboratory also employed a real-time PCR assay using primers directed against IS6110 followed by detection with fluorescent-labeled probes specific for MTBC (Amplitub-Rv, Syntol, Moscow, Russia) [10,11]. Culture-based DST was performed by either the BACTEC MGIT 960 [12], or the absolute concentration method on LJ medium [13] using standard critical concentrations of 1.0 mcg/ml and 40.0 mcg/ml for rifampicin, and critical concentrations of 0.1 mcg/ml and 0.2 mcg/ml for isoniazid, respectively.
The Xpert assay was performed according to the manufacturer’s instructions. Results were obtained using the Xpert MTB/RIF software, version 4.0.
At the CTRI laboratory only, in addition to the above procedures, a portion of the resuspended sediment (0.5 mL) was also tested with the TB-Biochip assay according to the manufacturer’s specifications. The presence and nature of mutations were determined by analysis of the fluorescence intensity pattern on the chip, using a fluorescence analyzer equipped with Imageware software (TB-Biochip-IMB, Moscow, Russia) [3,4].
Data analysis
Data were entered in an Epi Info version 3.3.5 (CDC, Atlanta, GA) database. Statistical analyses were performed using SAS software, version 9.1 (SAS Institute Inc., Cary, NC). Sensitivity and specificity of direct and concentrated Xpert, TB-Biochip, smear microscopy for MTBC detection were calculated using (1) culture positivity in the same specimen as the reference standard and (2) the patient’s final diagnosis (based on all available clinical, radiographic, as well as microbiologic information). These reference standards were applied to all specimens included in this evaluation. Sensitivity and specificity of direct and concentrated Xpert as well as TB-Biochip for detection of RIF resistance was calculated using as the reference standard: (1) the absolute concentration method on LJ, (2) BACTEC MGIT 960 result and (3) combined results of DST on LJ and MGIT when RIF resistance was defined as resistance by either LJ and/or MGIT DST methods, RIF susceptibility defined as susceptible by both LJ and MGIT methods or by either method when only results from one method were available.
Specimens from cases with non-tuberculous mycobacteria (NTM) were included in calculation of specificity of Xpert. Contaminated cultures were excluded for the Xpert assay and TB-Biochip sensitivity and specificity calculations.
Results
Specimens
A total of 238 specimens from 201 consecutive patients with presumptive TB were tested using the Xpert and conventional methods (134 in Vladimir; 104 at CTRI). Of the 238 specimens, 109 (45.8%) were culture-positive (liquid or solid) for MTBC, 65 were direct smear-positive, 93 were concentrated smear-positive, and NTM were isolated from 4 specimens (2 were M. kansasii, 1 - M. avium, and 1 - M. scrofulaceum). Of 109 culture-positive specimens, 67 (61.5%) were found to be rifampicin-resistant by phenotypic DST, and 64 of these (95.5%) were also resistant to isoniazid.
Comparison of direct Xpert assay with microscopy and culture results for detection of MTBC
In total, 230 of 238 (96.6%) specimens gave an interpretable result with the direct Xpert assay, 119 were positive, 111 were negative, and 8 (3.4%) were uninterpretable. All smear-positive, culture-positive specimens and 88.6% (39/44) of direct smear-negative, culture-positive specimens were identified by the direct Xpert as having MTBC DNA. A single direct Xpert test was more sensitive (95.3%, 102/107) than either direct (59.6%, 65/109) or concentrated (85.3%, 93/109) microscopy for detection of culture-positive M. tuberculosis complex (Table 1). Among 121 specimens with negative liquid and solid culture MTBC result and available direct Xpert result, 104 were MTB negative by direct Xpert, i.e. the specificity of direct Xpert was 86.0% (Table 1). Seventeen specimens that were culture-negative for MTBC, tested positive by the direct Xpert assay: 11 of these 17 specimens were from patients who were subsequently classified as TB patients (7 based on positive laboratory results from specimens collected outside of the project and 4 based on their final diagnosis). Of the remaining 6 patients, 3 were suspected of TB relapse, and TB was ruled out in the other 3 patients based on clinical and bacteriological criteria. The specificity of direct Xpert for excluding TB was 89.3% (50 of 56) when the final diagnosis was used as the reference standard. None of the 4 NTM specimens were positive in the Xpert assay.
Table 1.
Sensitivity and specificity in detection of MTBC by diagnostic test
| Diagnostic test | Positive culture as reference standard a | |||
|---|---|---|---|---|
| Sensitivity Culture-positive |
Specificity Culture-negative |
|||
| All culture- positive |
Smear- positive |
Smear- negative |
||
| N=109 | N=93 | N=16 | N=127 b | |
| Xpert, direct | ||||
| n/N (%) | 102/107c (95.3%) | 91/91c (100%) | 11/16 (68.8%) | 104/121d (86.0%) |
| 95% CI | 90.0–98.3% | 96.8–100% | 43.7–87.5% | 78.9–91.3% |
| Xpert, concentrated | ||||
| n/N (%) | 103/109 (94.5%) | 91/93 (97.9%) | 12/16 (75.0%) | 111/127 (87.4%) |
| 95% CI | 88.9–97.7% | 93.1–99.6% | 50.1–91.5% | 80.8–92.4% |
| Direct smear | ||||
| n/N (%) | 65/109 (59.6%) | 65/93 (69.9%) | 0/16 (0%) | 124/127 (97.6%) |
| 95% CI | 50.2–68.5% | 60.0–78.6% | 0–17.1% | 93.7–99.4% |
| Concentrated smear | ||||
| n/N (%) | 93/109 (85.3%) | 93/93 (100%) | 0/16 (0%) | 114/126g (90.5%) |
| 95% CI | 77.7–91.1% | 96.8–100% | 0–17.1% | 84.4–94.7% |
| LJ only | ||||
| n/N (%) | 84/104e (80.8%) | 77/89f (86.5%) | 7/15g (46.7%) | 125/125h (100%) |
| 95% CI | 72.4–87.5% | 78.2–92.5% | 21.5–68.1% | 97.6–100% |
| MGIT only | ||||
| n/N (%) | 107/109 (98.2%) | 92/92i (100%) | 15/15i (100%) | 116/116k (100%) |
| 95% CI | 94.1–99.7% | 96.8–100% | 81.9–100% | 97.5–100% |
| TB-Biochip (N=104)l | ||||
| n/N (%) | 69/71 (97.3%) | 64/64 (100%) | 5/7 (71.4%) | 25/32 (78.1%) |
| 95% CIi | 91.0–99.5% | 95.4–100% | 33.0–94.9% | 61.5–89.9% |
Note.
n=correct number of tests, compared to the reference standard.
N=total number of tests.
Reference standard: positive culture (either LJ and/or MGIT) in same specimen except as noted. For estimation of test performance, NTM were considered culture-negative.
One had both LJ and MGIT cultures contaminated; one culture result was missing.
Two were indeterminate.
Six were indeterminate.
Four cultures were contaminated, one culture result was missing.
Four cultures were contaminated.
One culture result was missing
Two culture results were missing.
One culture was contaminated.
Seven cultures were contaminated; four culture results were missing.
TB-Biochip was only performed at the CTRI laboratory on 104 specimens. A total of 71/104 specimens were MTBC culture-positive (64/71 smear-positive); 100/104 specimens were from patients with culture-confirmed or final diagnosis of TB and 4/104 specimens were from patients with ruled out TB.
The indeterminate rate (3.4%, 8/238) for direct Xpert was lower than the MGIT contamination rate (4.2%, 10/238) but higher than that of solid media (2.1%, 5/236).The overall sensitivity of Xpert (94.5%, 103/109) was not significantly affected when concentrated specimens were tested, but the indeterminate rate was reduced to zero. Median time from specimen receipt to report of MTBC from MGIT was 10 days (interquartile range [IQR]: 7, 15) and 24 days (IQR: 18, 34) from LJ culture compared to same day results using Xpert.
Comparison of direct Xpert results with phenotypic DST results for detection of RIF resistance
Xpert correctly detected rifampicin resistance-associated mutations in 55 of 56 (98.2%) specimens when compared with MGIT results, and in 45 of 50 (90.0%) specimens when compared with those obtained by LJ DST. RIF resistance-associated mutations were not identified by Xpert in 42 of 44 (95.5%) specimens that were RIF susceptibile by MGIT and in 28 of 30 (93.3%) that were RIF-susceptible by LJ (Table 2). Overall concordance between Xpert and phenotypic methods to detect RIF resistance was 97.0% (97/100) with MGIT and 91.3% (73/80) with LJ. The rate of isoniazid resistance in phenotypically RIF-resistant specimens was 95.5% (64/67). Given 98.2% sensitivity for detection of rifampicin resistance by Xpert compared to MGIT, 93.8% of MDR TB specimens were identified by the Xpert.
Table 2.
Sensitivity, specificity and predictive values of the Xpert and TB-Biochip for detection of RIF resistance with culture method as reference (N=109)
| Test method and DST reference standard |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| Xpert, direct a | ||||
| Reference standard: LJ b | ||||
| n/N (%) | 45/50 (90.0%) | 28/30 (93.3%) | 45/47 (95.7%) | 28/33 (84.9%) |
| 95% CI | 79.2–96.2% | 79.7–98.9% | 86.6–99.3% | 69.6–94.2% |
| Reference standard: MGIT c | ||||
| n/N (%) | 55/56 (98.2%) | 42/44 (95.5%) | 55/57 (96.5%) | 42/43 (97.7%) |
| 95% CI | 91.5–99.9% | 85.8–99.2% | 88.9–99.4% | 89.1–99.9% |
| Reference standard: LJ or MGIT d | ||||
| n/N (%) | 57/62 (91.9%) | 38/39 (97.4%) | 57/58 (98.3%) | 38/43 (88.4%) |
| 95% CI | 83.0–97.0% | 88.0–99.9% | 91.8–99.9% | 76.1–95.6% |
| Xpert, concentrated e | ||||
| Reference standard: LJ b | ||||
| n/N (%) | 45/50 (90.0%) | 27/29 (93.1%) | 45/47 (95.7%) | 27/32 (84.4%) |
| 95% CI | 79.2–96.2% | 79.0–98.8% | 86.6–99.3% | 68.7–94.0% |
| Reference standard: MGIT c | ||||
| n/N (%) | 55/56 (98.2%) | 40/42 (95.2%) | 55/57 (96.5%) | 40/41 (97.6%) |
| 95% CI | 91.5–99.9% | 85.2–99.2% | 88.9–99.4% | 88.6–99.9% |
| Reference standard: LJ or MGIT d | ||||
| n/N (%) | 57/62 (91.9%) | 36/37 (97.3%) | 57/58 (98.3%) | 36/41 (87.8%) |
| 95% CI | 83.0–97.0% | 87.4–99.9% | 91.8–99.9% | 75.0–95.4% |
| TB-Biochip (N=69)f | ||||
| Reference standard: LJ | ||||
| n/N (%) | 40/44 (90.9%) | 13/14 (92.9%) | 40/41 (97.6%) | 13/17 (92.9%) |
| 95% CI | 79.5–97.0% | 69.5–99.6% | 88.6–99.9% | 52.5–92.0% |
| Reference standard: MGIT | ||||
| n/N (%) | 50/50 (100%) | 18/19 (94.7%) | 50/51 (98.0%) | 18/18 (100%) |
| 95% CI | 94.2–100% | 76.7–99.7% | 90.7–99.9% | 84.7–100% |
| Reference standard: LJ or MGIT | ||||
| n/N (%) | 51/55 (92.7%) | 14/14 (100%) | 51/51 (100%) | 14/18 (77.8%) |
| 95% CI | 83.4–97.7% | 80.7–100.0% | 94.3–100.0% | 54.7–92.5% |
n=correct number of tests, compared to the reference standard.
N=total number of tests.
LJ = absolute concentration method on LJ medium
One hundred one specimens had direct Xpert result for RIF resistance: 58 with RIF resistance and 43 without RIF resistance.
Eighty four specimens had RIF DST result on LJ: 53 with RIF resistance and 31 with RIF susceptibility.
One hundred seven specimens had RIF DST result on MGIT: 61 with RIF resistance and 46 with RIF susceptibility.
RIF resistance defined as resistance by either LJ and/or MGIT DST methods. RIF susceptibility defined as susceptible by both LJ and MGIT methods or by either method when only results from one method were available. One hundred nine specimens had DST results by LJ and/or MGIT for RIF: 67 resistant, 42 susceptible.
Ninety nine specimens had concentrated Xpert results for RIF: 58 resistant, 41 without RIF resistance.
TB-Biochip was performed only at the CTRI laboratory. Sixty nine specimens had TB-Biochip results for RIF resistance: 51 resistant, 18 without RIF resistance.
Of 8 specimens with discordant RIF results between Xpert and phenotypic DST, 3 had mutations in the rpoB gene by Xpert and were susceptibile by either culture-based method (Table 3). Five specimens were found not to have rpoB gene mutations by Xpert but were phenotypically resistant. Six of these 8 discordant isolates were further tested by TB-Biochip and all results coincided with the Xpert result. Median time from specimen receipt to completion of phenotypic DST was 22 (IQR: 16, 30) and 52 days (IQR: 46, 64) for MGIT and LJ, respectively.
Table 3.
Comparison of rifampicin susceptibility testing by method of absolute concentrations on LJ, MGIT and Xpert (N=119)
| LJ result |
MGIT result |
Xpert result |
Direct Xpert n |
Concentrated Xpert n |
|---|---|---|---|---|
| R | R | R | 43 | 43 |
| R | S | R | 1 | 1 |
| S | R | R | 1 | 1 |
| S | S | R | 1 | 1 |
| R | R | S | 1 | 1 |
| R | S | S | 4 | 4 |
| S | S | S | 28 | 27 |
| N/A | R | R | 11 | 11 |
| N/A | S | S | 10 | 9 |
| R | N/A | R | 1 | 1 |
| N/A | N/A | S | 7 a | 6 b |
| N/A | N/A | R | 7 c | 4 d |
| N/A | S | N/A | 1 | 2 |
| S | S | N/A | 0 | 1 |
| N/A | N/A | N/A | 3 | 7 |
Note. R=resistant; S=susceptible; N/A=result not available (contamination on LJ or MGIT or “invalid”, “error” result on Xpert).
2 of 7 specimens were from cases with TB ruled out;
3 of 6 specimens were from cases with TB ruled out;
1 of 7 specimens from case with TB ruled out;
1 specimen from case with TB ruled out.
Performance of TB-Biochip
At the CTRI laboratory a portion of each concentrated specimen was also tested with the TB-Biochip assay. Of the 104 specimens tested, 71 (68.3%) were culture-positive for MTBC; 80.3% (57/71) were RIF-resistant and 87.3% (62/71) were INH-resistant by phenotypic DST. The TB-Biochip for detection of M. tuberculosis complex in culture-positive specimens had an overall sensitivity of 97.3% (69/71), (100% [64/64] for smear-positive, and 71.4% [5/7] for smear-negative specimens); specificity was 78.1% (25/32) in culture-negative specimens (Table 1).
For detection of rifampicin resistance, the sensitivity of TB-Biochip was 100% (50/50) when compared with MGIT DST results and 90.9% (40/44) compared with LJ results. Specificity was 94.7% (18/19) compared with MGIT and 92.9% (13/14) compared with LJ (Table 2). Of 54 phenotypically RIF-resistant, TB-Biochip mutation-positive specimens, the most frequent rpoB gene mutation detected by TB-Biochip was rpoB531 Ser→Leu (94.4%, 51/54); 3 specimens had concomitant mutations in rpoB533 Leu→Pro (n=2) or rpoB526_His→Pro (n=1). Two specimens had rpoB511_Leu→Pro mutation, and 1 specimen bore two mutations (rpoB516 Asp→Gly and rpoB 511 Leu→Pro). For detection of MTBC and RIF resistance, TB-Biochip results were 100% concordant with those obtained from direct and concentrated Xpert.
TB-Biochip detected mutations conferring isoniazid resistance in 98.2% (54/55) phenotypically INH-resistant specimens by MGIT and 97.8% (44/45) by LJ. TB-Biochip did not detect INH resistance-conferring mutations in 78.6% (11/14) of INH-susceptible specimens by MGIT and 84.6% (11/13) by LJ, respectively. Of 61 phenotypically INH-resistant, TB-Biochip mutation-positive specimens, a specific mutation in the katG gene (315 Ser→Thr) was detected in the majority of specimens (98.4%, 60/61) and 11/61 (18.0%) specimens had a concomitant mutation in the inhA gene (5 inhA15C→T and 6 inhA8T→G). A concomitant mutation in the intergenic region between ahpC and oxyR genes AhpC10 C→T was detected in 1 specimen. For one other specimen, the only mutation associated with resistance to INH was in the intergenic region between ahpC and oxyR genes AhpC10 C→T. There were no indeterminate test results for TB-Biochip.
Discussion
We validated the Xpert assay in two clinical laboratories in Russia where the rate of tuberculosis and proportion of drug resistance is relatively high, using single, pre-processed sputum specimens that were homogenized and split for testing by Xpert and conventional methods. For those specimens received at the CTRI laboratory, Xpert was also compared with TB-Biochip, an assay developed in Russia, using conventional laboratory methods as a reference standard.
A recent meta-analysis by Chang et al. [14], including results from 18 unique studies and 10,224 pulmonary specimens of TB suspects, estimated Xpert sensitivity for detection of M. tuberculosis complex to be 90.4% (95%CI 89.2%–91.4%), ranging from 73%–74% [15,16] to 98%–100% [5,17] across studies depending on the study population. In our project, using culture positivity (liquid or solid) as the reference standard, the sensitivity of Xpert was 95.3% for detection of MTBC when tests were performed directly from unprocessed sputum, and 94.5% when a concentrated portion of the same specimen was tested, similar to findings reported by Boehme et al. [5]. Interestingly, the indeterminate rate dropped from 3.4% to 0% when concentrated specimens were tested. Others have speculated that the higher viscosity of unprocessed sputum may contribute to the indeterminate results when these types of specimens are tested with Xpert [18]. The direct Xpert test was more sensitive (95.3%) than direct smear (59.6%), concentrated smear (85.3%) or LJ culture (80.8%), identifying 88.6% of direct smear-negative, culture-positive specimens.
In the same meta-analysis [14], the pooled specificity for MTBC detection was 98.4% (95%CI 98.0%–98.7%). In our project, the direct Xpert had lower specificity for excluding MTBC (86.0%) when compared with the culture result from the same specimen. Specificity of the direct Xpert for exclusion of TB, however, was 89.3% when compared with a combined reference standard that included all available microbiology and radiography results as well as the patient’s final diagnosis. Among the 6 specimens with direct Xpert-positive results from patients with active TB ruled out, 3 were from patients who completed treatment for TB in the past. The presence of residual DNA from nonviable organisms in the specimen or a sub-clinical relapse, in which the bacillary burden was very low, could explain the positive results [14]. Complete specimen homogeneity may not have been achieved after processing with glass beads and prior to removing a portion for direct Xpert, resulting in Xpert-positive, culture-negative findings. Furthermore, delays in transport, additional sample decontamination using NaOH or suboptimal culture conditions for resistant and/or unfit bacilli could have affected organism viability and growth in culture. False Xpert-positive results of the remaining 3 specimens are unexplained. It is well established that falsely positive NAAT results from amplicon contamination or cross-reaction with other species can occur [19,20]. However, these possible explanations are unlikely since Xpert has multiple features that may reduce false positive results such as its analytical specificity based on the molecular probe design, its closed cartridge system that minimizes specimen-to-specimen cross contamination and automated readout that minimizes user interpretation errors. Nevertheless, other possible causes such as specimen contamination at the time of collection, laboratory errors (mislabeling, pipetting or clerical errors) or cross contamination with a strongly positive specimen during the sample preparation process must be considered.
A principle advantage of the Xpert is rapid detection of DNA mutations associated with rifampicin resistance for timely initiation of treatment with second-line drugs and prevention of further transmission. This is especially important in Russia where nearly all TB patients are hospitalized at least for the intensive phase of treatment. Chang and colleagues [14] reported pooled sensitivity and specificity for detecting rifampicin resistance of 94.1% and 97.0%, respectively. However, the accuracy for detection of rifampicin resistance varied widely between reports, with ranges for sensitivity from 80% [21] to 97%–100% [5,6,15] and specificity from 72% [21] to 98%–100% [5,16,22]. In our project, concordance of direct Xpert for detection of mutations associated with RIF resistance, including sensitivity (98.2%) and specificity (95.5%), was higher with the MGIT DST method than with the absolute concentration method on LJ (sensitivity 90.0% and specificity 93.3%). The observed discordance between the two phenotypic DST methods re-emphasizes the complexities and limitations of conventional growth-based DST. Because of the high prevalence of rifampicin resistance in our sample (61.5%), the positive predictive value for RIF resistance by Xpert was >95% using phenotypic DST as a reference. Consistent with previous reports for high MDR TB prevalence settings [23,24], we found low rates of RIF resistance unaccompanied by resistance to isoniazid (5%), so RIF resistance identified by Xpert can be a proxy for MDR TB in such populations until DST for isoniazid is completed.
TB-Biochip performed well in detecting MTBC (sensitivity 97.3%, specificity 78.1%) compared with culture. Specificity for excluding TB was 100% when compared with a combined reference standard including final diagnosis. Concordance between Xpert and TB-Biochip was 100% for detection of MTBC. Sensitivity and specificity of TB-Biochip for RIF resistance was 100%, and 94.7%, respectively, when compared with MGIT DST. Perfect concordance between TB-Biochip and Xpert for RIF resistance-conferring mutations may be explained by overlapping nucleotide sequences of probes for the rpoB gene in both assays. Comparable to previously published data [3,25,26,27], we report sensitivity and specificity of 98.2% and 78.6% (compared to MGIT DST), respectively, for detection of INH resistance by TB-Biochip. However, TB-Biochip is a fully manual test and requires specialized molecular laboratory infrastructure and specialized expertise which limits use of this technology outside of central reference laboratories.
The incidence of tuberculosis is high and the prevalence of non-tuberculous mycobacteria within Russia is low. Consequently, few specimens from patients without active TB and with NTM were included in this study, thereby limiting calculations of specificity. Duplicate specimens were obtained from 37 patients; these were not analyzed separately with methods for correlated data. Since this validation project was restricted to sputum specimens of ≥5 ml, the results may not apply to patients with less productive or non-productive cough. HIV status of project patients was not collected, but HIV co-infection among TB patients in these two settings is relatively low (<5%).
Despite these limitations, our project has several strengths. All tests were performed on the same specimen which reduced variability caused by differences between specimens from the same patient, and all results were interpreted by staff members who were unaware of results from other tests included in the project. Xpert performance for detection of MTBC was assessed using culture results from the same specimen as well as its performance for the final diagnosis based on radiography, clinical judgment and culture results from other specimens concurrently collected.
Based on these favorable results, the regional laboratory in Vladimir has launched a pilot point-of-care testing program whereby Xpert testing has been implemented at three microscopy centers located within the region. Broadening the testing range beyond the low diagnostic capability of smear microscopy, currently available at these centers, to include Xpert testing may expedite diagnosis and treatment of MDR-TB and possibly become an important modality for improving patient care and outcomes within the region.
Conclusion
This project describes the results of an algorithm for validation of Xpert MTB/RIF used by two laboratories within Russia, and a comparison of a portion of those results with results obtained by TB-Biochip, a more technically demanding assay that requires both laboratory expertise and specialized facilities. Results demonstrate that a single unconcentrated sputum can be tested using Xpert with high sensitivity and specificity for detection of MTBC and RIF resistance, equivalent to those obtained with TB-Biochip. Coupled with its simplicity and speed, the Xpert assay addresses two major gaps in settings with a high incidence of TB, a high prevalence of MDR TB or limited laboratory infrastructure: rapid, accurate detection of MTBC and simultaneous identification of rifampicin-resistant tuberculosis. This tool is especially valuable in settings without the capacity for concentrating sputum, without high-level biosafety facilities, or highly skilled laboratory staff.
Acknowledgments
This work was supported by the United States Agency for International Development (USAID) country mission in Russia.
Footnotes
Disclaimer: The conclusions and interpretations of data presented in this report are solely those of the authors and do not necessarily represent an official position of CDC.
Conflicts of interest
No conflict of interest was reported for all authors.
References
- 1.World Health Organization. Geneva, Switzerland: 2011. Global tuberculosis control: WHO report 2011. [Google Scholar]
- 2.Institute of Medicine (US) Forum on Drug Discovery Development and Translation, Russian Academy of Medical Science. The New Profile of Drug-Resistant Tuberculosis in Russia: A Global and Local Perspective: Summary of a Joint Workshop. Washington DC: National Academy of Sciences; 2011. [PubMed] [Google Scholar]
- 3.Gryadunov D, Mikhailovich V, Lapa S, Roudinskii N, Donnikov M, et al. Evaluation of hybridisation on oligonucleotide microarrays for analysis of drug-resistant Mycobacterium tuberculosis. Clin Microbiol Infect. 2005;11:531–539. doi: 10.1111/j.1469-0691.2005.01183.x. [DOI] [PubMed] [Google Scholar]
- 4.Mikhailovich V, Lapa S, Gryadunov D, Sobolev A, Strizhkov B, et al. Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. J Clin Microbiol. 2001;39:2531–2540. doi: 10.1128/JCM.39.7.2531-2540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377:1495–1505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent PT, Kubica GP. Public health mycobacteriology: A guide for the level III laboratory. Atlanta, GA: Centers for Disease Control; 1985. [Google Scholar]
- 8.World Health Organization. Part 2: Microscopy. Geneva, Switzerland: 1998. Laboratory services in tuberculosis control. [Google Scholar]
- 9.Yu MC, Chen HY, Wu MH, Huang WL, Kuo YM, et al. Evaluation of the rapid MGIT TBc identification test for culture confirmation of Mycobacterium tuberculosis complex strain detection. J Clin Microbiol. 2011;49:802–807. doi: 10.1128/JCM.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, et al. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz M, Torres MJ, Llanos AC, Arroyo A, Palomares JC, et al. Direct detection of rifampin- and isoniazid-resistant Mycobacterium tuberculosis in auramine-rhodamine-positive sputum specimens by real-time PCR. J Clin Microbiol. 2004;42:1585–1589. doi: 10.1128/JCM.42.4.1585-1589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddiqi SH, Ruesch-Gerdes S. Becton Dickinson. Franklin Lakes, NJ: 2006. MGIT procedure manual for BACTEC MGIT 960 TB system. [Google Scholar]
- 13.Heifets LB. Drug susceptibility tests in the management of chemotherapy of tuberculosis. In: Heifets LB, editor. Drug Susceptibility in the Chemotherapy of Mycobacterial Infections. Boca Raton, Fla: CRC Press; 1991. pp. 90–121. [Google Scholar]
- 14.Chang K, Lu W, Wang J, Zhang K, Jia S, et al. Rapid and effective diagnosis of tuberculosis and rifampicin resistance with Xpert MTB/RIF assay: A meta-analysis. J Infect. 2012 doi: 10.1016/j.jinf.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Lawn SD, Brooks SV, Kranzer K, Nicol MP, Whitelaw A, et al. Screening for HIV-associated tuberculosis and rifampicin resistance before antiretroviral therapy using the Xpert MTB/RIF assay: a prospective study. PLoS Med. 2011;8:e1001067. doi: 10.1371/journal.pmed.1001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicol MP, Workman L, Isaacs W, Munro J, Black F, et al. Accuracy of the Xpert MTB/RIF test for the diagnosis of pulmonary tuberculosis in children admitted to hospital in Cape Town, South Africa: a descriptive study. Lancet Infect Dis. 2011;11:819–824. doi: 10.1016/S1473-3099(11)70167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marlowe EM, Novak-Weekley SM, Cumpio J, Sharp SE, Momeny MA, et al. Evaluation of the Cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Clin Microbiol. 2011;49:1621–1623. doi: 10.1128/JCM.02214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armand S, Vanhuls P, Delcroix G, Courcol R, Lemaitre N. Comparison of the Xpert MTB/RIF test with an IS6110-TaqMan real-time PCR assay for direct detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens. J Clin Microbiol. 2011;49:1772–1776. doi: 10.1128/JCM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apfalter P, Reischl U, Hammerschlag MR. In-house nucleic acid amplification assays in research: how much quality control is needed before one can rely upon the results? J Clin Microbiol. 2005;43:5835–5841. doi: 10.1128/JCM.43.12.5835-5841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noordhoek GT, van Embden JD, Kolk AH. Reliability of nucleic acid amplification for detection of Mycobacterium tuberculosis: an international collaborative quality control study among 30 laboratories. J Clin Microbiol. 1996;34:2522–2525. doi: 10.1128/jcm.34.10.2522-2525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott LE, McCarthy K, Gous N, Nduna M, Van Rie A, et al. Comparison of Xpert MTB/RIF with other nucleic acid technologies for diagnosing pulmonary tuberculosis in a high HIV prevalence setting: a prospective study. PLoS Med. 2011;8:e1001061. doi: 10.1371/journal.pmed.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theron G, Peter J, van Zyl-Smit R, Mishra H, Streicher E, et al. Evaluation of the Xpert MTB/RIF assay for the diagnosis of pulmonary tuberculosis in a high HIV prevalence setting. Am J Respir Crit Care Med. 2011;184:132–140. doi: 10.1164/rccm.201101-0056OC. [DOI] [PubMed] [Google Scholar]
- 23.Kurbatova EV, Cavanaugh JS, Shah NS, Wright A, Kim H, et al. Rifampicin-resistant Mycobacterium tuberculosis: susceptibility to isoniazid and other anti-tuberculosis drugs. Int J Tuberc Lung Dis. 2012;16:355–357. doi: 10.5588/ijtld.11.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SE, Kurbatova EV, Cavanaugh JS, Cegielski JP. Global isoniazid resistance patterns in rifampin-resistant and rifampin-susceptible tuberculosis. Int J Tuberc Lung Dis. 2012;16:203–205. doi: 10.5588/ijtld.11.0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caoili JC, Mayorova A, Sikes D, Hickman L, Plikaytis BB, et al. Evaluation of the TB-Biochip oligonucleotide microarray system for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:2378–2381. doi: 10.1128/JCM.00439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isakova Zh T. Fast identification of rifampicin-and isoniazid resistance of M. Tuberculosis strains by the "TB-biochip" test system. Georgian Med News. 2008:15–19. [PubMed] [Google Scholar]
- 27.Isakova Zh T. Practical value of the TB-biochip MDR test system in the rapid identification of multidrug-resistant M. tuberculosis strains. Klin Lab Diagn. 2009:50–51. [PubMed] [Google Scholar]