Abstract
A recently described fluorescence biosensor platform utilizes single-chain Fv (scFvs) that selectively bind and activate fluorogen molecules. In this report we investigated the display of tandem scFv biosensors at the surface of mammalian cells with the aim of advancing current fluorescence detection strategies. We initially screened different peptide linkers to separate each scFv unit, and discovered that tandem proteins joined by either flexible or α-helical linkers properly fold and display at the surface of mammalian cells. Accordingly, we performed a combinatorial scFv-dimer study and identified that fluorescence activation correlated with the cellular location (membrane distal versus proximal) and selections of the different scFvs. Furthermore, in vitro measurements showed that the stability of each scFv monomer unit influenced the folding and cell surface activities of tandem scFvs. Additionally, we investigated the absence or poor signals from some scFv-dimer combinations and discovered that intramolecular and intermolecular scFv chain mispairings led to protein misfolding and/or secretory-pathway-mediated degradation. Furthermore, when tandem scFvs were utilized as fluorescence reporter tags with surface receptors, the biosensor unit and target protein showed independent activities. Thus, the live cell application of tandem scFvs permitted advanced detection of target proteins via fluorescence signal amplification, Förster resonance energy transfer resulting in the increase of Stokes shift and multi-color vesicular traffic of surface receptors.
Keywords: biosensor, FAP, fluorescence, fluorogen, scFv
Introduction
Genetically, encoded fluorescence biosensors have radically advanced the fields of cellular and molecular biology. Currently, available protein-based reporters include fluorescent proteins (Shaner et al., 2005) or enzyme-tags that form covalent bonds with organic molecule fluorophores (Keppler et al., 2004; Hinner and Johnsson, 2010). A recently described next-generation platform, called fluorogen-activating proteins (FAPs), utilizes single-chain Fv (scFvs) selected against fluorogen molecules (intrinsically non-fluorescent) that results in the fluorescence activation of fluorogens (Szent-Gyorgyi et al., 2008). In this case, the scFv provides a high specificity interface that spatially restricts the fluorogen, and upon irradiation the fluorogen emits a fluorescence signal, instead of non-radiative decay to its ground state (Silva et al., 2007; Shank et al., 2013).
The scFv biosensor technology offers innovative strategies of fluorescence detection when compared with other protein-based biosensors. For example, the directed evolution of the scFv protein scaffold may allow for fluorogen activation in the cytoplasm of cells, as well as improved affinity binding resulting in lower concentration of fluorogen for fluorescence activation (Szent-Gyorgyi et al., 2008; Yates et al., 2013). Similarly, chemical modifications of fluorogens such as side group additions or charge alterations may result in properties of pH sensing, cellular permeability versus impermeability and spectra shifts (Ozhalici-Unal et al., 2008; Shank et al., 2009; Grover et al., 2012; Yushchenko et al., 2012). Additionally, the non-covalent interface of the scFv-ligand complex permits rapid fluorogen uncoupling and recoupling, as a result one may perform modular fluorescence detection and real-time signal exchanges (Gallo et al., 2014). Furthermore, the scFv platform permits sustained detection and application for time-lapse imaging due to its increased resistance to photobleaching, a result of the thermodynamically favorable exchange from oxidized fluorogens in the protein with unmodified fluorogens in the surrounding medium (Shank et al., 2009). Altogether, FAPs are bi-partite biosensors that allow the independent enhancement each constituent unit, scFv or fluorogen, for improved detection.
In order to further advance the scFv biosensor technology, in this report, we explored the expression of scFv multimers at the surface of mammalian cells. Such approach would offer several advantages: (i) user choice regarding fluorogen channel detection, (ii) simultaneous multi-color labeling of target proteins to facilitate detection of real signal versus background, (iii) fluorescence signal amplification to aid detection of low abundance proteins, (iv) opportunity for Förster resonance energy transfer (FRET) to increase the Stokes shift and (v) ability to perform studies of multi-color protein traffic. In the past two decades, scFv research showed rapid advancement in the generation of multivalent scFv molecules for achieving increased avidity, multi-specificity and improved renal retention in animals models (Hudson and Souriau, 2003; Holliger and Hudson, 2005). However, such research predominantly focuses on freely diffusing protein forms such as mini-bodies, diabodies, triabodies and derivatives thereof (Plückthun and Pack, 1997; Le Gall et al., 1999; Todorovska et al., 2001), and lacks validation for tethered cell surface display applications. In addition, synthesis of soluble scFv multimers frequently generates various byproducts species (Atwell et al., 1999; Xiong et al., 2006), and may form insoluble fractions that require denaturing and renaturing for their correct associations and solubility (Kurucz et al., 1995).
In order to utilize scFv multimers as protein reporters at the surface mammalian cells, we selected a tandem linear assembly of scFvs instead of freely diffusing proteins. In theory, this approach would facilitate folding in the highly restricted secretory compartments of eukaryotic cells, and limit incorrect variable light and heavy (VL and VH) pairings due to the sequential folding of a multipart protein. Thus, the aim of this report was to examine the tandem expression of scFv multimers at the surface of cells, and explore the possible challenges and requirements for the generation of functional biosensors. Additionally, here we also investigated tandem scFvs' novel properties of fluorescence detection for reporter-tag applications.
Materials and methods
Plasmid constructions
All transient transfections were performed using a mammalian expression pDisplaySacLac2 plasmid designed to target recombinant proteins to the surface of mammalian cells (Holleran et al., 2010). It consists of an N-terminal secretion signal sequence followed by a hemagglutinin (HA) sequence that is followed by an insert module flanked by two unique SfiI restriction sites, then followed by the transmembrane anchoring domain of platelet-derived growth factor receptor (PDGFR), and followed by a cytoplasmic insert module flanked by two unique PflMI restriction sites.
The original pDisplaySacLac2 plasmid was modified by ligating different cassette inserts at the SfiI restriction sites. Each cassette insert was generated using overlapped oligonucleotides with vector complementary ends that destroyed the original SfiI restriction sites when ligated to the vector, and resulted in a new domain insert with same as previous SfiI restriction sites followed by a linker (either flexible, α-helix, Z-peptide or proline-helix) and followed by a second domain insert flanked by two unique DraIII sites. The amino-acid sequence for the flexible linker consists of three tandem repeats of G4S, the α-helix is composed of four tandem repeats of EAAAK, the proline-helix consists of 15 prolines and the Z-peptide is composed of a three α-helix bundle with a sequence of VDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPK.
All scFvs used in this report were previously described (Ozhalici-Unal et al., 2008; Szent-Gyorgyi et al., 2008; Zanotti et al., 2011). The scFvs, eGFP and mRFP inserts were generated as previous (Holleran et al., 2010) and contained overhangs coding for the SfiI, DraIII or PflMI sites of the modified pDisplaySacLac2 plasmid; once the inserts were ligated to the vector the restriction sites were destroyed. For the construction of the tandem fluorescent protein plasmids, the eGFP insert was ligated into the insert domain flanked by SfiI sites, and the mRFP insert was ligated into the insert domain flanked by DraIII sites of the modified pDisplaySacLac2 plasmids (described earlier). The same ligation approach was performed for the generation of scFv-dimer plasmids. For the generation of scFv-dimer plasmids containing a cytoplasmic eGFP domain, the eGFP insert was ligated into the PflMI sites of the modified pDisplaySacLac2 plasmid that contained scFv dimers separated by a flexible linker. For the generation of plasmids containing trimeric scFvs, a scFv insert was first ligated into the DraIII sites of the modified pDisplaySacLac2 plasmid that contained a flexible linker; next a cassette insert that contained a flexible linker (described earlier) was ligated into the SfiI sites; following a scFv insert was ligated into the DraIII sites and another scFv insert was ligated into the SfiI sites.
The generation of adrenoreceptor-beta-2 (ADRB2) insert containing a stop codon and flanked by BsmI restriction sites was previously described (Fisher et al., 2010) and inserted into the BsmI site (before the PDGFR transmembrane domain) of the modified pDisplaySacLac2 plasmid. For expression of scFvs in Escherichia coli, a pET-21a plasmid was modified at NotI and XhoI restriction sites utilizing a cassette insert generated using overlapped oligonucleotides with NotI and XhoI complementary ends that resulted in two unique SfiI restriction sites followed by a stop codon. Subsequently, all scFv inserts were ligated into the SfiI sites.
Protein expression and purifications
Purified scFv proteins were obtained using a Rosetta-Gami E.coli strain (Novagen). The cells were induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside (RPIcorp), then lysed, and pelleted via high-speed centrifuging. The supernatant was used in nickel-nitrilotriacetic acid chromatography (Thermo-Fisher) according to the manufacturer's instructions. The eluted fractions were purified via gel-filtration chromatography, then pooled and concentrated using centrifugal-filter units (Millipore). The scFv protein purities were assessed via SDS–PAGE and concentrations were determined by spectroscopy at 280 nm wavelength using the Beer–Lambert equation. The protein samples were aliquoted and stored in phosphate buffer saline with 0.09% sodium azide at –20°C. Thawed samples were subsequently stored at 4°C for one month, and then discarded.
Optical spectroscopy
Different concentrations of guanidinium-chloride (Gm-Cl, Sigma-Aldrich) or urea (Sigma-Aldrich) were incubated with each scFv in phosphate-buffered saline (PBS) for 18 h at 4°C. Triplicate samples containing 0.5 μM protein and 0.5 μM cognate fluorogen in PBS were measured in an Infinite M1000 plate spectrometer (TECAN) using transparent, flat-bottom, 96-well microtiter plates (Corning). All measured values were corrected against fluorogen only samples.
The thermal denaturation/renaturation experiments were performed using 1 μM protein and 0.5 μM fluorogen in PBS. The samples were analyzed using a Varian Cary Eclipse fluorescence spectrometer (Varian Scientific Instruments), and measured every 1 min at a gradient of 2°C/1 min. The ramp-up and ramp-down temperature experiments were performed in the temperature range of 25–90°C and all measured values were corrected against fluorogen only samples. For all experiments, the excitation/emission wavelengths were 405/430 nm for OTB-SO3, 510/545 nm for TO1-2p fluorogen, 602/645 nm for DIR fluorogen and 630/660 nm for MG-2p fluorogen using a 5 nm band-pass filter.
Cell-culture conditions and transient transfections
HEK-293 cells were grown at 37°C, 5% CO2 in Dulbecco's modified Eagle's medium plus 10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin. All the transfections were performed using TransIT®-LT1 reagent (Mirus Bio) according to the manufacturer's instructions.
Flow cytometry
The cells were analyzed in PBS with propidium iodide (Sigma-Aldrich), used to gate out dead cells, in the presence of fluorogen with acquired live events >10 000 per sample. Data were collected with a FACS Vantage SE Flow Cytometer and FACS Diva option (Becton Dickinson) using a 405 nm laser with 450/20 nm filter, a 488 nm laser with 530/30 nm filter and a 633 nm laser with 685/35 nm filter. Quantitation was carried out using FACS Diva Software v5.0.2 (Becton Dickinson).
Fluorescence microscopy
Cells were imaged in PBS using 35-mm glass-bottom dishes (MatTek) in the presence of fluorogen. Images were acquired with a Carl Zeiss LSM 510 Meta/UV DuoScan inverted confocal microscope using a 405 nm laser and a 430–480 nm band-pass filter for OTB-SO3 fluorogen, a 488 nm laser and a 505–550 nm band-pass filter for TO1-2p fluorogen, a 561 nm laser and a 575 nm LP band-pass filter for DIR fluorogen, and a 633 nm laser and a 650 nm LP band-pass filter for MG-2p fluorogen. The acquired images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/).
FRET microscopy and quantification
For each sample, an image was acquired using donor only fluorogen (TO1-2p) or acceptor only fluorogen (DIR) in the medium; then, a same image was acquired using donor plus acceptor fluorogens (TO1-2p + DIR) in the medium after an incubation of 10 min. For data quantification, three independent experiments were performed with a summary total of 42 sample images and 100 regions of interest (ROIs). The mean pixel intensity values for each ROI were plotted for the donor and acceptor emission channels in the presence of TO1-2p only and TO1-2p + DIR fluorogens. To determine FRET efficiency, all ROI intensity values from the donor emission were averaged for each group (TO1-2p only and TO1-2p + DIR), and FRET efficiency was calculated as follows:
where E means FRET efficiency, I(DA) represents the average intensity (donor emission channel) from samples with both donor and acceptor fluorogens and I(D) represents the average intensity (donor emission channel) from samples with the donor fluorogen only. All samples were excited using the donor 488 nm laser (TO1-2p) or 561 nm laser (DIR), and measured at the donor channel of 505–550 nm band-pass filter, and the acceptor channel of 650 nm LP band-pass filter.
Cellular surface assays
Homo-scFv multimer constructs (1×, 2× and 3×) were transfected into HEK-293 cells, then labeled with anti-HA FITC (A01621, GenScript), and measured for fluorescence via flow cytometry in the presence of fluorogen. The receptor agonist stimulation assay was performed using HEK-293 cells transfected with a construct of A5/HL4, A5/dH6 or A5/dL5 fused to ADBR2. Initially, 100 nM MG-2p fluorogen was added to the cellular medium; after 5 min, the media was removed and replaced with PBS and 10 μM isoproterenol (Sigma-Aldrich). After a 30 min incubation at 37°C, the media was removed and replaced with PBS and 100 nM OTB-SO3 fluorogen. After 5 min, the cells were analyzed by microscopy.
Statistics
A Student's t-test was used to quantify statistical significance. In figures, the error bars denote the standard deviation of the mean, and stars represent significant differences (*P-value is <0.05, **P-value is <0.005, ***P-value is <0.0005). Also, we determined a Pearson correlation coefficient (cc) to assess the linear dependence between two different variables.
Results
Evaluating different peptide linkers between tandem proteins at the surface of mammalian cells
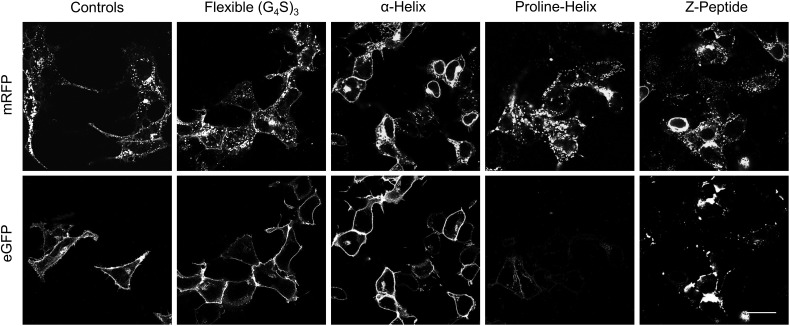
Recent antibody engineering technologies utilize complex arrangements of scFv multimers that result in multi-valency and -specificity (Hudson and Souriau, 2003). Such assemblies call for freely diffusing multipart protein arrangements generally expressed in prokaryotic systems. We chose to engineer less intricate constructs, based on a linear sequence of tandem scFvs to facilitate synthesis and folding of scFv multimers in the secretory compartments of mammalian cells. We selected four different peptide linkers to separate the different protein domains: a flexible linker composed of a trimer of four glycines and a serine (G4S)3 (Mack et al., 1995), a four turn α-helix linker (Arai et al., 2001), a proline-helix linker (Arora et al., 2002; Schuler et al., 2005) and a Z-domain linker composed of a bundle of three α-helices (Braisted and Wells, 1996; Nord et al., 1997). We separated two fluorescent proteins, eGFP and mRFP, with each linker type and assessed their surface activities in mammalian cells. After cellular transfection, the microscopy results revealed robust cell surface signal for the flexible or α-helix linker constructs; on the other hand, the proline-helix and Z-peptide linker constructs showed poor cell surface signals (Fig. 1). Additionally, all samples revealed intracellular fluorescence, particularly mRFP—a consequence of fluorescent protein folding and activity in the secretory compartments of mammalian cells. Here, however, the proline-helix and Z-peptide constructs showed large clusters of intracellular signal when compared with the flexible or α-helix linkers, and suggests protein aggregate formation resulting in their reduced cell surface traffic. Taken together, we find that either flexible or α-helix linkers result in the cell surface display of multipart protein assemblies in mammalian cells.
Fig. 1.
Assessing mammalian cell surface expression of tandem fluorescent proteins using different genetically encoded peptide linkers. The micrographs represent cells expressing surface tandem constructs of eGFP and mRFP separated using different linkers, and the control samples represent the cell surface expression of single fluorescent proteins. The scale bar indicates 30 μm for all samples.
Performing a large combinatorial screen of scFv dimers expressed at the surface of mammalian cells
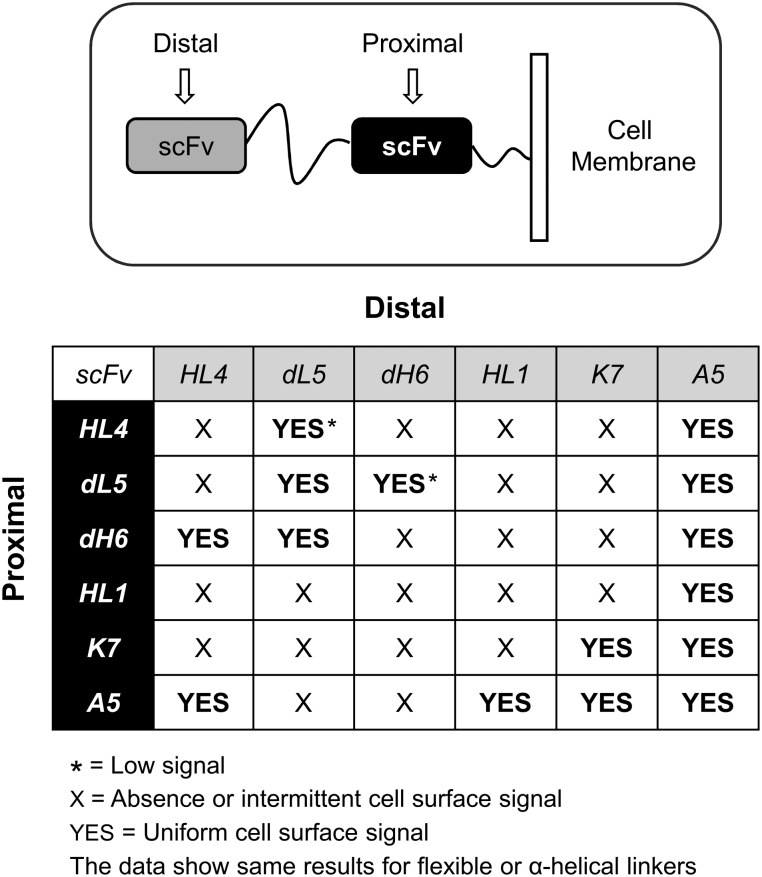
Next, we selected different scFv biosensors previously studied by our center with activation against different color cell-impermeant fluorogens (Supplementary Fig. S1). Because the selected blue emission scFvs A5, D10 and H10 lacked prior validation in mammalian cells, we transfected each one for cell surface expression and then measured for biosensor fluorescence. The micrograph data indicated abundant and uniform cell surface signal only for A5 (Supplementary Fig. S2); thus, we only utilized this scFv for all subsequent blue-channel experiments. Next, we asked whether tandem scFv assemblies, separated using flexible or α-helix linkers, would result in fluorogen activation at the cell surface. We performed a large combinatorial study of scFv dimers expressed in mammalian cells. Here, we observed varied results, such as, abundant cell surface signal for some scFvs, and poor cell surface activity—absence, dimness or intermittent signal—for others (Fig. 2, Supplementary Figs S4 and S5). Surprisingly, both linker types showed same data results for all scFv-dimer combinations and reveal their role as structural protein spacers rather than determinants for correct scFv folding and activities.
Fig. 2.
Summary graphic of microscopy images where different scFv-dimer configurations were expressed at the surface of cells. Both, flexible or α-helical linkers between the scFvs provided the same results.
Assessing the mechanisms for reduced cell surface activity of some scFv-dimer combinations
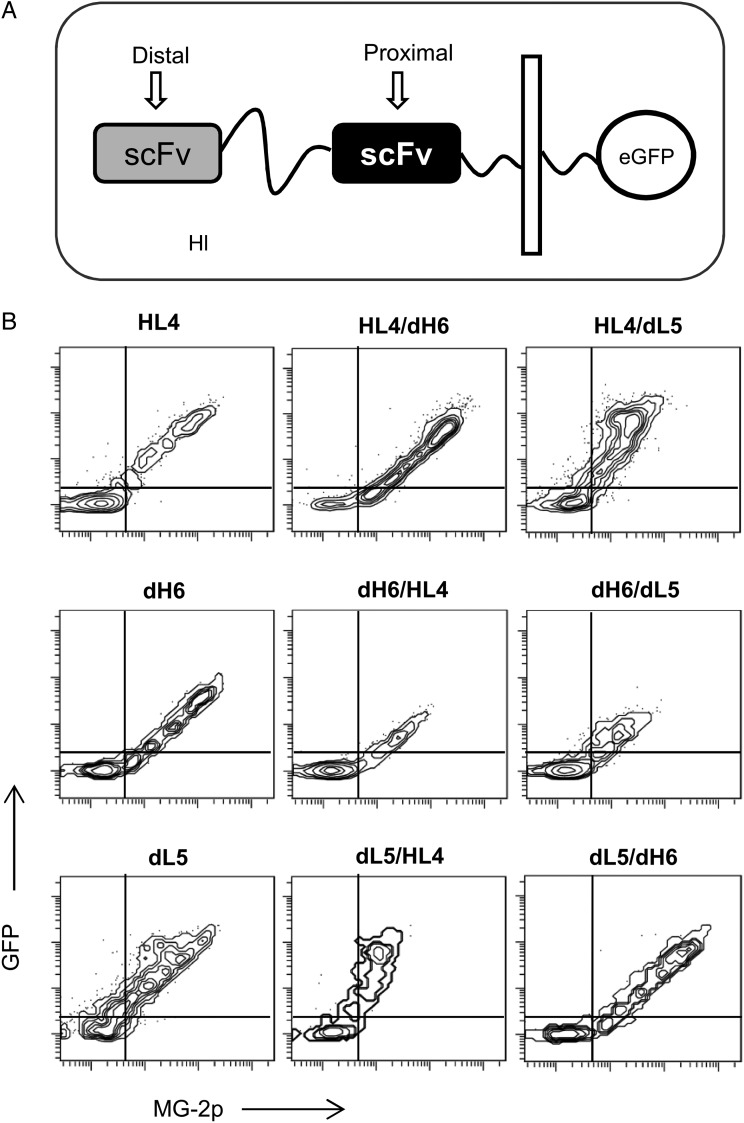
We further investigated the loss or reduced cell surface activity from scFv-dimer combinations. We genetically tethered a cytoplasmic eGFP domain to all pairwise combinations of MG-2p activating scFvs joined using a flexible linker (Fig. 3A) (we omitted the α-helix linker for all subsequent experiments since similar results are obtained). In such constructs, the GFP signal reports protein expression—intracellular and cell surface—while the cell impermeable MG-2p fluorogen reports only scFv cell surface activities. After cellular transfection, we analyzed a large population of cells (>10 000 events) via flow cytometry. For some cases, the data showed sustained GFP signal and decreased MG-2p fluorescence, such as, constructs HL4/dL5 and dL5/HL4 when compared with controls of single HL4 or dL5 (Fig. 3B). Here, the data suggest scFv misfolding (possibly incorrect scFv domain pairings) preventing fluorogen activation and/or cell surface traffic, yet sustained protein expression. On the other hand, when dH6 is placed distal to the cell membrane, as in the case of dH6/HL4 and dH6/dL5, both GFP and MG-2p signals decrease when compared with control of single dH6 (Fig. 3B). This observation hints of protein instability for which cellular quality control mechanisms target the tandem scFv protein for degradation (Meusser et al., 2005). Conversely, when dH6 is placed proximal to the cell membrane—as in the case of HL4/dH6 and dL5/dH6—the GFP and MG signals are preserved (Fig. 3B). This observation highlights that dH6 location (cell membrane distal versus proximal) influences correct folding and stability of scFv-dimer assemblies. In quick summary, here we propose two principal mechanisms for loss of biosensor activity at the surface of cells: (i) in some cases incorrect pairing of tandem scFv domains results in inactive scFv monomers and/or surface traffic of tandem scFvs and (ii) for other cases tandem scFvs form unstable assemblies that result in their degradation.
Fig. 3.
Fluorescence analysis using combinations of same color tandem scFvs and a cytoplasmic eGFP domain. (A) Graphical display of constructs at the cell surface. (B) Flow cytometry graphs from HEK-293 cells expressing each construct at the cell surface and measured in the presence of 150 nM MG-2p.
Understanding the role of scFv monomer stability for functional scFv tandem structures
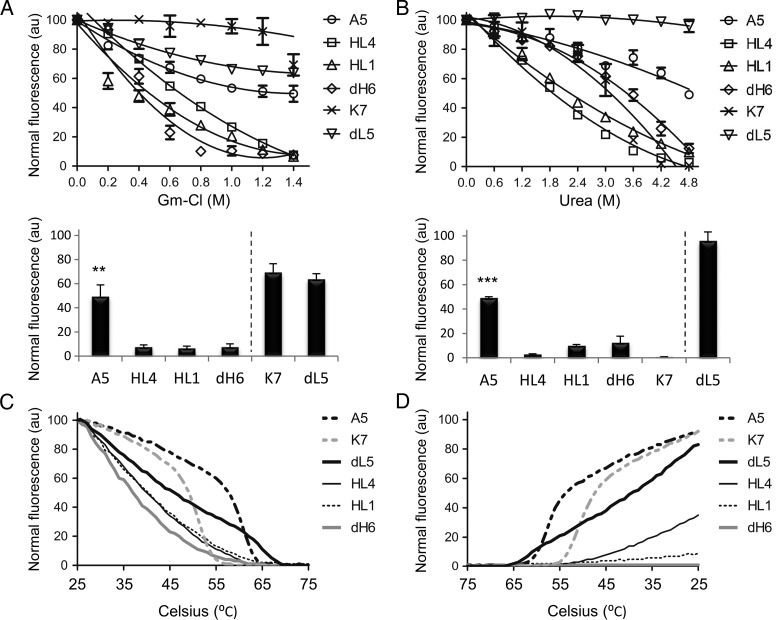
The scFv unit comprises of a pair of VH and VL chains held together by non-covalent forces, and the number of contacts and type of affinity interactions (i.e. ionic, van der Waals etc.) influence domain affinities and overall structure. Accordingly, we looked at the inherent stability of each constituent scFv monomer when subjected to chemical insults. We utilized two different chaotropic agents, Gm-Cl and urea, and measured for fluorogen activation at increasing concentrations of each reagent—note that chaotropic agent effects on fluorogens were undetected (Supplementary Fig. S6). The results showed A5 and dL5 as most resistant to chemical insults when compared with HL4, HL1, dH6 and K7 (Fig. 4A and B). Additionally, we observed chaotropic agent preferential effects, such as sensitivity of K7 to urea when compared with Gm-Cl (Fig. 4A and B). Such observation results from the molecular differences of each agent, such as the ionic nature of Gm-Cl versus uncharged urea (Rashid et al., 2005; England and Haran, 2011). More specifically, urea tends to unfold proteins through direct van der Waals forces, or indirect disruption of the water structure, which in turn weakens the solvation of hydrophobic groups (Bennion and Daggett, 2003; Canchi et al., 2010), while Gm-Cl tends to disrupt the protein structure via interference of electrostatic interactions (Monera et al., 1994). Thus, according to K7 sensitivity to urea we predict its stability to derive primarily from hydrophobic interactions.
Fig. 4.
Measuring in vitro protein stability from scFv monomers. (A) The top graph corresponds to the measured fluorescence intensity of each scFv in the presence of increased concentration of Gm-Cl, while the bottom bar graph shows the fluorescence intensities of each scFv at the highest concentration of Gm-Cl from the top graph. (B) Same as previous, but in the presence of increased urea concentration. (C) Graph of fluorescence intensities from each scFv at an increased thermal gradient. (D) Graph of fluorescence intensities from each scFv (post-thermal denaturation) at a decreased thermal gradient.
Because chemical insults may offer differential effects on protein stability results, we performed additional experiments involving thermal denaturation that uses enthalpy to indiscriminately disrupt bond interfaces in the protein. We exposed each scFv to an increased thermal gradient while measuring for fluorogen activation—note that thermal effects on fluorogens were undetected (Supplementary Fig. S7). Both A5 and K7 displayed a sigmoidal denaturation profile indicative of a cooperative thermal transition, and suggest their folded structure to be stabilized by energetically favorable interactions (Roy and Hecht, 2000). On the other hand, the rest of the scFvs showed non-cooperative denaturation profiles. Although dL5 maintained activity at high temperatures, A5 showed the most fluorescence activity at high temperatures (broadest shoulder), indicative of its high scFv stability (Fig. 4C). Next we performed the reverse assay; we denatured all the scFvs in a gradient fashion and followed by reversing the thermal gradient while measuring for fluorogen activation. Analysis showed initial refolding activity for dL5; however, as previously observed, A5 showed a sigmoidal transition profile with large measurements of fluorescence at high temperatures when compared with the rest of the scFvs (Fig. 4D). In contrast, dH6 and HL1 failed to regain activities when cooled, indicative of poor protein stability and/or ability to refold into correct VH and VL orientations (Fig. 4D). When analyzed together (chemical and thermal data), A5 shows high protein stability when compared with the other scFvs, while dH6 and HL1 show poor stabilities. In addition, the data stay congruent when compared with the microscopy data results (Fig. 2) and show that high stability scFvs generally form functional tandem assemblies. In the case of dL5, it displays high stability when exposed to chemical agents, yet moderate activity when assayed thermally with a non-cooperative denaturation profile. Accordingly, dL5 proves less stable than A5, which is also observed from the microscopy data (Fig. 2).
Determining fluorescence signal amplification using tandem scFvs at the surface of cells
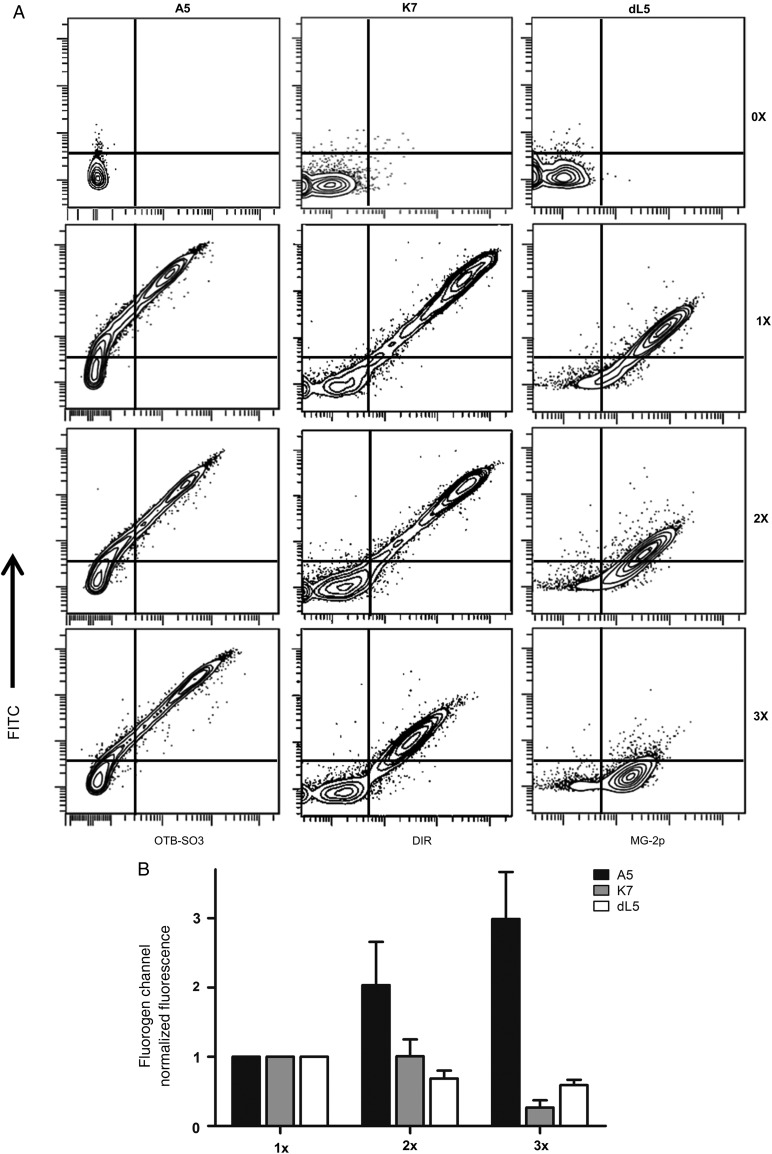
Because multimeric scFv biosensors increase the number of available scFv units, they may allow fluorescence signal amplification over single scFv reporters. For that reason, based on Fig. 2 we analyzed scFvs that showed fluorescence activity when paired with themselves—A5, dL5 and K7. We built scFv constructs containing mono-, di- and tri-meric scFvs separated by flexible linkers and transfected them into mammalian cells for surface expression. Each construct contained an N-terminal HA sequence, and after transfection the cells were labeled with anti-HA FITC antibody. After measuring a large population of cells for each group (>10 000 events), we only observed signal amplification for A5 (Fig. 5A). In the case of K7, each scFv unit addition resulted in decrease of fluorescence for both FITC and DIR, indicative of improper scFv assemblies with lower cell surface traffic and/or expression (Fig. 5A). Similarly, dL5 also showed lower signal for both FITC and MG-2p with each unit addition; however, we observed greater proportion of FITC signal loss than MG-2p—hinting at reduced antibody access to the N-terminal HA-epitope (Fig. 5A). This may occur because dL5 is a non-canonical scFv of same light-chain domain pairs (VL–VL) (Szent-Gyorgyi et al., 2013), and may form VL domain pairs among different scFv monomers that may bury the N-terminal HA-sequence for antibody access resulting in greater MG-2p activity than FITC. Additional quantitative analysis shows a linear increase of fluorescence activity for A5 that is proportional to the number of added scFv monomer units (Fig. 5B). On the other hand, fluorescence measurements for K7 and dL5 inform of decreased signal with each tandem unit increase (Fig. 5B), further validating the previous observations.
Fig. 5.
Determining fluorescence signal amplification of homo-scFv multimers. (A) Flow cytometry graphs from mammalian cells expressing a single (1×), double (2×) or triple (3×) homo-scFv multimers at the cell surface and co-labeled with anti-HA FITC antibody. (B) Summary graph of fluorogen fluorescence intensities from three independent flow cytometry experiments as presented above. All measurements were performed in the presence of 100 nM OTB-SO3, DIR or MG-2p fluorogens.
Assessing fluorescence energy transfer between chromophores in tandem scFv structures
Current protein fluorescent platforms show specific excitation and emission spectra with defined Stokes shifts. On the other hand, multimeric scFvs may offer opportunity for FRET between two different chromophore units via dipole–dipole intermolecular coupling (Mayor and Bilgrami, 2008). In our system, FRET would involve the transfer of excitation energy from a donor scFv–fluorogen complex separated by a flexible linker to an acceptor scFv–fluorogen complex. Such energy transfer would result in increased Stokes shift for the donor group resulting in fluorescence detection at longer wavelengths (acceptor channel). On the other hand, FRET also would allow the acceptor group to excite from multiple wavelength sources, either donor or acceptor excitations. As a result, FRET signal would provide a more flexible approach for fluorescence channel detection.
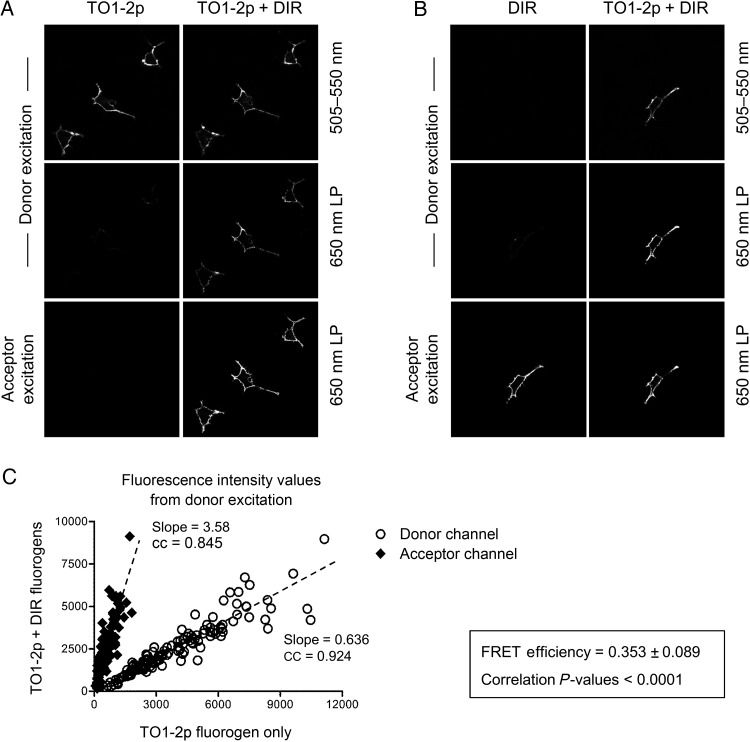
We utilized a tandem scFv dimer composed of A5 joined to K7 separated by a flexible linker. In this platform, scFv A5 would form a complex with TO1-2p fluorogen and would function as the donor unit (A5 also activates TO1-2p besides its cognate fluorogen, see Supplementary Fig. S3), and scFv K7 would form a complex with DIR fluorogen and would function as the acceptor unit. After the cell surface expression of tandem A5/K7, analysis showed that individual addition of TO1-2p or DIR fluorogens resulted in their single activation (Fig. 6A and B). On the other hand, when both fluorogens were present in the medium, the excitation of TO1-2p fluorogen resulted in DIR channel emission (Fig. 6A and B). Further quantitative analysis reveals a strong correlation (cc = 0.924) for the signal of the donor emission channel, i.e. when both fluorogens are present fluorescence signal diminishes resulting in a slope value of 0.636, <1 (Fig. 6C), indicative of energy transfer loss from donor to acceptor chromophore. Similarly, the signal for the acceptor emission channel also demonstrates strong correlation (cc = 0.845) with a sharp increase when both fluorogens are present and quantified by a slope value of 3.58, >1 (Fig. 6C). Overall, tandem scFv dimers offer opportunity for FRET signal detection and show 35.3% energy transfer efficiency.
Fig. 6.
Assessing FRET between tandem scFv biosensors. (A) Micrographs of mammalian cells expressing A5 and K7 joined by a flexible linker at the surface. Images were acquired using a donor (TO1-2p) excitation 488 nm laser or an acceptor (DIR) excitation 561 nm laser and measured using two different band-pass filters, 505–550 nm for the TO1-2p emission channel and 650 nm LP for the DIR emission channel. The cell sample was first measured using only TO1-2p fluorogen, and subsequently measured after the addition of acceptor DIR fluorogen into the medium. The scale bar represents 30 μm. (B) Same as A; however, the cell sample was first measured using only DIR fluorogen, and subsequently measured after the addition of donor TO1-2p fluorogen into the medium. (C) Graph of mean pixel intensity values for different ROIs (n = 100) from a total of 42 images from three independent experiments. The values were determined using the donor (TO1-2p) laser excitation and measured from the donor and acceptor emission channels. Each sample was analyzed using only TO1-2p fluorogen, and subsequently analyzed after the addition of acceptor DIR fluorogen into the medium. All data show strong correlation with P < 0.001 and cc close to 1. All the experiments were performed using 300 nM TO1-2p and/or 150 nM DIR fluorogens.
Validation of tandem scFv structures for live cell reporter applications
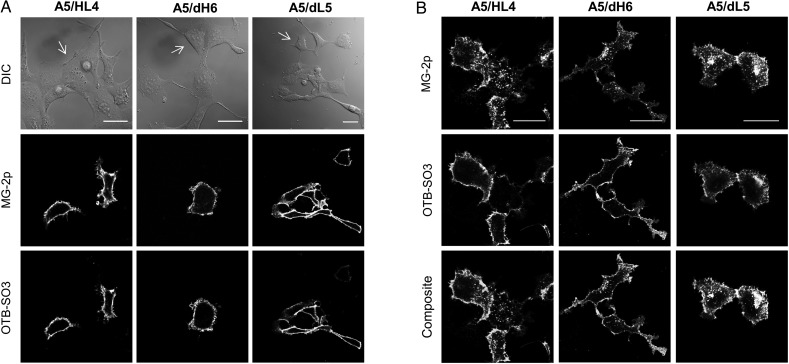
Next, we asked whether scFv dimers would result in functional reporters when fused to cell surface receptors. We genetically tagged a G-protein-coupled receptor, ADRB2, at its N-terminus with different tandem scFv dimers separated by a flexible linker (A5/HL4, A5/dH6 or A5/dL5). The microscopy images revealed multi-color fluorescence only for transfected cells, which showed biosensor activity from each scFv unit; while the untagged cells remained dark demonstrating minimal fluorogen background (Fig. 7A, Supplementary Fig. S8 color). Next, we asked whether tandem scFv reporters would interfere with the ADRB2 signaling response by measuring its attenuation (receptor internalization by endocytosis), a feature of G-protein-coupled receptors (Fisher et al., 2010). Accordingly, we initially introduced only MG-2p fluorogen to the cellular medium to label all the cell surface receptors with one channel. Next, we exchanged the medium to include an ADRB2 agonist ligand (isoproterenol) to induce signaling. After a short incubation, we washed the cells and exchanged the medium to include only OTB-SO3 fluorogen. Here, the microscopy results showed MG-2p channel signal in endosomal vesicles due to the rapid internalization response from ADRB2 to its ligand (Fig. 7B, Supplementary Fig. S8 color). After the removal of agonist from the cellular medium, we observed OTB-SO3 signal only at the cell surface, indicating that ADRB2 halted its vesicular internalization response (Fig. 7B, Supplementary Fig. S8 color). In conclusion, scFv-dimer biosensors form functional reporters-tags with cell surface proteins and permit the study of multi-color receptor vesicular traffic.
Fig. 7.
Determining biosensor activities of tandem scFv dimers when fused to a cell surface receptor. (A) Micrographs of mammalian cells expressing different tandem scFvs fused to ADRB2. Images were acquired using two different emission channels with corresponding excitation laser wavelengths. The white arrows on the DIC images indicate untransfected cells. (B) Same as A; however, the cells were initially labeled with MG-2p fluorogen, washed and incubated with an ADRB2 agonist, and then washed and replaced with medium containing only OTB-SO3 fluorogen. The MG-2p emission channel shows the ADRB2 agonist response of intracellular vesicular traffic. The OTB-SO3 emission channel only shows ADRB2 cell surface signal due to the removal of agonist from the medium. All the experiments were performed using 100 nM MG-2p and OTB-SO3 fluorogens, and the scale bar represents 30 μm.
Discussion
In this report, we set out to engineer scFv multimers expressed at the surface of mammalian cells with the aim of multi-color fluorescence and enhanced detection. We generated different tandem constructs separated using various peptide linkers. The initial cell surface display of dimer fluorescent proteins highlighted the influence of linker complexity for correct folding and cell surface protein traffic. For example, simple structural configurations based on flexible or α-helical linkers allow the proper folding, sorting and processing of tandem proteins in mammalian cells. Conversely, elaborate peptide linkers, such as the artificial proline-helix or the highly bundled Z-peptide, add folding complexities to multi-domain assemblies that limit their cell surface traffic.
A combinatorial study of protein assemblies offered insight into cell surface activities of tandem scFvs. That is, for some scFv combinations we observed uniform and bright signals, while for others we observed poor fluorescence activities—the absence, dimness or intermittent signals (Supplementary Figs S4 and S5). Here, we propose two independent models for the inactivity of tandem scFv biosensors: (i) the intramolecular interactions between the scFv monomers via VH and VL mispairings (De Jonge et al., 1995; Mack et al., 1995) may affect fluorogen activation, traffic and/or protein expression or (ii) the intermolecular interactions of scFv domains with neighboring scFvs (Szent-Gyorgyi et al., 2008, 2013; Senutovitch et al., 2012) may result in the observed clusters/intermittent fluorescence at the cell surface of some scFv tandem pairs.
Although the rate of scFv folding and molecular chaperones in the surrounding environment influence protein folding, in vitro stability measurements help discern favorable affinity interactions between VH and VL chains to fold into native conformations and circumvent alternate intramolecular/intermolecular arrangements. In this case, the magnitude of non-covalent forces and number of interactions between VH and VL domains direct the stability of each scFv monomer. As presented by our work, scFv monomer stabilities correlate with fluorescence activation (see Fig. 4 with 2). Furthermore, the cell surface activities from tandem scFvs depend on their cellular location (membrane distal versus proximal) and selections. Here, we see that because the N-terminus domain may function as a chaperone to facilitate folding of adjacent proteins (Harper and Speicher, 2011), the placement of high stability scFvs at this region (cell membrane distal) may result in functional scFv-dimer pairs (see Fig. 2 with 4, A5 data). Conversely, the placement of poor stability scFvs at the N-terminus may result in non-functional tandem scFvs (see Fig. 3 with 4, dH6 data). Accordingly, to facilitate future selections of functional scFv pairs, we advise a preliminary scFv stability screen prior labor intensive approaches of synthesis of scFv multimers and their subsequent live cell measurements. Additionally, poor stability scFvs may benefit from rational protein engineering strategies that increase their molecular stabilities, such as the introduction of point mutations and disulfide bonds between VL and VH domains (Wörn and Plückthun, 2001), as well as methods of loop grafting (Jung and Plückthun, 1997).
The ability to tether multiple scFv units into a single biosensor platform offers enhanced properties of fluorescence detection. For instance, stable homo-scFv multimers amplify signal due to the greater number of fluorescent units per reporter and may aid fluorescence sensitivity of tagged proteins that display low expression/abundance. Similarly, because a greater number of biosensor units increases the fluorescence lifetime of the reporter the susceptibility to signal loss due to photon-induced chemical damages decreases (Cooper et al., 2013). Thus, one may perform longer exposure experiments and time series microscopy reducing the risk of photobleaching effects. In addition, homo-scFv multimers may be utilized in conjunction with dyedron molecules, a novel class of fluorogens that stoichiometrically amplify the fluorescence output 5- to 10-fold per scFv unit (Szent-Gyorgyi et al., 2010). More specifically, a dyedron is composed of a single fluorogen conjugated with dendron-based cyanine dyes that increase the extinction coefficient of the excitation wavelength and allow more efficient excitation of the fluorogen via intramolecular energy transfer (Szent-Gyorgyi et al., 2010). As a result, the signal amplification of tandem scFvs forming complexes with dyedrons may theoretically surpass the fluorescence signal output of current protein reporters, and possibly fluorophore-based systems.
On the other hand, hetero-scFv multimers permit FRET signal detection where the energy from the excited state of a scFv–fluorogen unit is transferred to a different scFv–fluorogen unit. Thus, tandem scFv dimers must satisfy two crucial requirements for FRET: a short flexible linker that separates the scFvs to <10 nm and brings the chromophores to an optimal distance radius for energy transfer; and spectral overlap between the emission of the donor and absorbance of the acceptor fluorogen (Mayor and Bilgrami, 2008). Although FRET is commonly utilized in biological systems for resolving distances between molecules (Jares-Erijman and Jovin, 2006; Piston and Kremers, 2007) here we applied FRET to increase the Stokes shift of the donor scFv–fluorogen unit. In this case, the donor signal shifts to longer wavelengths in the red-channel emission where biological samples show reduced cellular background autofluorescence (Billinton and Knight, 2001). On the other hand, FRET signal allows multi-channel excitation for the acceptor scFv–fluorogen unit resulting in excitation flexibility, which favors settings of instrumentations with limited irradiation sources. We foresee future engineering approaches for the generation of tandem scFvs to utilize shorter peptide linkers between the scFv molecules to further improve proximity between the chromophores, and consequently increase the FRET efficiency (Berney and Danuser, 2003).
Protein reporters based on hetero-scFv multimers permit advanced multi-color protein traffic studies. Because fluorescence activation occurs only upon the presentation of cognate fluorogen in the cellular medium, the scFv biosensor technology offers a temporal manipulation of signal detection. In this case, a G-protein-coupled-receptor's response to its ligand may be resolved via the sequential addition of different cell-impermeant fluorogens (Fig. 7B). Thus, one may resolve receptor-mediated vesicular traffic and multi-color spatial resolution of cell surface versus intracellular tagged proteins. Furthermore, hetero-scFv multimers may permit co-localized signal analysis when co-labeled with different color fluorogens to facilitate the detection of real output signal for images with large background/noise. Likewise, the simultaneous labeling of hetero-scFv multimers using different color cell permeant versus impermeant fluorogens may allow the analysis of real-time protein synthesis (via measurement of the cell permeant fluorogen) and membrane receptor turnover/degradation (via measurement of the cell-impermeant fluorogen in cargo vesicles). Note, such approach currently proves difficult with conventional biosensors: fluorescent proteins with single color detection and intracellular background signals or fluorophore-labeled antibody systems that require timed incubations and fixed cells for intracellular labeling.
In conclusion, the fluorogen-activating technology facilitates understanding of scFv synthesis in live cells via standard methods of fluorescence detection. The signal measurements allowed us to comprehend the likely mechanisms for generating functional scFv multimers. Thus, scFv flurogen-activating technology offers the single-chain antibody research community a fluorescence assessment tool for dissecting scFv synthesis in cellular systems, such as correct folding, domain interferences and protein degradation. Additionally, we foresee future work based on the cell surface display of scFv multimers to focus on a combination of tethered and soluble scFvs to generate assemblies with increased complexities, such as diabody, triabody and tetrabody platforms (Iliades et al., 1997; Lawrence et al., 1998; Todorovska et al., 2001). Overall, the work presented here offers a glimpse on the possibilities and limitations for generating tandem scFv multimers at the surface of mammalian cells, and also expands on novel fluorescence detection properties for protein reporter applications.
Supplementary data
Funding
This work was supported by the National Institutes of Health [U54GM103529].
Supplementary Material
Acknowledgements
We would like to thank Yehuda Creeger and Haibing Teng for instrument assistance (flow cytometry and confocal microscopy, respectively). We also thank Brigitte Schmidt, Nathaniel Shank and Gloria Silva for the synthesis of all the fluorogens. Additionally, we are thankful to Kim Zanotti for providing the plasmids and sequences of the blue-channel scFvs used in this report.
References
- Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T. (2001) Protein Eng., 14, 529–532. [DOI] [PubMed] [Google Scholar]
- Arora P.S., Ansari A.Z., Best T.P., Ptashne M., Dervan P.B. (2002) J. Am. Chem. Soc., 124, 13067–13071. [DOI] [PubMed] [Google Scholar]
- Atwell J.L., Breheney K.A., Lawrence L.J., McCoy A.J., Kortt A.A., Hudson P.J. (1999) Protein Eng., 12, 597–604. [DOI] [PubMed] [Google Scholar]
- Bennion B.J., Daggett V. (2003) Proc. Natl Acad. Sci. U.S.A., 100, 5142–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney C., Danuser G. (2003) Biophys. J., 84, 3992–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billinton N., Knight A.W. (2001) Anal. Biochem., 291, 175–197. [DOI] [PubMed] [Google Scholar]
- Braisted A.C., Wells J.A. (1996) Proc. Natl Acad. Sci. U.S.A., 93, 5688–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchi D.R., Paschek D., García A.E. (2010) J. Am. Chem. Soc., 132, 2338–2344. [DOI] [PubMed] [Google Scholar]
- Cooper D., Uhm H., Tauzin L.J., Poddar N., Landes C.F. (2013) Chembiochem, 14, 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge J., Brissinck J., Heirman C., Demanet C., Leo O., Moser M., Thielemans K. (1995) Mol. Immunol., 32, 1405–1412. [DOI] [PubMed] [Google Scholar]
- England J.L., Haran G. (2011) Annu. Rev. Phys. Chem., 62, 257–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher G.W., Adler S.A., Fuhrman M.H., Waggoner A.S., Bruchez M.P., Jarvik J.W. (2010) J. Biomol. Screen., 15, 703–709. [DOI] [PubMed] [Google Scholar]
- Gallo E., Vasilev K.V., Jarvik J. (2014) Biotechnol. Bioeng., 111, 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A., Schmidt B.F., Salter R.D., Watkins S.C., Waggoner A.S., Bruchez M.P. (2012) Angew. Chem. Int. Ed. Engl., 51, 4838–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S., Speicher D.W. (2011) Methods Mol. Biol., 681, 259–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinner M.J., Johnsson K. (2010) Curr. Opin. Biotechnol., 21, 766–776. [DOI] [PubMed] [Google Scholar]
- Holleran J., Brown D., Fuhrman M.H., Adler S.A., Fisher G.W., Jarvik J.W. (2010) Cytometry A, 77, 776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliger P., Hudson P.J. (2005) Nat. Biotechnol., 23, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Hudson P., Souriau C. (2003) Nat. Med., 9, 129–134. [DOI] [PubMed] [Google Scholar]
- Iliades P., Kortt A.A., Hudson P.J. (1997) FEBS Lett., 409, 437–441. [DOI] [PubMed] [Google Scholar]
- Jares-Erijman E.A., Jovin T.M. (2006) Curr. Opin. Chem. Biol., 10, 409–416. [DOI] [PubMed] [Google Scholar]
- Jung S., Plückthun A. (1997) Protein Eng., 10, 959–966. [DOI] [PubMed] [Google Scholar]
- Keppler A., Pick H., Arrivoli C., Vogel H., Johnsson K. (2004) Proc. Natl Acad. Sci. U.S.A., 101, 9955–9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz I., Titus J.A., Jost C.R., Segal D.M. (1995) Mol. Immunol., 32, 1443–1452. [DOI] [PubMed] [Google Scholar]
- Lawrence L.J., Kortt A.A., Iliades P., Tulloch P.A., Hudson P.J. (1998) FEBS Lett., 425, 479–484. [DOI] [PubMed] [Google Scholar]
- Le Gall F., Kipriyanov S.M., Moldenhauer G., Little M. (1999) FEBS Lett., 453, 164–168. [DOI] [PubMed] [Google Scholar]
- Mack M., Riethmüller G., Kufer P. (1995) Proc. Natl Acad. Sci. U.S.A., 92, 7021–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Bilgrami S. (2008) Eval. Tech. Biochem. Res. Cell Press 43–49. [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol., 7, 766–772. [DOI] [PubMed] [Google Scholar]
- Monera O.D., Kay C.M., Hodges R.S. (1994) Protein Sci., 3, 1984–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord K., Gunneriusson E., Ringdahl J., Ståhl S., Uhlén M., Nygren P.A. (1997) Nat. Biotechnol., 15, 772–777. [DOI] [PubMed] [Google Scholar]
- Ozhalici-Unal H., Pow C.L., Marks S.A., Jesper L.D., Silva G.L., Shank N.I., Jones E.W., Burnette J.M., 3rd, Berget P.B., Armitage B.A. (2008) J. Am. Chem. Soc., 130, 12620–12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston D.W., Jan Kremers G. (2007) Trends Biochem. Sci., 32, 407–414. [DOI] [PubMed] [Google Scholar]
- Plückthun A., Pack P. (1997) Immunotechnology, 3, 83–105. [DOI] [PubMed] [Google Scholar]
- Rashid F., Sharma S., Bano B. (2005) Protein J., 24, 283–292. [DOI] [PubMed] [Google Scholar]
- Roy S., Hecht M.H. (2000) Biochemistry, 39, 4603–4607. [DOI] [PubMed] [Google Scholar]
- Schuler B., Lipman E.A., Steinbach P.J., Kumke M., Eaton W.A. (2005) Proc. Natl Acad. Sci. U.S.A., 102, 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senutovitch N., Stanfield R.L., Bhattacharyya S., Rule G.S., Wilson I.A., Armitage B.A., Waggoner A.S., Berget P.B. (2012) Biochemistry, 51, 2471–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Steinbach P.A., Tsien R.Y. (2005) Nat. Methods, 2, 905–909. [DOI] [PubMed] [Google Scholar]
- Shank N.I., Pham H.H., Waggoner A.S., Armitage B.A. (2013) J. Am. Chem. Soc., 135, 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank N.I., Zanotti K.J., Lanni F., Berget P.B., Armitage B.A. (2009) J. Am. Chem. Soc., 131, 12960–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G.L., Ediz V., Yaron D., Armitage B.A. (2007) J. Am. Chem. Soc., 129, 5710–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi C., Schmidt B.F., Creeger Y., et al. (2008) Nat. Biotechnol., 26, 235–240. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C., Schmidt B.F., Fitzpatrick J.A.J., Bruchez M.P. (2010) J. Am. Chem. Soc., 132, 11103–11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi C., Stanfield R.L., Andreko S., Dempsey A., Ahmed M., Capek S., Waggoner A., Wilson I.A., Bruchez M.P. (2013) J. Mol. Biol., 425, 4595–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovska A., Roovers R.C., Dolezal O., Kortt A.A., Hoogenboom H.R., Hudson P.J. (2001) J. Immunol. Methods, 248, 47–66. [DOI] [PubMed] [Google Scholar]
- Wörn A., Plückthun A. (2001) J. Mol. Biol., 305, 989–1010. [DOI] [PubMed] [Google Scholar]
- Xiong C.Y., Natarajan A., Shi X.B., Denardo G.L., Denardo S.J. (2006) Protein Eng. Des. Sel., 19, 359–367. [DOI] [PubMed] [Google Scholar]
- Yates B.P., Peck M.A., Berget P.B. (2013) Mol. Biotechnol., 54, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushchenko D.A., Zhang M., Yan Q., Waggoner A.S., Bruchez M.P. (2012) Chembiochem, 13, 1564–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti K.J., Silva G.L., Creeger Y., Robertson K.L., Waggoner A.S., Berget P.B., Armitage B.A. (2011) Org. Biomol. Chem., 9, 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.