Abstract
Display technologies such as yeast and phage display offer powerful alternatives to traditional immunization-based antibody discovery, but require conversion of displayed proteins into soluble form prior to downstream characterization. Here we utilize amber suppression to implement a yeast-based switchable display/secretion system that enables the immediate production of soluble, antibody-like reagents at the end of screening efforts. Model selections in the switchable format remain efficient, and library screening in the switchable format yields renewable sources of affinity reagents exhibiting nanomolar binding affinities. These results confirm that this system provides a seamless link between display-based screening and the production and evaluation of soluble forms of candidate binding proteins. Switchable display/secretion libraries provide a cloning-free, accessible approach to affinity reagent generation.
Keywords: amber suppression, antibody screening, phage display, synthetic antibody library, yeast display
Introduction
A number of immunization- and display-based methodologies enable the discovery of antibodies suitable for use in an array of applications (Langone, 1986; Winter et al., 1994; Hanly et al., 1995; Lipovsek and Plückthun, 2004; Gai and Wittrup, 2007; Boder et al., 2012; King et al., 2014). Immunization-based methods are generally reliable for the generation of polyclonal sera and, with hybridoma technology, renewable sources of monoclonal antibodies. However, polyclonal reagents generated with immunizations are nonrenewable, motivating the development of renewable cloned polyclonal antibody pools (Bradbury and Plückthun, 2015; Ferrara et al., 2015a).
Display methodologies offer versatile approaches to the generation of binding domains. However, lead candidates must be subcloned and expressed in soluble form for additional characterization, creating a bottleneck in the validation process. Different display platforms offer several approaches to convert, or switch, a displayed construct into soluble form, each with varying advantages and drawbacks. Some phage display constructs allow for the production of selected single-chain variable fragments (scFvs) or antigen-binding fragments (Fabs) in soluble form, but only after transforming clones of interest into separate bacterial expression strains (Winter et al., 1994; Andris-Widhopf et al., 2000). Similar methods have been applied in a combined phage/yeast display selection workflow, allowing the secretion of renewable polyclonal reagents in yeast after subcloning constructs of interest into a secretion vector (Ferrara et al., 2015a,b). Mammalian display technologies using alternative splicing have enabled the simultaneous display and secretion of antibodies (Horlick et al., 2013; Bowers et al., 2014). This approach is powerful, especially when coupled with somatic hypermutation, but the slow growth rates of mammalian cells limit screening to one round of sorting every 10–12 days. Yeast-based cell secretion and capture technologies are truly switchable (Rakestraw et al., 2006, 2011), but secreted binding proteins are not captured until after they have completely left the cell, leading to a potential loss of the genotype–phenotype linkage. Although these concerns can be partially alleviated by using viscous induction media and the avoidance of shaking during induction, these painstaking measures reduce the versatility of this approach. Thus, existing switchable display/secretion approaches require subcloning, an extended screening period, or careful handling to maintain the genotype–phenotype linkage.
Here, we present a yeast-based approach to switchable display/secretion that overcomes several of the challenges of existing systems. In our approach, we use amber suppression (Chin et al., 2003; Wang and Wang, 2008) to enable the display of an antibody-like protein on the yeast surface only when the media is supplemented with a noncanonical amino acid (ncAA). Thus, the ‘switch’ between display and secretion is mediated entirely by medium supplementation with a small molecule. Model selections in the switchable format remain efficient, and a naïve antibody library constructed in the format yields renewable binding reagents to multiple targets with affinities in the nanomolar range. This switchable screening format provides a rapid approach to the generation of soluble reagents for immediate downstream applications.
Materials and methods
Media preparation, plate preparation and yeast electroporation
Most procedures for preparing media, plates and electrocompetent yeast used here have been described previously (Van Deventer and Wittrup, 2014). Yeast minimal media prepared with individual amino acids (SD-SCAA and SG-SCAA) used the same buffer salts as conventionally prepared SD-CAA and SG-CAA and concentrations of individual amino acids described in the ‘SD-2XSCAA’ recipes used by Wittrup and Benig (1994). Unless otherwise noted, all SD-SCAA and SG-SCAA media used here were prepared without tryptophan (TRP), leucine (LEU) or uracil (URA).
Yeast strain construction
The strain RJY100 was constructed in order to eliminate expression of the amber codon-containing Trp1/YDR007W gene product coded in EBY100; amber suppression of this gene product would cause a loss of TRP sensitivity during amber suppression. Genomic DNA was isolated (Davis et al., 1980; Hoffman and Winston, 1987) from a TRP1 knockout strain and used as a template for PCR amplification of the TRP1 locus; this knockout encodes a selectable kanamycin resistance cassette in place of the TRP1 gene. The gel-purified PCR product was used to transform electrocompetent EBY100. Transformed yeast were diluted 1:1000 into YPD containing the kanamycin analog G418 (1000 μg/ml) and grown to saturation at 30°C. The cultures were diluted 1:1000 and plated on SD-SCAA solid media lacking histidine but containing TRP and LEU. Five colonies were used to inoculate YPD media containing G418 and grown to saturation. To confirm the complete knockout of TRP1, the candidate knockout strains were inoculated in SD-CAA; no growth was observed. Isolated genomic DNA was used as the basis for PCR assays confirming the presence of the properly sized insert and the kanamycin resistance marker. One of the strains exhibiting the proper genomic and phenotypic characteristics was termed RJY100, stocked in 15% glycerol at −80°C, and used for all subsequent experiments.
Plasmid construction
To construct a suppression plasmid containing a LEU marker, the SacI–PstI fragment of pSNR-OmeRS (Wang and Wang, 2008) encoding a constitutively expressed variant of an Escherichia coli tyrosyl-tRNA synthetase (TyrRS) that aminoacylates O-methyl-l-tyrosine (OmeY) and a constitutively expressed E. coli tyrosyl tRNACUA was ligated into the similarly digested LEU marker plasmid pRS315 and transformed into competent E. coli. Transformants capable of growing on LB plates supplemented with ampicillin were sequence verified, resulting in the plasmid pRS315-OmeRS. All sequencing described here was performed by the Koch Institute Biopolymers Facility or by Macrogen.
Plasmids encoding antibody-like scFv-Fcs (Shu and Qi, 1993; Wu et al., 2001), fused at their C-termini to the display anchor Aga2p, were assembled in several steps. The CH2 and CH3 domains of the human IgG1 antibody 4m5.3 (Sazinsky et al., 2008) with codon optimization for improved expression in yeast were amplified in 5′ and 3′ pieces in order to eliminate an internal BamHI sequence, with the 3′ fragment being amplified to encode either a CAG codon or TAG codon at the end of the constant region. After gel purification, the 5′ and 3′ fragments were assembled, amplified and extended with PCR, and subsequently gel purified. These inserts and the vector pCHA4GBamHI → NcoI (Mata-Fink et al., 2013) were doubly digested with the enzymes BamHI and NheI. After gel purifying the vector backbone and column purifying the inserts, the backbone and inserts were ligated, transformed into chemically competent XL1 blue E. coli, rescued, and plated on LB plates containing ampicillin. Colonies were picked and grown in liquid LB media containing ampicillin in order to enable plasmid extraction. The resulting plasmids were sequenced, and plasmids with the correct sequences were named pCHA-FcSup-CAG and pCHA-FcSup-TAG.
The scFvs used in model selection experiments were isolated from a conventional yeast display library with a pCTCON2 backbone (Chao et al., 2006) containing the same antibody diversity used here (see below) and standard isolation approaches. These scFvs, recognizing human fibroblast activation protein (hFAP) and lysozyme, were used in the pCTCON2 backbone (conventional format) or PCR amplified and homologously recombined with linearized pCHA-FcSup-CAG or pCHA-FcSup-TAG (linearized as described below) to create constructs in the scFv-Fc-Aga2p or scFv-Fc-TAG-Aga2p formats, respectively. Sequencing (Koch Institute Biopolymers Facility) was used to confirm the correct recombination of all of these constructs.
Phage library construction and characterization
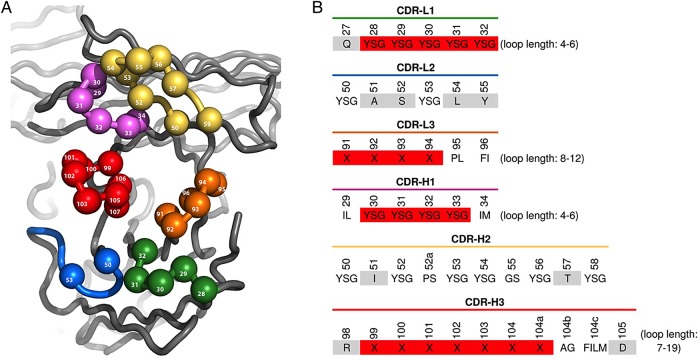
The phage-displayed scFv library ‘G’ was constructed using methods similar to the those used for the construction of previously described synthetic Fab libraries ‘D’ and ‘F’ (Fellouse et al., 2007; Rajan and Sidhu, 2012; Persson et al., 2013). Briefly, the Fab template was converted to an scFv format that was cloned into an IPTG-inducible phage-mid designed for the secretion and phage display of scFvs fused to the C-terminal domain of the gene-3 minor coat protein. TAA (“ochre”) stop codons were inserted into the regions encoding complementarity determining regions (CDRs) L1, L2, H1 and H2 prior to randomization to prevent display of the parental sequence. These four CDRs were randomized and subjected to two rounds of phage selection with protein A in order to enrich for properly folded and displayed scFvs. The output of these selections was used as a template for introducing diversity into CDRs L3 and H3. The diversity design is described in Fig. 3, and library size was estimated by colony counts. Several hundred individual clones from the naïve library were sequenced in order to validate the phage library construction (Supplementary Fig. S2).
Fig. 3.
Design of scFv-phage library G. (A) Structural representation of CDR diversification. Residue numbering is based on the Fab structure 1FVC (Eigenbrot et al., 1993), and residues that were diversified are shown as spheres. (B) CDR diversity design. At diversified positions, amino acids that were allowed are shown in the single-letter code. ‘X’ indicates the following amino acid mixture: Y (25%); S (20%); G (20%); A (10%); F, W, H, P, V (5% each). Nondiversified positions are shaded light gray. Shading of randomized positions indicates positions that were replaced by random loops of all possible varying lengths, as indicated.
Yeast library construction
Homologous recombination in RJY100 was used to construct a billion member library of synthetic antibodies with a pCHA-FcSup-TAG backbone using diversity from library G. Phage DNA encoding the library was extracted from intact phage using a QIAprep Spin M13 Kit (Qiagen), PCR amplified and gel extracted. The plasmid pCHA-FcSup-TAG was linearized in two steps: first, overnight incubation with NdeI was performed, followed by a second overnight incubation after adding the enzymes NheI and XmaI to the digestion mixture. This digested mixture was combined directly with the gel-purified scFv PCR products and transformed into electrocompetent RJY100 containing pRS315-OmeRS. Cells were prepared for electroporation using previously described methods (Van Deventer and Wittrup, 2014) with the following modifications: (i) prior to the final outgrowth in YPD on the day of electroporation, the RJY100 cells containing the suppressor plasmid were propagated in SD-SCAA lacking LEU and URA (but containing TRP); (ii) each individual electroporation (using the standard amount of 5 × 108 cells) was conducted with 12-µg insert DNA, 4-µg linearized vector and 4-µg additional pRS315-OmeRS (intact vector). Transformed cultures were recovered in SD-SCAA; small portions of transformed cells were plated on SD-SCAA plates to confirm the final reported library size of 1 × 109. The library was expanded in SD-SCAA, pelleted, resuspended in 15% glycerol and stored in aliquots containing 1010 yeast. Library quality was assessed by sequencing a small sampling of the library diversity after DNA isolation with the Zymoprep Yeast Plasmid Miniprep Kit II (Zymo Research), selective removal of the suppression plasmid by digestion with restriction enzymes HpaI and NotIHF (NEB) overnight and transformation into E. coli. All clones sequenced from the naive library were found to be in the expected format. A minority (∼5–10%) of unselected and selected clones have been found to contain diversity not coded by the randomization scheme; this diversity was likely introduced during the construction of the original library or during propagation in phage.
Induction of yeast display
Yeast cultures, single colonies or thawed library aliquots were grown to saturation at 30°C in SD-SCAA and diluted to an OD600 of 1 in fresh media. The diluted cultures were then grown to mid-logarithmic phase (OD600 of 2–5; reached in 4–6 h) at 30°C, pelleted and resuspended to an OD600 of 1 in induction media (SG-SCAA) and incubated at 20°C overnight. In yeast populations containing the pCHA-FcSup-TAG plasmid, induction media were supplemented with 1 mM OmeY (Chem-Impex International) to facilitate read-through of the amber stop codon and display of the scFv-Fc construct.
Antigen production
Fc fusions of hFAP and murine interleukin-2 (bFcIL-2) were produced in-house using transient transfections of suspension-phase HEK293 cells in Freestyle medium (Life Technologies) according to the recommendations of the manufacturer. The murine antibody TA99 was produced from a stably transfected HEK293 cell line. These antigens were purified using protein A columns and buffer exchanged into 1× phosphate-buffered saline, pH 7.4 (PBS; Corning). The biotin acceptor peptide on hFAP was subjected to enzymatic biotinylation with biotin ligase (Avidity). Biotinylated versions of bFcIL-2 and TA99 were prepared using EZ-Link NHS-LC-Biotin (Thermo Scientific) with a target biotinylation level of 1–2 biotins/protein. All antigens were buffer exchanged into PBS after biotinylation.
Flow cytometric analysis and sorting
All flow cytometry experiments reported here were performed using standard protocols (Van Deventer and Wittrup, 2014). Initial experiments to assess display levels utilized simultaneous detection of the N- and C-termini of the constructs via the HA tag (primary label: 1:200 16B12 mouse anti-HA (Covance); secondary label: 1:100 goat anti-mouse Alexa Fluor 488 (Life Technologies)) and c-Myc tag (primary label: 1:250 chicken anti-c-Myc (Gallus); secondary label: 1:100 goat anti-chicken Alexa Fluor 647 (Life Technologies)), respectively. In antigen-binding experiments, the c-Myc tag was detected as described above, and the antigen was detected using appropriate concentrations of antigen during primary labeling and 1:100 goat anti-mouse Alexa Fluor 488 during secondary labeling.
Model selections
Mixtures (1:10 000) of antigen-binding (anti-hFAP) and irrelevant (anti-lysozyme) scFvs displayed in different formats (Aga2p-scFv, scFv-Fc-Aga2p and scFv-Fc-TAG-Aga2p) constructed as described above were used in model selections. Cultures of each construct in the various formats were grown and induced separately, with OD600 values used to enable ‘spiking’ of the antigen-binding cells within a culture of irrelevant antigen binders in the corresponding format. The populations were then washed twice in PBSA (1× PBS + 0.1% (w/v) bovine serum albumin (BSA)), split into three equal subpopulations (2 billion cells/subpopulation), and each added to 10 µl of previously prepared hFAP-coated beads. Incubation, selection and recovery were performed according to previously described methods (Van Deventer and Wittrup, 2014). After the populations reached saturation, they were diluted and induced as described above and subjected to flow cytometric detection of hFAP binding. The fraction of displaying cells binding to hFAP was used to calculate the enrichment; values reported here are the result of triplicate experiments.
Library screening and clone isolation
All sorts were performed using coverages of at least 10 times the expected library or population size using Dynabeads biotin binder beads (Life Technologies). Each round of bead sorting consisted of two negative sorts using TA99-coated beads followed by a positive enrichment against the target of interest. Sorting progress was monitored using flow cytometry and deemed to be completed after five rounds for both hFAP and bFcIL-2. The final populations were zymoprepped, subjected to restriction enzyme digestion as described above to eliminate the suppression plasmid from the DNA prep, transformed into E. coli and plated on solid media containing ampicillin. Colonies were picked, miniprepped and sequenced; individual clones were transformed into RJY100 containing pRS315-OmeRS using the Frozen-EZ Yeast Transformation II Kit (Zymo Research) and plated on selective solid media.
Affinity characterizations on the yeast surface
Individual colonies containing clones recognizing bFcIL-2 were inoculated in 5 ml SD-SCAA cultures, grown to saturation and induced in media containing 1 mM OmeY as described above. To determine antibody affinity, assays were conducted in 96-well plates containing 15 000 cells per well. Induced yeast were incubated with anti-c-Myc antibody (1:1000 dilution) and a concentration of bFcIL-2 ranging from 1 µM to <1 nM overnight with agitation. To ensure that antigen remained in excess when concentrations approached the effective concentration of display antibody, nondisplaying cells were added to reduce the total number of scFv-Fc constructs present (Hackel et al., 2008). After primary incubation, cells were pelleted and washed three times with 200 µl ice-cold PBSA. Secondary labeling was performed for a minimum of 10 min on ice with detection antibodies added to a final dilution of 1:200. C-Myc labeling was performed as described above, and antigen binding was detected via the murine Fc using goat anti-mouse Alexa Fluor 488 secondary (Life Technologies). Cells were pelleted, washed two times in ice-cold PBSA, and analyzed on an LSR II HTS (BD Biosciences) at the Koch Institute Flow Cytometry Core. The normalized median fluorescence intensity as a function of bFcIL-2 concentration was used to establish Kd values for all clones of interest. Values reported here are the result of triplicate measurements.
scFv-Fc secretion and purification
Yeast polyclonal or monoclonal populations were inoculated at 107 cells/ml (OD600 of 1) and grown to an OD600 of 6–8 in 50 ml SD-SCAA overnight at 30°C. The saturated cultures were pelleted and resuspended in 100 ml induction media (YPG + 0.1% (w/v) BSA). To expand polyclonal populations, 0.5 ml of the saturated SD-SCAA culture was used to seed a new 50 ml SD-SCAA culture (100× expansion; 10 000× expansions were generated by repeating the expansion a second time). Cultures in induction media were grown for 4 days at 20°C with shaking. Upon completion of the incubation period, the cultures were pelleted at 10 000 × g for 15 min and the supernatant was filtered using a 0.2 µM filter (Thermo). The pH of the filtrate was adjusted to pH 7.4 with the addition of 10× PBS, pH 7.4 (Corning) to a final concentration of 1× and passed twice over a pre-equilibrated protein A column containing 1 ml resin (Genscript). Resin containing bound scFv-Fc was washed three times using 10 ml 1× PBS. ScFv-Fc was eluted from the column using 5 ml 100 mM glycine, pH 3.0, followed by immediate neutralization with 500 µl 1 M Tris, pH 8.0. Neutralized eluant was concentrated and buffer exchanged into 1× PBS using centrifugal filtration units (Millipore, 30 kDa molecular weight cut-off).
ScFv-Fc yield was quantified by A280 measurements on a Nanodrop spectrophotometer (Thermo Scientific), and purity was assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using 4–12% gels (Life Technologies). Proteins were visualized using Coomassie SimplyBlue SafeStain (Life Technologies). For analysis of deglycosylated constructs, PNGase F (NEB) was used according to the manufacturer's protocols prior to SDS–PAGE analysis. Protein purity was assessed via densitometry analysis using ImageJ. For western blot analysis, gels were transferred to a nitrocellulose membrane using an iBlot Dry Blotting System (Life Technologies). Membranes were blocked overnight with 5% nonfat dry milk followed by staining for presence of human IgG (primary: 1:500 mouse anti-human IgG Alexa Fluor 488, Life Technologies), c-Myc (primary 1:1000 chicken anti-c-Myc, secondary 1:1000 goat anti-chicken Alexa Fluor 647) or HA (primary 1:1000 16B12 mouse anti-HA, secondary 1:1000 goat anti-mouse Alexa Fluor 488).
Enzyme-linked immunosorbent assay quantitation of secretion
To directly assess secretion yield from clarified supernatant, an enzyme-linked immunosorbent assay (ELISA) was used with anti-HA capture and anti-human Fc secondary detection. Plates were nonspecifically coated with mouse anti-HA antibody (Clone 16B12, Covance, 1:50 dilution) overnight followed by blocking with 5% nonfat dry milk and subsequent incubation with a 100× dilution of clarified supernatant for 1 h. Plates were washed, incubated with goat anti-human Fc horse radish peroxidase conjugate (Sigma, 1:100 000 dilution) for 1 h, and binding was detected after incubation with 1-Step Ultra TMB substrate (Life Technologies).
Affinity characterizations on the mammalian cell surface
All scFv-Fc polyclonal populations (original, 100× expanded and 10 000× expanded) or individual clones recognizing hFAP were titrated on the surface of cells from the CCD-18Co cell line (American Type Culture Collection), a human colon-derived fibroblast line that expresses hFAP under tissue culture conditions. Assays were conducted in 96-well plates containing 20 000 trypsinized cells per well in PBSA. Cell suspensions were incubated with scFv-Fc clones of interest at concentrations ranging from 1 µM to <1 nM for 3 h at 25°C, or in PBSA only (secondary control). The cells were then pelleted and washed three times with 200 µl ice-cold PBSA. ScFv-Fc binding was detected by incubating the cells with mouse anti-human Alexa Fluor 647-labeled secondary antibody (Life Technologies) added to a final concentration of 1:200 in ice-cold PBSA for a minimum of 10 min. Cells were pelleted, washed two times in ice-cold PBSA, and analyzed on an LSR II HTS at the Koch Institute Flow Cytometry Core. The normalized median fluorescence intensity as a function of scFv-Fc concentration was used to establish Kd values for all clones and populations of interest. Values reported here are the result of triplicate measurements.
Statistics
All errors reported in the current work refer to standard errors of three replicate experiments. For comparison of data between two groups, a two-tailed Student's t-test was performed. Statistical analysis was completed with the aid of Graphpad Prism 6.
Results
Switchable display/secretion platform
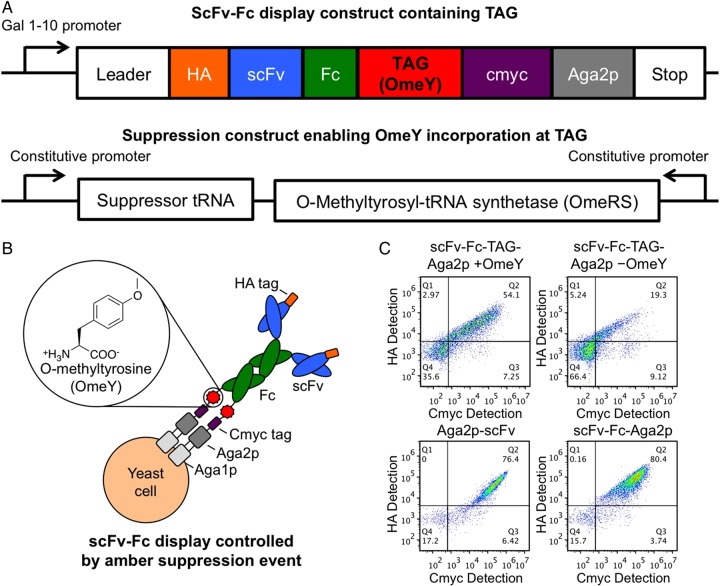
We used a modular, two-plasmid approach to encode a switchable display/secretion system in yeast (Fig. 1A). The display construct encodes a galactose-inducible, HA-tagged antibody-like scFv-Fc upstream of an amber codon, a c-Myc epitope tag and the Aga2p display anchor (Boder and Wittrup, 1997; Chao et al., 2006). The suppression construct encodes a constitutively expressed E. coli tyrosyl tRNA with an amber anticodon and a TyrRS variant (O-methyltyrosyl-tRNA synthetase, OmeRS) that comprise a previously described orthogonal pair (Chin et al., 2003). OmeRS activates the ncAA OmeY, but not any of the 20 canonical amino acids. Figure 1B schematically depicts the expected scFv-Fc display product upon induction of scFv-Fc expression in the presence of OmeY. The ncAA is incorporated at the amber codon, resulting in the display of the full-length scFv-Fc on the yeast surface. Flow cytometry results depicted in Fig. 1C demonstrate the OmeY-mediated control of protein display. Cells encoding the switchable system induced in the presence of OmeY (scFv-Fc-TAG-Aga2p + OmeY) exhibit c-Myc and HA fluorescence levels one to two orders of magnitude above the levels of nondisplaying cells, while cells induced in the absence of OmeY (scFv-Fc-TAG-Aga2p − OmeY) exhibit lower overall fluorescence levels, with a minority of cells exhibiting c-Myc and HA detectable above background (see also Supplementary Table SI; this scFv-Fc display is attributed to aminoacylation of E. coli tyrosyl tRNA with a canonical amino acid). Although a portion of the induced scFv-Fc-TAG-Aga2p − OmeY population displays some scFv-Fc, this aberrant expression will not introduce growth biases (Daugherty et al., 1999) during selections or screens because populations are propagated in an uninduced state and always induced in the presence of OmeY. Display levels of the scFv-Fc-TAG-Aga2p + OmeY population are somewhat reduced compared with the display levels in a conventional display format (Aga2p-scFv) or an scFv-Fc display framework lacking the stop codon of the switchable system (scFv-Fc-Aga2p).
Fig. 1.
Switchable system construction and display levels. (A) Schematic representation of scFv-Fc display construct (scFv-Fc-TAG-Aga2p) and amber suppression construct. (B) Insertion of OmeY in response to the amber stop codon results in the display of full-length scFv-Fcs on the yeast surface. (C) Yeast display levels of switchable (scFv-Fc-TAG-Aga2p) and conventional (Aga2p-scFv (pCTCON2 backbone); scFv-Fc-Aga2p (stop codon from switchable format converted to glutamine) constructs. The identical scFv was used to construct all of the formats interrogated for display levels here. The scFv-Fc-TAG-Aga2p format was assayed for display levels in the presence (+OmeY) and absence (−OmeY) of ncAA in the induction media. The N- and C-termini of the constructs were detected via HA and c-Myc epitope tags, respectively.
Model selections
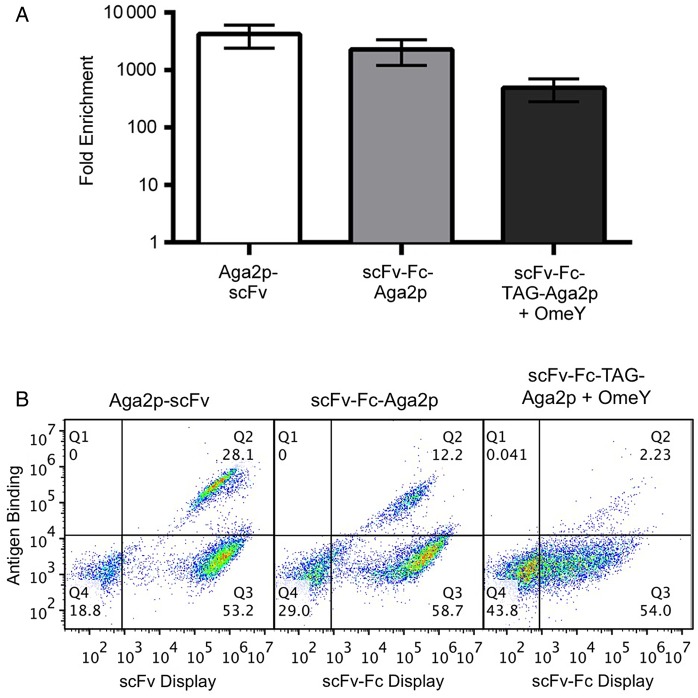
Model selections were used to investigate selection efficiencies in conventional and switchable formats. In these experiments, we employed the exact same antigen-binding and irrelevant scFvs in conventional (Aga2p-scFv), scFv-Fc-Aga2p and switchable (scFv-Fc-TAG-Aga2p) formats. Figure 2 depicts the results of single-pass, bead-based enrichments performed with 1:10 000 mixtures of antigen-binding and irrelevant clones in each format (see Supplementary Fig. S1 and Table SII for raw data). The Aga2p-scFv and scFv-Fc-Aga2p selection outputs were enriched by 4200 ± 1800-fold and 2300 ± 1100-fold, respectively, indicating that these two formats lead to statistically indistinguishable (Student's t-test, P > 0.05) enrichments in this model system. On the other hand, the selection performed in switchable format yielded a 490 ± 210-fold enrichment, statistically lower (Student's t-test, P < 0.05 compared with each other format) than the formats lacking stop codons, although improved considerably over previously reported secretion-and-capture approaches (Rakestraw et al., 2006, 2011), suggesting that this format is suitable for library selections.
Fig. 2.
Model enrichments in Aga2p-scFv, scFv-Fc-Aga2p and scFv-Fc-TAG-Aga2p formats. (A) Enrichments after a single round of bead-based selections. A starting mixture of 1:10 000 antigen-specific: irrelevant binders in the indicated formats was enriched using antigen-coated magnetic beads. Selection outputs were assayed by labeling the recovered and induced populations with antigen. These experiments were performed in triplicate. (B) Representative flow cytometry data assessing the percentage of antigen-binding proteins in enriched populations.
Library construction and screening
A two-step approach was used to construct a high-quality, phage-displayed synthetic scFv library in which diversity was generated in all six CDRs using strategies similar to those published previously (Fellouse et al., 2007; Rajan and Sidhu, 2012; Persson et al., 2013). In the first step, a phage display template containing stop codons in CDRs L1, L2, H1 and H2 was used as template for combinatorial mutagenesis reactions with degenerate oligonucleotides that simultaneously repaired the stop codons and incorporated designed diversity at key positions within these CDRs. Because the heavy chain framework binds to protein A, the resulting phage library was subjected to two rounds of binding selections with this immobilized super-antigen to select for phage particles displaying properly folded scFvs. The resulting enriched phage pool was used as the template for subsequent combinatorial mutagenesis reactions with highly diverse degenerate oligonucleotides designed to mutate CDRs L3 and H3. The resulting library ‘G’ (Fig. 3) contained 1011 unique members. DNA sequencing of several hundred clones confirmed >90% mutagenesis efficiency in all CDRs, and 100% mutagenesis efficiency in CDRs L1, H1 and H2. Moreover, all designed length variations were observed in CDRs L1, L3, H1 and H3, although there was a slight bias in favor of shorter lengths in CDRs L1, L3 and H1 (Supplementary Fig. S2).
Plasmid DNA representing library G was used as the template for PCRs that amplified the scFv open reading frames in a format suitable for transfer into a yeast display vector using previously described methods (Chao et al., 2006; Van Deventer and Wittrup, 2014). Following homologous recombination-based construction in yeast, the yeast display version of library G was estimated to contain 109 unique members. DNA sequencing of individual clones (40 total) confirmed proper assembly of scFv-Fcs (data not shown). To investigate the functionality of the library, we performed screens using Fc fusions of hFAP and murine interleukin-2 (bFcIL-2). In each of these screening campaigns, we were successful in isolating populations of binding proteins using several rounds of magnetic bead-based selections. Characterization of these screen outputs is presented below. Standard flow cytometric enrichments have also proved successful in the switchable format (Supplementary Fig. S3).
Polyclonal reagent production and binding characteristics
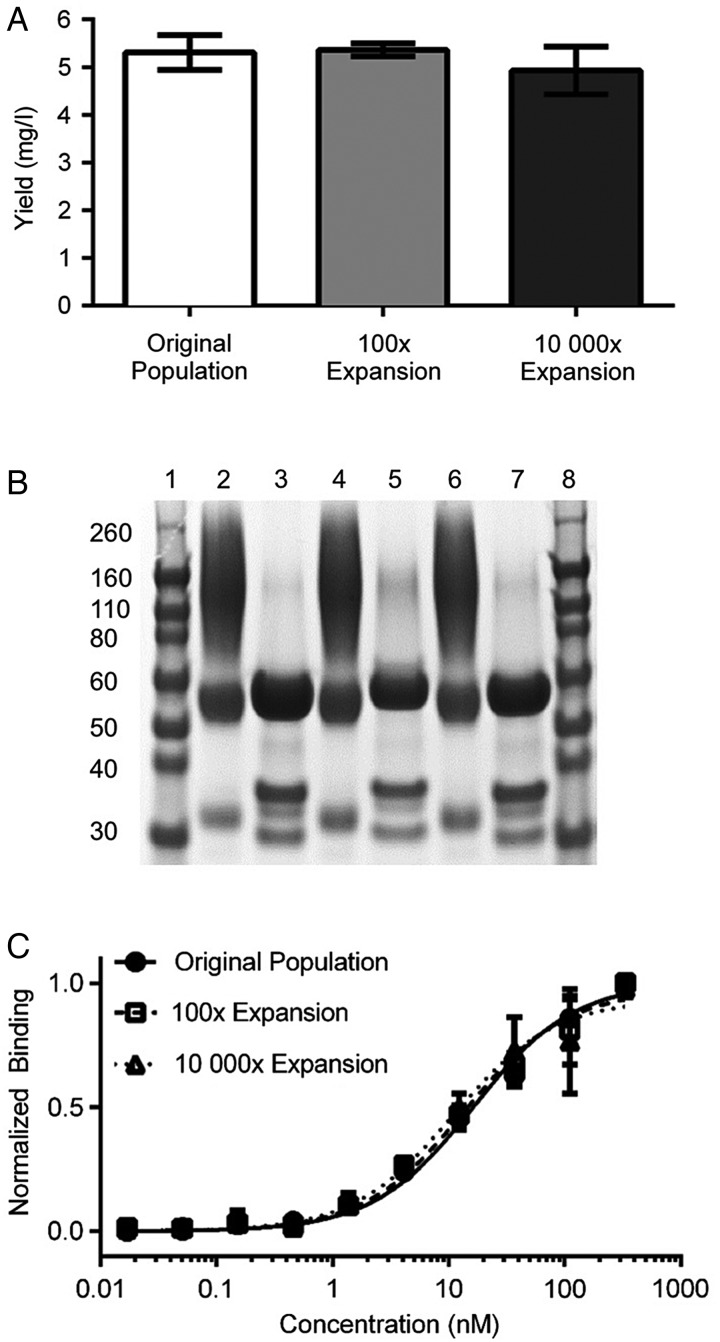
The isolated population of clones recognizing hFAP was assessed for its ability to provide a consistent, renewable source of binding reagents directly after screening. The original, a 100× expansion and a 10 000× expansion of the populations were used to produce soluble scFv-Fcs in yeast culture by simply omitting OmeY from the secretion media. After purification using protein A, the purified polyclonal reagent yields were found to be indistinguishable among the original and expanded populations (Fig. 4A; original: 5.3 ± 0.4 mg/l, 100×: 5.4 ± 0.1 mg/l and 10 000×: 4.9 ± 0.5 mg/l). As noted above, some induced cells display detectable levels of scFv-Fc even in the absence of OmeY. However, this phenomenon is unlikely to affect the yield or clonal diversity of secreted clones. We have observed that the presence of OmeY in the secretion media does not change scFv-Fc secretion yield (Supplementary Fig. S4). Moreover, any growth biases resulting from residual display are anticipated to be minimal, as cells double only three times during the secretion period. SDS–PAGE (Fig. 4B) revealed that scFv-Fc quality remains consistent between original and expanded populations. In unreduced samples (lanes 2, 4 and 6 representing products purified from the original, 100× and 10 000× expanded populations, respectively), the major bands appear at 120 and 55 kDa, representing dimeric and monomeric scFv-Fc protein products, respectively. We attribute the minor band at ∼30 kDa to the product of a cleaved scFv-Fc, with weight corresponding to an scFv. Densitometry indicated that 70 ± 1% of the product is an scFv-Fc dimer, 21 ± 1% is an scFv-Fc monomer and 8 ± 0.4% appears to be an scFv. In reduced, deglycosylated samples (lanes 3, 5 and 7), the major band appears at ∼55 kDa (82 ± 1% purity), consistent with the expected molecular weight of a monomeric scFv-Fc protein product. Western blot detection of human IgG, the N-terminal HA tag or the C-terminal c-Myc tag (Supplementary Fig. S5) further confirms the identity of the full-length products and 30-kDa fragment. The binding properties of the polyclonal binding populations were assessed using titrations on the CCD-18Co cell line (Fig. 4C), a human fibroblast line that expresses hFAP in culture. The data and associated Kd values (Supplementary Table SIII) indicate that the binding behaviors of each propagated polyclonal population are indistinguishable.
Fig. 4.
Polyclonal anti-hFAP scFv-Fc production, purification and characterization. (A) Expression yields after scFv-Fc purification. The bead-selected scFv-Fc population recognizing hFAP was used for secretion directly (original population) or expanded in selective media 100× or 10 000× prior to induction and purification. (B) SDS–PAGE analysis of purified scFv-Fcs. Lanes 1 and 8: protein ladders. Lanes 2, 4 and 6: original, 100× and 10 000× expanded populations, respectively, loaded under nonreducing conditions. Lanes 3, 5 and 7: original, 100× and 10 000× expanded populations, respectively, loaded after reduction and PNGase F treatment. Gel was stained using Coomassie stain. (C) Titration of the purified original, 100× expanded and 10 000× expanded populations on CCD-18Co cells.
Characteristics of isolated clones
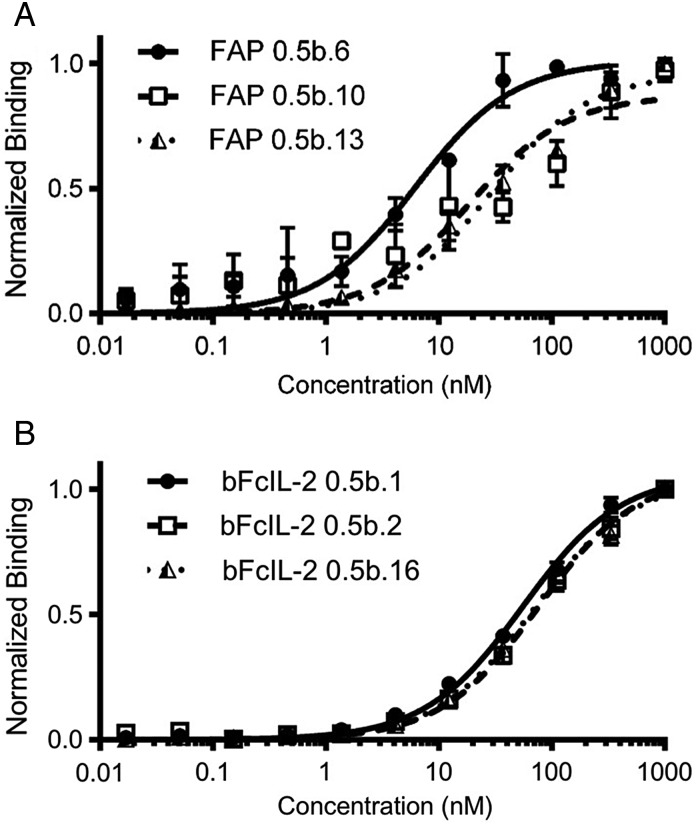
Individual clones recognizing hFAP and bFcIL-2 were isolated, sequenced and assayed in binding titration experiments. Figure 5 depicts a subset of the binding titration curves, and Table I and Supplementary Table SIII summarize sequence and Kd information for all of the unique clones characterized in this work. Clones recognizing hFAP (18 unique) were secreted, protein A purified and titrated on the surface of CCD-18Cos (Fig. 5A); these clones exhibited a range of Kd values from ∼6 nM to >1 μM. ScFv-Fcs recognizing bFcIL-2 (10 unique clones selected for full titration analysis) were displayed on the yeast surface and titrated with soluble antigen (Fig. 5B), exhibiting Kd values ranging from 55 to 138 nM. These Kd values are consistent with values measured in the scFv-Fc-Aga2p display format (Supplementary Fig. S6). Thus, screens of a naïve library in switchable format yielded multiple nanomolar affinity reagents.
Fig. 5.
Titrations of selected clones. (A) Titrations of soluble anti-hFAP scFv-Fcs on the surface of CCD-18Cos. (B) Titrations of anti-bFcIL-2 clones displayed on the surface of yeast with soluble bFcIL-2.
Table I.
Sequences and Kd values for selected clones
| Clone name | CDR sequences |
Kda | |||||
|---|---|---|---|---|---|---|---|
| VL |
VH |
||||||
| L1 | L2 | L3 | H1 | H2 | H3 | ||
| FAP0.5b.6 | QYYSGV | GASYLY | QYSSSLFT | NIYGSSMH | ASISPSYSSTYY | RGGSYSHGGAFD | 6.15 ± 2.28 |
| FAP0.5b.10 | QGSGSV | YASYLY | QSSYSLIT | NLSSGGSYMH | ASIYSYGSGTYY | RSHPSGWYSGFD | 18.29 ± 12.21 |
| FAP0.5b.13 | QYSGSV | SASYLY | QGSALIT | NLGGYYMH | ASIYPGSGGTYY | RGWSSAMD | 28.41 ± 7.29 |
| FcIL-20.5.1 | QYGYYPV | SASYLY | QSSYSLIT | NLGGGYIH | ASISPYYSSTYY | RTVRGSKKPYFSGWAMD | 55.1 ± 5.5 |
| FcIL-20.5.2 | QSYGYSGV | GASYLY | QSSSYSSLIT | NLGYYSMH | AGIYSYGGSTYY | RAGHYSGMD | 76.0 ± 9.1 |
| FcIL-20.5.16 | QGYGSV | GASYLY | QSSYSLIT | NIYGSYIH | ASISSGGSSTYY | RGGWWGASAFD | 68.9 ± 8.5 |
aFAP clones were produced in soluble scFv-Fc form and titrated on the surface of CCD-18Cos. FcIL-2 clones were displayed on the surface of yeast and titrated with soluble bFcIL-2. All values reported here are the result of triplicate experiments and are reported as Kd fit values plus or minus the 95% confidence interval.
Discussion
The switchable display/secretion system described here exploits amber suppression for the purpose of immediately producing soluble reagents at the end of a yeast display screening campaign. The use of a yeast-based system harnesses the advantages of protein expression with eukaryotic expression machinery (Shusta et al., 1999) with minimal recovery times in between screening rounds. Moreover, the amber suppression event that links the scFv-Fc to the cell surface occurs intracellularly, preventing the loss of the genotype–phenotype linkage that can occur with yeast-based secretion-and-capture approaches to switchable system construction. One drawback of the amber suppression-mediated switchable system is the lower display levels observed compared with conventional display. In the model selections performed here, the reduction in display levels reduced enrichment efficiencies by approximately one order of magnitude. Recent findings suggest that aminoacyl-tRNA synthetases with poor catalytic properties are at least partially responsible for the low expression yields observed with suppression methodologies (Guo et al., 2014). The modular implementation of display and secretion used here will facilitate high-throughput screening and identification of improved orthogonal pairs, which in turn should improve the display levels and screening efficiency achievable in switchable format.
Our screening results suggest that the switchable approach will be useful for the rapid generation of polyclonal binding reagents. With hFAP and bFcIL-2, we were able to isolate collections of antigen-specific clones using bead-based enrichments on the benchtop. Our flow cytometric analyses and sorting data with hFAP indicate that fluorescence-activated cell sorting is also efficient in the switchable format. Upon completion of sorting, the soluble expression and protein A purification of scFv-Fcs yields a reasonable 5 mg/l of polyclonal reagent. This amount is sufficient for most routine biological assays such as cell binding, western blots and ELISAs. Moreover, polyclonal yeast cultures can be expanded greatly without loss of productivity or function, facilitating large-scale production of reagents as necessary. Affinity measurements on individual clones indicate that isolated scFv-Fcs exhibit affinities within the nanomolar range, comparable with the binding strengths of many commercially available affinity reagents. Moreover, in cases where polyclonal reagents can be used directly at the end of screening, no cloning steps are required at any point of the discovery or production process, making this entire approach (bead-based screening, polyclonal population amplification, production and purification) more accessible to laboratories without specialized library screening expertise.
Combining yeast display with amber codon suppression enhances the versatility of this already powerful display technique. The integration of display-based screening and soluble reagent production simplifies the process of producing and characterizing both polyclonal and monoclonal affinity reagents, and future improvements will continue to extend the reach of this approach. For example, incorporating fluorescent proteins into displayed/secreted affinity reagents could lead to the end-to-end identification and production of reagents enabling direct, one-step detection of target antigens.
Supplementary data
Funding
This work was supported by seed money from the Koch Institute. J.A.V. was supported by a Ruth L. Kirschstein National Research Service Award [grant number F32CA168057]; R.L.K. was supported by a graduate fellowship from the National Institute of General Medical Sciences Interdepartmental Biotechnology Training Program at the National Institutes of Health [grant number T32 GM008334-25].
Supplementary Material
Acknowledgements
We would like to acknowledge the Koch Institute Flow Cytometry Core for assistance.
References
- Andris-Widhopf J., Rader C., Steinberger P., Fuller R., Barbas C.F. III (2000) J. Immunol. Methods, 242, 159–181. [DOI] [PubMed] [Google Scholar]
- Boder E., Wittrup K. (1997) Nat. Biotechnol., 15, 553–557. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Raeeszadeh-Sarmazdeh M., Price J.V. (2012) Arch. Biochem. Biophys., 526, 99–106. [DOI] [PubMed] [Google Scholar]
- Bowers P.M., Horlick R.A., Kehry M.R. et al. (2014) Methods, 65, 44–56. [DOI] [PubMed] [Google Scholar]
- Bradbury A., Plückthun A. (2015) Nature, 518, 27–29. [DOI] [PubMed] [Google Scholar]
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M., Wittrup K.D. (2006) Nat. Protoc., 1, 755–768. [DOI] [PubMed] [Google Scholar]
- Chin J.W., Cropp T.A., Anderson J.C., Mukherji M., Zhang Z., Schultz P.G. (2003) Science, 301, 964–967. [DOI] [PubMed] [Google Scholar]
- Daugherty P.S., Olsen M.J., Iverson B.L., Georgiou G. (1999) Protein Eng. Des. Sel., 12, 613–621. [DOI] [PubMed] [Google Scholar]
- Davis R., Thomas M., Cameron J. (1980) Methods Enzymol., 65, 404–411. [DOI] [PubMed] [Google Scholar]
- Eigenbrot C., Randal M., Presta L., Carter P., Kossiakoff A.A. (1993) J. Mol. Biol., 229, 969–995. [DOI] [PubMed] [Google Scholar]
- Fellouse F.A., Esaki K., Birtalan S. et al. (2007) J. Mol. Biol., 373, 924–940. [DOI] [PubMed] [Google Scholar]
- Ferrara F., D'Angelo S., Gaiotto T. et al. (2015a) mAbs, 7, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara F., Kim C.Y., Naranjo L.A., Bradbury A.R.M. (2015b) mAbs, 7, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai S.A., Wittrup K.D. (2007) Curr. Opin. Struct. Biol., 17, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L.T., Wang Y.S., Nakamura A. et al. (2014) Proc. Natl Acad. Sci. U.S.A., 111, 16724–16729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B.J., Kapila A., Wittrup K.D. (2008) J. Mol. Biol., 381, 1238–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanly W.C., Artwohl J.E., Bennett B.T. (1995) ILAR J., 37, 93–118. [DOI] [PubMed] [Google Scholar]
- Hoffman C.S., Winston F. (1987) Gene, 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Horlick R.A., Macomber J.L., Bowers P.M., Neben T.Y., Tomlinson G.L., Krapf I.P., Dalton J.L., Verdino P., King D.J. (2013) J. Biol. Chem., 288, 19861–19869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D., Bowers P., Kehry M., Horlick R. (2014) Curr. Drug Discov. Technol., 11, 56–64. [DOI] [PubMed] [Google Scholar]
- Langone J.J. (1986) Immunochemical Techniques Part I: Hybridoma Technology and Monoclonal Antibodies. Academic Press, San Diego. [PubMed] [Google Scholar]
- Lipovsek D., Plückthun A. (2004) J. Immunol. Methods, 290, 51–67. [DOI] [PubMed] [Google Scholar]
- Mata-Fink J., Kriegsman B., Yu H.X., Zhu H., Hanson M.C., Irvine D.J., Wittrup K.D. (2013) J. Mol. Biol., 425, 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H., Ye W., Wernimont A., Adams J.J., Koide A., Koide S., Lam R., Sidhu S.S. (2013) J. Mol. Biol., 425, 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan S., Sidhu S.S. (2012) In Wittrup K.D., Verdine G.L. (eds), Protein Engineering for Therapeutics. Elsevier Inc., San Diego, CA, 3–23. [Google Scholar]
- Rakestraw J.A., Baskaran A.R., Wittrup K.D. (2006) Biotechnol. Prog., 22, 1200–1208. [DOI] [PubMed] [Google Scholar]
- Rakestraw J.A., Aird D., Aha P.M., Baynes B.M., Lipovsek D. (2011) Protein Eng. Des. Sel., 24, 525–530. [DOI] [PubMed] [Google Scholar]
- Sazinsky S.L., Ott R.G., Silver N.W., Tidor B., Ravetch J.V., Wittrup K.D. (2008) Proc. Natl Acad. Sci. U.S.A., 105, 20167–20172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L., Qi C. (1993) Proc. Natl Acad. Sci. U.S.A., 90, 7995–7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shusta E.V., Kieke M.C., Parke E., Kranz D.M., Wittrup K.D. (1999) J. Mol. Biol., 292, 949–956. [DOI] [PubMed] [Google Scholar]
- Van Deventer J.A., Wittrup K.D. (2014) In Ossipow V., Fischer N. (eds), Monoclonal Antibodies. Humana Press, Totowa, NJ, 151–181. [Google Scholar]
- Wang Q., Wang L. (2008) J. Am. Chem. Soc., 130, 6066–6067. [DOI] [PubMed] [Google Scholar]
- Winter G., Griffiths A., Hawkins R., Hoogenboom H. (1994) Annu. Rev. Immunol., 12, 433–455. [DOI] [PubMed] [Google Scholar]
- Wittrup K., Benig V. (1994) Biotechnol. Tech., 8, 161–166. [Google Scholar]
- Wu A.M., Tan G.J., Sherman M.A., Clarke P., Olafsen T., Forman S.J., Raubitschek A.A. (2001) Protein Eng. Des. Sel., 14, 1025–1033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.