Abstract
The plant WRKY gene family represents an ancient and complex class of zinc-finger transcription factors (TFs) that are involved in the regulation of various physiological processes, such as development and senescence, and in plant response to many biotic and abiotic stresses. Despite the growing number of studies on the genomic organisation of WRKY gene family in different species, little information is available about this family in grapevine (Vitis vinifera L.). In the present study, a total number of 59 putative grapevine WRKY transcription factors (VvWRKYs) were identified based on the analysis of various genomic and proteomic grapevine databases. According to their structural and phylogentic features, the identified grapevine WRKY transcription factors were classified into three main groups. In order to shed light into their regulatory roles in growth and development as well as in response to biotic and abiotic stress in grapevine, the VvWRKYs expression profiles were examined in publicly available microarray data. Bioinformatics analysis of these data revealed distinct temporal and spatial expression patterns of VvWRKYs in various tissues, organs and developmental stages, as well as in response to biotic and abiotic stresses. To also extend our analysis to situations not covered by the arrays and to validate our results, the expression profiles of selected VvWRKYs in response to drought stress, Erysiphe necator (powdery mildew) infection, and hormone treatments (salicilic acid and ethylene), were investigated by quantitative real-time reverse transcription PCR (qRT-PCR). The present study provides a foundation for further comparative genomics and functional studies of this important class of transcriptional regulators in grapevine.
Introduction
Plants have evolved unique strategies to adapt to and cope with biotic and abiotic stresses due to their inability to escape predation or adverse environmental conditions. Plant response to these stresses is mediated by a complex signal transduction network that results in changes in biochemistry, physiology and morphology.1–4 These adaptive responses are associated with distinct changes in gene expression, mediated by the action of transcription factors. 5–7
WRKY proteins represent an important class of transcriptional regulators in higher plants and are found troughout the whole green lineage (green algae and land plants).6 The majority of studies on WRKY transcription factors (TFs) showed that numerous members of this multigene family are involved in the response to biotic stresses and are central components of many aspects of the innate plant immune system. Many loss- and gain-of-function studies in Arabidopsis thaliana, which are extensively described in Rushton et al.,8 demonstrated that these regulatory proteins act in a complex defense response network as both positive and negative regulators of the two partly interconnected branches of plant innate immunity: the microbe/pathogen-triggered immunity and the effector-triggered immunity. Pathogen-triggered immunity and effector-triggered immunity activate both local and systemic defence responses, which are mainly modulated by the signalling pathways related to the jasmonic acid (JA),9,10 generally associated to necrotrophic pathogens, chewing insects attack and salicilic acid (SA), more related to defenses triggered by biotrophic pathogenes. JA and SA signalling often act antagonistically, but synergisms between these two phytohormones have also been observed.11 In Arabidopsis, many WRKY TFs appear to be involved in fine regulation of the balance between SA- and JA-dependent defense pathways. For example, AtWRKY70, a common regulatory component of SA- and JA-dependent defence signalling, mediates the cross-talk between these antagonistic pathways and is a positive regulator of R-gene-mediated resistance and systemic defense responses.12,13 In 2008, Lai et al.14 demonstrated that two closely related WRKY TFs, AtWRKY3 and AtWRKY4, play positive role in plant resistance toward necrotrophic pathogens, as Atwrky4, Arwrky3 and Atwrky3 wrky4 mutants showed increased susceptibility toward the fungus B. cinerea, wheras the AtWRKY4 overexpression enanched the plant susceptibility to the biotrophic bacterium Psudomonas siringae. Many other WRKY TFs were found to act as negative regulators of defence signalling, including AtWRKY7, −38, −48 and −62.15–18 The role and complexity of WRKY TFs in plant defense signalling have been demonstrated not only in Arabidopsis, but also in other plants, such as rice (Oryza sativa),19,20 barley (Hordeum vulgare)21 and tobacco (Nicotiana attenuata).22 In rice, a recent study reavealed that the pair of allelic genes OsWRKY45-1 and OsWRKY45-2, which encode proteins with a 10-amino acid difference, play opposite roles in rice resistance against bacterial pathogens.19 Despite both alleles positively regulate resistance to the rice fungal pathogen Magnaporte grisea, they differentially regulate resistance to Xanthomonas oryzae since the former appears to be a negative regulator of the defense response, whereas the latter acts as a positive regulator. Again, the opposite roles of the two allelic genes seem to be due to their mediation of different JA and SA signalling defense pathways.
Altought research on the role of WRKY TFs in plant response to abiotic stresses has lagged behind that on biotic stresses, a growing number of evidences are unrevealing the important role of WRKYs in the regulation of abiotic stress-induced pathways. Many examples illustrates that WRKY TFs form part of signalling processes associated with transcriptional reprogramming when plant encounters high salt, heat, osmotic stress, high CO2 levels, high ozone concentrations, cold or drought.19,23–25 Moreover, recently, scientists have focussed on additional roles of WRKY factors in other important plant processes, such as seed dormancy and germination,24,26,27 plant development28,29 and leaf senescence.30–33
One of the defining features of WRKY TFs is their DNA binding region, also called WRKY domain, that consists of about 60 amino acids,34,35 characterized by the highly conserved WRKY signature at the N-terminus adjacent to an atypical, either Cx4–5Cx22–23HXH or Cx7Cx23HXC, zinc-finger motif at the C-terminus.8 WRKY genes have been classified into three major groups based on the number of WRKY domains present.35 Group I members are characterized by two WRKY domains containing a C2H2 zinc-finger motif. Group II WRKY genes contain only one WRKY domain, characterized by a C2H2 zinc-finger motif. This group has been further divided into five subgroups (II-a, b, c, d and e, respectively). Group III consists of a small number of genes characterized by a single WRKY domain with a C2HC zinc-finger motif. In a few WRKY proteins, the WRKY amino-acid sequences have been replaced by WRRY, WSKY, WKRY, WVKY or WKKY.36 The conservation of the WRKY domain is mirrored by a remarkable conservation of its cognate binding site, the W box (TTGACC/T).34,35 Both bioinformatic-based and functional studies of plant promoters have found clusters of W boxes in stress-inducible promoters.37 In some cases, the multiple W boxes appear to have a synergistic effect on transcriprion, as observed in parsley (Petroselinum crispum).5,38
Grapevine (Vitis vinifera L.) is a major economic crop species with world production approaching 70 million tonnes and a harvest area of over seven million hectares. It is cultivated for table grapes, juice, raisins and wine production.39 Over the course of the last few decades, there has been a great increase in the economic competitiveness of grape production. The competitive challenge is associated with the need to maintain quality and productivity, and the need to adapt production to new geographical areas. Climate changes, the advent of new diseases and increasing market demand have led to a remarkable increase in studies related to the of physiology and pathology studies of this species.
Only six individual WRKYs have been characterized in grapevine thus far. Among them, VvWRKY1 appears to be involved in plant response to downy mildew (Plasmopara viticola) infection through a JA-mediated transcriptional reprogramming.40 VvWRKY2 plays a role in regulating lignification, possibly in response to biotic or abiotic stresses,41 and VvWRKY11 has an important role in the response to water stress.42 The other three WRKYs, VpWRKY1, VpWRKY2 and VpWRKY3, were cloned from a wild Chinese species, Vitis pseudoreticulata W. T. Wang ‘Baihe-35-1’, and all of them have been shown to be involved in the resistance to both biotic and abiotic stresses.43
The availability of the genome sequences of a highly homozygous clone PN40024 of Pinot Noir44 and the cultivated Pinot Noir clone ENTAV 11545 represents a strong foundation for a vast range of genetic and functional studies of entire gene families. In the present study, a total of 59 VvWRKY genes were identified and manually annotated from the grapevine genome. Detailed analysis, including gene classification, annotation, phylogenetic evaluation, conserved motif determination and expression profiling based on previously published microarray data were performed on all members of the family. Additionally, the expression patterns of Group II-a and III WRKY transcription factors (VvWRKYs) in response to abiotic and biotic stresses were further investigated using quantitative real-time reverse transcription PCR (qRT-PCR). Our results provide a foundation for further comparative genomics and functional studies of this important class of transcriptional regulators in grapevine.
Materials and methods
Identification of putative WRKY proteins in grapevine
To identify a complete list of grapevine WRKY genes, annotated grapevine proteins were downloaded from three public databases: the National Centre for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), the Grapevine Genome Browser (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/) and the Grapevine Genome CRIBI Biotech website (http://genomes.cribi.unipd.it/). Putative grapevine WRKY proteins were identified as described by Wu et al.46 The HMM profile of the WRKY DNA-binding domain (PF03106) was downloaded from the Pfam protein families database (http://pfam.janelia.org) and used to survey all grapevine proteins in the 12× coverage assembly of the V. vinifera PN40024 genome. All non-redundant WRKY protein sequences were considered and used in further analysis. Length of sequences, molecular weights and isoelectric points of deduced polypeptides were calculated by using tools provided at the ExPasy website (http://web.expasy.org/protparam/). Finally, manual annotation was performed to resolve any discrepancy between incorrectly predicted genes and the actual chromosomal locations of involved genes in question. The Arabidopsis WRKY gene family database was downloaded from the Database of Arabidopsis Transcription Factorshttp://datf.cbi.pku.edu.cn/) and used for comparative analysis.
Phylogenetic analysis and classification of the grapevine WRKY family
All identified grapevine WRKY genes (VvWRKYs) were classified into different groups based on the AtWRKY classification scheme and the alignment of VvWRKY and AtWRKY DNA-binding domains using Clustal X 2.147 with default settings. The phylogenetic trees were inferred using MEGA 5.048 with the neighbour-joining method. Bootstrap values were calculated for 1000 iterations.
Chromosomal location of VvWRKYs
All VvWRKY genes were mapped to grapevine chromosomes based on information available at the Grape Genome CRIBI website (http://genomes.cribi.unipd.it/). The map was drafted using MapInspect software (http://www.plantbreeding.wur.nl/uk/software-mapinspect.html). Tandem duplicated VvWRKY genes in the grape genome were identified by checking their physical locations on individual chromosomes and were defined as adjacent paralogous on a grape chromosome, with no more than one intervening gene.49 MCScanX software (http://chibba.pgml.uga.edu/mcscan2/) was used to detect the gene duplication events, with the E-value set below 1×10−5. Physical chromosomal locations were graphically represented by scaling the 19 chromosomes.
Analysis and distribution of conserved motifs and exon–intron structures
The exon–intron organisation of VvWRKY genes was determined by comparing predicted coding sequences with their corresponding genomic sequences using the GSDS software (http://gsds.cbi.pku.edu.cn). The Fancy Gene online tool (http://bio.ieo.eu/fancygene/) was used to illustrate the exon–intron structure of VvWRKYs.50 The Expectation Maximization for Motif Elicitation online program (http://meme.sdsc.edu/meme/intro.html)51 for DNA and protein sequence analysis was used to identify conserved motifs in the 59 VvWRKY-deduced proteins. The optimized parameters of Expectation Maximization for Motif Elicitation were set as follows: the number of repetitions, any; the maximum number of motifs, 50; and the optimum width of each motif, between 6 and 300 residues. The online program 2ZIP (http://2zip.molgen.mpg.de/index.html) was used for predicting Leu zipper motifs, while HARF, LXXLL and LXLXLX motifs were identified by manual inspection.
Microarray data analysis
The expression profiles of VvWRKY genes predicted from an analysis of the grapevine genome releases was analysed in a V. vinifera cv ‘Corvina’ (clone 48) gene expression atlas of different organs at various developmental stages. Microarray data were downloaded from Gene Expression Omnibus under the series entry GSE36128 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lfcrxesyci qgsjoandacc=GSE36128).52 Expression analyses in response to abiotic and biotic stresses were based on microarray data (series matrix accession numbers GSE31594, GSE31677, GSE6404, GSE12842 and GSE31660) downloaded from the NCBI gene expression omnibus (GEO) datasets. Data were analysed and graphically represented using the mean of Multi Experiment Viewer software.53 Apart from data obtained from the V. vinifera cv. ‘Corvina’ expression atlas, which were normalized based on the mean expression value of each gene in all tissues/organs analysed, all other expression data were calculated as log2 fold change in treated vs. untreated samples.
Plant material and treatments
In vitro grapevine plants (V. vinifera, genotype PN40024) were kindly provided by Dr Anne-Françoise Adam-Blondon (INRA, Castanet Tolosan cedex France) and maintained in vitro on half-strength MS medium supplemented with 0.3 mg L−1 indole 3-butyric acid (Sigma Chemical Co., St. Louis, MO, USA), at 25 °C in a culture room under 16 h light/8 h dark photoperiod cycle. To validate VvWRKYs microarray expression data obtained from the Corvina expression atlas in different tissues and developmental stages,52 three biological replicates of roots, stems, leaves and shoot tips, were independently sampled from 4-week-old in vitro subcultured explants. To validate VvWRKYs microarray data in response to various stresses, Group II-a and Group III VvWRKY genes were selected for analysis using qRT-PCR. For drought stress treatment, five-week old in vitro PN40024 plants were transplanted into pots and acclimatized in a growth incubator until about 40 cm tall with 14 leaves at 23±2 °C, a 16/8 h dark photoperiod, and 70%–80% relative humidity. Plants grown in pots were first kept well-watered and then water was withheld to impose a water stress. The sixth leaves were harvested at 0, 4, 8 and 12 days after water was withheld and immediately frozen in liquid nitrogen and stored at −70 °C until further analysis was conducted. Control plants were maintained in well-watered conditions. For pathogen challenge experiments, a local strain of E. necator (powdery mildew) was maintained on ‘Pinot Noir’ plants in a greenhouse. Similar-sized E. necator-infected leaves were pressed onto in vitro leaves of PN40024 for inoculation. Leaves were collected at 0, 4, 12 and 24 h after inoculation. All treated tissue samples were immediately frozen in liquid nitrogen and stored at −70 °C for subsequent analysis. Pressing similar-sized, non-infected leaves onto in vitro leaves of PN40024 served as a mock control. Five-week-old in vitro grapevine plants were used for the following hormone treatments. In vitro plants were sprayed with 5 mM SA or 0.5 g L−1 ET and leaves were sampled at 0, 4, 12 and 24 h after treatment. Treatment with deionized water served as a control. Three biological replicates were used in all the described experiments.
qRT-PCR
qRT-PCR was conducted on an ABI 7300 Real Time PCR System (Applied Biosystems, Foster City, CA USA) using the SYBR-Green (Takara, Dalian, China) method. The oligonucleotide primers were designed based on the identified 3′-untranslated region and the 3′ terminal sequences of the predicted coding region using Beacon designer software (version 2) and were listed in Supplementary Table S8. Primers were checked using BLAST tool of NCBI and dissociation curve was analysed after the PCR reaction for their specificity. In addition, the specificity of PCR products was verified by cloning the relative amplicons in the pMD19-T Vector (Takara), sequencing and aligning them onto the reference genome. Each reaction was carried out with a volume of 20 μl, which contained 10 μL SYBR, 8.6 μL ddH2O, 1 μL diluted template (1 μL of the generated first-strand cDNA diluted by 9 μL ddH2O) and 0.2 μL of each of two gene specific primers. The following program was used conditions: 94 °C for 30 s (pre-denaturation) followed by 40 cycles at 94 °C for 30 s (denaturation), 60 °C for 20 s (primer annealing) and 72 °C for 43 s (extension and gathering the fluorescent signal). At the end, the melting curve analysis was executed for verifying the specificity of the primer with the following program: 95 °C/15 s, 60 °C/1 min and 95 °C/15 s. The baseline and threshold cycles (Ct) were automatically determined by the own software of the system. Three technical replicates were taken in each biological replicate. The actin-101-like gene (VIT_12S0178g00200) whose expression levels remained nearly constant under all experimental conditions was used as an internal control. Transcript levels were normalized against the average expression of the actin gene. The relative expression level for all selected genes was calculated using the formula method, where .54
Statistical analyses
Statistical analyses were performed using Dunnett’s two-tailed t-test. Mean values±s.d. of at least three replicates are presented, and significant differences relative to controls are given at *p≤0.05 and **p≤0.01.
Results
Identification and annotation of WRKY genes in the grapevine genome
A total of 59 full-length genes encoding putative WRKY proteins were identified in the grapevine genome. Among them, 58 gene sequences were obtained from the 12X V1 prediction of the PN40024 grape genome sequence which is available at the CRIBI Biotech website (http://genomes.cribi.unipd.it/). One additional gene, corresponding to accession XP_002277383.1 in the NCBI GenBank database, was found in the old version of the PN40024 genotype representing the 8× coverage of the grape genome (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/). A homologous sequence (CAN70150) was also identified in the PN ENTAV 115 grape genome sequence, indicating that this WRKY member is real and was most likely missed in the PN40024 12X V1 prediction of proteins. Three WRKY genes identified as VIT_11s0052g00450, VIT_15s0021g01310 and VIT_06s0004g00230 in the 12X V1 prediction, were found to have truncated coding sequences and manual annotation was performed to correct their sequences. All 59 identified WRKY genes were named as VvWRKY1 to VvWRKY59 based on their chromosome location according to Vannozzi et al.55 The parameters used to describe the VvWRKY proteins are listed in Table 1 and included the deduced protein length, the molecular weight, the isoelectric point, the aliphatic index and the grand average of hydropathicity. The deduced length of VvWRKY proteins ranged from a minimum of 101 amino acids (VvWRKY12) to a maximum of 612 residues (VvWRKY59), whereas the PI ranged from 5.02 (VvWRKY38) to 9.91 (VvWRKY34). This range of variability implies that different VvWRKY proteins might operate in different microenvironments.
Table 1. Grapevine WRKY genes identified in the PN40024.
| Proposed name | PN40024 12× V1 ID | Chr | ORF (aa) | MW (kDa) | pI | Ai | GRAVY | Group | Conserved motif | Domain pattern | Zinc finger |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VvWRKY01 | VIT_01s0011g00720 | 1 | 305 | 33.3585 | 5.62 | 47.67 | −0.882 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY02 | VIT_01s0026g01730 | 1 | 594 | 64.3392 | 6.02 | 57.24 | −0.689 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY03 | VIT_01s0010g03930 | 1 | 189 | 21.2588 | 9.13 | 54.13 | −0.723 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY04 | VIT_02s0025g00420 | 2 | 329 | 36.3666 | 5.71 | 60.52 | −0.602 | IIe | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY05 | VIT_02s0025g01280 | 2 | 342 | 38.5618 | 6.05 | 55.03 | −0.793 | III | WRKYGQK | C–X7–C–X23–H–X–H | C2HC |
| VvWRKY06 | VIT_04s0008g01470 | 4 | 166 | 18.9167 | 5.2 | 48.19 | −1.122 | IIc | WRKYGKK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY07 | VIT_04s0008g05750 | 4 | 261 | 28.8756 | 9.1 | 65.4 | −0.716 | IIa | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY08 | VIT_04s0008g05760 | 4 | 317 | 35.2637 | 8.22 | 73.19 | −0.718 | IIa | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY09 | VIT_04s0008g06600 | 4 | 491 | 54.3129 | 8.88 | 57.8 | −0.837 | I | 2×[WRKYGQK] | C–X4–C–X23–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY10 | VIT_04s0069g00920 | 4 | 338 | 36.5692 | 9.49 | 63.20 | −0.501 | IId | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY11 | VIT_04s0069g00970 | 4 | 156 | 17.4673 | 5.92 | 55.58 | −0.75 | IIc | WRKYGKK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY12 | VIT_04s0069g00980 | 4 | 101 | 12.0977 | 9.8 | 54.95 | −1.067 | IIc | WRKYGKK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY13 | VIT_04s0023g00470 | 4 | 502 | 54.8737 | 7.07 | 57.71 | −0.806 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY14 | VIT_05s0077g00730 | 5 | 309 | 34.4082 | 5.72 | 54.05 | −0.825 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY15 | VIT_06s0004g00230 | 6 | 129 | 14.989 | 8.89 | 67.36 | −0.512 | NG | WKKYGQK | C–X4–C–X23H–X–H | C2H2 |
| VvWRKY16 | VIT_06s0004g07500 | 6 | 603 | 66.2845 | 6.42 | 47.06 | −0.915 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY17 | VIT_07s0141g00680 | 7 | 340 | 38.0411 | 9.84 | 64.24 | −0.826 | IId | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY18 | VIT_07s0005g01520 | 7 | 242 | 27.5485 | 5.86 | 50.7 | −0.933 | IIe | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY19 | VIT_07s0005g01710 | 7 | 302 | 33.7616 | 6.74 | 54.9 | −0.831 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY20 | VIT_07s0005g02570 | 7 | 506 | 54.6601 | 8.03 | 58.32 | −0.646 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY21 | VIT_07s0031g00080 | 7 | 336 | 36.6164 | 9.57 | 63.54 | −0.624 | IId | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY22 | VIT_07s0031g01710 | 7 | 180 | 19.8731 | 6.3 | 54.17 | −0.652 | IIc | WRKYGKK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY23 | VIT_07s0031g01840 | 7 | 226 | 25.7181 | 9.08 | 65.18 | −0.681 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY24 | VIT_08s0058g00690 | 8 | 552 | 61.0009 | 7.29 | 44.71 | −0.895 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY25 | VIT_08s0058g01390 | 8 | 357 | 40.4237 | 5.73 | 62.55 | −0.797 | III | WRKYGQK | C–X7–C–X23–H–X–H | C2HC |
| VvWRKY26 | VIT_08s0040g03070 | 8 | 477 | 52.3037 | 8.84 | 52.91 | 52.91 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY27 | VIT_08s0007g00570 | 8 | 299 | 33.1578 | 5.19 | 61.34 | −0.752 | NG | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY28 | VIT_09s0018g00240 | 9 | 311 | 34.3754 | 8.71 | 65.85 | −0.764 | IIa | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY29 | VIT_10s0116g01200 | 10 | 535 | 57.4599 | 6.04 | 64.67 | −0.57 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY30 | VIT_10s0003g01600 | 10 | 278 | 30.9392 | 5.14 | 42.81 | −0.845 | IIe | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY31 | VIT_10s0003g02810 | 10 | 319 | 35.0008 | 6.76 | 50.78 | −0.782 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY32 | VIT_10s0003g05740 | 10 | 312 | 33.8314 | 7.56 | 51.67 | −0.821 | IIe | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY33 | VIT_11s0037g00150 | 11 | 450 | 53.8146 | 6.12 | 55.91 | −0.822 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY34 | VIT_11s0052g00450 | 11 | 245 | 26.7143 | 9.91 | 68.49 | −0.586 | IId | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY35 | VIT_12s0028g00270 | 12 | 311 | 34.7884 | 6.92 | 42.67 | −0.949 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY36 | XP_002277383.1 | 12 | 244 | 26.3864 | 5.41 | 62.79 | −0.517 | IIe | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY37 | VIT_12s0059g00880 | 12 | 593 | 64.1152 | 6.48 | 60.46 | −0.665 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY38 | VIT_12s0057g00550 | 12 | 403 | 44.3906 | 5.02 | 63.33 | −0.917 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY39 | VIT_12s0055g00340 | 12 | 487 | 53.6434 | 5.21 | 63.92 | −0.823 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY40 | VIT_13s0067g03130 | 13 | 364 | 40.288 | 5.97 | 55.8 | −0.729 | III | WRKYGQK | C–X7–C–X23–H–X–H | C2HC |
| VvWRKY41 | VIT_13s0067g03140 | 13 | 313 | 35.3091 | 5.45 | 60.1 | −0.738 | III | WRKYGQK | C–X7–C–X23–H–X–H | C2HC |
| VvWRKY42 | VIT_14s0081g00560 | 14 | 362 | 41.2639 | 9.68 | 64.59 | −0.801 | IId | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY43 | VIT_14s0068g01770 | 14 | 182 | 20.7873 | 9.41 | 52.42 | −0.884 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY44 | VIT_14s0108g00120 | 14 | 502 | 53.9169 | 6.48 | 64.36 | −0.582 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY45 | VIT_14s0108g01280 | 14 | 527 | 57.4979 | 8.12 | 59.41 | −0.788 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY46 | VIT_15s0021g01310 | 15 | 202 | 22.8743 | 9.13 | 49.16 | −1 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY47 | VIT_15s0046g01140 | 15 | 323 | 36.1682 | 5.14 | 58.24 | −0.622 | III | WRKYGQK | C–X7–C–X23–H–X–H | C2HC |
| VvWRKY48 | VIT_15s0046g02150 | 15 | 201 | 22.6178 | 9.28 | 72.29 | −0.624 | IIc | WRKYGQK | C–X4–C–X23–H–X–H | C2H2 |
| VvWRKY49 | VIT_15s0046g02190 | 15 | 348 | 37.99 | 5.73 | 55.32 | −0.633 | IIe | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY50 | VIT_16s0050g01480 | 16 | 182 | 20.5055 | 5.28 | 48.9 | −1.057 | IIe | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY51 | VIT_16s0050g02510 | 16 | 364 | 40.0434 | 5.45 | 57.66 | −0.704 | III | WRKYGQK | C–X7–C–X23–H–X–H | C2HC |
| VvWRKY52 | VIT_17s0000g01280 | 17 | 151 | 17.7131 | 9.67 | 49.07 | −1.043 | IIc | WRKYGQK | C–X4–C-X23–H–X–H | C2H2 |
| VvWRKY53 | VIT_17s0000g05810 | 17 | 605 | 65.1789 | 7.55 | 52.66 | −0.746 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY54 | VIT_18s0001g10030 | 18 | 319 | 34.7564 | 9.16 | 67.55 | −0.502 | IId | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY55 | VIT_19s0090g00840 | 19 | 551 | 59.2499 | 9.24 | 57.75 | −0.646 | IIb | WRKYGQK | C–X5–C–X23–H–X–H | C2H2 |
| VvWRKY56 | VIT_19s0090g01720 | 19 | 590 | 64.3486 | 6.27 | 65.88 | −0.658 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY57 | VIT_19s0015g01870 | 19 | 573 | 62.016 | 5.04 | 59.76 | −0.709 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| VvWRKY58 | VIT_01s0011g00220 | RND | 502 | 54.6082 | 7.66 | 51.53 | −0.846 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) | |||||||||||
| VvWRKY59 | VIT_00s0463g00010 | UN | 612 | 66.5023 | 6.1 | 65.15 | −0.72 | I | 2×[WRKYGQK] | C–X4–C–X22–H–X–H(N) | C2H2 |
| C–X4–C–X23–H–X–H(C) |
Abbreviations: Ai, aliphatic index; Chr, chromosome numbers; GRAVY, grand average of hydropathicity; MW, molecular weight; NG, no chromosomal group identified; ORF, open reading frame; pI, isoelectric point; RDM, random chromosome; UN, unknown chromosome.
The variants of the conserved WRKYGQK peptide are shown in bold.
Chromosomal distribution and exon–intron organisation of VvWRKY genes
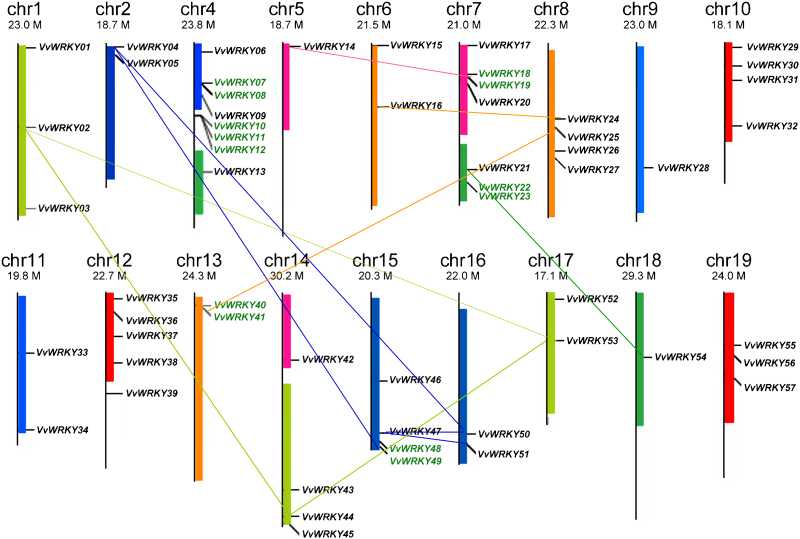
Using TblastN, 57 out of the 59 VvWRKY genes could be mapped to 18 out of 19 grapevine chromosomes (chr), with the exclusion of chr 3, indicating a comprehensive distribution of VvWRKYs within the grapevine genome (Figure 1). VvWRKY58 and 59, which mapped to chr1_random and chrUn, respectively, are not showed. The paralogous chromosome segments resulting from whole-genome duplication or fusion events are illustrated in the same colour as in Jaillon et al.44 Chromosome 4, with eight VvWRKY genes, possesses the largest number followed by chr 7 with six genes.
Figure 1.
Chromosomal distribution of VvWRKY genes. Chromosome numbers are provided at the top of each chromosome together with the approximate size. Paralogous chromosome segments resulting from whole-genome duplication or fusion events are shown in the same colour. Genes belonging to clusters are indicated in green font. The 11 synteny blocks are linked by lines. VvWRKY58 and VvWRKY59, which were not assigned to any chromosome bacause on the PN40024 12X V1 prediction, are not reported in the figure.
According to the definition of a gene cluster provided by Holub,56 13 VvWRKY genes could be placed in six clusters. Among the six clusters, two were found on chromosomes 4 and 7. While only one cluster was located on chromosomes 13 and 15. Tandem and segmental duplications have been suggested to be two of the main causes for gene family expansion in plants.57 We identified two WRKY tandem duplication cluster (VvWRKY11/VvWRKY12 and VvWRKY40/VvWRKY41) on grapevine chromosome 4 (Figure 1). We then examined the synteny block within the grape genome and found that 16 VvWRKY genes were located in 11 pairs of duplicated genome regions (Figure 1). In summary, 20 of 59 VvWRKY genes were associated in either tandem and segmental duplication events.
The exon–intron structure of all the identified VvWRKY genes was analysed in order to gain more insight into the evolution of the WRKY family in grapevine. As results indicated that a large number VvWRKY genes contain two to four introns (Figure 2), twenty-seven VvWRKY genes were found to possess two introns, eight members had three introns and eleven had four introns. Five VvWRKY genes had five introns, one had six introns and seven VvWRKY genes had only one intron. The intron located in the conserved WRKY domain can be classified as either an R-type intron or V-type intron, similar to what has been reported in rice.46 The R-type intron, a phase-2 intron, is spliced prior to an arginine residue (R). However, the V-type intron, a phase-0 intron, is spliced before a valine residue, six amino acids after the second cysteine residue of the zinc finger C2H2 motif46 (Supplementary Fig. S1). The V-type intron was only observed in the WRKY domains of genes belonging to groups II-a and II-b, whereas the R-type intron is extensively distributed in all the other VvWRKY groups (I-C, II-c, II-d, II-e and III) (Supplementary Fig. S1). No introns were found in the sequence encoding the Group I N-terminal WRKY domains.
Figure 2.
Structures of VvWRKY genes. Exon–intron composition of VvWRKY genes. Names of genes are indicated on the left. Exons, represented by black or red boxes, were drawn to scale. Dashed lines connecting two exons represent an intron. Intron phases 0, 1 and 2 are indicated by numbers 0, 1 and 2, respectively. WRKY domains in VvWRKY proteins are marked in red.
Phylogenetic analysis, classification and motifs of the grapevine WRKY gene family
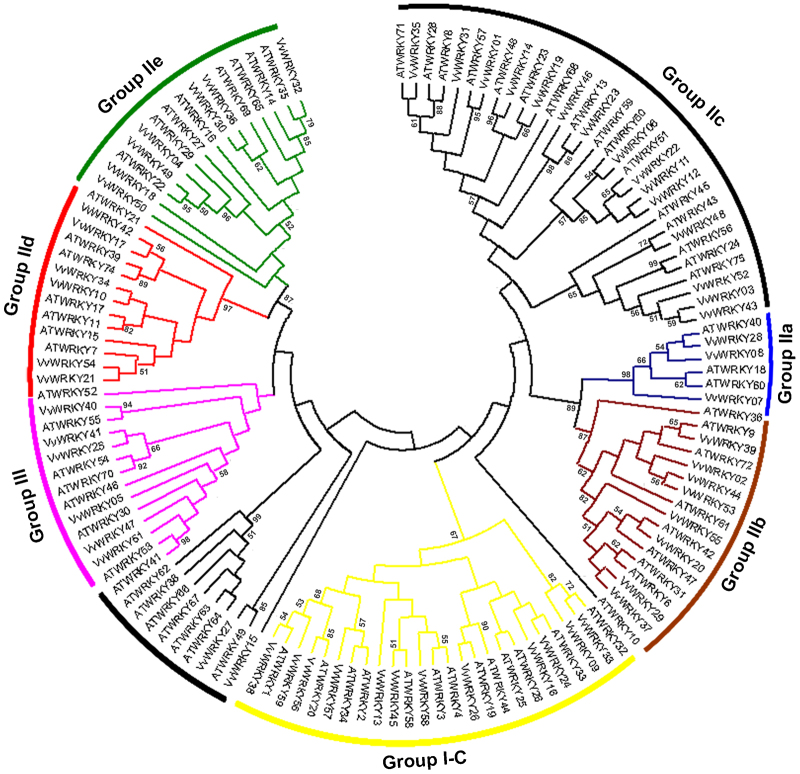
In order to examine the phylogenetic relationship between the predicted WRKY DNA-binding domains in grapevine and Arabidopsis, an unrooted phylogenetic tree was inferred based on the alignment of 131 WRKY domains from the two species (excluding N-terminal domains of Group I) (Figure 3). Based on AtWRKY classification, VvWRKYs were classified into three major groups. Group I is comprised of 12 proteins, each containing two WRKY domains and the C2H2-type zinc-finger motifs (C–X4–C–X22–23–H–X1–H). The only exception within this group is VvWRKY57, which does not possess any zinc-finger motifs at the C-terminus. Thirty-nine VvWRKY proteins possessing a single WRKY domain were assigned to Group II in which the C2H2-type zinc-finger structure is C–X5–C–X23–H–X1–H. Group II was further divided into five subgroups based on their primary amino-acid sequence and was comprised of Groups II-a, -b, -c, -d and -e with 3, 8, 15, 6 and 7 members, respectively. Finally, six VvWRKYs, each with a single WRKY domain, were assigned to Group III, differing from Group II VvWRKYs based on their C2HC zinc-finger structure (C–X4–C–X23–H–X1–C). VvWRKY15 and 27 exhibit sequence divergence in the unique WRKY domain; therefore, they were not classified into any group. The grouping of VvWRKY proteins produced the following pairwise relationships between grapevine and Arabidopsis: VvWRKY24 and AtWRKY33 in Group I; VvWRKY22 and AtWRKY51 in Group II-c; and VvWRKY49 and AtWRKY22 in Group II-e. A strong relationship among some of the grapevine WRKY proteins may imply the occurrence of gene duplication. For example, VvWRKY11 and 12 belong to the same cluster, and VvWRKY44, 53 and 02 were located within a synteny block (Figure 1). A consensus phylogenetic tree obtained by aligning all the deduced grapevine WRKY domains (including both N-terminal and C-terminal domains) is shown in Figure 4, while the details of the alignment of domains are reported in Supplementary Figure S1. Out of a total number of 71 WRKY domains, 54 contain WRKYGQK domains that were perfectly conserved, while the other domains differ in one amino acid in the conserved WRKY signature. The WRKY domain sequence in VvWRKY6, 11, 12 and 22 all belong to the Group II-c. While the WRKY domain sequence in VvWRKY15, which does not fall into one of the three WRKY groups, is WKKYGQK.
Figure 3.
Phylogenetic tree of WRKY domains based on an alignment of grapevine and Arabidopsis. The consensus unrooted phylogenetic tree generated after an alignment of deduced grapevine and Arabidopsis WRKY domains at N-terminus. The phylogenetic tree was generated with ClustalX 2.1 and using the NJ method. The phylogenetic tree was inferred using MEGA 5.0 software. Reliability of the predicted tree was tested using bootstrapping with 1000 replicates. Numbers at the nodes indicate how often the group to the right appeared among bootstrap replicates. Branch lines of subtrees are coloured indicating different WRKY subgroups. NJ, neighbour-joining.
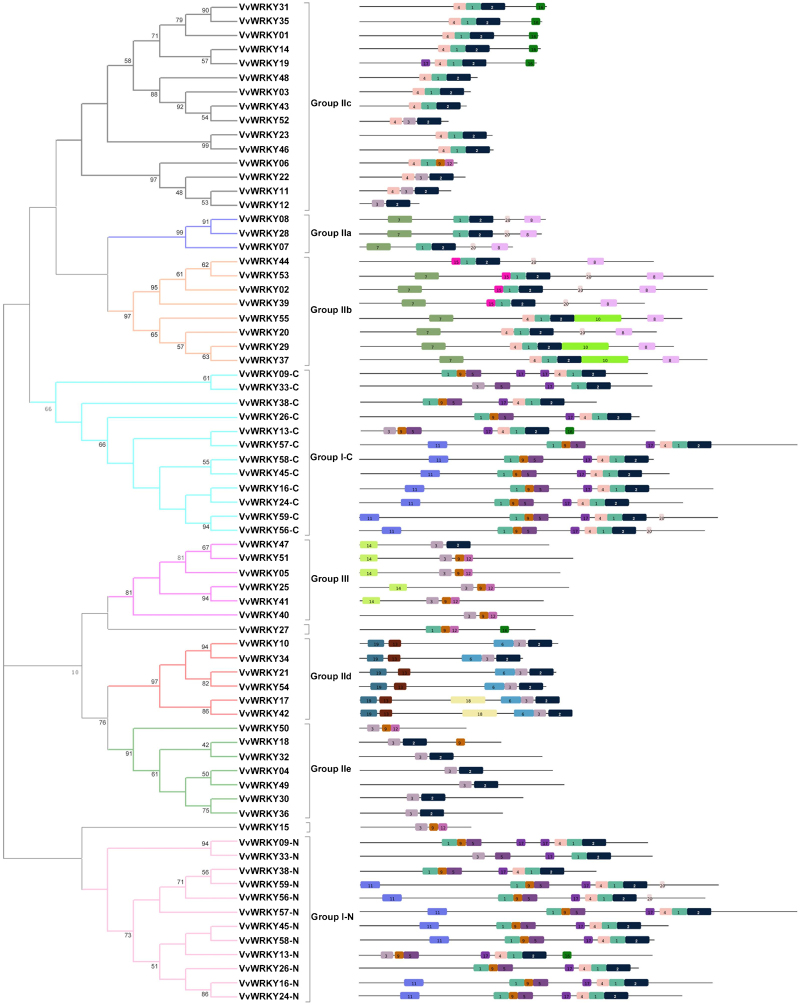
Figure 4.
Phylogenetic tree of deduced VvWRKY domains associated with the motif compositions in the amino-acid sequences. Consensus phylogenetic tree generated after alignment of deduced VvWRKY domains (both N-terminal and C-terminal domains) with ClustalX 2.1 using the NJ method. The phylogenetic trees were constructed using MEGA 5 software. Reliability of the predicted tree was tested using bootstrapping with 1000 replicates. Numbers at the nodes indicate how often the group to the right appeared among bootstrap replicates. Subtrees branch lines are coloured indicating different WRKY subgroups. Group I N-terminal WRKY domains are indicated with the suffix N after the name, whereas Group I C-terminal WRKY domains are indicated with the suffix C. The motif composition related to each VvWRKY protein is displayed on the right-hand side. The motifs, numbered 1–20, are displayed in different coloured boxes. The sequence information for each motif is provided in Table 2. NJ, neighbour-joining.
Conserved motifs other than the WRKY domain were detected manually in the VvWRKY proteins (Supplementary Table S1). A ‘Leucine zipper’ motif, present in Group II-a and II-b WRKY proteins of Arabidopsis and rice,35,36 was detected in VvWRKY07 belonging to Group II-a. Four members (VvWRKY10, 21, 34 and 54) of Group II-d contain the novel HARF (RTGHARFRR [A/G] P) motif, whose function has not been clearly determined. The LxLxLx repressor and the LxxLL co-activator motifs58,59 were found in seven (VvWRKY08, 14, 21, 34, 41, 48 and 54) and ten (VvWRKY02, 07, 08, 15, 25, 34, 39, 44, 53 and 55) VvWRKY proteins, respectively. Interestingly, VvWRKY08 contains both co-activator and active repressor motifs.
Twenty other distinct motifs (Table 2 and Figure 4) were identified by using the Multiple Expectation Maximization for Motif Elicitation online tool (http://meme.sdsc.edu/meme/intro.html). As illustrated in Figure 4, other than motifs 1 and 3 which are the WRKY domains widely distributed in all 59 proteins, VvWRKY members within the same subgroups were generally found to share a common motif composition, as illustrated in Figure 4. For instance, motif 14 is unique to Group III, whereas motifs 13 and 19 are specific to Group II-d. Group II-a and II-b both contain motifs 7 and 8 except for VvWRKY44 which possesses only motifs 8. As observed for the amino-acid motifs detected in VvWRKY deduced proteins, WRKY genes within the same group generally also possess similar exon–intron arrangements.
Table 2. Analysis and distribution of conserved motifs in grapevine VvWRKYs.
| Motif | E-value | Width | Best possible match |
|---|---|---|---|
| 1 | 1.6×10−994 | 26 | DGYRWRKYGQKVVKGNP[FN]PRSYYRCT |
| 2 | 3.8×10−954 | 41 | [SA]AGCP[VA][RK]K[QHR]V[EQ]R[ASC][SA]EDPS[IM][VL][IV][TV]TYEG[EK]HNH[PD]LPAARx[AS]xAS |
| 3 | 1.4×10−359 | 21 | [YFW][SA]WRKYGQKPI[KL]GS[PK][YH]PR[GS]YY |
| 4 | 1.3×10−359 | 21 | E[KR][TK][IV]R[EK]PR[VF][AV][VF]QT[RK]SEVDILD |
| 5 | 3.1×10−105 | 26 | GQ[IV]TEI[IV]YKGTH[ND]HPKPQPN[RK]RS[AKS]LG |
| 6 | 1.0×10−083 | 34 | SGRCHCSK[RK]RKLRVKR[STV][IV][RK]VPAIS[NS]K[IM]ADIP[PA]D[ED] |
| 7 | 2.5×10−083 | 41 | [EK][LTV][EGV][VGS][LA][QKV][AEV]E[LM][EGN][RE][MV][RN][AEK]EN[EKQR][KR]L[RT][ES]ML[ENS][QI][IMV][TCM][KNE][ND]Y[NS][ADT]LQM[HQ][LFV]V[ET][LI]M |
| 8 | 1.4×10−081 | 28 | [TL][VI][EA][AT]AT[AK][AS][IL]T[AS]DP[NS]F[TR]AALAAAI[TS]SIIG |
| 9 | 3.3×10−069 | 15 | HPNCP[AV]KK[KQ]V[EQ]RSH[DE] |
| 10 | 5.9×10−043 | 80 | TTS[AS]AA[NRT]MLLSGSM[PS]S[AS]DG[LI]M[NS][SP][NS]F[LH][AS]RT[IMV][LF]PCS[SP][SN][ML]ATISASAPFPT[VI]TLDLT[HQ][NST]P[NS][PL]LQ[FHY]QRP[NPT][SA]QF[HPY]VP[FS][APQ][NA][PL][APT]Q[NS][FL] |
| 11 | 3.4×10−036 | 33 | [LMN][PRS][PI][SA][KP][LP]P[IV][SA][RP]S[PS][YC][FL][TS]IP[PS]GLSP[ST]SLL[DE]SP[VG][LF][LF]S |
| 12 | 1.5×10−025 | 15 | [DEN]PSM[FY][IEK][ITV]TY[IER]G[DK]HT[CH] |
| 13 | 1.8×10−023 | 21 | [CL][RM][AE][EIV][TA]DE[AT]VSKFK[RK]V[IV]S[LI]LNR |
| 14 | 8.9×10−019 | 30 | [MPW][EDPS][EMNT][AGKMR][LGIR][SEK][WGT][DEQ][QRH][KN][RTN][LAV][IV][NDG]EL[TLV][QHNR]G[RM][EDS][LFM][AT][KN]QL[KQR][INS][HLV]L |
| 15 | 4.7×10−017 | 15 | [AC]RVSVRARC[DQ]T[PAL]TMN |
| 16 | 1.7×10−016 | 15 | [DP][EHY]GLL[QE]D[ILM]VPS[FM][IM]R[KN] |
| 17 | 2.8×10−016 | 15 | [ED][DE][DE][EDG]D[ED]P[ED][SP]KRRKKE |
| 18 | 1.2×10−014— | 59 | QQMK[HY]QAD[MT]MY[RS]RSNSGINL[KN]FD[GS]SSCTPTMSSTRSFISSLS[MV]DGSVANLDGN[AS]FHLIG |
| 19 | 5.0×10−013 | 28 | [MQ]EA[IV][QE]EA[AN]SA[GA][LV][EK]S[CMV]E[KH][LV][IL]RLLS[HQ]xQ[DQ]Q |
| 20 | 2.2×10−012 | 8 | [PD]T[IV][TS]LDL[TG] |
Numbers correspond to the motifs in Figure 4.
The number of VvWRKY genes belonging to each subgroup was compared with the number in other plant species where this gene family has been fully characterized, i.e., Arabidopsis, rice, castor bean, poplar and tomato (Table 3). The number of genes in Group I WRKY family is similar in grapevine, Arabidopsis, rice and tomato, but has undergone a significant expansion in poplar. The numbers of members belonging to the different Group II subclades are also comparable between all the considered species, although an obvious expansion was detected in poplar Group II-d and tomato group II-e WRKYs. Lastly, apart from Castor bean, grapevine showed a lower number of members belonging to Group III if compared to the other species, especially rice, which has the largest number of WRKY proteins in this subfamily.
Table 3. Size of the WRKY groups and sub-groups in different plant species.
VvWRKYs expression in different organs, tissues and developmental stages
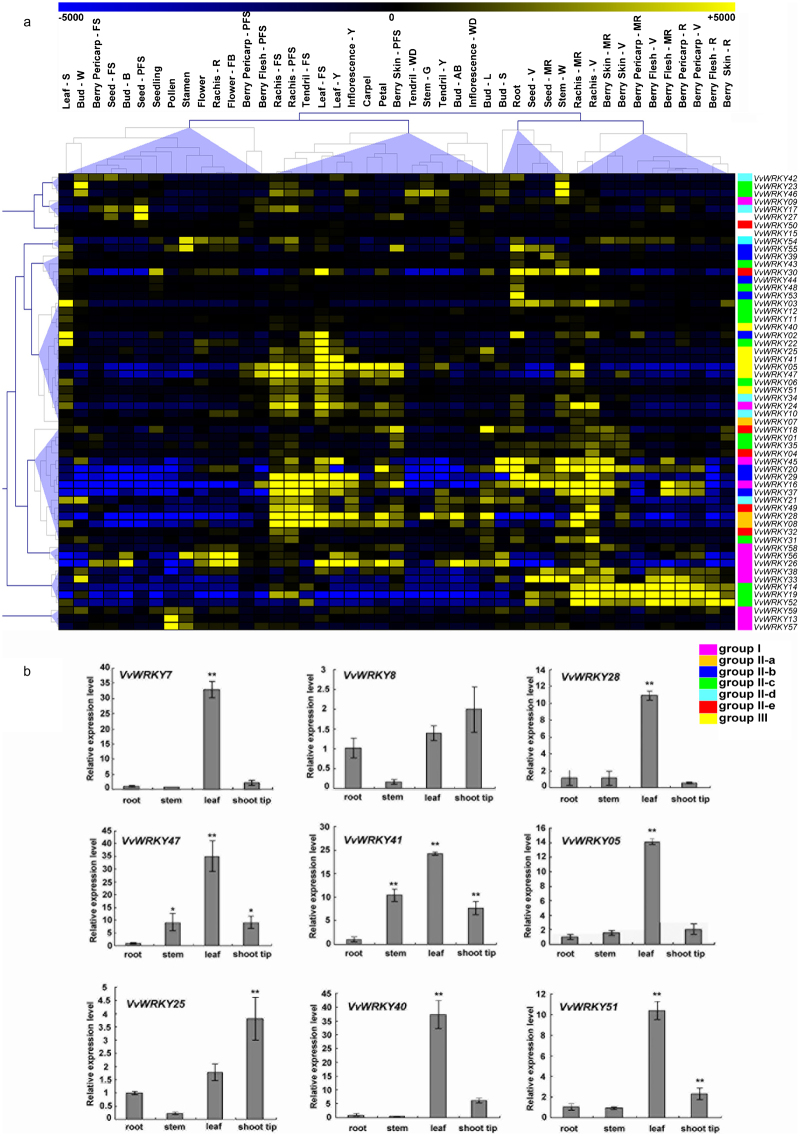
The expression patterns of VvWRKY genes were analysed in the V. vinifera cv. Corvina global gene expression atlas, which consists of 45 different organs/tissues at various developmental stages obtained by microarray analysis (Supplementary Table S2).52 All VvWRKY genes, except VvWRKY36, had corresponding probes on the NimbleGen array. Figure 5a presents a graphical representation of the expression pattern of each VvWRKY gene in which both genes and samples were ordered based on a hierarchical clustering analysis.
Figure 5.
Expression profiles of the grapevine VvWRKY genes in different grapevine organs, tissues and developmental stages. (a) Expression of VvWRKY genes in the V. vinifera cv ‘Corvina’ atlas (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lfcrxesyciqgsjoandacc=GSE36128). Data were normalized based on the mean expression value of each gene in all tissues analysed. Genes were hierarchically clustered based on average Pearson’s distance metric and ‘average linkage’ method. Yellow and blue boxes indicate high and low expression levels, respectively, for each gene. (b) qRT-PCR validation of VvWRKY expression in roots, stems, leaves and shoot tips obtained from 4-week-old in vitro explants of the V. vinifera PN40024 genotype. Transcripts were normalized to the expression of the actin gene. The mean±s.d. of three biological replicates are presented. Asterisks indicate that the corresponding gene was significantly up- or downregulated in a given organ (*p≤0.05, **p≤0.01). Bud-AB, bud after burst; Bud-B, Bud burst; Bud-W, winter bud; Bud-L, latent bud; Bud-S, bud swell; Flower-F, flowering; Flower-FB, flowering begins; FS, fruit set; Inflorescence-Y, young inflorescence with single flowers separated; Inflorescence-WD, well-developed inflorescence; Leaf-FS, mature leaf; Leaf-S, senescing leaf; Leaf-Y, young leaf; MR, mid-ripening; R, ripening; PFS, post fruit set; Stem-G, green stem; Stem-W, woody stem; Tendril-FS, mature tendril; Tendril-WD, well-developed tendril; Tendril-Y, young tendril; V, véraison.
This analysis indicates that only some members within the same group share a similar expression profile in grapevine organs/tissues during development (Figure 5a). For example, VvWRKY14, 19 and 52, belonging to Group II-c, are highly expressed in berries and rachis during or following véraison (V, MR and R). Other members of this group, such as VvWRKY23 and 46, exhibit high levels of expression in overwinter organs tissues, such as buds (Bud-W) and stems (Stem-W), suggesting an involvement of Group II-c members in berry ripening and cold acclimation.
Members of Group III VvWRKYs, including VvWRKY05, 47, 41 and 25, and Group II-a, including VvWRKY07, 08 and 28, are characterized by an high levels of expression in a heterogeneous group of tissues including inflorescence, leaf, rachis, tendril and berry in pre-véraison phases and are less expressed (especially VvWRKY05 and 47) in berries and the rachis after véraison phase. The expression pattern of these genes was also evaluated by qRT-PCR in selected organs of the V. vinifera PN40024 genotype, confirming their high level of expression in leaves (Figure 5b). Low modulation in the levels of expression were detected for VvWRKY40 and 51 (Group III) in all tissues in the Corvina atlas. However, results on expression levels obtained by qRT-PCR revealed high transcript accumulation in leaves (Figure 5b). Another interesting gene cluster in Group I, composed of VvWRKY13, 59 and 57, was all characterized by a high level of expression that was restricted to stamen and pollen tissues. A high level of modulation was observed in some VvWRKYs in senescing leaves (S). These include VvWRKY22 (Group II-c), VvWRKY 02 (Group II-b) and VvWRKY03 (Group II-c). Although the upregulation of VvWRKY22 and 02 was also present in previous leaf developmental stages (Y and FS), high levels of expression of VvWRKY03 were restricted to the senescing stage of leaves. VvWRKY17 (Group II-d) and VvWRKY27 (NG) were induced in the very early phases of seed development, especially at post fruit-set, while VvWRKY44, 48 and 53 were upregulated in roots.
Expression patterns of VvWRKY genes in response to biotic and abiotic stresses
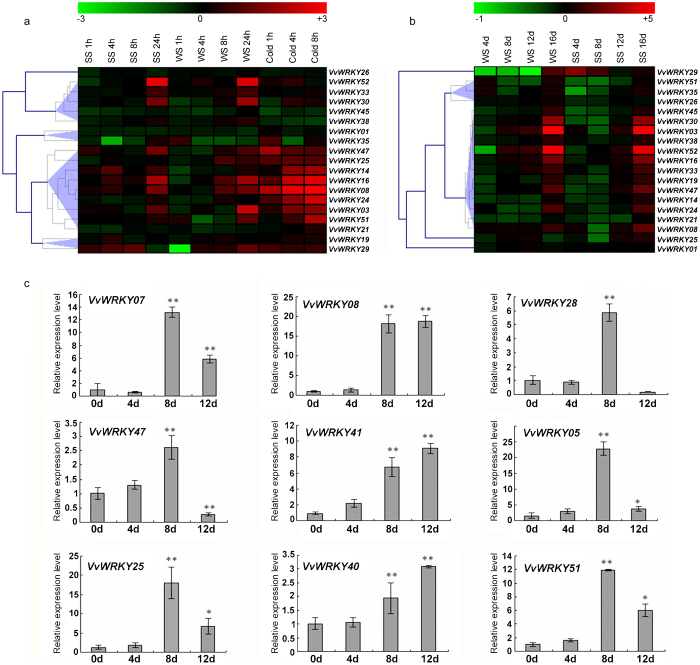
The expression of VvWRKY genes in response to both biotic and abiotic stresses was investigated using microarray data from several previously published papers. Regarding abiotic stresses, expression datasets were obtained from two studies (GSE31594 and GSE31677) conducted on transcriptomic response of leaves of V. vinifera cv ‘Cabernet Sauvignon’ to short-term salt, water and cold stress, and long-term water and salt stress. Since the Affymetix array used in these analysis was based on the few VvWRKYs sequences (cDNA and ESTs) known at the time, the expression of only a limited number of VvWRKY genes could be determined (Supplementary Tables S3 and S4). In general, cold stress induced the most rapid upregulation of VvWRKY genes of all the abiotic stress treatments (Figure 6a). In fact, a relatively large group of VvWRKY genes, including VvWRKY08, 14, 16 and 24, was induced since the first hour after the imposition of the stress, whereas the majority of genes induced by salt (SS) and water (WS) stresses, were upregulated after 24 h. Anyway, the quicker transcriptional response observed upon cold treatment could be related to a more rapid percepition of the cold compared to the gradual imposition of the water stress. More in detail, VvWRKY24 was found to be a cold-specific responsive gene since it was induced at all time points of the cold treatment, but was not in response to either the water or salt stresses. Conversely, VvWRKY30 and 52 were both strongly induced 24 h after water and salt stress, but not in response to cold stress. VvWRKY29 appeared to be weakly induced since in the first hours following salt and cold treatments, but interestingly was strongly repressed 1 h after the water stress treatment. The data presented in Figure 6b also confirmed the putative involvement of VvWRKY30 and 52 in water and salt stress responses together other genes such as VvWRKY03 and 16. Interestingly, all the VvWRKY genes that were induced within 24 h in response to short-term water and salt stress (Figure 6a) appeared to be more highly induced in the last point of the long-term stress treatments (16 days after treatment, Figure 6b). VvWRKY29 was again downregulation in almost all time points of the water stress treatment, whereas it appeared to be slightly induced in response to salt stress (Figure 6b).
Figure 6.
Expression profiles of VvWRKY genes in response to abiotic stresses. Expression of VvWRKY genes in the V. vinifera cv ‘Cabernet Sauvignon’ (microarray) or 4-week-old in vitro explants of V. vinifera PN40024 genotype (qRT-PCR) in response to cold (cold), drought (WS) and salt stress (SS). Microarray analysis presented in a and b. qRT-PCR data presented in c. Microarray data were downloaded from the NCBI GEO datasets (GSE31594 and GSE31677), processed as log2 of the ratio between treated and untreated samples and graphically represented with MeV software. (a) V. vinifera cv ‘Cabernet Sauvignon’ plants grown in a hydroponic drip system were treated with 120 mM salt (SS), polyethylene glycol (PEG), cold (5 °C) or left untreated. Shoots with leaves were collected at 0, 1, 4 and 8 h for all treatments, and at 24 h for all treatments except cold (GEO series GSE31594). (b) Potted V. vinifera cv ‘Cabernet Sauvignon’ vines in the greenhouse were exposed to a water-deficit stress (WC) by withholding water or a salt stress (SS) by watering plants with a saline solution for 16 days. Non-stressed, normally watered plants served as the control for both treatments. Shoot tips were harvested every 4 days (0, 4, 8, 12 and 16 days) (GEO series GSE31677). (c) qRT-PCR expression analysis of VvWRKYs in V. vinifera PN40024 genotype subjected to a drought treatment. Transcripts were normalized to the actin gene expression. The mean±s.d. of three biological replicates are presented. Asterisks indicate that the corresponding gene was significantly up- or downregulated in a given organ (*p≤0.05, **p≤0.01). MeV, Multi Experiment Viewer.
In order to validate the results obtained from the analysis of the microarray data, qRT-PCR was used to determine the expression profiles of VvWRKY08, 25, 47 and 51 genes in leaves of V. vinifera clone PN40024 plants subjected to drought stresses (Figure 6c).
The expression profiles of selected mumbers belonging to Groups II-a (VvWRKY07 and 28) and Group III (VvWRKY05, 40 and 41) were examined (Figure 6c). In Arabidopsis, these two groups include most of the stress-responding WRKY genes. The majority of the tested VvWRKY transcripts exhibited a high level of accumulation at 8-day after the onset of the stress treatment, approximately increasing up to 22-fold for VvWRKY05 (Figure 6c).
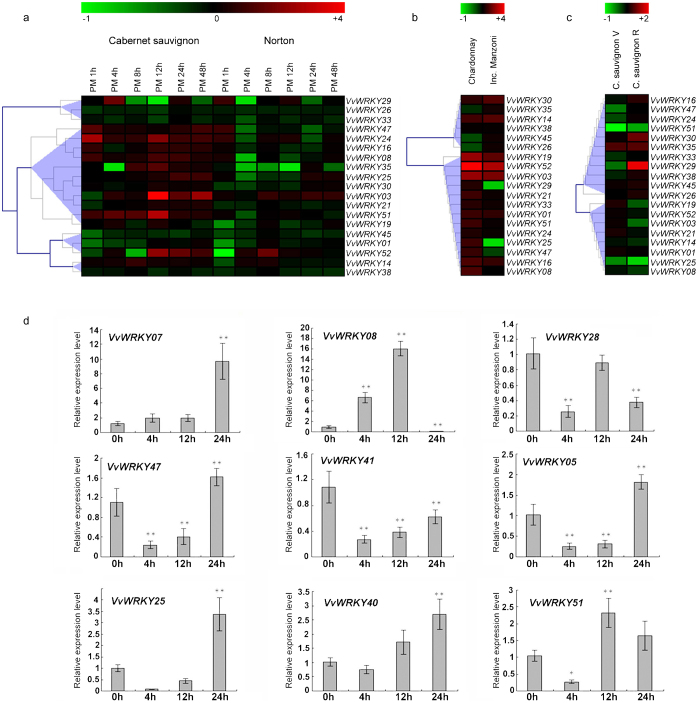
In regards to VvWRKY expression in response to biotic stresses, expression datasets obtained from three different host–pathogen interaction experiments were examined, including: the inoculation of Erysiphe necator on leaves of the susceptible V. vinifera cv. Cabernet sauvignon and the tolerant V. aestivalis cv. Norton60 (Supplementary Table S5); infection of V. Vinifera cv. Chardonnay and cv. Incrocio Manzoni infection with the Bois Noir phytoplasma61 (Supplementary Table S6); and the infection of V. vinifera cv. Cabernet Sauvignon with grapevine leaf-roll-associated virus-3 (GLRaV-3) during véraison and ripening stages of berry development62 (Supplementary Table S7).
The effect E. necator infection on VvWRKY response appeared to be much stronger in the susceptible cv. Cabernet sauvignon than in the resistant cv. Norton (Figure 7a). Several WRKY genes were upregulated in Cabernet sauvignon, including five that were previously reported to be upregulated by Fung et al.60 VvWRKY47, 24, 16, 08 and 51 were induced after 1 hour after inoculation and remained upregulated during the entire sampling period. A peak in the expression of VvWRKY51, 35, 03 and 52 was observed at 12 h after inoculation, whereas a weak, late induction was observed for VvWRKY29. Phytoplasma infection led to a general induction of the majority of VvWRKY genes in susceptible V. Vinifera cv. Chardonnay compared to the tolerant cv. Incrocio Manzoni (Figure 7b). VvWRKY51, 25, 47 and 08 were found to be also upregulated in the susceptible cultivar noted in the previous experiment (Figure 7b). Finally, the expression profiles of VvWRKY genes in response of grapevine to GLRaV-3 infection in both vèraison and ripening phases revealed a general repression of many members of the VvWRKY family (Figure 7c). However, a weak-upregulation, limited to VvWRKY30, 35 and 29, specifically induced during the ripening phase.
Figure 7.
Expression profiles of VvWRKY genes in response to biotic stresses. (a) V. vinifera cv ‘Cabernet sauvignon’ and V. aestivalis cv ‘Norton’ plants were grown in an environmental chamber and inoculated with Erysiphe necator conidiospores (PM). Inoculated leaves were harvested at 0, 4, 8, 12, 24 and 48 h after inoculation (GEO series GSE6404). (b) Field-grown plants of V. vinifera cv ‘Chardonnay’ and ‘Incrocio Manzoni’ naturally infected with Bois Noir phytoplasma (BN), compared to healthy samples (GEO series GSE12842). (c) V. vinifera cv ‘Cabernet Sauvignon’ was infected with GLRaV-3 during véraison and the ripening stages of berry development (GEO series GSE31660). (d) qRT-PCR expression analysis of VvWRKY genes in Pinot PN40024 genotype infected with powdery mildew. Transcripts were normalized to actin gene expression. The means±s.d. of three biological replicates are presented. Asterisks indicate that the gene was significantly up- or downregulated in a given organ (*p≤0.05, **p≤0.01). GLRaV-3, grapevine leaf roll-associated virus-3.
qRT-PCR were performed on PN40024 leaves infected by E. necator in order to validate and integrate microarray data (Figure 7d). In general, qRT-PCR results for VvWRKY08, 25, 47 and 51 corroborated transcriptomic data obtained from the microarrays. VvWRKY07 and VvWRKY08, belonging to Group IIa, exhibited a much higher level of induction compared to genes belonging to group III (VvWRKY 47, 41, 05, 25, 40 and 51). Interestingly, VvWRKY07 exhibited a peak in induction at 24 h, while VvWRKY08 exhibited a peak at 12 h.
It is well established that the plant hormones SA and ethylene mediate plant responses to biotic stresses.63 To investigate their effect on VvWRKY expression profiles, qRT-PCR, using a select number of VvWRKY genes, was conducted on Pinot PN40024 leaves after treatment with these two hormones. Excluding the response of VvWRKY25 to ethylene, all the VvWRKYs analysed by qRT-PCR were significantly modulated upon treatment with both hormones. In general, the VvWRKY genes examined were upregulated to a higher levels by SA than by ethylene (Supplementary Fig. S2). VvWRKY07, 08 and 25 were strongly (≥10-fold change) induced by SA whereas VvWRKY07 appeared higly induced by ethylene.
Discussion
Organisation of the grapevine VvWRKY gene family
WRKY genes encode a class of transcription factors that are ubiquitous to all plant species and have been surveyed in several species whose genomes have been sequenced, such as Arabidopsis,35 rice,46 cucumber,64 poplar,65 tomato,66 sunflower67 and coffee.68 In the current study, a search for WRKY genes in the last and previous versions of the genome assembly of the grape PN40024 genotype resulted in the identification of 59 members, which were designated VvWRKY01 through VvWRKY59 on the basis of their chromosomal location.
The structure of the phylogentic tree obtained from an alignment of the deduced WRKY domains of grapevine and Arabidopsis indicated that the 59 VvWRKY genes identified in grapevine can be divided into the three major groups (I, II and III) as previously described in other plant species.35 Members within the same group, or subgroup within group II, shared a similar gene structure (intron/exon organisation), length and amino-acid motif composition, indicating their close evolutionary relationship.
Group I VvWRKYs, which are characterized by the presence of two WRKY domains, contain approximately 20% of the entire VvWRKY family and is comparable in size to Group I in Arabidopsis, rice, tomato and castor bean (Table 3). This group has undergone a significant expansion in poplar, where it contains approximately 50% of the entire PtWRKY family. Group II subgroups in Arabidopsis, rice and grapevine were comparable in size, whereas a sensible expansion was detected for poplar Group II-d and tomato group II-e. In grapevine, the members of Group II-a and II-b are characterized by the presence of motifs 7 and 8, whereas Group II-d contains motifs 13 and 19 and the HARF sequence. WRKY Group III, containing the motif 14, was found to be smaller in number than in Arabidopsis or rice, with only 10% of the entire VvWRKY family falling in this clade. Similar percentages were detected for castor bean, poplar and tomato. Gene orthology may represent a starting point for genetical studies which aim to define the function of a candidate gene. Some of the predicted WRKY domains from grapevine and Arabidopsis showed pairwise relationships, such as the those between VvWRKY22 and AtWRKY51, VvWRKY24 and AtWRKY33, and VvWRKY49 and AtWRKY22. Indeed, the function of these Arabidopisis genes was previously inferred. AtWRKY51 proteins appear to be involved in the SA-dependent and low oleic acid-dependent repression of JA signaling pathway.69 AtWRKY33 plays a role in Arabidopsis tolerance to high concentrations of NaCl concentration,24 while AtWRKY22 appears to be involved in the regulation of dark-induced leaf senescence in Arabidopsis.70
Gene duplication and divergence events have been suggested to be the main contributors to evolutionary momentum.71,72 An analysis of gene duplication in the current study indicated that 20 of 59 (33.89%) VvWRKY genes have arisen through either tandem or segmental duplication. Two pair of the WRKY genes, VvWRKY11/VvWRKY12 and VvWRKY40/VvWRKY41, appeared to have undergone tandem duplication (Figure 4). Due to the genome of grapevine originated from a hexaploidisation,44 several synteny blocks would be expected to exist in this genome. Among the 11 collinearity relationships, only three of them, VvWRKY2/44/53 were identified as triplicates (Figure 1). Taken together, this implies that low tandem and high segmental duplications existed in VvWRKY genes, which is in consistent with the results in Arabidopsis.57 Segmental duplicates may be more often retained due to subfunctionalisation, without increasing the likelihood of gene rearrangment.73,74
VvWRKY genes involved in grapevine growth and development
Patterns of expression of all of the predicted coding members of the VvWRKY family in different grapevine tissues and at different developmental stages were examined using an expression atlas of V. vinifera cv Corvina.52 The analysis revealed that many VvWRKY genes can be grouped together based on their abundant expression in specific grapevine tissues, possibly reflecting their involvement in a common metabolic and/or developmental process. In addition to the well-known response of WRKY genes to biotic and abiotic stresses, a number of studies have provided evidence of a role for WRKY TFs in developmental processes, such as seed and trichome development,28,75,76 dormancy and germination,75,77 and senescence.30,32,33,78
The expression analysis revealed that VvWRKY23 and 46, belonging to Group II-c, were highly expressed in ‘woody’ tissues, suggesting a putative role in the regulation of lignin biosynthesis. Involvement of WRKY genes in the lignin synthesis has been previously reported for one grapevine WRKY within the same group (VvWRKY01).41 In tobacco, VvWRKY01 was able to induce lignin accumulation by regulating lignin biosynthetic genes when overexpressed.
VvWRKY02 of Group II-b was found to be specifically expressed in leaf tissues and highly accumulated during senescence. In Arabidopsis, AtWRKY6, belonging to the same group as VvWRKY02, was strongly upregulated during leaf senescence and targets a senescence-specific receptor-like kinase (SIRK/FRK1).32 Several other Arabidopsis WRKY genes were also upregulated in the Arabidopsis leaf transcriptome during senescence.79 VvWRKY03, another gene belonging to Group II-c, was specifically induced during senescence and would be an interesting candidate for functional characterisation.
AtWRKY53 and AtWRKY70 play a critical roles in leaf senescence;31,80,81 however, none of the grapevine VvWRKY genes belonging to the same group (III) were determined to be highly expressed in senescencing leaves. Both VvWRKY13, 59 and 57, belonging to Group I, were higly expressed in stamen and pollen tissues and are closely related to Arabidopsis AtWRKY34, which was shown to have a role in coordinating gene expression during the formation of the tapetum and microspores.82
Stress induced VvWRKY expression in grapevine
The involvement of WRKY TFs in the regulation of plant response to biotic and abiotic stresses has been described in many species.8 Recent studies have indicated that hormones involved in plant defence signalling pathways83 are mediated by the induction of WRKY TFs.7,19,84 The transcriptomic databases generated in previous studies of grapevine subjected to biotic and abiotic stresses, together with our qRT-PCR analysis, allowed us to identify VvWRKY genes putatively involved in stress response. Since Arabidopsis WRKYs involved in stress response mainly belong to Groups II-a and III, the current focus is also on VvWRKYs within the same groups.
All members of Group II-a, the least abundant subgroup in both grapevine and Arabidopsis, are associated with stress response. In particular, VvWRKY07, 08 and 28, had the strongest upregulation in response to drought stress. Water stress induces the accumulation of abscisic acid (ABA) which triggers closing of stomata, thus reducing water loss.84 In Arabidopsis, AtWRKY18, 40 and 60 of Group II-a exhibited a complex pattern of expression in plant responses to ABA and abiotic stresses.85 Although an ABA treatment was not included in our survey, VvWRKY08, which is closely related to these Arabidopsis WRKYs, was induced by cold, salt and drought stresses, and by SA and ethylene treatments. VvWRKY08 shares 99% amino-acid sequence identity with VpWRKY3, a Group II-a member of V. pseudoreticulata, which has been shown to be involved in the ABA signalling pathway, in response to salt stress, and to increase resistance to Ralstonia solanacearum when overexpressed in tobacco.39 The similar expression response of VvWRKY08 and VpWRKY3 in response to drought, SA and ethylene treatments, and E. necator (powdery mildew) infection suggests a putative role for VvWRKY08 in grapevine plant stress tolerance that deserves further investigation.
Regarding biotic stresses, the low response of WRKY genes detected in resistant varieties compared to susceptible genotypes is not surprising since overall changes in the global transcriptome were generally lower in the resistant genotypes.60,61 In the case of E. necator infection, it is important to note that despite showing a low fold change in infected vs. mock-inoculated leaves in ‘Norton’ (resistant genotype), the majority of VvWRKY genes showed a higher level of constitutive expression in the mock-inoculated leaves compared to what was detected in ‘Cabernet sauvignon’ (susceptible genotype) mock-inoculated leaves. This indicates that, although a significant induction of VvWRKYs did not occur in response to infection, the baseline levels of these transcript are always higher in the resistant genotype than in the susceptible genotype.60
VvWRKY07 and VvWRKY08, two members of Group IIa, are significantly upregulated in response to E. necator infection, suggesting that these two genes may be involved in the pathogen response pathway. The involvement of Group II-a WRKY genes in biotic responses has been previously in Arabidopsis where AtWRKY18 and AtWRKY40 play a significant role in controlling the basal defense.86 All members of Group III VvWRKYs were induced by both abiotic and biotic stresses, except for VvWRKY41. In Arabidopsis, AtWRKY70 and AtWRKY54, also in Group III, play an important role in plant defence against E. amylovora and in osmotic stress signaling.87,88 Interestingly, VvWRKY25, which is ortholog to AtWRKY54, is significantly induced by cold and drought treatments, and by E. necator infection. VvWRKY25 shares 88.66% amino-acid sequence identity with VpWRKY1 of V. pseudoreticulata was also reported to be rapidly induced by SA treatment and E. necator infection.43 This suggests that VvWRKY25 may also play a role in mediating plant defense response in grapevine V. vinifera.
Despite many recent advances in functional studies of WRKYs in grapevine, the biological function of most VvWRKY genes in physiological and developmental processes and plant defence still needs to be elucidated. The bioinformatic analysis and expression patterns of the VvWRKY gene family conducted in the present study provide an overall picture of the composition and expression of WRKY genes in grapevine that will facilitate selecting candidate genes for cloning and further functional characterisation.
Conclusions
Fifty-nine WRKY genes in grapevine were bioinformatically identified and characterized. The VvWRKY family genes were classified into three main groups, with high similar exon–intron structures and motif compositions within the same groups and subgroups. VvWRKY genes play important roles in grapevine growth and development as indicated by their spatial and temporal expression profiles. Group II-a and Group III genes exhibited different patterns of expression in response to different stresses. Most members in Group II-a and Group III were expressed under drought stress, and represent good candidate genes for exploring the role of WRKYs in grapevine stress response pathways.
Acknowledgments
This work was supported by the Priority Academic Program Development of Modern Horticultural Science in Jiangsu Province.
The authors declare no conflict of interest.
References
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 2002; 5: 325–331. [DOI] [PubMed] [Google Scholar]
- Singh K, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 2002; 5: 430–436. [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Gomez-Buitrago A, Eckardt N et al. Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 2003; 4: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F. A global view of defense gene expression regulation—a highly interconnected signaling network. Curr Opin Plant Biol 2004; 7: 506–511. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J 1999; 18: 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 2004; 7: 491–498. [DOI] [PubMed] [Google Scholar]
- van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol 2008; 146: 1983–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci 2010; 15: 247–258. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol 2004; 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 2005; 43: 545–580. [DOI] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Atzorn R, Miersch O, Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 2006; 140: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2006; 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth C, Ringler J, Dangl JL, Eulgem T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact 2007; 20: 120–128. [DOI] [PubMed] [Google Scholar]
- Lai Z, Vinod K, Zheng Z, Fan B, Chen Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol 2008; 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Fan BF, Chen ZX. Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol 2006; 142: 1180–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai ZB, Fan BF, Chen ZX. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 2008; 20: 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Duan MR, Wei CH, Li Y. WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol 2007; 48: 833–842. [DOI] [PubMed] [Google Scholar]
- Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant 2008; 1: 459–470. [DOI] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D et al. A pair of allelic WRKY genes play opposite roles in rice–bacteria interactions. Plant Physiol 2009; 151: 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A et al. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 2007; 19: 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007; 315: 1098–1103. [DOI] [PubMed] [Google Scholar]
- Skibbe M, Qu N, Galis I, Baldwin IT. Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 2008; 20: 1984–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep 2009; 28: 21–30. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 2009; 69: 91–105. [DOI] [PubMed] [Google Scholar]
- Li S, Fu Q, Huang W, Yu D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep 2009; 28: 683–693. [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Hanzlik S, Cook E, Shen QJ. Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol Biol 2007; 64: 293–303. [DOI] [PubMed] [Google Scholar]
- Zou X, Neuman D, Shen QJ. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol 2008; 148: 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002; 14: 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 2007; 19: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J 2001; 28: 123–133. [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 2004; 55: 853–867. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 2002; 16: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing SJ, Zhou X, Song Y, Yu DQ. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul 2009; 58: 181–190. [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 1996; 15: 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci 2000; 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 2005; 137: 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T et al. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 2000; 26: 403–410. [DOI] [PubMed] [Google Scholar]
- Mare C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L. Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol 2004; 55: 399–416. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Shi J, Cao J, He M, Wang Y. VpWRKY3, a biotic and abiotic stress-related transcription factor from the Chinese wild Vitis pseudoreticulata. Plant Cell Rep 2012; 31: 2109–2120. [DOI] [PubMed] [Google Scholar]
- Marchive C, Mzid R, Deluc L et al. Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J Exp Bot 2007; 58: 1999–2010. [DOI] [PubMed] [Google Scholar]
- Guillaumie S, Mzid R, Mechin V et al. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol 2010; 72: 215–234. [DOI] [PubMed] [Google Scholar]
- Liu H, Yang W, Liu D, Han Y, Zhang A, Li S. Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol Biol Rep 2011; 38: 417–427. [DOI] [PubMed] [Google Scholar]
- Li H, Xu Y, Xiao Y et al. Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 2010; 232: 1325–1337. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B et al. The grapevine genome sequence suggests ancestral hexaploidization in major Angiosperm phyla. Nature 2007; 449: 463–467. [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2007; 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 2005; 12: 9–26. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mao L, Wang H et al. Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS ONE 2012; 7: e32153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics 2009; 25: 2281–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009; 37: W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M, Dal Santo S, Zenoni S et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 2012; 24: 3489–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC et al. TM4 microarray software suite. Methods Enzymol 2006; 411: 134–193. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics 2006; 7: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M. Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol 2012; 12: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet 2001; 2: 516–527. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 2004; 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004; 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, Mcknight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 1988; 240: 1759–1764. [DOI] [PubMed] [Google Scholar]
- Fung RW, Gonzalo M, Fekete C et al. Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol 2008; 146: 236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertazzi G, Milc J, Caffagni A et al. Gene expression in grapevine cultivars in response to Bois Noir phytoplasma infection. Plant Science 2009; 176: 792–804. [Google Scholar]
- Vega A, Gutierrez RA, Pena-Neira A, Cramer GR, Arce-Johnson P. Compatible GLRaV-3 viral infections affect berry ripening decreasing sugar accumulation and anthocyanin biosynthesis in Vitis vinifera. Plant Mol Biol 2011; 77: 261–274. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 2012; 159: 266–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Jiang W, Zhang Y et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 2011; 12: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Dong Q, Shao Y et al. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep 2012; 31: 1199–1217. [DOI] [PubMed] [Google Scholar]
- Huang S, Gao Y, Liu J et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Genet Genomics 2012; 287: 495–513. [DOI] [PubMed] [Google Scholar]
- Giacomelli JI, Weigel D, Chan RL, Manavella PA. Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol 2012; 195: 766–773. [DOI] [PubMed] [Google Scholar]
- Ramiro D, Jalloul A, Petitot AS, de sa MFG, Maluf M, Fernandez D. Identification of coffee WRKY transcription factor genes and expression profiling in resistance responses to pathogens. Tree Genet Genomes 2010; 6: 767–781. [Google Scholar]
- Gao QM, Venugopal S, Navarre D, Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 2011; 155: 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 2011; 31: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Wolf U, Atkin NB. Evolution from fish to mammals by gene duplication. Hereditas 1968; 59: 169–187. [DOI] [PubMed] [Google Scholar]
- Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science 2003; 300: 1701–1703. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science 2000; 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary demography of duplicate genes. J Struct Funct Genomics 2003; 3: 35–44. [PubMed] [Google Scholar]
- Jiang W, Yu D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol 2009; 9: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA 2005; 102: 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 1995; 29: 691–702. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J 2001; 28: 123–133. [DOI] [PubMed] [Google Scholar]
- Guo S, Zhang Z, Tong T. Cloning and characterization of cellular senescence-associated genes in human fibroblasts by suppression subtractive hybridization. Exp Cell Res 2004; 298: 465–472. [DOI] [PubMed] [Google Scholar]
- Ulker B, Shahid Mukhtar M, Somssich IE. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 2007; 226: 125–137. [DOI] [PubMed] [Google Scholar]
- Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J 2009; 58: 333–346. [DOI] [PubMed] [Google Scholar]
- Honys D, Oh SA, Renak D et al. Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC Plant Biol 2006; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol 2009; 69: 473–488. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 2001; 410: 327–330. [DOI] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 2010; 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Roccaro M, Schon M, Logemann E, Somssich IE. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J 2010; 64: 912–923. [DOI] [PubMed] [Google Scholar]
- Moreau M, Degrave A, Vedel R et al. EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora. Mol Plant Microbe Interact 2012; 25: 421–430. [DOI] [PubMed] [Google Scholar]
- Li J, Besseau S, Toronen P, et al. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol 2013; 200: 457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Zhang LB, Guo D, Li CZ, Peng SQ. Identification and expression profiles of the WRKY transcription factor family in Ricinus communis. Gene 2012; 503: 248–253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.