Abstract
Date palm (Phoenix dactylifera) is an economically important fruit tree in the Middle East and North Africa and is characterized by large cultivar diversity, making it a good model for studies on fruit development and other important traits. Here in gel comparative proteomics combined with tandem mass spectrometry were used to study date fruit development and ripening. Total proteins were extracted using a phenol-based protocol. A total of 189 protein spots were differentially regulated (p≤0.05). The identified proteins were classified into 14 functional categories. The categories with the most proteins were ‘disease and defense’ (16.5%) and ‘metabolism’ (15.4%). Twenty-nine proteins have not previously been identified in other fleshy fruits and 64 showed contrasting expression patterns in other fruits. Abundance of most proteins with a role in abiotic stress responses increased during ripening with the exception of heat shock proteins. Proteins with a role in anthocyanin biosynthesis, glycolysis, tricarboxylic acid cycle and cell wall degradation were upregulated particularly from the onset of ripening and during ripening. In contrast, expression of pentose phosphate- and photosynthesis-related proteins decreased during fruit maturation. Although date palm is considered a climacteric species, the analysis revealed downregulation of two enzymes involved in ethylene biosynthesis, suggesting an ethylene-independent ripening of ‘Barhi’ fruits. In summary, this proteomics study provides insights into physiological processes during date fruit development and ripening at the systems level and offers a reference proteome for the study of regulatory mechanisms that can inform molecular and biotechnological approaches to further improvements of horticultural traits including fruit quality and yield.
Introduction
Date palm (Phoenix dactylifera) is a perennial, dioecious monocot that is highly heterozygous.1 Dates have been an agriculturally and economically important fruit crop for centuries in the Middle East and Northern Africa.2 They are not only a staple for millions of people,3 but they also have potential health benefits due to their high nutrient content and bioactive compounds including polyphenols such as flavonoids, tannins and phenolics.4–6 The date industry generated about 7.5 million tons worldwide in 2012 and Saudi Arabia, the third major producer after Egypt and Iran, grows over 400 cultivars.7 ‘Sukkary’ and ‘Barhi’ are the two most popular cultivars;8,9 however, their productivity is irregular. Therefore, breeding strategies, including the use of molecular tools, offer the potential to contribute towards improving productivity and quality.
Like other major commercial fruits such as apples (Malus × domestica) and bananas (Musa acuminata), dates are classified as climacteric fruits as the ripening processes are associated with a concomitant burst of ethylene and a peak in respiration.10,11 Fruit development and ripening consist of complex biological processes associated with major metabolic pathways and cellular processes undergoing gradual switches from cell division to energy, nutrient and metabolite storage during cell expansion and eventually starch degradation during ripening.12–14 Understanding date palm fruit development biology at the molecular level is still in its infancy, but the recent release of the date genome enables a more systematic investigation of fruit development.9,15–17 Proteomics in combination with phenotypical and physiological data offers a promising approach to further characterize fruit development and quality traits. Comparative proteomics has furthered our understanding of biological processes during fruit development, ripening, postharvest and senescence in apple,18 tomato (Solanum lycopersicum),19,20 strawberry (Fragaria × ananassa),21 grape (Vitis vinifera),22–24 peach (Prunus persica)25,26 and apricot (Prunus armeniaca).27 In date palm, comparative proteomics has been applied only to the analysis of somatic and zygotic embryos,28 but not to date fruit tissues. The objective of this study was to generate comparative proteomic profiles for inferring of date hypanthium protein changes during development and ripening with a view to provide new insights into date fruit biology and support efforts to improve date horticulture.
Materials and methods
Plant material
Date palm fruits (cultivar ‘Barhi’) were collected at Thuwal, Province of Makkah, on the shores of the Red Sea in Saudi Arabia at four developmental stages: 25 days after pollination (DAP; S1), 70 DAP (MD), 110 DAP (NTR) and 125 DAP (RIPE-Khalal stage). The optimal date of harvest, designated as S1, range from 1 to 25 April 2011 and 2012. This was based on fruit size, shape, color and the historical harvest date for this cultivar. Fruits were immediately rinsed with sterile distilled water, frozen in liquid nitrogen, transported to the laboratory and stored at −80 °C. For comparative proteomics, three biological repetitions comprising 10-pooled fruits harvested over two consecutive years for each stage were used.
Protein extraction
Proteins were extracted from date hypanthia by phenol extraction as described previously,29 with some modifications. Fruit skin and innermost layers including the seed were removed prior to extraction. Approximately 5 g of frozen tissue was ground twice for 5 s each to a fine powder in 5 mL of cold extraction buffer (1% (w/v) polyvinylpolypyrrolidone, 0.7 M sucrose, 0.1 M KCl, 0.5 M Tris-HCl pH 7.5, 500 mM ethylenediaminetetraacetic acid, 1 mM phenylmethanesulfonylfluoride, 2% (v/v) β-mercaptoethanol) on ice using a PowerGen™ Model 125 homogenizer (Fisher Scientific, Waltham, MA, USA). Volume was adjusted to 15 mL and the mixture homogenized for 30 min at 4 °C. The mixture was homogenized for a further 30 min at 4 °C after addition of an equal volume of 0.5 M Tris-HCl pH 7.5-saturated phenol and then centrifuged at 10 000g for 30 min at 4 °C. The upper phenol phase was used to extract proteins twice in extraction buffer. The final phenol phase was precipitated with five volumes of saturated ammonium acetate in 100% (v/v) methanol overnight at −20 °C. Proteins were pelleted by centrifugation at 10 000g for 30 min at 4 °C and washed once in 100% (v/v) ice-cold methanol and subsequently three times in 100% (v/v) ice-cold acetone. Each wash was followed by centrifugation at 10 000g for 10 min at 4 °C to collect proteins, which were finally air-dried and solubilized in isoelectric focusing buffer (7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 1% (v/v) IPG buffer pH 3–10). Proteins were quantified by Bradford30 using Quick Start Bradford reagent (Bio-Rad, Hercules, CA, USA) and bovine serum albumin as standard. Quality was assessed by one-dimensional gel electrophoresis.
Comparative two-dimensional gel electrophoresis (2-DE) and protein identification
Proteins (50 μg) were used to rehydrate a 7-cm-long linear IPG strip, pH range 4–7 (GE Healthcare, Pittsburgh, PA, USA). 2-DE, SYPRO® Ruby (Molecular Probes, Eugene, OR, USA) staining and imaging were performed as detailed previously.31 Image analysis with Delta 2D v4.2 (DECODON, Greifswald, Germany) and trypsin digestion of differentially expressed spots were carried out as reported previously.32 Digested peptides were dried and stored at −20 °C. Peptides were analyzed with an LTQ-Orbitrap Velos (Thermo-Scientific, Bremen, Germany) coupled to nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark) for liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously.32 Raw data files were converted to mgf using Proteome Discoverer v1.2 (Thermo-Scientific). All spectra were submitted to a local MASCOT (Matrix Science, London, UK) server and searched against Phoenix dactylifera dataset downloaded from http://qatar-weill.cornell.edu/research/datepalmGenome/download.html (version 3) using a precursor mass tolerance of ±10 ppm, a fragment ion mass tolerance of ±0.6 Da, and strict trypsin specificity allowing up to one missed cleavage, carbamidomethyl on cysteine residues as fixed modification, and oxidation of methionine residues and phosphorylation of serine, threonine and tyrosine residues as variable modifications. Proteins were considered positive if the Mascot score was over the 95% confidence limit (≥26 for plant). Data was validated with Scaffold v4 (Proteome Software, Portland, OR, USA). The dataset presented here only include proteins and peptides with ≥95% probability each and ≤1% false discovery rate. Scaffold PTM (Proteome Software) was used to identify and position phosphorylation and oxidation posttranslational modifications (PTMs).
Bioinformatics analysis for functional enrichment and protein classification
Blast2GO (http://www.blast2go.org; v2.6.6)33 was used for gene ontology (GO) functional enrichment analysis. Blast-p search of identified protein sequences was performed against the NCBI non-redundant database with a minimum expectation value of 1×10−3. Annotations were made with default parameters: pre-eValue-Hit-Filter at 1×10−6, cut-off was set at 55 and GO weight at 5. Annotation was augmented using Annotation Expander (ANNEX) and the addition of GO terms associated with functional domains derived from scanning the InterPro database. Functional classification was performed as described previously.34
Results and discussion
Comparative 2-DE analysis and protein identification
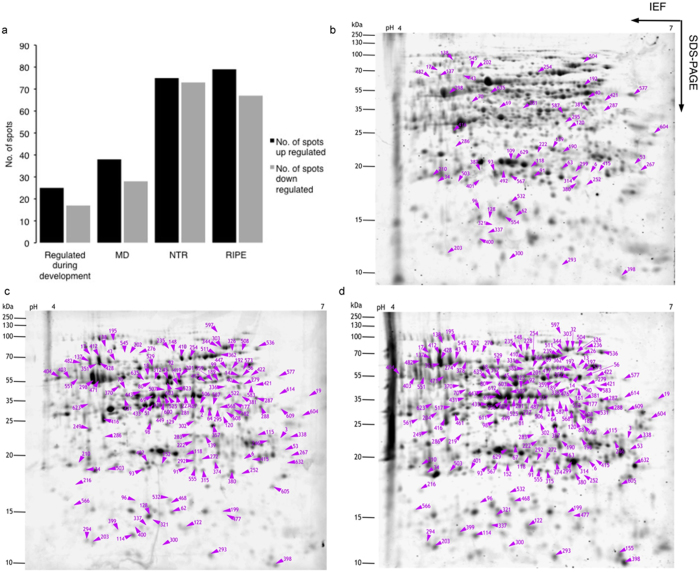
‘Barhi’ fruits, when harvested at RIPE (Khalal stage) before over-ripening (Rutab stage), are firm, yellow and contain about 30% moisture.35 A phenol-based extraction method yielded a protein fraction that produced high-quality 2-DE maps with minimal streaking in the acidic pH range and well-resolved protein spots (Figure 1). Reference gel images displayed distinct dissimilarities in terms of spot abundance and distribution among the four developmental stages (S1, MD, NTR and RIPE). Differences were observed particularly over pI range 4–5. The principal component analysis showed a closer correlation between biological replicates than between developmental stages (Supplementary File 1). The Delta 2D analysis revealed an average of 960 unique spots resolved on individual 2-DE and this is comparable to recently reported fruit proteome profiles.27,36–41
Figure 1.
2-DE proteome maps of the date hypanthium showing differentially expressed spots. The relative molecular mass and pI are indicated on the left and at the top of the gels. Total soluble proteins (∼50 μg) from date hypanthium collected at different developmental stages were separated in the first dimension on immobilized linear pH 4–7 gradient and then on 12% acrylamide gels for the second dimension. Gels were stained with SYPRO® Ruby and comparatively analyzed with Delta 2D (Decodon). a shows number of up- and downregulated spots overall and at each stage in comparison to S1. Differentially expressed spot numbers are shown at MD (b), NTR (c) and RIPE (d) in comparison to S1.
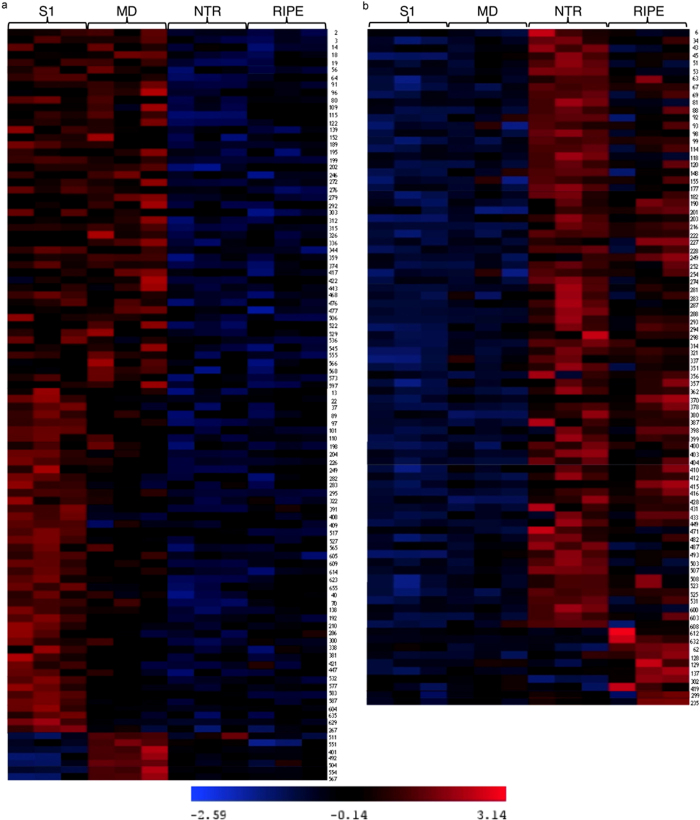
The abundance of 189 protein spots was significantly altered (p≤0.01) with a minimum fold change of ±1.5 during the four developmental stages and these spots were selected for further analyses (Figure 1). Heat maps revealed major changes in protein expression over the course of date development and ripening. A first subset of spots was downregulated from NTR stage (Figure 2a), while a second subset was upregulated (Figure 2b). Of the 189 significantly altered spots, 171 were positively identified by LC-MS/MS corresponding to 193 unique proteins (Supplementary Files 2 and 3), while 18 spots (6, 51, 118, 139, 148, 152, 161, 204, 207, 219, 236, 276, 293, 315, 321, 399, 505 and 517) remained unidentified (Figure 1). Of the 171 identified spots, 68 (representing 82 proteins) and 78 (representing 96 proteins) were up- or downregulated at different times. A further 25 spots (representing 36 proteins) showed differential accumulation throughout development (Supplementary File 2).
Figure 2.
Heat maps of differentially expressed protein spots during fruit development and ripening. Clustered spots show different expression during fruit development according to the normalized spot pixel intensity in each replicate. a shows spots mainly downregulated from NTR and b shows spots mainly upregulated at NTR and RIPE.
GO analysis of differentially expressed proteins
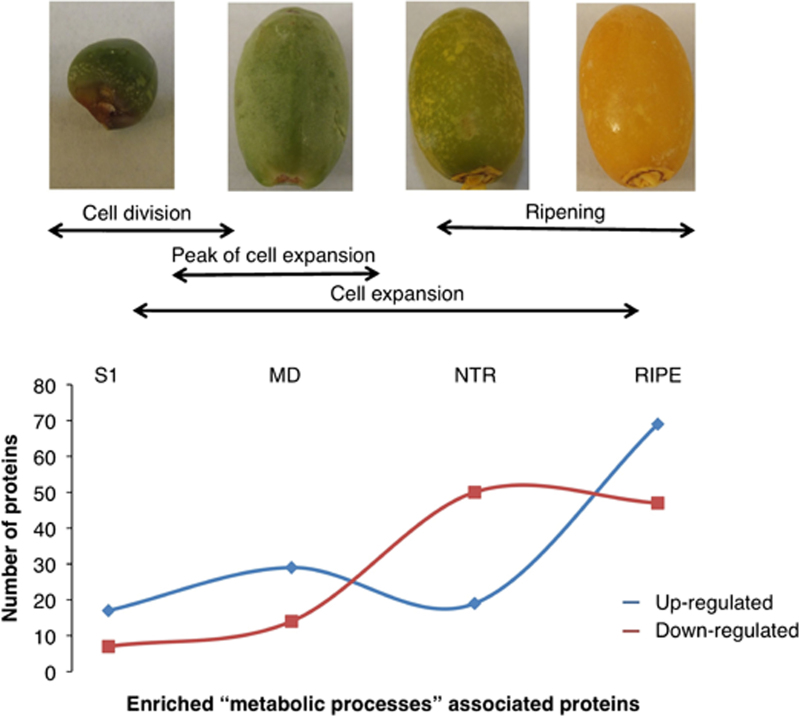
GO analysis using Blast2GO facilitated classification of the 193 identified proteins. The GO analysis is a tool that allows inferring function based on enrichments in comparison to the total proteome. Among the 82 upregulated and 96 downregulated proteins during fruit development, the most enriched biological processes (p≤0.05) are ‘metabolic’ and ‘cellular processes’, ‘response to stimulus’ and ‘biological regulation’ (Supplementary File 4). The most enriched molecular functions (p≤0.05) included ‘catalytic activity’ and ‘binding’. Enrichments were also detected in cellular compartments ‘vesicle’, ‘fumarate reductase complex’ and ‘respiratory chain complex II’ (Supplementary File 4). GO terms detected were most markedly enriched at RIPE stage, suggesting occurrence of a distinct metabolic phase that could possibly serve as an indicator of developmental reprogramming (Figure 3).
Figure 3.
Analysis of enriched ‘metabolic processes’-associated proteins against each of the four developmental stages investigated.
The most represented GO term was ‘metabolic processes’ with 134 proteins differentially regulated during fruit development and ripening (Figure 3 and Supplementary File 4). In this category, abundance of 17 proteins consistently increased throughout development and ripening, while 29, 19 and 69 proteins were transiently upregulated only at MD, NTR and RIPE, respectively (Figure 3). In the ‘response to stress’ category, 22 proteins were upregulated throughout the entire development and ripening stages, while 29 proteins were transiently downregulated at NTR and RIPE (Supplementary File 4). The GO analysis of the date fruit-dependent proteome revealed that proteins from specific metabolic and cellular processes are induced at particular developmental stages suggesting maturation specific roles.
Date-specific hypanthium proteins and their expression
Of the 193 identified proteins that were significantly differentially expressed (p≤0.01), we noted that 29 were specific to date fruit and, according to Bevan et al.,34 belong to 10 functional categories (Table 1). To date, these proteins have not been identified in any other fleshy fruits, possibly due to limited proteomic data available from fleshy fruits of monocot plants.40,41 However, identification of this set of proteins could also be due to the different stages at which fruits were collected and characterized. For example banana fruits were grouped into two stages, pre- and post-climacteric,40 while in the current study, four stages were considered, two before ripening (S1 and MD) and two after the onset of ripening (NTR and RIPE). An additional five proteins, proline iminopeptidase (spot 129), aspartyl-tRNA synthetase (spot 274), GDP dissociation inhibitor (spot 370), disproportionating enzyme (spot 385) and ornithine carbamoyltransferase (spot 566), which have been detected in other fleshy fruits, but not characterized as differentially expressed during development or ripening, were identified in this study (Table 1).
Table 1. Date specific proteins identified as differentially regulated during fruit development and ripening.
| Spot no. | Identified protein | Protein probability | Accession no. | Theoretical MW (Da)/pI | Observed MW (Da)/pI | Sequence coverage | NUP | FC S1/MD | FC S1/NTR | FC S1/R |
|---|---|---|---|---|---|---|---|---|---|---|
| 01 | Metabolism | |||||||||
| 197 | Furostanol glycoside 26-O-β-glucosidase | 100.00% | 30s65509133g003 | 64132/5.34 | 61000/6.1 | 13.30% | 7 | ns | ns | −7.28 |
| 227 | Dolichyl-phosphate-mannose-glycolipid α-mannosyltransferase | 98.50% | 30s785981g002 | 64116/9.91 | 39000/5.6 | 1.44% | 1 | ns | 4.20 | 6.00 |
| 235 | Lysosomal α-subunit | 100.00% | 30s945081g001 | 202855/7.02 | 82000/5.35 | 1.44% | 3 | ns | −3.61 | 1.99 |
| 385 | Disproportionating enzymea | 99.70% | 30s1194551g005 | 52152/5.41 | 61000/5.0 | 3.70% | 1 | ns | ns | −1.91 |
| 482 | AMP-dependent | 100.00% | 30s993441g001 | 29388/5.16 | 66000/4.5 | 12.50% | 3 | −2.02 | 4.57 | 5.56 |
| 566 | Ornithine carbamoyltransferasea | 99.50% | 30s699641g001 | 42893/7.66 | 16000/4.3 | 3.08% | 1 | ns | −2.03 | −2.99 |
| 02 | Energy | |||||||||
| 477 | UMP6_mitochondrial flags: precursor | 100.00% | 30s1009951g003 | 16907/5.11 | 15000/5.95 | 13.60% | 2 | ns | −2.16 | −2.70 |
| 03 | Cell growth/division | |||||||||
| 366 | Growth regulator | 98.10% | 30s797691g002 | 19927/10.01 | 62000/5.6 | 6.29% | 1 | ns | ns | −3.21 |
| 387 | KU P80 DNA | 99.50% | 30s862811g001 | 75649/6.15 | 20000/4.9 | 1.48% | 1 | 2.75 | ns | ns |
| 04 | Transcription | |||||||||
| 302 | Gata transcription factor 25 | 99.70% | 30s1133641g001 | 80728/9.32 | 25000/5.65 | 1.37% | 1 | ns | 2.14 | −1.96 |
| 302 | Inducer of cbf expression 1 DNA binding transcription activator transcription factor | 99.70% | 30s953031g001 | 38914/10.25 | 25000/5.65 | 2.51% | 1 | ns | 2.14 | −1.96 |
| 05 | Protein synthesis | |||||||||
| 249 | Gag-pol polyprotein | 99.70% | 30s811431g002 | 49976/7.27 | 24000/4.5 | 2.04% | 1 | ns | 1.65 | 1.72 |
| 274 | Aspartyl-tRNA synthetasea | 100.00% | 30s6550950g017 | 66976/5.84 | 65000/4.75 | 19.30% | 7 | ns | ns | 3.30 |
| 06 | Protein destination and storage | |||||||||
| 129 | Proline iminopeptidasea | 100.00% | 30s775611g009 | 40654/5.64 | 31000/5.45 | 4.57% | 2 | ns | 2.22 | ns |
| 210 | Proteasome subunit β type 7-A | 100.00% | 30s1126271g001 | 29520/6.79 | 19000/4.4 | 4.40% | 1 | −2.20 | −4.21 | −4.79 |
| 300 | Nuclear transport | 99.20% | 30s1197741g004 | 16883/5.77 | 11000/5.25 | 6.80% | 1 | −2.91 | −3.50 | −1.66 |
| 370 | GDP dissociation inhibitora | 100.00% | 30s1179061g005 | 49697/5.83 | 47000/4.8 | 4.50% | 2 | ns | 1.74 | 2.60 |
| 531 | Chaperonin containing t-complex protein epsilon | 100.00% | 30s1179991g005 | 59114/5.67 | 30000/6.15 | 7.32% | 4 | ns | ns | 1.64 |
| 08 | Intracellular traffic | |||||||||
| 417 | Protein binding structural molecule | 99.90% | 30s784571g001 | 33386/5.26 | 50000/5.75 | 6.89% | 1 | ns | −1.93 | −2.51 |
| 11 | Disease/defence | |||||||||
| 303 | Heat shock protein 82 | 100.00% | 30s947641g003 | 80204/5.04 | 82000/5.9 | 10.70% | 2 | ns | −2.50 | −1.57 |
| 303 | Heat shock protein 82 | 100.00% | 30s722381g008 | 89708/5.18 | 82000/5.9 | 12.40% | 3 | ns | −2.50 | −1.57 |
| 344 | Heat shock protein 82 | 99.60% | 30s1082281g001, 30s722381g008, 30s947641g003 | 89708/4.66 | 80000/5.85 | 3.17% | 1 | ns | −2.73 | −1.57 |
| 352 | Heat shock protein 82 | 99.70% | 30s1082281g001, 30s722381g008, 30s947641g003 | 89708/4.66 | 54000/5.9 | 3.17% | 1 | ns | ns | −2.00 |
| 401 | Aldose reductase | 100.00% | 30s1006721g002 | 34679/6.01 | 19000/5.0 | 14.10% | 5 | 2.13 | ns | 1.78 |
| 12 | Unclear classification | |||||||||
| 91 | Riboflavin α- subunit | 99.00% | 30s703111g002 | 28760/9.09 | 19000/5.5 | 5.22% | 2 | 2.09 | −1.58 | −2.61 |
| 199 | Nucleic acid binding | 99.80% | 30s1050381g022 | 36142/4.99 | 22000/5.88 | 6.98% | 1 | ns | −2.95 | −2.56 |
| 477 | Nucleic acid binding | 100.00% | 30s1050381g022 | 14374/4.99 | 15000/5.95 | 10.90% | 2 | ns | −2.16 | −2.70 |
| 507 | JHL06P13.3-like proteinb | 99.80% | 30s1042771g010 | 50733/5.46 | 43000/5.3 | 2.22% | 1 | ns | 3.96 | 9.99 |
| 508 | JHL06P13.3-like proteinb | 100.00% | 30s1042771g010 | 50733/5.46 | 55000/6.26 | 4.44% | 2 | ns | 1.73 | ns |
| 561 | Mitochondrial glycoprotein | 99.20% | 30s796661g001 | 33383/5.55 | 26000/4.35 | 4.05% | 1 | ns | ns | −2.03 |
| 20 | Secondary metabolism | |||||||||
| 431 | Glutamate 1-semialdehyde aminotransferase | 100.00% | 30s1039641g001 | 50533/5.98 | 39000/4.8 | 5.29% | 2 | ns | 4.54 | 5.18 |
| 603 | Glutamate 1-semialdehyde aminotransferase | 99.60% | 30s1039641g001 | 50533/5.98 | 55000/5.33 | 2.54% | 1 | ns | 4.07 | 3.70 |
| 623 | Formamidase-like protein | 99.50% | 30s702741g005 | 48663/5.73 | 34000/4.5 | 2.47% | 1 | ns | −1.73 | −3.02 |
| 635 | Formamidase-like protein | 99.50% | 30s702741g005 | 48663/5.73 | 54000/5.1 | 2.47% | 1 | −1.66 | −3.63 | −4.27 |
Abbreviations: ns, not significant; NUP, no. of unique peptides.
Proteins identified in other fruit studies but not shown to be linked with fruit development or ripening.
Highly homologous to Barbados nut (Jatropha curcas) and involved in ATP-binding in addition to having ATP-dependent helicase activity.
Sixty-four differentially expressed date proteins (Table 2) showed opposite transient accumulation to previously reported expression changes in fruits such as tomato,19 strawberry21 and apple.14 These proteins include 30s ribosomal protein s1 (spot 287), whose expression increased in date during development and ripening, particularly at NTR and RIPE, and aldo/keto reductase (spots 274, 378, 523 and 525) that was upregulated at NTR and RIPE. These proteins were reported to decrease in tomato,19 strawberry21 and apple14 during fruit development and ripening. Likewise, proteins involved in metabolism such as glucan endo-1,4-β-glucosidase (spot 192), energy metabolism-related proteins like transaldolase-like protein (spot 577) and transketolase 1 (spot 202), signal transduction proteins such as zinc finger (spot 587) and disease/defense protein, E3 ubiquitin ligase (spot 583) were downregulated at NTR and RIPE in the date hypanthium, but upregulated in papaya (Carica papaya),42 peach,43 mango (Mangifera indica)44 and grapes45 during fruit ripening. The different protein expression patterns noted here may indicate date specific development patterns and/or result from differing plant interactions with their respective environments.46
Table 2. Differentially regulated proteins in the date hypanthium showing contrasting transient patterns compared to other fruits.
| Spot no. | Identified protein | Protein probability | Accession no. | Theoretical MW (Da)/pI | Observed MW (Da) /pI | Sequence coverage | NUP | FC S1/MD | FC S1/NTR | FC S1/R | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | Metabolism | |
|
|||||||||
| 91 | Isoamyl acetate-hydrolyzing esterase | 100.00% | 30s696051g001 | 36883/5.86 | 19000/5.5 | 4.79% | 2 | 2.09 | −1.58 |
−2.61 |
||
| 192 | Glucan endo-1,4-β-glucosidase | 100.00% | 30s896091g001 | 16069/6.54 | 60000/6.1 | 14.50% | 3 | −1.90 | −2.96 |
−15.84 |
||
| 196 | Cytochrome p450 | 99.70% | 30s6550951g018 | 24174/4.39 | 23000/6.1 | 7.37% | 1 | ns | ns |
−2.09 |
||
| 202 | Transketolase 1 | 99.00% | 30s656601g005 | 84589/5.66 | 95000/4.9 | 1.42% | 1 | 1.86 | −8.33 |
−8.45 |
||
| 312 | UDP-glucose pyrophosphorylase | 100.00% | 30s904561g011 | 34207/6.1 | 54000/5.25 | 7.47% | 2 | ns | −2.28 |
−1.65 |
||
| 317 | Sorbitol dehydrogenase | 100.00% | 30s830031g004 | 38361/5.53 | 45000/5.6 | 21.30% | 7 | ns | ns |
−1.56 |
||
| 443 | S-adenosylmethionine synthetase | 100.00% | 30s1183641g001 | 43368/5.68 | 43000/5.1 | 11.20% | 3 | ns | −2.25 |
−4.48 |
||
| 443 | S-adenosylmethionine synthetase | 100.00% | 30s1207141g006 | 43344/5.76 | 43000/5.1 | 8.08% | 1 | ns | −2.25 |
−4.48 |
||
| 471 | Aldehyde dehydrogenase | 100.00% | 30s693451g001 | 59067/6.48 | 55000/4.65 | 18.80% | 11 | ns | 2.18 |
−2.17 |
||
| 476 | S-adenosyl-l-homocysteine hydrolase | 100.00% | 30s1132381g003, 30s825931g001 | 38838/6.72 | 45000/6.0 | 13.00% | 6 | ns | ns |
−2.83 |
||
| 476 | Aldehyde dehydrogenase | 100.00% | 30s693451g001 | 59067/6.98 | 45000/6.0 | 12.20% | 5 | ns | ns |
−2.83 |
||
| 522 | Reversibly glycosylated polypeptide | 100.00% | 30s969461g002 | 41051/6.12 | 37000/5.9 | 3.59% | 2 | ns | −2.27 |
−3.20 |
||
| 573 | Pyruvate decarboxylase | 99.70% | 30s919171g003 | 38883/8.66 | 60000/6.2 | 3.46% | 1 | ns | −2.33 |
−5.94 |
||
| 660 | S-adenosylmethionine synthetase | 99.60% | 30s1183641g001, 30s1207141g006, 30s799761g004, 30s933141g001 | 43344/5.68 | 25000/6.1 | 3.28% | 1 | ns | ns |
−2.64 |
||
| 02 | Energy | |
|
|||||||||
| 2 | Pyruvate dehydrogenase E1 component β-subunit | 99.90% | 30s852391g002 | 40148/5.46 | 34000/5.74 | 6.43% | 1 | ns | −3.12 |
−2.73 |
||
| 18 | Enolase | 100.00% | 30s663761g002 | 47828/5.91 | 55000/5.15 | 7.64% | 3 | ns | −1.90 |
−1.50 |
||
| 403 | Citrate synthase | 99.50% | 30s1040901g002 | 30711/6.25 | 55000/4.3 | 6.64% | 1 | ns | 2.52 |
3.23 |
||
| 421 | Malate dehydrogenase | 100.00% | 30s892681g001 | 48314/7.11 | 49000/6.25 | 6.11% | 1 | −1.64 | −7.02 |
−4.54 |
||
| 461 | Malate dehydrogenase | 100.00% | 30s903851g004, 30s946431g012 | 26083/8.76 | 30000/4.7 | 4.39% | 2 | ns | ns |
−2.09 |
||
| 506 | Pyruvate dehydrogenase E1 component β-subunit | 100.00% | 30s855801g002 | 22740/5.7 | 35000/5.31 | 13.00% | 2 | ns | −1.60 |
−1.70 |
||
| 531 | Bisphosphoglycerate-independent phosphoglycerate mutase | 100.00% | 30s661041g002, 30s693071g003 | 43543/5.55 | 30000/6.15 | 5.60% | 1 | ns | ns |
1.64 |
||
| 566 | Fructose-bisphosphate aldolase | 100.00% | 30s1148281g010 | 40222/7.55 | 16000/4.4 | 12.10% | 4 | ns | −2.03 |
−2.99 |
||
| 577 | Transaldolase-like protein | 100.00% | 30s1065691g004 | 43557/5.05 | 55000/6.5 | 4.88% | 2 | −1.79 | −7.48 |
−4.49 |
||
| 03 | Cell growth/division | |
|
|||||||||
| 45 | Condensin complex subunit 1 | 99.30% | 30s702551g004 | 127265/5.78 | 51000/5.34 | 1.47% | 1 | ns | 2.53 |
1.95 |
||
| 252 | Enhancer of polycomb-like protein | 99.70% | 30s827291g002 | 51710/9.99 | 18000/6.2 | 5.79% | 1 | 4.51 | 4.99 |
2.11 |
||
| 415 | 14-3-3-like protein | 99.70% | 30s819041g002 | 17032/5.09 | 19000/6.24 | 7.95% | 1 | 2.71 | 2.55 |
1.72 |
||
| 04 | Transcription | |
|
|||||||||
| 416 | Transcription factor IIA small subunit | 99.70% | 30s656261g002, 30s760301g006 | 12093/6.08 | 27000/4.7 | 10.40% | 1 | 2.52 | 3.51 |
2.46 |
||
| 05 | Protein synthesis | |
|
|||||||||
| 45 | Translation initiation factor (eif-4a) | 99.90% | 30s724051g001, 30s997411g005 | 47064/5.39 | 51000/5.34 | 2.42% | 1 | ns | 2.53 |
1.95 |
||
| 53 | Elongation factor 1 | 99.20% | 30s667161g002 | 16163/4.26 | 21000/6.6 | 11.90% | 1 | −1.84 | −1.79 |
1.55 |
||
| 287 | 30s ribosomal protein s1 | 97.70% | 30s783011g009 | 45242/4.95 | 35000/6.25 | 2.15% | 1 | 1.58 | 8.73 |
11.49 |
||
| 416 | Peptide chain release factor, putative | 99.70% | 30s682511g003 | 9163/4.85 | 27000/4.7 | 13.30% | 1 | 2.52 | 3.51 |
2.46 |
||
| 504 | 60s ribosomal protein l23a | 99.70% | 30s1035671g001 | 9453/11.22 | 88000/6.0 | 11.00% | 1 | 1.62 | ns |
−3.63 |
||
| 525 | Translation initiation factor (eif-4a) | 100.00% | 30s724051g001, 30s997411g005 | 47064/5.39 | 39000/5.34 | 9.93% | 4 | ns | 3.98 |
5.12 |
||
| 06 | Protein destination and storage | |
|
|||||||||
| 3 | Cysteine protease | 98.80% | 30s790241g001 | 36042/4.87 | 22000/6.55 | 4.98% | 1 | ns | −2.48 |
−1.80 |
||
| 32 | Luminal binding protein | 100.00% | 30s685511g001 | 56109/8.42 | 82000/5.95 | 7.24% | 2 | ns | ns |
−2.52 |
||
| 92 | Mitochondrial processing peptidase | 100.00% | 30s927641g002 | 59451/5.76 | 55000/5.48 | 6.08% | 3 | ns | 1.92 |
1.88 |
||
| 239 | Luminal binding protein | 100.00% | 30s685511g001 | 56109/8.42 | 70000/5.8 | 18.00% | 8 | ns | ns |
−1.62 |
||
| 288 | Cysteine protease | 99.70% | 30s790241g001 | 36042/4.87 | 32000/6.3 | 4.98% | 1 | ns | 2.43 |
2.24 |
||
| 303 | Subtilisin-like serine proteinase | 100.00% | 30s808251g002 | 48672/4.79 | 82000/5.9 | 5.76% | 2 | ns | −2.50 |
−1.57 |
||
| 419 | Mitochondrial processing peptidase | 100.00% | 30s927641g002 | 59451/5.76 | 55000/5.5 | 4.60% | 1 | ns | 2.78 |
ns |
||
| 609 | Cysteine protease | 99.60% | 30s790241g001 | 36042/4.87 | 31000/6.48 | 4.98% | 1 | ns | −1.86 |
−1.99 |
||
| 611 | Cysteine protease | 99.40% | 30s790241g001 | 36042/4.87 | 29000/5.15 | 4.98% | 1 | ns | ns |
−2.35 |
||
| 07 | Transporters | |
|
|||||||||
| 99 | Vacuolar ATP synthase subunit v- proton pump b v-ATPase 57 kDa | 100.00% | 30s735571g002, 30s837971g002 | 58290/5.43 | 58000/5.9 | 3.08% | 2 | ns | 2.13 |
ns |
||
| 312 | ATP synthase β chain | 100.00% | 30s884401g004 | 55268/5.31 | 54000/5.25 | 5.10% | 2 | ns | −2.28 |
−1.65 |
||
| 419 | ATP synthase β chain | 100.00% | 30s884401g004 | 55268/5.31 | 55000/5.5 | 7.25% | 3 | ns | 2.78 |
ns |
||
| 507 | ATP synthase β chain | 99.50% | 30s884401g004 | 55268/5.31 | 43000/5.3 | 2.75% | 1 | ns | 3.96 |
9.99 |
||
| 403 | CMP-sialic acid | 99.90% | 30s808171g003 | 23646/9.71 | 55000/4.3 | 9.52% | 1 | ns | 2.52 |
3.23 |
||
| 468 | Mitochondrial deoxynucleotide carrier, putative | 99.70% | 30s656531g003 | 36808/9.09 | 16000/5.35 | 2.69% | 1 | ns | −2.26 |
−2.47 |
||
| 09 | Cell structure | |
|
|||||||||
| 3 | Fibrillin-like protein | 98.80% | 30s724791g001, 30s837971g003 | 39091/9.45 | 22000/6.55 | 4.19% | 1 | ns | −2.48 |
−1.80 |
||
| 19 | Plastid-lipid associated protein 3 | 98.30% | 30s761251g002 | 20316/6.28 | 37000/6.8 | 5.35% | 1 | ns | −1.61 |
−2.37 |
||
| 53 | Fibrillin-like protein | 99.20% | 30s724791g001, 30s837971g003 | 39091/9.45 | 21000/6.6 | 4.19% | 1 | −1.84 | −1.79 |
1.55 |
||
| 317 | Actin | 100.00% | 30s1070141g012, 30s717671g011, 30s844721g006, 30s951221g005 | 41610/5.31 | 45000/5.6 | 22.30% | 3 | ns | ns |
−1.56 |
||
| 356 | Tubulin β-2 | 100.00% | 30s837411g003, 30s965611g001 | 50234/4.72 | 34000/5.2 | 9.60% | 1 | ns | ns |
2.39 |
||
| 356 | Tubulin β-chain | 100.00% | 30s1088541g002, 30s697331g005, 30s708731g001 | 50095/4.77 | 34000/5.2 | 9.60% | 1 | ns | ns |
2.39 |
||
| 10 | Signal transduction |
|||||||||||
| 34 | Germin-like protein | 98.10% | 30s1014231g001 | 13337/5.63 | 18000/4.5 | 17.60% | 1 | 2.71 |
5.01 |
4.50 | ||
| 96 | Nucleoside diphosphate kinase | 100.00% | 30s835621g002 | 12145/9.0 | 16000/4.9 | 28.80% | 3 | 1.74 |
−1.50 |
−1.62 | ||
| 314 | IN2-1 protein | 99.80% | 30s696261g003 | 37485/6.46 | 19000/6.0 | 2.73% | 1 | −1.83 |
ns |
6.10 | ||
| 587 | Zinc finger | 100.00% | 30s796661g001 | 41956/5.55 | 35000/5.8 | 5.15% | 2 | −1.85 |
−8.66 |
−6.63 | ||
| 11 | Disease/defence | |
|
|||||||||
| 13 | Leucine rich repeat-containing | 100.00% | 30s857351g006 | 49345/4.81 | 64000/6 | 9.64% | 3 | ns |
ns |
−1.95 | ||
| 32 | Heat shock protein 70 | 100.00% | 30s941391g004 | 71057/5.13 | 82000/5.95 | 10.90% | 3 | ns |
ns |
−2.52 | ||
| 81 | Glutathione transferase | 100.00% | 30s806101g003 | 37154/5.57 | 30000/5.4 | 8.00% | 2 | ns |
ns |
1.67 | ||
| 239 | Heat shock protein 70 | 100.00% | 30s795061g002 | 71222/5.17 | 70000/5.8 | 19.40% | 1 | ns |
ns |
−1.62 | ||
| 239 | Heat shock protein 70 | 100.00% | 30s941391g005 | 61596/5.11 | 70000/5.8 | 18.10% | 3 | ns |
ns |
−1.62 | ||
| 239 | Heat shock protein 70 | 100.00% | 30s941391g004 | 71057/5.13 | 70000/5.8 | 15.10% | 1 | ns |
ns |
−1.62 | ||
| 239 | Heat shock cognate 70 kDa expressed | 99.80% | 30s1034811g003 | 13813/4.45 | 70000/5.8 | 17.60% | 1 | ns |
ns |
−1.62 | ||
| 246 | ABA-hypersensitive germination 2 nucleic acid binding ribonuclease | 98.70% | 30s1120391g003 | 79218/5.98 | 22000/6.37 | 1.44% | 1 | ns |
−1.50 |
−2.15 | ||
| 274 | Aldo/keto reductase | 100.00% | 30s1183861g001 | 38732/5.97 | 65000/4.75 | 10.50% | 3 | ns |
ns |
3.30 | ||
| 357 | ATGLX1 (glyoxalase i homolog) | 99.70% | 30s1166451g001 | 16766/5.94 | 24000/5.78 | 8.97% | 1 | ns |
1.75 |
1.91 | ||
| 362 | Heat shock protein 70 | 100.00% | 30s941391g005 | 61596/5.11 | 70000/5.9 | 5.90% | 2 | ns |
1.83 |
−3.04 | ||
| 362 | Heat shock protein 70 | 100.00% | 30s795061g002 | 71222/5.17 | 70000/5.9 | 6.02% | 3 | ns |
1.83 |
−3.04 | ||
| 378 | Aldo/keto reductase | 99.80% | 30s1183861g001 | 38732/5.97 | 65000/4.85 | 2.27% | 1 | ns |
ns |
3.09 | ||
| 403 | Catalase | 100.00% | 30s1080231g005 | 55425/6.62 | 55000/4.3 | 10.70% | 5 | ns |
2.52 |
3.23 | ||
| 404 | Catalase | 100.00% | 30s1080231g005 | 55425/6.62 | 55000/4.25 | 4.62% | 2 | ns |
3.79 |
3.82 | ||
| 503 | Glutathione s-transferase GSTF2 | 100.00% | 30s987821g001 | 13597/6.89 | 19000/4.7 | 14.90% | 2 | 2.27 |
2.55 |
2.52 | ||
| 511 | Heat shock protein 70 | 99.80% | 30s941391g005 | 61596/5.11 | 70000/5.85 | 3.76% | 1 | ns |
2.18 |
−2.14 | ||
| 523 | Aldo/keto reductase | 100.00% | 30s1183861g001 | 38732/5.97 | 43000/5.45 | 11.90% | 3 | ns |
1.84 |
2.64 | ||
| 525 | Aldo/keto reductase | 100.00% | 30s1183861g001 | 38732/5.97 | 39000/5.34 | 9.07% | 2 | ns |
3.98 |
5.12 | ||
| 583 | E3 ubiquitin ligase | 99.70% | 30s1065691g004 | 121028/6.79 | 46000/6.25 | 0.91% | 1 | ns |
−5.11 |
−6.36 | ||
| 20 | Secondary metabolism | |
|
|||||||||
| 63 | Isopentenyl diphosphate isomerase | 100.00% | 30s802901g001 | 32219/7.16 | 20000/5.8 | 6.86% | 2 | 1.75 |
ns |
−2.50 | ||
| 63 | Isopentenyl pyrophosphate isomerase | 100.00% | 30s762371g001 | 17552/5.91 | 20000/5.8 | 13.10% | 2 | 1.75 |
ns |
−2.50 | ||
| 91 | Chalcone isomerase | 100.00% | 30s1070141g002 | 24742/5.29 | 19000/5.5 | 4.27% | 1 | 2.09 |
−1.58 |
−2.61 | ||
| 272 | Chalcone isomerase | 100.00% | 30s1070141g002 | 24742/5.29 | 20000/5.7 | 24.80% | 6 | ns |
−5.09 |
−4.71 | ||
| 274 | Chalcone isomerase | 100.00% | 30s1070141g002 | 24742/5.29 | 65000/4.75 | 14.50% | 3 | ns |
ns |
3.30 | ||
| 286 | Anthocyanidin synthase | 100.00% | 30s1125611g001 | 38960/5.99 | 23000/4.7 | 5.51% | 4 | −1.63 | −2.44 | −1.62 | ||
Abbreviations: ns, not significant; NUP, no. of unique peptides.
Classification of development and ripening-specific proteins
Based on UniProt (http://www.uniprot.org/), TAIR (http://www.arabidopsis.org/) and published literature, differentially expressed proteins were assigned to 14 categories previously described:34 ‘disease and defense’ (16.5%), ‘metabolism’ (15.4%), ‘unclassified’ (15.1%), ‘protein destination and storage’ (10.7%), ‘energy’ (9.9%), ‘cellular structure’ (7.0%), ‘secondary metabolism’ (5.5%), ‘signal transduction’ (5.1%), ‘protein synthesis’ (5.1%), ‘unclear classification’ (2.6%), ‘transporters’ (2.6%), ‘transcription’ (2.2%), ‘cell growth/division’ (1.8%) and ‘intracellular traffic’ (0.4%) (Figure 4 and Supplementary File 2). Some proteins have dual roles including ‘response to abiotic stress’ and this will be discussed in the sections below.
Figure 4.
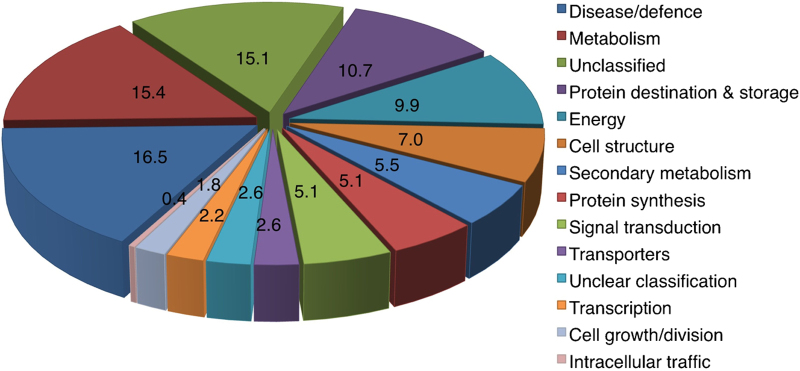
Classification of the differentially expressed proteins. Following 2-DE, protein spots were visualized by SYPRO® Ruby staining and comparatively analyzed with Delta 2D (Decodon). Differentially expressed spots were identified using LC-MS/MS and classified into functional categories according to Ref. 34.
Response to abiotic stress
In this category, nine proteins were identified as date hypanthium-specific, 29 proteins showed contrasting accumulation patterns compared to other fruits and seven had similar patterns. Of the nine date hypanthium-specific proteins, expression of two transcription-related proteins, GATA transcription factor 25 (spot 302) and inducer of C-repeat-binding factor expression 1 DNA-binding transcription activator (spot 302) (Table 1), was reduced at NTR and later increased at RIPE in comparison to S1 stage. C-repeat-binding factor expression 1 is a transcription factor regulating expression of a number of genes with a role in abiotic stress responses including responses to cold,47,48 drought, high temperature and salt stress.49,50 The transient upregulation of C-repeat-binding factor expression 1 at NTR suggests that it might play a role in abiotic stress responses at the onset of date hypanthium ripening, a stage characterized by changes in metabolic activities and fruit physiology.
Two date-specific proteins involved in protein synthesis, group antigen polymerase (Gag-pol) polyprotein (spot 249) and aspartyl-tRNA synthetase (spot 274), were upregulated during ripening (Table 1). Aspartyl-tRNA synthetase, essential during the first step of translation, has been implicated in processes including response to cadmium ion and aspartyl-tRNA aminoacylation, protein maturation and response to salt stress. Its expression was also detected in apple and strawberry fruit extracts.51 In addition to Gag-pol polyprotein and aspartyl-tRNA synthetase, another protein identified in this category, elongation factor thermo unstable (EF-Tu; spots 431 and 433; Supplementary File 2) has been previously detected in maturing fruits. This protein belongs to the GTP binding EF-Tu family and plays an essential role in translational elongation. In plants, EF-Tu is involved in chaperone activity to protect protein aggregation in response to environmental stress, thereby facilitating degradation of N-terminally blocked proteins by the proteasome and triggering resistance to pathogenic bacteria.52 Over-expression of an EF-Tu gene has been shown to improve heat tolerance in maize (Zea mays).53 Similar to the upregulation observed in the current study, it was also detected as increasing during grape ripening.24
Further, five stress-responsive proteins classified in the category ‘protein destination and storage’ showed differential accumulation during development. Three proteins were detected as upregulated, proline iminopeptidase (spot 129) at NTR only, GDP dissociation inhibitor (spot 370) from the onset of ripening, and the ε-subunit chaperonin containing tailless complex protein (spot 531) at RIPE. Expression of two other proteins, proteasome subunit β type 7-A (spot 210) and nuclear transport (spot 300), decreased at all stages (Table 1). GDP dissociation inhibitor is involved in signaling on the plasma membrane and may be essential for actin reorganization.54 The protein was detected in young satsuma mandarins (Citrus unshiu)55 and its expression increased in peach fruits following post-harvest heat treatment.43 In the latter study, GDP dissociation inhibitor was proposed to play a vital role in maintenance of cell integrity during fruit ripening and senescence, and its upregulation at NTR and RIPE suggests a similar role in the date hypanthium.
Expression of aldose reductase (spot 401), a disease/defense-related protein and a member of the aldo-keto reductase family, increased at both MD and RIPE. Unlike other members of the family (e.g., aldo-keto reductase), this protein has not been reported as differentially expressed during development in other fruits. Aldose reductase catalyzes the Nicotinamide adenine dinucleotide phosphate-dependent reduction of carbonyl and aldehyde metabolites like the reduction of glucose to sorbitol.56 In rice (Oryza sativa), expression of aldose reductase increased throughout seed development57 and it was also induced by exogenous application of abscisic acid and abiotic stress such as water deficiency and salinity in vegetative tissues.58 Other proteins identified from this family included aldo-keto reductases (spots 70, 274, 378, 461, 522, 523, 525, 529, 566 and 567). The aldo-keto reductase family protein is involved in tolerance to abiotic stress like heavy metals and drought57,59 and in scavenging reactive oxygen species and their toxic counterparts generated due to environmental stress.59 The transient abundance changes of aldose reductase and aldo-keto reductases in dates might suggest a role in fruit development and/or in abiotic stress tolerance.
Heat shock proteins (HSPs) facilitate refolding of denatured proteins in response to high light intensity, heat, hydrogen peroxide and/or pathogenic attack.60,61 HSPs also act as molecular chaperones assisting and regulating maturation of target proteins. In this study, three HSP82 (spots 303, 344 and 352) were identified as downregulated at NTR and RIPE, and these were date response specific proteins. The HSP82 gene is strongly induced by temperatures around 37–42 °C in maize, particularly in embryos and tassels.62 In other fruits like peach and apple, differential accumulation of only the small HSPs has been reported after heat stress treatment during ripening and storage.18,26 The comparative analysis in this study did not identify small HSPs (molecular mass of 15–30 kDa) but other HSPs including HSP70, also reported in other fruits (Supplementary File 2). Accumulation of nine HSPs (from seven spots: 26, 32, 40, 239, 303, 326 and 352) decreased at NTR and RIPE and two HSP70 (spots 362 and 511) increased transiently at NTR. This result is evidence for the complex role HSPs and suggests that different HSP isoforms might be regulated at various stages during fruit development.
Other stress-related proteins were detected as differentially expressed and included universal-stress protein (USP, spot 660) and E3 ubiquitin ligase (spot 380 and 583) (Supplementary File 2). USP is ubiquitously induced during defense to protect cellular components from heat stress, oxidants or DNA-damaging agents.63 In rice, USPs are involved in ethylene-mediated stress adaptation64 and it was proposed that they have a similar role in tomato fruit as expression increased simultaneously with ethylene-related enzymes during development and ripening.19 In contrast, in dates, USP was downregulated at RIPE, which correlated with the decrease in the ethylene-related enzymes identified. E3 ubiquitin ligase catalyzes the transfer of ubiquitin to target proteins, resulting in their labeling for degradation.65 Ubiquitination also plays a crucial role in abiotic and biotic stress responses,65 as well as in the regulation of plant immune signaling (for review see, Ref. 66). In the current study, E3 ubiquitin ligase (spot 380) accumulation increased at all stages in comparison to S1, while expression of spot 583 decreased at NTR and RIPE. The expected mass of E3 ubiquitin ligase (121 kDa) differed from the observed mass of spots 380 (18 kDa) and 583 (46 kDa), suggesting that these spots might represent either subunits of E3 ubiquitin ligase or truncated version of their full length.38
Proteins with a role in primary metabolism
Spot abundance of most proteins in the category ‘primary metabolism’ declined from the onset of ripening, and six of them were specific to the date hypanthium. The majority of these proteins have a role in sugar and polysaccharide metabolism (Supplementary File 2). In this category, five spots were identified as sorbitol dehydrogenase (SDH; spots 281, 317, 603, 629 and 635), an enzyme that catalyzes the conversion of sorbitol to fructose, the first step of sorbitol utilization.67 In apples, SDH expression was observed throughout fruit development and was proposed to be critical in sugar metabolism during early seed and cortex development.68 Rising SDH accumulation was reported during the transition from cell division to cell expansion, and also at ripening in some apple cultivars.69,70 Of the five date spots identified as SDH, spot 281 was the most highly upregulated, increasing from 1.83-fold at MD to 11.43-fold at NTR and 23.71-fold at RIPE. Accumulation of spot 603 increased at NTR and RIPE, while the other three spots decreased at all stages (Supplementary File 2). Overall increase in SDH levels is consistent with an increase in fructose and NADH levels during ripening.
A number of UDP-glucose pyrophosphorylase (UGPase) isoforms were also identified as differentially regulated. This enzyme catalyzes the reversible conversion between glucose-1-phosphate and UDP glucose required for sucrose biosynthesis in plants. In banana, transcripts of UGPase increased in response to exogenous ethylene treatment, and this was more severe in the pulp of unripe fruits than in ripe ones,71 showing that UGPase expression was ethylene-inducible. Exogenous sucrose and fructose have been reported to increase UGPase abundance in leaves and fruit pulp.71 In dates, accumulation of UGPase spots (92, 201 and 508) increased during ripening with the exception of spot 312. This correlated with the increase in SDH levels, and may imply that, similarly to banana, the enzyme is induced by sucrose and fructose but not ethylene since accumulation of two biosynthesis enzymes identified in this study, S-adenosylmethionine synthetase (SAMS) and S-adenosyl-l-homocysteine hydrolase (SAHH), decreased (Supplementary File 2).
Δ-1-pyrroline-5-carboxylate dehydrogenase (spot 298) and glutamate dehydrogenase (GDH; spot 298), both involved in glutamate biosynthesis, were upregulated at MD and NTR. Δ-1-pyrroline-5-carboxylate dehydrogenase catalyzes the conversion of (S)-1-pyrroline-5-carboxylase to L-glutamate, and GDH converts L-glutamate to glutamate γ-aminobutyric acid.72 Previous studies have also depicted a GDH increase in tomato fruits during cell enlargement and a subsequent decline at ripening.73,74 Upregulation of GDH andΔ-1-pyrroline-5-carboxylate dehydrogenase was also observed during fruit development in Chinese barberry (Myrica rubra).75 Like in Chinese barberry, upregulation of both proteins in dates suggests an increase in amino acid metabolism during MD and NTR, the main stages of cell expansion (Figure 3).
Aldehyde dehydrogenase (ALDH) is an enzyme catalyzing the oxidation of various (toxic) aldehydes to their corresponding carboxylic acids in the presence of NAD or NADP as cofactors.76 In Arabidopsis, transgenic lines overexpressing Ath-ALDH3 showed improved tolerance to salt, dehydration, heavy metals and oxidative stress-inducing compounds like H2O2.77 In apple, ALDH was reported as induced by ethylene during fruit development but not at the ripening stage.69 In our study, isoforms of ALDH (spots 298, 471 and 554) were upregulated at MD and NTR, and spots 471, 476 and 568 were downregulated at RIPE. The decline of the latter three isoforms of ALDH correlates with the decreased accumulation of ethylene-related proteins, particularly at RIPE, and may suggest a link between ALDH accumulation and the ethylene pathway.69
Role of ethylene in date ripening
The ethylene biosynthesis pathway starts with the conversion of methionine to S-adenosylmethionine (SAM) by SAMS. SAM is then converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase, the rate-limiting step. Finally, ACC is converted to ethylene in the presence of oxygen by ACC oxidase.78 SAM can also be demethylated to form S-adenosyl-l-homocysteine and catabolized to homocysteine and adenosine by action of SAHH. SAMS has also been linked to the biosynthesis of polyamines that are necessary for cell growth and division,79,80 possibly explaining the increased accumulation of SAMS during early fruit development when cell division reaches its maximum. Recent work on papaya reported that increased SAMS and methionine synthase during fruit ripening was a possible indication of their prerequisite for the ethylene burst in climacteric fruits.36 In dates, a small peak in ethylene production and respiration rate at early ripening in cultivar ‘Negros’ has been reported,81,82 while in another cultivar like ‘Helali’, fruit ripening was inhibited following application of the ethylene inhibitor Ethrel, implying a putative role for ethylene in fruit ripening.83 Furthermore, ethylene was detected from 91 DAP in ‘Hillawi’ dates, then increased until Khalal stage before declining sharply at harvest.11 However, no change was observed in yellow ‘Barhi’ fruits exposed to 100 ppm ethylene for 48 h at 20 °C and 85%–90% relative humidity.35 Expression of SAHH was reported to increase during ripening of climacteric fruits.84,85 Results in olive (Olea europaea) drupe suggested that decreased expression of SAMS and SAHH is specific to non-climacteric fruits.80 In the current study, expression of SAMS (spots 443 and 660) and SAHH (spot 476) was detected as declining at RIPE and this could retard ‘Barhi’ fruit ripening. Although date palm has been classified as a climacteric plant, accumulation patterns of SAMS and SAHH are similar to that of olive drupes, a non-climacteric fruit. In total, about 34 proteins (corresponding to 55 spots; Supplementary File 2) that have been characterized as specific to non-climacteric ripening and only six proteins (corresponding to 17 spots) to climacteric ripening86 were identified in the present study, suggesting that cultivar ‘Barhi’ may follow an ethylene-independent ripening process.
Proteins associated with secondary metabolism
Proteins with a role in secondary metabolism may also play a pivotal role in environmental stress adaptation.87 In this category, 14 proteins were identified and two of them, glutamate 1-semialdehyde aminotransferase (spots 431 and 603) and formamidase-like protein (spots 623 and 635), have not been previously reported in fruits (Table 1). The remaining proteins, including chalcone isomerase isoforms (spots 91, 272, 274 and 374), chalcone synthase (spot 70) and anthocyanidin synthase (spots 69 and 286), have previously been characterized in other fruits. Anthocyanins are pigments commonly induced under stress conditions.88 They have also been shown to slow down over-ripening in tomato fruits.89 Interestingly, the two spots identified as anthocyanidin synthase, abundance of spot 69 increased at all stages, especially at ripening (21-fold). In contrast, expression of spot 286 declined throughout development, particularly at NTR (−2.44-fold). The marked increase of spot 69 at NTR and RIPE might cause anthocyanidin accumulation and synthesis of anthocyanins.
The correlation between anthocyanins and photosynthesis might be controlled by ethylene since an increase in ethylene was shown to negatively regulate anthocyanin synthesis, while an increase in anthocyanin inhibited photosynthesis.90 This suggests that the decrease in ethylene biosynthesis enzymes observed here might have promoted anthocyanin production and indirectly reduced photosynthesis (in correlation with the downregulation of Rubisco ∝-subunit, spots 13 and 14, observed during fruit maturation). Furthermore, increased accumulation of anthocyanins may suggest a response to light stress since their synthesis has also been linked to high light exposure90 and protection from photo-oxidative damages.91,92 For example, as fruit bearing branches grow heavier and bend downwards away from the protective leaves, direct light exposure to the fruit may increase.
Proteins associated with energy production
A total of 20 proteins involved in energy generation were identified. Expression of UMP6 mitochondrial precursor (spot 477), detected as decreasing during ripening, has not previously been detected in other fruits (Table 1). This protein is located in the mitochondria and thylakoid and is involved in organelle organization, DNA metabolic and cell cycle processes. Additionally, nine proteins showed contrasting expression patterns in relation to other fruit species (Table 2). Amongst them, a transaldolase-like protein, transketolase 1, chloroplast oxygen evolving enhancer protein, isoforms of Rubisco α-subunit and malate dehydrogenase (MDH) were identified (Supplementary File 2). The MDH enzyme catalyzes the reversible oxidation of malate to oxaloacetate with concomitant reduction of NAD+. It is involved in the TCA cycle and facilitates exchange of metabolites between the cytoplasm and organelles.93 In higher plants, four isoforms of MDH were detected, three are NAD-dependent and located in mitochondria, cytoplasm and microbodies, while the fourth one is NADP-dependent and was detected in chloroplasts.94 In grapes, expression of MDH increases throughout ripening.95 Here, accumulation of spot 421 decreased at all stages, particularly at NTR. Accumulation of spot 482 decreased at MD and then increased at NTR and RIPE, while spot 461 decreased at RIPE. Other spots identified as MDH (spots 129, 137, 274, 412 and 428) were upregulated at different stages of fruit development (Supplementary File 2). Overall significant changes in TCA cycle enzymes suggest increasing energy generation throughout the process of fruit development and ripening and this is consistent with previous findings.24
Green fruits contain photosynthetically active chloroplasts that contribute to energy metabolism.96 The decline of Rubisco α-subunit isoforms (spots 13 and 14) and chloroplast oxygen evolving enhancer protein (spots 283 and 555) during ripening processes has also been reported in tomatoes,19 apricots27 and grapes.24,97 Reduced expression of the two proteins correlated with a change in skin color from green to light yellow (Figure 3) similarly to apples.18 Decrease in Rubisco and chloroplast oxygen evolving enhancer proteins is a potential indicator of chlorophyll degradation, marking the transition of chloroplasts to chromoplasts.20 Further, proteins involved in other photosynthesis-related pathways such as the pentose phosphate pathway (PPP) and the Calvin cycle were identified. PPP is an important process that generates Nicotinamide adenine dinucleotide phosphates and pentoses that are the substrates for the Calvin cycle, the dark reactions of photosynthesis. Transketolase 1 is involved in both the non-oxidative phase of PPP and the Calvin cycle. In the former, transketolase 1 is involved in both up- and down-stream of transaldolase-like protein that catalyzes the transfer of a 3-C dihydroxyacetone moiety from sedoheptulose-7-phosphate to glyceraldehyde-3-phosphate. In the Calvin cycle, transketolase 1 converts sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate to aldose ribose-5-phosphate and ketose D-xylulose-5-phosphate.98 In the current study, transaldolase-like protein (spot 577) and transketolase 1 (spot 202) significantly decreased at all stages, and more prominently so at NTR and RIPE, suggesting a downregulation of non-oxidative phase of PPP and the Calvin cycle.
The reduction of proteins associated with photosynthesis correlated with the reduction of pyruvate dehydrogenase (PDH) E1 component (spots 2 and 506) and the increase of mitochondrial pyruvate dehydrogenase kinase isoform 1 (mPDK, spot 431, also classified as a signaling protein). PDH E1 component is a key enzyme involved in the decarboxylation of pyruvate and reductive acetylation of lipoic acid to form acetyl-CoA, while mPDK regulates activity of the PDH complex by phosphorylating PDH E1 component.99 In grapes, the mPDK isoform was induced in a ripening-specific manner.100 In the current study, decrease in PDH E1 component and increase in mPDK accumulation both occurred in a ripening-specific manner (i.e., at NTR and RIPE). This may indicate that while accumulation of PDH E1 component decreased, the increased abundance of mPDK could promote inactivation of the remaining PDH E1 components of PDH complex.
Enzymes involved in the glycolysis pathway were also identified including fructose-bisphosphate aldolase (spot 566), triosephosphate isomerase (spot 503), phosphoglycerate kinase (spot 431), phosphoglycerate mutase (spot 531) and enolase (spot 18). Triosephosphate isomerase was upregulated at all stages, and similarly to apples, possibly enhancing respiration and catabolic metabolism.18 Expression of phosphoglycerate kinase increased at NTR and RIPE, and phosphoglycerate mutase at RIPE, while that of enolase decreased at NTR and RIPE. Increases in phosphoglycerate kinase and phosphoglycerate mutase suggest enhanced energy production, particularly since this correlated with the upregulation of ATP synthase β-chain (spot 312, 419 and 507) of the ATP biosynthesis pathway. ATP synthase β-chain is involved with fuelling cellular energy, and has recently been demonstrated as a key regulator of fruit ripening and senescence.101 In litchi (Litchi chinensis) fruit pericarp, ATP synthase β-chain accumulation significantly increased during fruit development and early post-harvest, while alternative oxidase 1 significantly increased two days post-harvest.101 Exogenous application of ATP induced a reduction of alternative oxidase 1 expression, enhancing antioxidant systems and delayed pericarp browning in litchi101–103 and ‘Conference’ pears.104 Additionally, elevated ATP and energy levels decrease membrane permeability and production of reactive oxygen species during ripening.104,105 Here, expression of ATP synthase β-chain, spot 507, increased at RIPE (9.99-fold), spot 419 transiently increased at NTR (2.78-fold), while spot 312 decreased at NTR (−2.28-fold) and RIPE (−1.65-fold, Supplementary File 2). The overall increase of this ATP-generating enzyme particularly at NTR and RIPE could help maintain membrane integrity and promote antioxidant capacity, which could in turn protect dates against oxidative stress and senescence.102,103
Proteins implicated in cellular structures
Decline in fruit firmness during ripening and senescence is partially controlled by changes in the expression of cytoskeleton-related proteins and cell wall-degrading enzymes.38,42 These can lead to biochemical and physical/structural alterations in cells and their walls. Of the proteins identified in this category, actin (spot 317), fibrillin (spot 3 and 53), plastid-lipid associated protein 3 (spot 19) and tubulin β-chain (spot 356) showed contrasting differential accumulation in dates compared to other fruits such as grapes (Table 2 and Supplementary File 2). Actins are involved in cytoplasmic streaming, cell division, organelle movement and cell shape maintenance and, in grapes, their abundance remained elevated throughout fruit development.22,106 In this study, actin was identified in four spots (129, 317, 400 and 487). Spot 129 was upregulated at NTR, spot 400 increased at MD and NTR, and spot 487 increased at NTR and RIPE, while spot 317 decreased at RIPE (Table 2 and Supplementary File 2). Besides actins, tubulins, which are cytoskeletal proteins implicated in cellular processes including the preservation of cell structure and intracellular transport, were also identified. Increased tubulin levels have been reported in the early phase of cucumber (Cucumis sativus) development107 and elevated expression of actins and tubulins has been observed during rapid cell enlargement, a phase characterized by extensive cytoskeleton rearrangement.23 Here, eight spots (14, 56, 189, 197, 356, 359, 417 and 422) identified as tubulins were downregulated at NTR and RIPE, with the exception of spot 356 that increased at RIPE. Similarly to the present study, expression of tubulins was reported to decline during grape berry development and was very limited during ripening. Additionally, xylose isomerase (spot 507 and 508), which is involved in a reversible conversion of D-xylose to xylulose, was upregulated at ripening. D-xylose is the primary constituent of xylans, which are involved in cell wall stabilization.108
Posttranslational modifications
The current study highlights interactions of various cellular and metabolic processes linked to fruit development and ripening, as observed by changes in protein accumulation. A number of proteins undergo PTMs for acquiring stability, subcellular localization, (de)-activation and/or protein–protein interactions. Previous studies reported carbonylation of several proteins during senescence in apple109 and peach, 110 initiating PTM investigations in fruit proteomics. In the present study, a total of 362 methionine oxidation sites positioned on 204 proteins and 443 phosphosites on 328 proteins were detected using Scaffold PTM (Supplementary File 5). These PTMs potentially have important roles in regulating fruit development and ripening and merit a detailed follow-up study.
Conclusions
Here, we provide the first detailed comparative proteome of the date fruit, a fruit that is of particular horticultural interest in the Middle East and North Africa. Proteomics afforded four major insights in differential protein expression during fruit development and ripening. The first is an increase in the accumulation of abiotic stress-responsive proteins during ripening with the exception of HSPs and universal stress protein, which decreased, suggesting intricate metabolic systems for stress response and, in particular, the ability to withstand high temperatures. The second is an overall increase of glycolysis and TCA cycle-related proteins in contrast to pentose phosphate and photosynthesis from ripening onset. This indicates a shift in energy supply between the fruit development stages markedly S1-MD and NTR-RIPE. The third is a decrease of two enzymes involved in ethylene biosynthesis, S-adenosylmethionine synthetase and S-adenosyl-l-homocysteine hydrolase, which are among the 34 identified proteins proposed to be specific to non-climacteric ripening. This implies that the cultivar ‘Barhi’ might follow an ethylene-independent ripening process. The fourth is a differential accumulation of enzymes involved in secondary metabolism, notably the sharp increase in anthocyanidin synthase, from the anthocyanin biosynthesis pathway, during ripening. Anthocyanins act as natural antioxidants and play a crucial protective role in response to light. The results from this investigation will further our understanding of the role and regulation of proteins and pathways during fruit ripening, and will contribute to the development of innovative practices for fruit quality improvements.
Acknowledgments
The authors express their gratitude to Natalia Serrano, Cléa Marondedze and Frederick Kinyua for their technical assistance and help with the sampling, and Fouad Lemtiri-Chlieh and Lee Gehring for critical reading of the manuscript.
The authors declare no conflict of interest.
References
- Saker MM, Moursy HA. Molecular characterisation of Egyptian date palm cultivars: RAPD fingerprints. Arab J Biotechnol 1999; 2: 71–78. [Google Scholar]
- Morton J. Date. In: Morton J (ed). Fruits of Warm Climates. West Lafayette: Purdue University Center for New Crops and Plants Products, 1987: 5–11. [Google Scholar]
- Vayalil PK. Date Fruits (Phoenix dactylifera Linn): an emerging medicinal food. Crit Rev Food Sci 2012; 52: 249–271. [DOI] [PubMed] [Google Scholar]
- Maier VP, Metzler DM. Quantitative changes in date polyphenols and their relation to browning. J Food Sci 1965; 30: 80–84. [Google Scholar]
- Kikuchi N, Miki T. Separation of date (Phoenix dactylifera) sterols by liquid-chromatography. Mikrochim Acta 1978; 1: 89–96. [Google Scholar]
- Al-Farsi M, Alasalvar C, Morris A, Baron M, Shahidi F. Comparison of antioxidant activity, anthocyanins, carotenoids, and phenolics of three native fresh and sun-dried date (Phoenix dactylifera L.) varieties grown in Oman. J Agric Food Chem 2005; 53: 7592–7599. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organisation of the United Nations Statistical Databases (FAOSTAT), 2012. Available at http://faostat.fao.org/site/339/default.aspx (accessed April 2014).
- El-Rayes DA. Characterization of three date palm cultivars based on RAPD finger prints and fruit chemical composition. Environ Arid Land Agric Sci 2009; 20: 3–20. [Google Scholar]
- Yang M, Zhang XW, Liu GMet al. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS ONE 2010; 5: e12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qurashi AD, Awad MA. 5-Aminolevulinc acid increases tree yield and improves fruit quality of ‘Rabia’ and ‘Sukkariat-Yanbo’ date palm cultivars under hot arid climate. Sci Hort 2011; 129: 441–448. [Google Scholar]
- Abbas MF, Ibrahim MA. The role of ethylene in the regulation of fruit ripening in the hillawi date palm (Phoenix dactylifera L). J Sci Food Agric 1996; 72: 306–308. [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. Fruits: a developmental perspective. Plant Cell 1993; 5: 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 2001; 52: 725–749. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Thodey K, Schaffer RJet al. Global gene expression analysis of apple fruit development from the floral bud to ripe fruit. BMC Plant Biol 2008; 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dous EK, George B, Al-Mahmoud MEet al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat Biotechnol 2011; 29: 521–584. [DOI] [PubMed] [Google Scholar]
- Al-Mssallem IS, Hu SN, Zhang XWet al. Genome sequence of the date palm Phoenix dactylifera L. Nat Commun 2013; 4: 2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Zhang X, Fang Yet al. High-throughput sequencing-based gene profiling on multi-staged fruit development of date palm (Phoenix dactylifera L.). Plant Mol Biol 2012; 78: 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Jiang L, Zhang L, Kang R, Yu Z. Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage. Hort Res 2014; 1: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurobert M, Mihr C, Bertin Net al. Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol 2007; 143: 1327–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco M, D’Ambrosio C, Arena S, Faurobert M, Scaloni A, Marra M. Proteomic analysis of tomato fruits from two ecotypes during ripening. Proteomics 2006; 6: 3781–3791. [DOI] [PubMed] [Google Scholar]
- Bianco L, Lopez L, Scalone AGet al. Strawberry proteome characterization and its regulation during fruit ripening and in different genotypes. J Proteomics 2009; 72: 586–607. [DOI] [PubMed] [Google Scholar]
- Giribaldi M, Perugini I, Sauvage FX, Schubert A. Analysis of protein changes during grape berry ripening by 2-DE and MALDI–TOF. Proteomics 2007; 7: 3154–3170. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ma H, Feng J, Zeng L, Wang Z, Chen S. Grape berry plasma membrane proteome analysis and its differential expression during ripening. J Exp Bot 2008; 59: 2979–2990. [DOI] [PubMed] [Google Scholar]
- Martinez-Esteso MJ, Selles-Marchart S, Lijavetzky D, Pedreno MA, Bru-Martinez R. A DIGE-based quantitative proteomic analysis of grape berry flesh development and ripening reveals key events in sugar and organic acid metabolism. J Exp Bot 2011; 62: 2521–2569. [DOI] [PubMed] [Google Scholar]
- Hu H, Liu Y, Shi GLet al. Proteomic analysis of peach endocarp and mesocarp during early fruit development. Physiol Plantarum 2011; 142: 390–406. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu Z, Jiang L, Jiang J, Luo H, Fu L. Effect of post-harvest heat treatment on proteome change of peach fruit during ripening. J Proteomics 2011; 74: 1135–1149. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Arena S, Rocco Met al. Proteomic analysis of apricot fruit during ripening. J Proteomics 2013; 78: 39–57. [DOI] [PubMed] [Google Scholar]
- Sghaier-Hammami B, Drira N, Jorrin-Novo JV. Comparative 2-DE proteomic analysis of date palm (Phoenix dactylifera L.) somatic and zygotic embryos. J Proteomics 2009; 73: 161–177. [DOI] [PubMed] [Google Scholar]
- Saravanan RS, Rose JK. A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 2004; 4: 2522–2532. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Marondedze C, Lilley K, Thomas L. Comparative gel-based phosphoproteomics in response to signaling molecules. In: Gehring C (ed.) Cyclic Nucleotide Signaling in Plants: Methods and Protocols. New York: Humana Press, 2013: 1016, 139–154. [DOI] [PubMed] [Google Scholar]
- Thomas L, Marondedze C, Ederli L, Pasqualini S, Gehring C. Proteomic signatures implicate cAMP in light and temperature responses in Arabidopsis thaliana. J Proteomics 2013; 83: 47–59. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005; 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent Eet al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998; 391: 485–488. [DOI] [PubMed] [Google Scholar]
- Kader AA, Hussein A. Harvesting and Postharvest Handling of Dates. Aleppo: ICARDA; 2009. [Google Scholar]
- Nogueira SB, Labate CA, Gozzo FC, Pilau EJ, Lajolo FM, Oliveira do Nascimento JRProteomic analysis of papaya fruit ripening using 2DE-DIGE. J Proteomics 2012; 75: 1428–1439. [DOI] [PubMed] [Google Scholar]
- Marondedze C, Thomas LA. Insights into fruit function from the proteome of the hypanthium. J Plant Physiol 2011; 169: 12–19. [DOI] [PubMed] [Google Scholar]
- Marondedze C, Thomas LA. Apple hypanthium firmness: new insights from comparative proteomics. Appl Biochem Biotechnol 2012; 168: 306–326. [DOI] [PubMed] [Google Scholar]
- Xu J, Pascual L, Aurand Ret al. An extensive proteome map of tomato (Solanum lycopersicum) fruit pericarp. Proteomics 2013; 13: 3059–3063. [DOI] [PubMed] [Google Scholar]
- Toledo TT, Nogueira SB, Cordenunsi BRet al. Proteomic analysis of banana fruit reveals proteins that are differentially accumulated during ripening. Postharvest Biol Technol 2012; 70: 51–58. [Google Scholar]
- Loei H, Lim J, Tan Met al. Proteomic analysis of the oil palm fruit mesocarp reveals elevated oxidative phosphorylation activity is critical for increased storage oil production. J Proteome Res 2013; 12: 5096–5109. [DOI] [PubMed] [Google Scholar]
- Lazan H, Ng SY, Goh LY, Ali ZM. Papaya beta-galactosidase/galactanase isoforms in differential cell wall hydrolysis and fruit softening during ripening. Plant Physiol Biochem 2004; 42: 847–853. [DOI] [PubMed] [Google Scholar]
- Zhang B, Xi WP, Wei WW, Shen JY, Ferguson I, Chen KS. Changes in aroma-related volatiles and gene expression during low temperature storage and subsequent shelf-life of peach fruit. Postharvest Biol Technol 2011; 60: 7–16. [Google Scholar]
- Andrade Jde M, Toledo TT, Nogueira SB, Cordenunsi BR, Lajolo FM, do Nascimento JR. 2D-DIGE analysis of mango (Mangifera indica L.) fruit reveals major proteomic changes associated with ripening. J Proteomics 2012; 75: 3331–3341. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Suzuki S, Tanzawa F, Takayanagi T. Low expression of flavonoid 3′,5′-hydrolase (F3′,5′H) associated with cyanidin-based anthocyanins in grape leaf. Am J Enol Viticul 2009; 60: 362–367. [Google Scholar]
- Ungerer MC, Halldorsdottir SS, Purugganan MA, Mackay TF. Genotype–environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 2003; 165: 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature or high-salt stress. Plant Cell 1994; 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Medina J, Rodriguez-Franco M, Neuhaus G, Salinas J. Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. J Exp Bot 2012; 63: 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Matus-Cqdiz M, Pozniak C, Cattivelli L, Fowler DB. Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol Genet Genomics 2009; 282: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Li XP, Chen W, Lu W, Mao J, Liu T. Molecular cloning characterization and expression analysis of CpCBF2 gene in harvested papaya fruit under temperature stresses. Electron J Biotechnol 2013; 16: 1–10. [Google Scholar]
- Zheng Q, Song J, Doncaster K, Rowland E, Byers DM. Qualitative and quantitative evaluation of protein extraction protocols for apple and strawberry fruit suitable for two-dimensional electrophoresis and mass spectrometry analysis. J Agric Food Chem 2007; 55: 1663–1673. [DOI] [PubMed] [Google Scholar]
- Fu J, Momcilovic I, Prasad PV. Roles of protein synthesis elongation factor EF-Tu in heat tolerance in plants. J Bot 2012; 2012: 1–8. [Google Scholar]
- Ristic Z, Gifford DJ, Cass DD. Heat shock proteins in two lines of Zea mays L. that differ in drought and heat resistance. Plant Physiol 1991; 97: 1430–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson RM, Gandhi PN, Tong Xet al. An activating mutant of Cdc42 that fails to interact with Rho GDP-dissociation inhibitor localizes to the plasma membrane and mediates actin reorganization. Exp Cell Res 2004; 301: 211–222. [DOI] [PubMed] [Google Scholar]
- Boo KH, Kim DW, Cho SKet al. Construction and profiling of a cDNA library from young fruit satsuma mandarin. J Plant Biol 2007; 50: 403–409. [Google Scholar]
- Petrash JM. All in the family: aldose reductase and closely related aldo-keto reductases. Cell Mol Life Sci 2004; 61: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuna Sree B, Rajendrakumar CS, Reddy AR. Aldose reductase in rice (Oryza sativa L.): stress response and developmental specificity. Plant Sci 2000; 160: 149–157. [DOI] [PubMed] [Google Scholar]
- Turoczy Z, Kis P, Torok Ket al. Overproduction of a rice aldo-keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol Biol 2011; 75: 399–412. [DOI] [PubMed] [Google Scholar]
- Oberschall A, Deak M, Torok Ket al. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J 2000; 24: 437–446. [DOI] [PubMed] [Google Scholar]
- Almoguera C, Coca MA, Jordano J. Differential accumulation of sunflower tetraubiquitin mRNAs during zygotic embryogenesis and developmental regulation of their heat-shock response. Plant Physiol 1995; 107: 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehat A, Lurie S, Weiss D. Expression of small heat-shock proteins at low temperatures. A possible role in protecting against chilling injuries. Plant Physiol 1998; 117: 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA, Casey ES, Capitant SAet al. Characterization of two maize HSP90 heat shock protein genes: expression during heat shock, embryogenesis, and pollen development. Dev Genetics 1993; 14: 27–41. [DOI] [PubMed] [Google Scholar]
- Tkaczuk KL, Shumilin IA, Chruszcz M, Evdokimova E, Savchenko A, Minor W. Structural and functional insight into the universal stress protein family. Evol Appl 2013; 6: 434–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M, Rzewuski G, Marwedel T, Lorbiecke R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J Exp Bot 2002; 53: 2325–2331. [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E, Belloni S, Marone Det al. The E3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics 2006; 7: 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan V, Rivas S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci 2014; 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguayo MF, Ampuero D, Mandujano Pet al. Sorbitol dehydrogenase is a cytosolic protein required for sorbitol metabolism in Arabidopsis thaliana. Plant Sci 2013; 205: 63–75. [DOI] [PubMed] [Google Scholar]
- Nosarzewski M, Archbold DD. Tissue-specific expression of sorbitol dehydrogenase in apple fruit during early development. J Exp Bot 2007; 58: 1863–1872. [DOI] [PubMed] [Google Scholar]
- Yamada K, Mori H, Yamaki S. Identification and cDNA cloning of a protein abundantly expressed during apple fruit development. Plant Cell Physiol 1999; 40: 198–204. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Kanayama Y. Changes in the amounts of the NAD-dependent sorbitol dehydrogenase and its involvement in the development of apple fruit. J Am Soc Hort Sci 1996; 121: 848–852. [Google Scholar]
- Pua EC, Lim SS, Liu P, Liu JZ. Expression of a UDPglucose pyrophosphorylase cDNA during fruit ripening of banana (Musa acuminata). Aust J Plant Physiol 2000; 27: 1151–1159. [Google Scholar]
- Gallardo F, Canton FR, Garcia-Gutierrez A, Canovas M. Changes in photorespiratory enzymes and glutamate synthases in ripening tomatoes. Plant Physiol Biochem 1993; 31: 189–196. [Google Scholar]
- Gallego PP, Whotton L, Picton S, Grierson D, Gray JE. A role for glutamate-decarboxylase during tomato ripening—the characterization of a cDNA-encoding a putative glutamate-decarboxylase with a calmodulin-binding site. Plant Mol Biol 1995; 27: 1143–1151. [DOI] [PubMed] [Google Scholar]
- Rolin D, Baldet P, Just D, Chevalier C, Biran M, Raymond P. NMR study of low subcellular pH during the development of cherry tomato fruit. Aust J Plant Physiol 2000; 27: 61–69. [Google Scholar]
- Feng C, Chen M, Xu CJet al. Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genomics 2012; 13: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozich J, Nicholas H, Wang BC, Lindahl R, Hempel J. Relationships within the aldehyde dehydrogenase extended family. Protein Sci 1999; 8: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Bartels D, Kirch HH. Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J 2003; 35: 452–464. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher-plants. Annu Rev Plant Physiol Plant Mol Biol 1984; 35: 155–189. [Google Scholar]
- Liu JH, Honda C, Moriguchi T. Involvement of polyamine in floral and fruit development. Jarq-Jpn Agric Res Q 2006; 40: 51–58. [Google Scholar]
- Bianco L, Alagna F, Baldoni L, Finnie C, Svensson B, Perrotta G. Proteome regulation during Olea europaea fruit development. PLoS ONE 2013; 8: e53563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Pretel MT, Botella MA, Amoros A. Physicochemical changes during date ripening related to ethylene production. Food Sci Technol Int 2001; 7: 31–36. [Google Scholar]
- Lelievre JM, Latche A, Jones B, Bouzayen M, Pech JC. Ethylene and fruit ripening. Physiol Plantarum 1997; 101: 727–739. [Google Scholar]
- Awad MA. Increasing the rate of ripening of date palm fruit (Phoenix dactylifera L.) cv. Helali by preharvest and postharvest treatments. Postharvest Biol Technol 2007; 43: 121–127. [Google Scholar]
- Medina-Suarez R, Manning K, Fletcher J, Aked J, Bird CR, Seymour GB. Gene expression in the pulp of ripening bananas—two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis of in vitro translation products and cDNA cloning of 25 different ripening-related mRNAs. Plant Physiol 1997; 115: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield KA, Dang T, Guis M, Pech JC, Bouzayen M, Bennett AB. Characterization of ripening-regulated cDNAs and their expression in ethylene-suppressed Charentais melon fruit. Plant Physiol 2000; 122: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molassiotis A, Tanou G, Filippou P, Fotopoulos V. Proteomics in the fruit tree science arena: New insights into fruit defense, development, and ripening. Proteomics 2013; 13: 1871–1884. [DOI] [PubMed] [Google Scholar]
- Nascimento NC, Fett-Neto AG. Plant secondary metabolism and challenges in modifying its operation: an overview. Methods Mol Biol 2010; 643: 1–13. [DOI] [PubMed] [Google Scholar]
- Gould KS. Nature’s Swiss army knife: the diverse protective roles of anthocyanins in leaves. J Biomed Biotechnol 2004; 2004: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, de Stefano Ret al. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol 2013; 23: 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PK, Geul B, Choi SB, Yoo SD, Park YI. Photosynthesis-dependent anthocyanin pigmentation in Arabidopsis. Plant Signal Behav 2011; 6: 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie RM, Hetherington SE. Photoabatement by anthocyanin shields photosynthetic systems from light stress. Photosynthetica 1999; 36: 451–463. [Google Scholar]
- Merzlyak MN, Chivkunova OB. Light-stress pigmanet changes and evidence for anthocyanin photoprotection in apples. J Photochem Photobiol B 2000; 55: 155–163. [DOI] [PubMed] [Google Scholar]
- Musrati RA, Kollarova M, Mernik N, Mikulasova D. Malate dehydrogenase: distribution, function and properties. Gen Physiol Biophys 1998; 17: 193–210. [PubMed] [Google Scholar]
- Poeydomenge O, Marolda M, Boudet A, Grima-Pettenati J. Nucleotide sequence of a cDNA encoding mitochondrial malate dehydrogenase from eucalyptus. Plant Physiol 1995; 107: 1455–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taureilles-Saurel C, Romieu CG, Robin JP, Flanzy C. Grape (Vitis vinifera L.) malate dehydrogenase. I. Intracellular compartmentation of the isoforms. Am J Enol Viticulture 1995; 46: 22–28. [Google Scholar]
- Tanaka A, Fujita K, Kikuchi K. Nutrio-physiological studies on the tomato plant. III. Photosynthetic rate on individual leaves in relation to dry matter production of plants. Soil Sci Plant Nutr 1974; 20: 173–183. [Google Scholar]
- Terrier N, Ageorges A, Abbal P, Romieu C. Generation of ESTs from grape berry at various developmental stages. J Plant Physiol 2001; 158: 1575–1583. [Google Scholar]
- Kruger NJ, von Schaewen A. The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 2003; 6: 236–246. [DOI] [PubMed] [Google Scholar]
- Tovar-Mendez A, Miernyk JA, Randall DD. Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur J Biochem 2003; 270: 1043–1049. [DOI] [PubMed] [Google Scholar]
- Pilati S, Perazzolli M, Malossini Aet al. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at veraison. BMC Genomics 2007; 8: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qian Z, Ma Set al. Energy status of ripening and postharvest senescent fruit of litchi (Litchi chinensis Sonn.). BMC Plant Biol 2013; 13: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Qu HX, Jiang YMet al. ATP-induced changes in energy status and membrane integrity of harvested litchi fruit and its relation to pathogen resistance. J Phytopathol 2008; 156: 365–371. [Google Scholar]
- Yi C, Jiang YM, Shi Jet al. ATP-regulation of antioxidant properties and phenolics in litchi fruit during browning and pathogen infection process. Food Chem 2010; 118: 42–47. [Google Scholar]
- Saquet AA, Streif J, Bangerth F. Energy metabolism and membrane lipid alterations in relation to brown heart development in ‘Conference’ pears during delayed controlled atmosphere storage. Postharvest Biol Technol 2003; 30: 123–132. [Google Scholar]
- Duan XW, Jiang YM, Su XGet al. Role of pure oxygen treatment in browning of litchi fruit after harvest. Plant Sci 2004; 167: 665–668. [Google Scholar]
- Sarry JE, Sommerer N, Sauvage FXet al. Grape berry biochemistry revisited upon proteomic analysis of the mesocarp. Proteomics 2004; 4: 201–215. [DOI] [PubMed] [Google Scholar]
- Ando K, Carr KM, Grumet R. Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genomics 2012; 13: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Teng Y, Yin Q, Chen W, Zhao F. Identification, expression, and characterization of the highly conserved D-xylose isomerase in animals. Acta Biochim Biophys Sin 2009; 41: 116–122. [DOI] [PubMed] [Google Scholar]
- Qin G, Wang Q, Liu J, Li B, Tian S. Proteomic analysis of changes in mitochondrial protein expression during fruit senescence. Proteomics 2009; 9: 4241–4253. [DOI] [PubMed] [Google Scholar]
- Qin G, Meng X, Wang Q, Tian S. Oxidative damage of mitochondrial proteins contributes to fruit senescence: a redox proteomics analysis. J Proteome Res 2009; 8: 2449–2462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.