Abstract
Since stem elongation is a gibberellic acid (GA) response, GA inhibitors are commonly used to control plant height in the production of potted ornamentals and bedding plants. In this study, we investigated interfering with GA signaling by using molecular techniques as an alternative approach. We isolated three putative GID1 genes (PhGID1A, PhGID1B and PhGID1C) encoding GA receptors from petunia. Virus-induced gene silencing (VIGS) of these genes results in stunted growth, dark-green leaves and late-flowering. We also isolated the gai mutant gene (gai-1) from Arabidopsis. We have generated transgenic petunia plants in which the gai mutant protein is over-expressed under the control of a dexamethasone-inducible promoter. This system permits induction of the dominant Arabidopsis gai mutant gene at a desired stage of plant development in petunia plants by the application of dexamethasone (Dex). The induction of gai in Dex-treated T1 petunia seedlings caused dramatic growth retardation with short internodes.
Introduction
Gibberellic acid (GA), a plant hormone, regulates many crucial growth and developmental processes, including seed germination, leaf expansion, induction of flowering and stem elongation.1 A common problem in the production of ornamental potted plants is undesirably tall growth, so inhibitors of GA biosynthesis including A-rest (ancymidol), B-nine (daminozide), Bonzi (paclobutrazol), Cycocel (chloromequat chloride) and Sumagic (uniconazole), are commonly used to control plant height.2,3 To provide an alternative strategy for managing plant architecture and preventing postharvest ‘stretching’, we propose to investigate genetic manipulation of the GA response pathway.
In the current model of GA signaling, GA binds to a soluble GID1 receptor, which in turn binds to the DELLA repressor protein. The bound DELLA protein is then targeted for degradation by the 26S proteasome, thus relieving DELLA-mediated repression of GA-dependent growth processes.4,5 The genes encoding the GA response cascade have been identified using dwarf mutants of Arabidopsis, wheat and rice.6,7 A soluble GA receptor was identified as the basis of the rice GA-insensitive dwarf1 (GID1) mutant.8 In Arabidopsis, there are three GID1 orthologs (AtGID1a, AtGID1b and AtGID1c); the gid1a/gid1b/gid1c triple mutant was severely dwarfed 9 and showed high levels of RGA (REPRESSOR OF GA1-3) and GAI (GA-INSENSITIVE) proteins.10 These proteins, characterized by the conserved DELLA domain at their N termini, function as repressors in GA signalling.11,12 Loss-of-function mutants such as rice slr1,13 and Arabidopsis gai-t6 and rga-24,14 are DELLA deficient, and are taller and earlier flowering than wild-type plants. Conversely, DELLA gain-of-function mutants or transgenic plants are dwarfed, and flower late. Such a mutant, the DELLA protein mutant gai-1 from Arabidopsis has a 17-amino acid deletion in the conserved DELLA domain.11
Previous researchers showed that heterologous expression of the Arabidopsis gai mutant gene reduced plant height and altered GA response in transgenic rice,15 tobacco,16 chrysanthemum17 and apple.18 However, the native or constitutive promoters used in these studies resulted in permanent inhibition of GA responses, which resulted in severe dwarfing and other undesirable phenotypes. To use this approach in practice would require that expression of the mutant gene be coupled to an inducible system,19 such as the dexamethazone-inducible promoter20 or the alcohol-inducible promoter,21 which permits the expression of transgenes to be turned on or off at desired stages of development of an organism or tissue.
This study tested the hypothesis that interfering with GA signalling by silencing GID1-like receptor genes using the virus-induced gene silencing (VIGS), with GA signal transduction by over-expression of the Arabidopsis gai mutant gene under the control of the dexamethasone (Dex)-inducible promoter, would modulate plant growth and architecture in petunia.
Materials and methods
Plant material and growth conditions
Petunia (Petunia x hybrida cv. Primetime Blue) seeds were obtained from Goldsmith Seeds (Gilroy, CA, USA). Plants were grown from seed in growth chambers under a 16-h photoperiod (ca 350 µmol m−2 s−1 PPFD) with a day/night temperature regime of 22°C/18°C. VIGS experiments used the purple-flowered ‘Primetime Blue’ cultivar, but studies on stable transformants used white-flowered cultivar ‘Mitchell Diploid’.
Isolation of GID1-like genes from petunia
Total RNA was extracted from petunia tissues using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and was treated with RNase-free DNase (Promega, Madison, WI, USA) to remove any contaminating genomic DNA. First strand cDNA was synthesized using 2 µg total RNA, oligo d(T) primer, random hexamer, and then replicated by superscript reverse transcriptase (Invitrogen), as previously described.3,22 GID1-like gene fragments were amplified from the cDNA by RT-PCR using degenerate primers designed from the three GID1 receptor gene sequences of Arabidopsis or partial EST sequences of petunia. The full-length sequences of GID1-like genes were isolated by standard techniques using 3′ or 5′ rapid amplification of cDNA end (RACE) with the Clontech kit (Clontech, Mountain View, CA, USA), and following the manufacturer's instructions. The sequences were analysed by the sequencing service of the College of Biological Science at UC Davis, and are deposited in Genbank as accession numbers JX501238 (PhGID1A), JX501239 (PhGID1B), JX501240 (PhGID1C). BLAST and CLUSTALW analysis tools were used to compare the nucleotide and deduced amino acid sequences of PhGID1-like genes with those of GID1 genes from other organisms.
Expression analysis of PhGID1-like genes from petunia
Total RNA was extracted from different plant tissues including young leaves, mature leaves, stem, root, pollen, petal and stigma using TRIzol Reagent (Invitrogen). The isolated RNA was treated with RNase-free DNase (Promega) to remove any contaminating genomic DNA. First-strand cDNA was then synthesized from 2 µg total RNA, oligo d(T) primer and random hexamers using Superscript III reverse transcription kit (Invitrogen) according to the manufacturer's protocol. This cDNA was used as template for semi-quantitative PCR using primers (Supplementary Table S1) for PhGID1A (1526 bp, 5′-TCT ATG GCA AGA AAT AAT GAA GCT G-3′ and 5′-GAA GCA AAC ATA GTT CTA TAT AA-3′), PhGID1B (1432 bp, 5′-ACC AGT CAA ACT TGG TCA AAC TC-3′ and 5′-CAA GTG CCA ATT CCA CAA ATT AC-3′) and PhGID1C (1079 bp, 5′-TTG TGT AAT AGT CAT GGC TGG TG-3′ and 5′-GCT GCT TGT ATA TGA TGT TAA AG-3′). The abundance of 26S ribosomal RNA was used as an internal control and the amplification primers were 5′-AGC TCG TTT GAT TCT GAT TTC CAG-3′ and 5′-GAT AGG AAG AGC CGA CAT CGA AGG-3′ (185 bp).
VIGS
The TRV1 and TRV2 VIGS vectors were kindly provided by Dinesh-Kumar, Yale University, and have been described in detail previously.3,22,23
To silence all three GID1-like genes in petunia, a 199 bp fragment of the PhGID1 gene was amplified from total petunia leaf cDNA using the primers listed in Supplementary Table S1. The resulting product was cloned into the pGEM-T Easy vector (Promega) for amplification, sequencing and subcloning. The fragment was excised from this plasmid by Sac I and Xho I digestion, then sub-cloned in the antisense orientation into a modified TRV2 vector with the CHS fragment (TRV2/CHS) constructed by Chen et al.23 as a visual reporter to generate TRV2/CHS/GID1 in a tandem manner. The constructs, TRV1, TRV2, TRV2/CHS and TRV2/CHS/GID1 were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Agroinfection of petunia plants was then performed as described by Chen et al.23 Cultures of Agrobacterium transformed with pTRV1 or the relevant pTRV2 construct were grown separately to an optical density of 2.0 at 600 nm, then mixed. Primary leaves of petunia seedlings (infected when the plants had two true leaves) were infected with the mixed culture using a 1 mL disposable syringe without a needle. Infected plants were grown under controlled conditions (22°C day/18°C night with a 16 h photoperiod). For the evaluation of plant growth, the phenotypic appearance was observed every day and the length of three branches from each infected plant was recorded.
To evaluate silencing efficacy, RT-PCR analysis was performed to measure the relative transcript levels of all three GID1-like genes. Total RNA was extracted from petunia leaves. Total cDNA was synthesized as previously described.24 The cDNA was then used as a template for semiquantitative PCR using primers for a different region of each target gene than the region used for gene silencing.
The primers for amplifying PhGID1 transcripts were designed outside the region targeted for gene silencing to avoid amplification of RNA from the TRV2/CHS/GID1construct. The VIGS primers (Supplementary Table S1) used for PhGID1A were 5′-CTT AGC TGG AGA TAG CTC TGG TG-3′ and 5′-GAA GCA AAC ATA GTT CTA TAT AA-3′ (957 bp); for PhGID1B were 5′-ATA GAG TCT ATC GAC ATG CTC CC-3′ and 5′-ACC AAA CAT TGG ATG AAG ATG G-3′ (434 bp); for PhGID1C were 5′-TTG TGT AAT AGT CAT GGC TGG TG-3′ and 5′-GCT GCT TGT ATA TGA TGT TAA AG-3′ (1079 bp).
Generation of plants overexpressing gai
The gai mutant gene (gai-1) was isolated from Arabidopsis. We confirmed a 17 amino acid deletion in the conserved DELLA domain of the putative GAI protein, and used the full-length sequence together with the GVG inducible system,20 to generate a GVG::gai construct. This construct was then used to transform into ‘Mitchell diploid’ petunia plants by UC Davis plant transformation facility. Seeds were collected from self-pollinated transgenic plants and T1 seedlings containing the transgene were selected on MS medium containing 20 mg L−1 hygromycin. Three T1 transgenic lines were selected and sprayed with 30 µM dexamethazone (the GVG inducer) for 10 days and the resulting plant architecture was compared with sprayed wild-type and H2O-sprayed transgenic plants; three mature leaves of each seedling were selected for the measurement of leaf size. Total RNA was extracted from leaf tissues and semiquantitative RT-PCR was used to compare the abundance of each gai transcript using primers (Supplementary Table S1) forward 5′-GTG TAA GCT GGC TCA TTT AGC TG-3′ and reverse 5′-GTT GAA CAG AGC CAA AAG CAT AC-3′ (535 bp).
Results
Identification of GID1-like genes in petunia
The sequences of the three GID1 receptor genes from Arabidopsis and the GID1-like sequences from other species were used to identify consensus region and to design degenerate primers based on the conserved regions of the GID1 gene. Multiple fragments were amplified by PCR from petunia, and their identity was confirmed by sequencing. The 3′ and 5′ RACE PCR was used to extend both 5′ and 3′ ends of the genes to obtain full-length sequences of all petunia GID1-like genes.
Three putative GID1 genes were identified in petunia and named PhGID1A, PhGID1B and PhGID1C (Supplementary Figure S1). The deduced amino acid sequences were aligned with three Arabidopsis GID1 genes; one of the GID1-like sequences shares 72% identity with both AtGID1A (accession number NP_187163) and AtGID1C (accession number NP_198084) and the other two petunia GID1-like proteins share 74% and 77% identities with AtGID1B (accession number NP_191860) (Supplementary Figure S2). The nucleotide sequences similarities are 81% between PhGID1B and PhGID1C; 64% between PhGID1A and PhGID1C; and 48% between PhGID1A and PhGID1B.
To determine the tissue distribution of the three PhGID1 transcripts, semiquantitative RT-PCR was performed using gene-specific primers. With the exception of pollen, the three PhGID1 transcripts were detected at moderate abundance in all tested petunia tissues, including stems, leaves, roots and flowers. Transcripts of PhGID1A and PhGID1B were barely detected in pollen (Figure 1).
Figure 1.
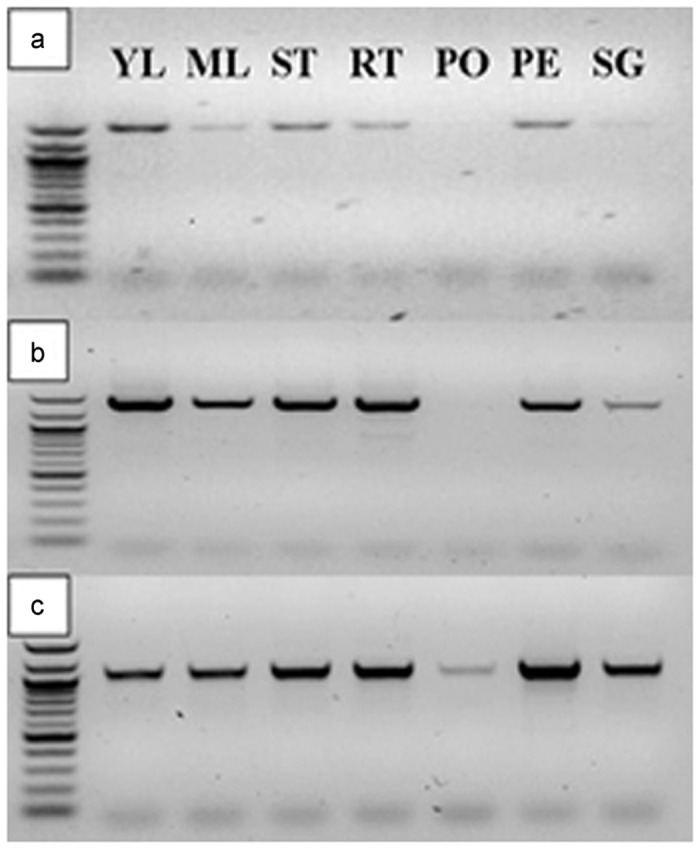
Abundance of transcripts of three putative PhGID1 genes in different petunia tissues. (a) PhGID1A; (b) PhGID1B; (b) PhGID1C. Total RNA was extracted from tissues of young leaves (YL), mature leaves (ML), stem (ST), root (RT), pollen (PO), petal (PE) and stigma (SG) and cDNA was synthesized for evaluation of transcript abundance. 26S rRNA abundance determined by real-time PCR was used to normalize loading for each cDNA sample.
VIGS of GID1-like genes in petunia
To examine the function of PhGID1 in petunia, we used VIGS to silence expression of these genes. We cloned different PhGID1 fragments into the TRV CHS vector (silencing of CHS was used as a visual reporter for cosilencing the GID1 target), and infected young petunia seedlings with Agrobacterium transformed with silencing constructs containing single or multiple GID1-like genes.
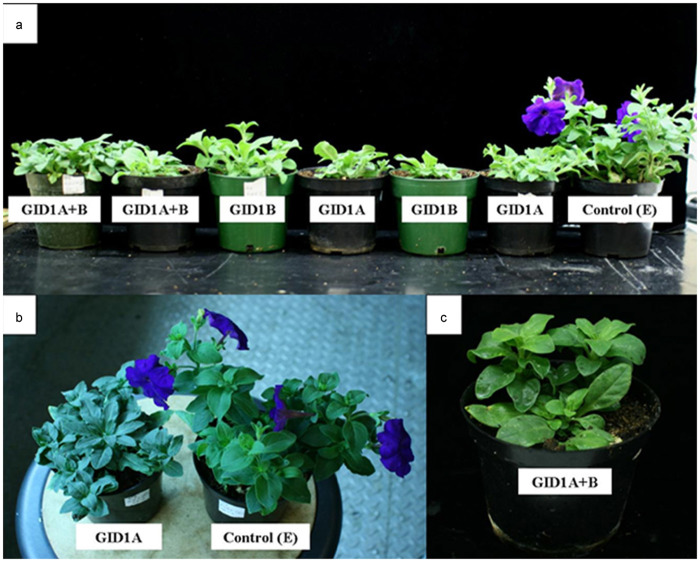
Silencing of PhGID1 had a strong impact on plant architecture, resulting in plants with short branches and internodes, small, dark leaves and late flowering (Figures 2 and 3). A range of dwarf phenotypes was observed from single or double silencing of GID1-like genes (Figure 2) and the frequency of silenced phenotype was relatively low (data not shown). We therefore tested the effects of simultaneously silencing all three PhGID1 genes. A severely dwarfed phenotype was observed (Figure 3b–3d) in these plants and the silenced branches were significantly shorter than the controls (Table 1).
Figure 2.
Effect of silencing GID1-like genes in petunia. The phenotypes resulting from silencing one (PhGID1A or PhGID1B) or two (PhGID1A+B) GID1 genes is shown in comparison with control (E) plants transformed with the empty vector.
Figure 3.
Effect of silencing three GID1-like genes in petunia. (a) The phenotype resulting from silencing with a TRV vector containing fragments of all three PhGID1 genes and the CHS reporter gene (left) is compared with that of the CHS and empty vector controls. (b–d) The severely dwarfed phenotype of plants silenced with all PhGID1 gene fragments.
Table 1. Effect of silencing PhGID1 on the branch length in petunia. Data collected at 60 days after infection.
| Branch length (cm) |
|||
|---|---|---|---|
| PhGID1 | PhCHS | H2O | |
| Experiment 1 | 18.7±8.2*** | 27.0±2.6 | 28.7±2.4 |
| (n=15) | (n=10) | (n=10) | |
| Experiment 2 | 13.1±8.3*** | 21.3±3.3 | 22.4±3.7 |
| (n=30) | (n=10) | (n=10) | |
Data are means±s.d.; n represents 10–30 plant replicates of each treatment.
indicates significant differences (P<0.05) analyzed by Tukey test.
A semiquantitative RT-PCR analysis confirmed that the abundance of PhGID1A, PhGID1B and PhGID1C transcripts in infected plants were all strongly reduced (Figure 4). The severely dwarfed phenotypes observed in plants where all three PhGID1 genes were silenced (Figure 3b and 3d) were correlated with reduction in abundance of transcripts of the three PhGID1 genes (samples 4 and 5 of Figure 4), particularly PhGID1A (Figure 4a).
Figure 4.

Virus-induced gene silencing of GID1-like genes in petunia. (a) PhGID1A; (b) PhGID1B; (c) PhGID1C; (d) 26S ribosomal RNA; cDNA samples 1–5 were synthesized from total RNA extracted from leaf tissues of five independently silenced plants. WT, wild-type.
Overexpression of the gai mutant gene from Arabidopsis in petunia
A GA-insensitive mutant gene (gai) isolated from Arabidopsis was cloned into a GVG inducible system, using Dex as an inducer, to generate 10 independent lines of GVG::gai transgenic plants (T0). The presence of the gai transgene was confirmed by PCR in all T0 lines. Since a phenotype of growth retardation was observed in some T0 lines under normal growth conditions, gai expression was examined in all lines, and three lines for induction experiments, including two normal lines (#1 and #4—showing no gai expression) and one leaky line (#6—showing gai expression in the absence of the Dex inducer).
GVG::gai T1 petunia seedlings were sprayed daily with 30 µM dexamethasone for 10 days. The resulting over-expression of gai caused a dramatic retardation of plant growth in the treated seedlings (Figure 5c) and result in smaller leaf size in line 4 and 6, but there was no significant difference in line 1 (Table 2). In the control plants (no Dex-induction), plant growth rate and leaf size were similar to non-transformed controls except for the leaky line 6 (Table 2). After Dex spraying was discontinued, line 4 plants recovered gradually from the dwarf phenotype but the leaky line 6 plants continued to grow slower than wild-type control plants (Figure 6b).
Figure 5.

Induction of gai expression and the phenotype of GVG::gai T1 petunia after 10 days of spray treatment with 30 µM dexamethasone. (a) Abundance of gai transcripts (WT: wild-type; D1, D4 and D6: Dex-treated plants; C1, C4 and C6: no Dex-treated plants); (b) 26S rRNA transcripts; (c) Phenotype of GVG::gai T1 petunia lines 4 and 6 after 10-days treatment with Dex compared with that of a wild-type control plant.
Table 2. Effect of overexpression of the Arabidopsis gai on leaf size in GVG-gai T1 petunia plants. Data were collected after 10 days of Dex treatment.
| WT | Line 1 | Line 4 | Line 6 | |
|---|---|---|---|---|
| Leaf length (cm) | ||||
| Dex | 7.6±0.7 a | 7.1±0.7 a | 5.4±0.9 b | 4.8±1.2 b |
| Non-Dex | 7.4±0.8 a | 7.2±1.0 a | 7.5±0.5 a | 5.5±0.7 b |
| Leaf width (cm) | ||||
| Dex | 2.6±0.2 a | 2.5±0.2 ab | 2.1±0.3 b | 2.2±0.2 b |
| Non-Dex | 2.5±0.1 a | 2.4±0.2 a | 2.5±0.1 a | 2.3±0.2 a |
Data are means±s.d. (n=9). Different letters indicate significant differences (P<0.05) analyzed by Tukey test.
Figure 6.

Recovery of the gai overexpressed phenotype following Dex-induction in GVG::gai petunia. The photograph was taken 70 days after termination of Dex treatments (b). Abundance of gai transcripts was determined using RT-PCR on cDNA prepared from total RNA extracted from tissues at the same time (a).
To confirm that the dwarf phenotype was caused by gai overexpression, the abundance of gai transcripts was measured from GVG::gai plants with or without Dex induction. The expression of gai was strongly induced in treated plants of line 4 and 6 (Figure 5, D4a, D4b, D6a and D6b), compared to the treated wild-type plants (WT) and the non-treated transgenic plants (C1, C4, C6a and C6c) (Figure 5a). However, gai transcripts were not induced in line 1 plants (D1a and D1c) by Dex treatment (Figure 5a). In correlation with recovery from the dwarf phenotype, gai transcripts were barely detected in line 4 after spraying ceased, but were still abundant in line 6 (Figure 6a).
Discussion
Interfering with GA signal transduction through downregulation of GID1-like genes in petunia
Three GID1-like genes, named PhGID1A, PhGID1B and PhGID1C, were identified in petunia (Supplementary Figure S1). In Arabidopsis, the three GID1 proteins have redundant function as GA receptors; gid1 double mutants are partially impaired in GA responses but gid1 triple mutants are totally insensitive to GA.10 The sequences of AtGID1A and AtGID1C (A/C type) are highly similar with 73% identity but AtGID1B (B type) is quite different (about 40% identity with the A/C type); gid1a/gid1c double mutants show a strong dwarf phenotype than the other two double mutants.10 In petunia, the sequences of PhGID1B and PhGID1C are much closer to AtGID1B (above 74% identity) and have 81% similarity between them (Supplementary Figure S2). It seems that there are two B-types of GID1 receptors existing in petunia. VIGS-induced silencing of single or double PhGID1 genes resulted in only modest dwarfing and a low silencing frequency (data not shown). Silencing of all three PhGID1 genes showed a severely dwarfed phenotype (Figures 3 and 4).
All three PhGID1 genes are expressed throughout the plants except for in pollen (Figure 1). Although the genes are expressed in flower organs, downregulation had no visible effect on floral development (data not shown). Future experiments will examine the phenotype in transgenic petunia plants transformed with antisense GID1 under the control of an inducible promoter. Such plants could be treated at the desired stage so that inhibition of GID1 expression would reduce stem elongation without negative effects on other aspects and stages of plant growth and development.
Interfering with GA signal transduction by the over-expression of the Arabidopsis gai in petunia
This study demonstrated that heterologous expression of the mutant Arabidopsis gai inhibited plant growth in petunia. One line of transgenic GVG::gai petunia seedlings (line 4) showed the anticipated phenotype of stunted growth only when gai expression was induced by dexamethasone. No gai expression was detected in line 4 seedlings not treated with Dex, and they had the same growth rate as non-transgenic wild-type petunia. Line 1, although it contained the transgene, showed no phenotypic response or gai expression in response to Dex. This may be the result of loss of GVG control of the transgene during transformation, or could reflect epigenetic interference. Line 6 GVG::gai seedlings appeared to be ‘leaky’, expressing gai even before the Dex-induction, and showing a slower growth rate. After Dex-induction, line 6 seedlings showed strong gai expression and a dramatic retardation of seedling growth. Therefore, it suggested that the phenotype of growth retardation was caused by induced gai expression (Figure 5).
It took about 2 months after the end of the Dex treatment for petunia plants to return to their normal growth rate (Figure 6). This could be the effect of residual inducing Dex on the leaves or in the soil. The question of how the induced mutant gai proteins are degraded remains to be answered. The dwarfed plants of line 6 had shorter internodes but no difference in number of nodes (Table 3) compared to the wild-type plants. The data confirmed that GA plays an important role on stem elongation. It is worth mentioning that we detected the gai expression in the absence of the dexamethasone inducer in some transgenic lines. This is most likely caused by integration sites of the transgene in the genome of the petunia, leading to the leaky expression of the gai.
Table 3. Effects of GVG::gai induction on branch length and internode number in the leaky line (line 6). Data were collected 80 days after Dex induction.
| WT | Line 6 | |
|---|---|---|
| Branch length (cm) | 56.9±6.0 | 26.0±3.3*** |
| Node numbers | 14.7±2.1 | 14.7±1.8 |
Data are means±s.d..
indicates significant differences (P<0.05) analyzed by Tukey test.
This study showed the potential for commercial application of biotechnology to prevent plant stretching. An inducible promoter system could provide flexible control of plant height that would allow grower to inhibit GA signalling during finishing but avoid adverse effects on growth and flowering during production. An ethanol-inducible system was used to control gai expression in Arabidopsis,21 and this could be the basis for a practical system to make use of the effects of GID1 silencing or gai over-expression in petunia and other ornamentals. Control of plant architecture using these molecular strategies could be of considerable benefit to commercial producers by reducing costs and environmental contamination, and permitting height control only when desired.
Acknowledgments
This study was supported in part by funds from United States Department of Agriculture (USDA) Floriculture Initiative (5306-21000-019-00D and 5306-13210-001-02S). We thank Linda Donnelly and Alejandro Estrada for their assistance.
The authors declare no conflict of interest.
References
- Phillips AL. Gibberellins in Arabidopsis. Plant Physiol Biochem 1998; 36: 115–124. [Google Scholar]
- Rademacher W. Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 2000; 51: 501–531. [DOI] [PubMed] [Google Scholar]
- Liu YL, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J 2002; 31: 777–786. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. Understanding gibberellic acid signaling—are we there yet? Curr Opin Plant Biol 2008; 11(1): 9–15. [DOI] [PubMed] [Google Scholar]
- Hirano K, Ueguchi-Tanaka M, Matsuoka M. GID1-mediated gibberellin signaling in plants. Trends Plant Sci 2008; 13: 192–199. [DOI] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-Ali T, Harberd NP. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 2001; 52: 67–88. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J. Genes controlling plant architecture. Curr Opin Biotechnol 2006; 17: 123–129. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M et al. Gibberellin insensitive dwarf1 encodes a soluble receptor for gibberellin. Nature 2005; 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006; 18: 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 2007; 19: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 1997; 11: 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Fu X. GA action: turning on de-DELLA repressing signaling. Curr Opin Plant Biol 2007; 10: 461–465. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 2001; 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 2001; 159: 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Sudhakar D, Peng J, Richards DE, Christou P, Harberd NP. Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 2001; 13: 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes LW, Peng J, Richards DE, Harberd NP. Transgenic expression of the Arabidopsis DELLA proteins GAI and gai confers altered gibberellin response in tobacco. Transgen Res 2003; 12: 707–714. [DOI] [PubMed] [Google Scholar]
- Petty LM, Harberd NP, Carre IA, Thomas B, Jackson SD. Expression of the Arabidopsis gai gene under its own promoter causes a reduction in plant height in chrysanthemum by attenuation of the gibberellin response. Plant Sci 2003; 164: 175–182. [Google Scholar]
- Zhu L, Li X, Welander M. Overexpression of the Arabidopsis gai gene in apple significantly reduces plant size. Plant Cell Rep 2008; 27: 289–296. [DOI] [PubMed] [Google Scholar]
- Dutt M, Dhekney SA, Soriano L, Kandel R, Grosser JW. Temporal and spatial control of gene expression in horticultural crops. Hort Res 2014; 1: 14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 1997; 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Ait-ali T, Rands C, Harberd NP. Flexible control of plant architecture and yield via switchable expression of Arabidopsis gai. Plant Biotechnol J 2003; 1: 337–343. [DOI] [PubMed] [Google Scholar]
- Jiang CZ, Lu F, Imsabai W, Meir S, Reid MS. Silencing polygalacturonase expression inhibits tomato petiole abscission. J Exp Botany 2008; 59: 973–979. [DOI] [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Gookin TE, Hunter DA, Clark DG, Reid MS. Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol Biol 2004; 55: 521–530. [DOI] [PubMed] [Google Scholar]
- Singh A, Liang YC, Kumar P, Jiang CZ, Reid MS. Co-silencing of the mirabilis antiviral protein (MAP) permits virus-induced gene silencing (VIGS) of other genes in Four O′Clock plants (Mirabilis jalapa). J Hort Sci Biotechnol 2012; 87: 334–340. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.