Abstract
Gene co-expression analysis has been widely used for predicting gene functions because genes within modules of a co-expression network may be involved in similar biological processes and exhibit similar biological functions. To detect gene relationships in the grapevine genome, we constructed a grapevine gene co-expression network (GGCN) by compiling a total of 374 publically available grapevine microarray datasets. The GGCN consisted of 557 modules containing a total of 3834 nodes with 13 479 edges. The functions of the subnetwork modules were inferred by Gene ontology (GO) enrichment analysis. In 127 of the 557 modules containing two or more GO terms, 38 modules exhibited the most significantly enriched GO terms, including ‘protein catabolism process’, ‘photosynthesis’, ‘cell biosynthesis process’, ‘biosynthesis of plant cell wall’, ‘stress response’ and other important biological processes. The ‘response to heat’ GO term was highly represented in module 17, which is composed of many heat shock proteins. To further determine the potential functions of genes in module 17, we performed a Pearson correlation coefficient test, analyzed orthologous relationships with Arabidopsis genes and established gene expression correlations with real-time quantitative reverse transcriptase PCR (qRT-PCR). Our results indicated that many genes in module 17 were upregulated during the heat shock and recovery processes and downregulated in response to low temperature. Furthermore, two putative genes, Vit_07s0185g00040 and Vit_02s0025g04060, were highly expressed in response to heat shock and recovery. This study provides insight into GGCN gene modules and offers important references for gene functions and the discovery of new genes at the module level.
Introduction
The rapid accumulation of genome sequences and high-throughput microarray data provides rich materials for research on gene function and regulation at the system level.1 However, integrating and exploiting these data sets has been challenging. Biological networks constructed by bioinformatic methods can help ‘put the function in genomics,2 and allow researchers to understand how biomolecules interact with one another at the system level to perform specific biological functions in living plant cells.3,4
The molecular interaction network is a type of biological network in which a node represents a gene, gene product or metabolite, and a link or edge refers to an interaction between them.4 A gene co-expression network, in which nodes and links represent genes and indicate their co-expression relationships, can characterize such topological properties as small-world, hierarchically modular and scale-free.5 A gene co-expression network can be divided into several substructures, including motifs, modules and pathways. Its substructure exhibits topological properties described by specific terms, such as network density, degree distribution, clustering coefficient and betweenness.3
Co-expression network analysis is a powerful method to extract functional modules of co-expressed genes, analyze their biological meanings and identify important novel genes. In recent studies, several plant gene co-expression networks have been built and many functional modules have been inferred or identified.6–13 For instance, Mao and colleagues7 constructed an Arabidopsis gene-expression network and identified many functional modules associated with photosynthesis, protein biosynthesis, cell cycle, defense response and others, and these modules revealed new insights into gene function organization. The expression of genes related to the same metabolic function may show co-expression patterns.14 Wang and colleagues employed co-expression network analysis to identify related cell wall genes in Arabidopsis.11 Gene modules were extracted in response to drought in rice by network-based analysis, and many hub genes clustered in some rice chromosomes have been found to significantly associate with quantitative trait loci (QTLs) for drought tolerance.12
Microarray datasets and genome sequences provide an excellent opportunity to understand gene relationships and biological functions in the grapevine.15,16 In this report, we constructed a GGCN by using 374 high quality microarrays (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL1320). Qcut,17 a graph portioning algorithm, was applied to identify subnetwork modules from the gene co-expression network. The functions represented by the extracted modules were evaluated by GO enrichment analysis.18 Next, we validated module 17 by examining gene expression by qRT-PCR and inferred that two putative uncharacterized proteins might be potentially related to heat stress.
Materials and methods
Raw expression data
The grapevine microarray data set for the construction of the co-expression network was obtained from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL1320) (platform accession number GPL1320). The platform consists of experimental samples using Affymetrix GeneChip Grapevine Genome Array. A total of 374 CEL files of samples from platform GPL1320 were used to construct the network and involved three treatment types (biotic stress, development, abiotic stress) and 13 series. The grapevine and Arabidopsis genome sequences were downloaded from Phytozome (http://www.phytozome.net).15
Annotation of probe sets and homolog search
A total of 16 436 probe sets from the Affymetrix Grapevine GeneChip were mapped to the grapevine gene loci in CRIBI (http://genomes.cribi.unipd.it/) using BlastN. If more than six probes from the set aligned perfectly to a gene, the probe set was assigned to that gene. Arabidopsis protein sequences and gene information were obtained from the Arabidopsis Information Resource release 10 (http://www.arabidopsis.org/). Grapevine protein sequences were used to search complete Arabidopsis protein sequences using BlastP with an e-value cutoff of 1e−4, and the best hits were selected as Arabidopsis orthologs.
Construction of GGCN
The construction of a gene co-expression network involves the measuring gene expression similarity, visualizing gene expression data, and identifying modular structures. To measure the similarity of gene expression, we utilized the Pearson correlation coefficient (PCC) between pairwise genes. The 374 arrays from Gene Expression Omnibus were normalized by the justRMA function in R/BioConductor.19 Gene co-expression data were calculated in ATTED-II and applied to the PCC calculation (http://atted.jp/help/coex_cal.shtml).
To determine the PCC cutoff threshold for network construction, the numbers of probe sets, edges, and network density (ND) were calculated along with the PCC cutoffs. The network density was calculated according to where m was the observed number of edges in the network and n was the number of nodes in the network. Co-expressed genes are selected at a certain PCC cutoff threshold, and a co-expression network was constructed and visualized by Cytoscape software20 (http://www.cytoscape.org/).
The algorithm Qcut, which identifies statistically significant graph partitions in a biological network,17 was applied to identify sub-network modules from the co-expression network (http://www.mybiosoftware.com/pathway-analysis/12211).
GO enrichment analysis of modules in GGCN
GO annotations of grapevine genes were downloaded from agriGO (http://bioinfo.cau.edu.cn/agriGO/download.php). The GO enrichment was performed within each module using BiNGO 2.4.18 The statistical significance of GO term enrichment was measured by a hypergeometric test21 using the genes in a whole co-expression network as the back ground. A Bonferroni correction22 was used to control the false positive rate in the multiple testing problems, and a GO term in a module was considered significantly enriched in the given module if the family-wise error rate (FWER) corrected p value was less than 0.05.
Validation of expression genes in module 17 by qRT-PCR
Pinot Noir PN40024 (the genotype deriving the reference genome sequence) was subcultured in vitro on 3/4 Murashige and Skoog medium23 at 22 °C with a 16-h/8-h photoperiod and an illumination intensity of 150 μmol m−2 s−1 for 6 weeks. Young leaves, including second and third expanding leaves, were sampled for gene expression analysis.
To analyze the response of module 17 genes to continuous heat shock stress, whole plants were treated at 40 °C for 0.5, 1, 2, 3 or 6 h in the plant growth chamber. Meanwhile, to analyze the heat shock recovery response, a fraction of the plants that were heat-shocked for 1 h was placed under the original temperature (22 °C) for 2 h and 5 h (the third hour or sixth hour from the beginning of heat shock). The plants without heat shock treatment were used as the controls and handled in an identical manner. To analyze their responses to low temperature, a set of plants was placed in a plant growth chamber at 4 °C for 1 h. All the plant samples were then frozen in liquid nitrogen before total RNA extraction and first strand cDNA synthesis by the reported method.24
We designed 29 pairs of oligonucleotide primers (Supplementary Table 1) in module 17 with Primer 5.0 (http://www.premierbiosoft.com/crm/jsp/com/pbi/crm/clientside/ProductList.jsp) according to the putative cDNA sequences of the grapevine genome. PCR amplification was carried out in a 25 μL reaction solution consisting of 20 ng template cDNA, 2.0 mM MgCl2, 2.5 μL 10× PCR buffer, 200 μM dNTP, 0.2 pM of each primer and 0.25 U Taq DNA polymerase. To validate the specificity of PCR products, the amplicons were cloned into a pMD19-T vector (Takara, Dalian, China), sequenced at Shanghai Invitrogen Biotechnology Co., Ltd (2715 Longwu Road, Shanghai 200231, China) according to the protocol24 and aligned onto the grapevine reference genome. The qRT-PCR oligonucleotide primers (Table 1) targeting the expressed grapevine genes in module 17 (response to environmental stress) were designed with Beacon Designer 7.0 (http://www.premierbiosoft.com/molecular_beacons/). Because of high homology and some unknown gene information, all primers were blasted against the grapevine reference genome sequences. Each primer differs from non-target genes by at least three nucleotides, and at least one nucleotide at the 3′-end.25
Table 1. qRT-PCR primer sequences of genes in module 17.
| Gene number | Grapevine gene | Forward primers (5′ to 3′) | Reverse primers (5′ to 3′) |
|---|---|---|---|
| 1 | Vit_10s0003g00260 | TCAACATCAAGTTTCCAACAAGG | ACAGTCGCACATCATTAGCC |
| 2 | Vit_07s0185g00040 | AGGATGCGAGAGGATGAGAC | ACAAGAGAAACACCAGACAAGG |
| 3 | Vit_13s0019g03160 | AGTTCCTTCGTCGGTTCAG | GCCTTCACCTCAGCCTTC |
| 4 | Vit_18s0041g01230 | GTCAACAACCCAAACTATCAAGG | GCACCATCATATCATATACACTCC |
| 5 | Vit_02s0025g04060 | TTGATAGTATGTCTGAGTTATGGAG | CCTTGGGTGTGAAACAAATGG |
| 6 | Vit_04s0008g01590 | TTGAGGTGAAGGTTGCTTGAG | CATACTGACTTGGGAGACATCG |
| 7 | Vit_06s0004g04470 | CATAAGAAGGATATTAGCGGAAGT | GTTGTGTAGAAATCAATACCATCGA |
| 9 | Vit_16s0050g01150 | GACCTTGTGATGCTCCTATATG | ATCTTGCTCTCCTCATTGCC |
| 11 | Vit_01s0010g02290 | GTATGACCAAGGATGATGTGAAG | ACTCCATCTTTGACCTCTGC |
| 12 | Vit_16s0098g01060 | TGGAGGATGACTTGCTTGTG | CTCTACCTTGGTCTTAGGAATGG |
| 13 | Vit_11s0016g04080 | GTGAACAAGGCTATCCGGTC | TCATCTTCTTCTCCAACCTCG |
| 14 | Vit_07s0005g01980 | GGGGTTTGTCACGGTTAG | GTATGACTGGAAGTAATTTGCC |
| 15 | Vit_17s0000g07190 | TAGATGCGGGAGTGTCAGG | CCTCTTCGTCTTCTATTTCTTCG |
| 19 | Vit_19s0085g01050 | GAGTTCAAGAGTCAAGACACAG | ACCTCCAGTTTCACCTCATTC |
| 20 | Vit_06s0004g06010 | GCTATTATAGAAGGCGGCATTAC | GACCCAGGAGTGAGAGACC |
| 22 | Vit_13s0019g00860 | AAGGTGGAGATAGAAGATGGAAAC | TGGAACAACGATGGTGAGAAC |
| 23 | Vit_08s0007g00130 | GATTGAGGATGCCATTGAGC | TCTTTGCTATGATGGGGTTG |
| 24 | Vit_16s0022g00510 | AGATACAGCAGCAGAATTGATTTG | TCAGTCCTCTCCTCTTCCTTCAG |
| 26 | Vit_06s0004g05770 | GTTCTTACTGTTACTGTTCCTAAGAAG | CGCTGATATATGATATGATGGTCTC |
There were 41 nodes (probes) in module 17. Among them, 29 probes were matched with grapevine genes annotated by CRIBI Genomics, University of Padua (http://genomes.cribi.unipd.it/). However, the genes numbered 8, 10, 16, 17, 18, 21, 25, 27, 28 and 29 in module 17 did not express in response to heat shock or cold treatment stress and were therefore not cloned (listed in Table 1).
The qRT-PCR reaction was carried out in a 20 μL reaction solution consisting of 10 μL SYBR (Takara), 8.7 μL ddH2O, 1 μL cDNA diluted 10-fold and 0.15 μL of each specific primer. qRT-PCR amplifications were performed with the following procedure: 94 °C for 4 min and 40 cycles of 94 °C for 20 s, 60 °C for 20 s and 72 °C for 43 s. The qRT-PCR data were analyzed as previously described.25 Each treatment data point represents three biological replicates (individual plants) with three technical replicates each. The actin-101-like gene (VIT_12S0178g00200) was used as an internal reference. The expression ratio was calculated by the formula , as previously described.16,25
Goodness of fit test of gene expression in module 17
To test the goodness of fit of all gene expression values between each two time points treated with heat shock and recovery, we employed ‘LOESS’, locally weighted scatterplot smoothing,26 and ‘Linear’, a unitary linear regression, to add a fit line and calculate R2, the coefficient of determination,27 with SPSS 19.0 software.28 Firstly, a matrix scatter was created between the variables ‘gene expression value’ and ‘treatment time point’ following the steps Graphs→Legacy Dialogs→Scatter/Dot→Matrix Scatter. Next, a fit line was added in the matrix scatterplot by ‘LOESS’ with parameters 95% individual confidence intervals, 30% percentage of points to fit and Epanechnikov kernel function. Secondly, ‘Linear’ was performed with 95% individual confidence intervals following the steps Graphs→Legacy Dialogs→Scatter/Dot→Matrix Scatter→Linear. R2 between the dependent and independent variables ‘gene expression value’ and ‘treatment time point’ in the linear regression were obtained for goodness of fit analysis.27,28
Results
Construction of GGCN
The raw microarray data could be divided into the following three categories: biotic stress, development, and abiotic stress. The array accession and the experiment conditions are listed in Table 2. After normalization of gene expression values, the PCC was calculated between each pair within the 16,436 genes. An appropriate PCC cutoff value is necessary to construct a co-expression network. Figure 1 reveals a negative correlation between the network density and PCC cutoff values. At approximately 0.78, the network density approached the minimal value and then increased gradually. The PCC cutoff value of 0.78 was then chosen to screen significant co-expression correlation from a large-scale expression data set (Figure 1). At the PCC cutoff value of 0.78, the network contained 3834 nodes (probe sets) with 13 479 edges (Figure 2 and Supplementary Table 2) and a network density of 0.001856078. The GGCN view was created by the Cytoscape software package.20
Table 2. Microarray data used to construct the grapevine co-expression network.
| Condition | Series ID | Number of gene chips | Experimental conditions |
|---|---|---|---|
| Biotic stress | GSE6404 | 72 | Erysiphe necator conidiospores infection |
| GSE11857 | 12 | Downy mildew infection | |
| GSE12842 | 10 | Bois noir infection | |
| GSE31660 | 14 | Viral diseases in berry | |
| Development | GSE31674 | 27 | Berry transcriptome during ripening |
| GSE31664 | 12 | Skin transcriptome in the berries | |
| GSE31662 | 8 | Grape skin transcriptome in the berries | |
| GSE11406 | 32 | Berries during ripening initiation | |
| GSE17502 | 84 | Photoperiod regulation of bud dormancy | |
| Abiotic stress | GSE31677 | 39 | Salt and water stress |
| GSE31675 | 12 | High temperature | |
| GSE31594 | 48 | Short term abiotic stress | |
| GSE27180 | 4 | Micropropagated plants were transferred to ex vitro conditions |
Figure 1.
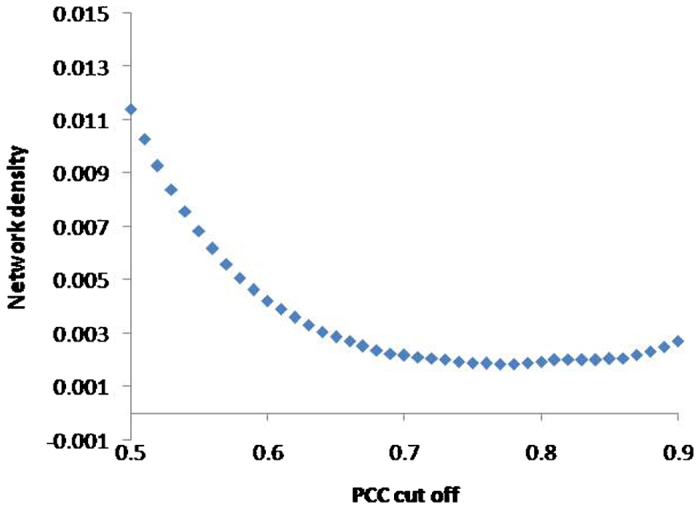
Relationship between network densities and PCC cutoff values.
Figure 2.
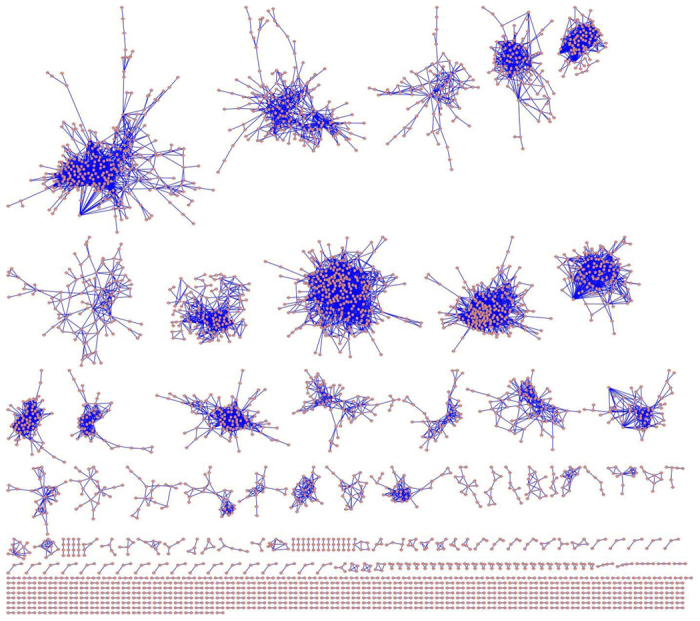
The co-expression network of grapevine genes. A red dot represents a node, and a blue line connecting two nodes represents an edge.
Modules in GGCN
In the 3834 nodes, a partitioning analysis was performed to detect 557 modules with a Q value of 0.78, demonstrating a strong modular structure. The modular structure, one of the important features of the biological network, indicates the interaction of biomolecules at the system level. However, all modules in the GGCN were completely independent and represented by different sizes (Figure 2 and Supplementary Table 2). For instance, the two largest modules, module 1 and module 2, each contained 312 nodes in their network, but with 1521 and 2284 edges, respectively, and the smallest modules had only two nodes (Supplementary Table 2).
BiNGO 2.4,18 a Cytoscape plugin, was used to perform GO term enrichment analysis of biological processes. A total of 127 modules that contained more than two nodes were analyzed using the 1256 probes with a biological process GO term as the custom reference set. As a result, 15 modules were identified with significantly over-represented GO terms with a FWER-adjusted p<0.01 from the hypergeometric test.21 Table 3 lists the most significantly enriched functional categories and the GO term number in a module and in the grapevine gene co-expression network. Because the biotic or abiotic stress response and its regulation are important biological processes in plants, we highlight the details of one interesting module here, module 17, which responds to environmental stresses Figure 3 and Table 4.
Table 3. Significantly enriched GO terms in 38 modules.
| Module | GO term description | GO term | p value |
|---|---|---|---|
| 1 | Protein catabolic process | 13/30 | 2.1×10−5 |
| 2 | Ribonucleoprotein complex biogenesis | 152/207 | 3.0×10−90 |
| 3 | Photosynthesis | 54/69 | 1.0×10−40 |
| 4 | Cellular amine metabolic process | 18/82 | 2.6×10−2 |
| 5 | Response to salicylic acid stimulus | 5/8 | 2.1×10−4 |
| 7 | Carbohydrate metabolic process | 18/102 | 2.4×10−5 |
| 11 | DNA metabolic process | 21/40 | 5.7×10−19 |
| 12 | ATP synthesis coupled electron transport | 9/16 | 1.5×10−8 |
| 15 | Cellular biosynthetic process | 34/408 | 4.4×10−7 |
| 17 | Response to heat | 11/31 | 3.5×10−10 |
| 20 | Plant-type cell wall biogenesis | 6/7 | 1.5×10−9 |
| 24 | Response to auxin stimulus | 3/10 | 2.8×10−2 |
| 25 | Phenylpropanoid biosynthetic process | 9/28 | 6.7×10−11 |
| 26 | ATP metabolic process | 5/14 | 1.6×10−5 |
| 29 | Protein folding | 6/57 | 1.0×10−5 |
| 30 | Lipid transport | 3/14 | 2.1×10−2 |
| 31 | Flavonoid biosynthetic process | 6/8 | 6.2×10−11 |
| 34 | Response to wounding | 3/10 | 3.5×10−5 |
| 35 | Carboxylic acid metabolic process | 6/141 | 3.4×10−4 |
| 36 | Response to biotic stimulus | 5/37 | 6.1×10−6 |
| 37 | Protein ubiquitination | 2/14 | 5.9×10−3 |
| 38 | Acyl-carrier-protein biosynthetic process | 4/25 | 1.1×10−4 |
| 42 | Metal ion transport | 3/18 | 9.9×10−5 |
| 48 | Modification-dependent protein catabolic process | 4/24 | 2.1×10−6 |
| 51 | Nucleic acid metabolic process | 4/96 | 2.5×10−3 |
| 57 | Cell redox homeostasis | 3/15 | 1.3×10−4 |
| 75 | Fatty acid biosynthetic process | 3/21 | 8.9×10−5 |
| 79 | Water homeostasis | 1/1 | 2.1×10−2 |
| 83 | One-carbon metabolic process | 3/9 | 7.9×10−6 |
| 87 | Xylulose metabolic process | 1/1 | 3.6×10−2 |
| 96 | Regulation of cell cycle | 2/6 | 1.6×10−3 |
| 101 | Nucleosome assembly | 2/25 | 4.6×10−2 |
| 105 | D-xylose metabolic process | 3/3 | 9.1×10−8 |
| 107 | Oligosaccharide metabolic process | 2/29 | 3.4×10−2 |
| 112 | Ketone biosynthetic process | 3/13 | 3.1×10−5 |
| 115 | Chitin catabolic process | 3/9 | 5.1×10−6 |
| 124 | Lipid transport | 3/14 | 1.8×10−5 |
| 139 | Response to chlorate | 3/3 | 5.5×10−8 |
A GO term indicates numerical values of the same GO term in one module and the grapevine gene co-expression network.
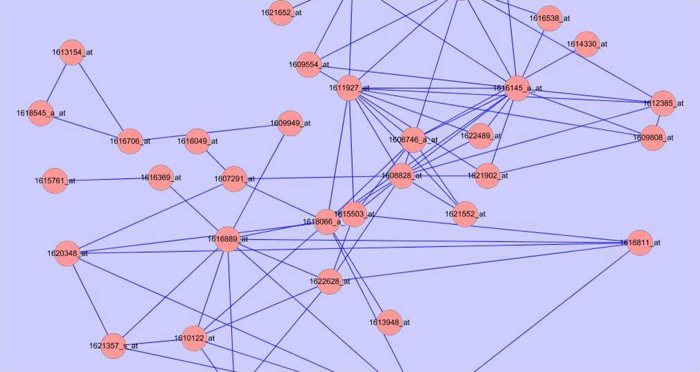
Figure 3.
The fraction of module 17 enriched with the GO term ‘in response to heat stress’. Red circles represent nodes, the blue lines represent edges, and the numbers in the red circles represent gene chip probes.
Table 4. Gene ontology enrichment analysis in module 17.
| GO ID | p value (FWER corrected) | Number of GO terms in module 17 in−1 GGCN | Description |
|---|---|---|---|
| 6950 | 4.0537×10−18 | 26/183 | Response to stress |
| 50896 | 1.0848×10−13 | 26/267 | Response to stimulus |
| 9408 | 3.5017×10−10 | 11/31 | Response to heat |
| 9266 | 4.5005×10−8 | 11/46 | Response to temperature stimulus |
| 9644 | 3.2480×10−7 | 6/9 | Response to high light intensity |
| 9642 | 3.4062×10−6 | 6/12 | Response to light intensity |
| 9628 | 9.9960×10−6 | 12/92 | Response to abiotic stimulus |
| 42542 | 1.7589×10−5 | 6/15 | Response to hydrogen peroxide |
| 10035 | 2.7093×10−5 | 7/25 | Response to inorganic substance |
| 302 | 1.2576×10−4 | 20/29 | Response to reactive oxygen species |
| 6979 | 3.4874×10−3 | 6/34 | Response to oxidative stress |
| 9416 | 6.7133×10−3 | 6/38 | Response to light stimulus |
| 9314 | 6.7133×10−3 | 6/38 | Response to radiation |
| 6986 | 2.3696×10−2 | 2/2 | Response to unfolded protein |
| 43335 | 2.3696×10−2 | 2/2 | Protein unfolding |
| 35966 | 2.3696×10−2 | 2/2 | Response to topologically incorrect protein |
Module 17, a module in response to environmental stresses
We examined one module, module 17, in detail because we are interested in stress responses, as module 17 was found to be enriched with GO terms relating to environment stresses. Module 17 contained 41 nodes (genes) and 89 edges and was significantly enriched with 16 GO terms (p<2.3696×10–2) (Figure 3 and Table 4). The over-expressed GO terms include ‘response to stimulus’, ‘response to high light intensity’, ‘response to abiotic stimulus’, ‘response to oxidative stress’, ‘response to hydrogen peroxide’ and particularly ‘response to heat’ (GO: 0009408) (p=3.5017×10−10). A total of 19 genes in module 17 encode for heat shock proteins (HSPs), including members of the HSP20, HSP40, HSP70, HSP90 and HSP100 families (Table 5).
Table 5. Homologous genes between 29 grapevine genes in module 17 and those in Arabidopsis thaliana.
| Gene number | Grapevine gene | Probe number | Homologs in Arabidopsis thaliana | Information of gene classification and function |
|---|---|---|---|---|
| 1 | Vit_10s0003g00260 | 1616811_at | AT2G20560 | DNAJ heat shock protein |
| 2 | Vit_07s0185g00040 | 1621759_s_at | AT3G07150 | Unknown protein |
| 3 | Vit_13s0019g03160 | 1616145_a_at | AT1G53540 | HSP17.6C-CI |
| 4 | Vit_18s0041g01230 | 1616369_at | AT5G49910 | Chloroplast HSP70−2; ATP binding |
| 5 | Vit_02s0025g04060 | 1611927_at | AT4G11740 | Unknown protein |
| 6 | Vit_04s0008g01590 | 1611192_at | AT5G12020 | HSP17.6II |
| 7 | Vit_06s0004g04470 | 1621357_s_at | AT5G02500 | HSC70−1; ATP binding |
| 8 | Vit_04s0008g01490 | 1614330_at | AT5G12020 | HSP17.6II |
| 9 | Vit_16s0050g01150 | 1618066_a_at | AT5G52640 | HSP90.1; ATP binding |
| 10 | Vit_08s0007g00740 | 1613948_at | AT3G09350 | Armadillo/beta-catenin repeat family protein |
| 11 | Vit_01s0010g02290 | 1608828_at | AT4G27670 | HSP21 |
| 12 | Vit_16s0098g01060 | 1620985_at | AT4G27670 | HSP21 |
| 13 | Vit_11s0016g04080 | 1621552_at | AT3G24500 | MBF1C |
| 14 | Vit_07s0005g01980 | 1609808_at | AT2G47180 | GolS1 |
| 15 | Vit_17s0000g07190 | 1615503_at | AT1G74310 | HSP101; ATP binding |
| 16 | Vit_17s0000g00070 | 1611931_at | AT5G07330 | Unknown protein |
| 17 | Vit_13s0047g00110 | 1606746_a_at | AT4G02450 | Glycine-rich protein |
| 18 | Vit_11s0078g00260 | 1608348_a_at | AT5G35320 | Unknown protein |
| 19 | Vit_19s0085g01050 | 1616538_at | AT1G53540 | HSP17.6C-CI |
| 20 | Vit_06s0004g06010 | 1615761_at | AT1G07350 | Arginine-rich ribonucleoprotein |
| 21 | Vit_05s0020g03330 | 1621709_at | AT2G32120 | HSP70T−2; ATP binding |
| 22 | Vit_13s0019g00860 | 1622489_at | AT5G37670 | HSP15.7−CI |
| 23 | Vit_08s0007g00130 | 1609949_at | AT3G12580 | HSP70; ATP binding |
| 24 | Vit_16s0022g00510 | 1616889_at | AT4G25200 | Mitochondrion-localized HSP23.6 |
| 25 | Vit_08s0217g00090 | 1611195_at | AT3G08970 | Endoplasmic reticulum-localized J protein |
| 26 | Vit_06s0004g05770 | 1621652_at | AT1G07400 | HSP17.8−CI |
| 27 | Vit_02s0154g00480 | 1620348_at | AT4G25200 | Mitochondrion-localized HSP23.6 |
| 28 | Vit_12s0035g01910 | 1613858_at | AT4G10250 | HSP22.0 |
| 29 | Vit_18s0089g01270 | 1609222_at | AT4G10250 | HSP22.0 |
Module 17 contains 41 nodes (probes). Among them, 12 probe sets were not matched with grapevine genes annotated by CRIBI Genomics, University of Padua (http://genomes.cribi.unipd.it/) (listed in Supplementary Table 2). These probe sets were 1609554_at, 1615503_at, 1607291_at, 1610779_at, 1613154_at, 1622489_at, 1616706_at, 1611195_at, 1621902_at, 1610122_at, 1616049_at and 1618545_a_at. Therefore, 29 grapevine genes are listed in this table.
Plants respond to various stresses in a similar manner—by producing HSPs that protect cells against many stresses.29 The accumulation of HSPs plays a key role in acquired heat tolerance during heat stress.30 MBF1C (Vit_11s0016g04080) is an important transcription factor that responds to stresses,31 and as a key regulator of heat tolerance in Arabidopsis thaliana, the MBF1C protein accumulates rapidly during heat stress. The inositol galactoside (GolS2) enzyme (Vit_07s0005g01980) is a key synthase that regulates the drought and cold responses.32 Liu et al.33 inferred that galactinol synthase may be important for grapevine heat tolerance. The endoplasmic reticulum-localized J protein Vit_08s0217g00090 is an important molecular chaperone of HSP70.34 In addition, four putative uncharacterized proteins in module 17, Vit_07s0185g00040, Vit_02s0025g04060, Vit_17s0000g00070 and Vit_11s0078g00260, are clearly interrelated to other nodes and edges involved in the stress response, but no information about their domain and homologous alignments is available. Therefore, we considered these four putative genes to have unknown functions in the stress response.
Expression patterns of genes in module 17 at different time points after heat shock and recovery
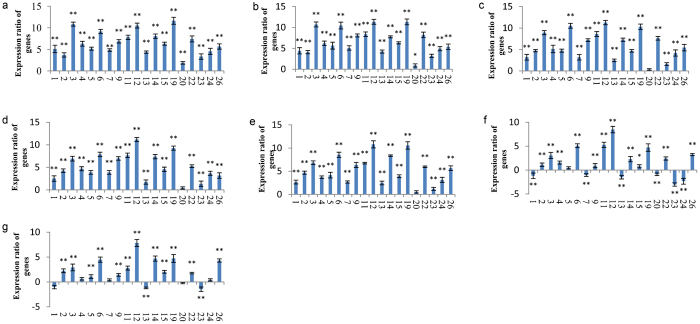
We tested module 17 in response to heat shock, one environmental stress. When grapevine plants were treated with heat shock at 40 °C for 6 h, 19 of 29 genes in module 17 were upregulated and their expression quantities exhibited variable regulation from low-level to high-level, ranging from 1.86- to 11.63-fold (Figure 4a−4e). However, some gene expression quantities maintained a high level from 0.5 h to 6 h, ranging from 6.85- to 11.63-fold (p<0.01). These included Vit_13s0019g03160, Vit_04s0008g01590, Vit_16s0098g01060, Vit_07s0005g01980 and Vit_19s0085g01050, which encode HSP17.6, HSP17.6, HSP21, galactinol synthase 1 and HSP17.6, respectively, in which galactinol synthase 1 (GolS1) is a heat shock factor target gene responsible for the heat-induced synthesis of the raffinose family of oligosaccharides in Arabidopsis.35
Figure 4.
Gene expression patterns in module 17 treated with heat shock and recovery at different time points. a–e: heat shock for 0.5, 1, 2, 3 and 6 h, respectively. f–g: heat shock recovery for 2 and 5 h after plants were treated at 40 °C for 1 h, respectively. The value in the Y-axis is −ΔΔCt. The expression ratio of a gene was considered significant if *p<0.05. Expression ratio of genes was significant if **p<0.01. The numbers from 1 to 26 on the X-axis represent the grapevine genes listed under ‘gene number and grapevine gene’ in Table 1.
Moreover, 12 of 19 genes were still upregulated significantly (p<0.01) after 2 h and 5 h of recovery. After 2 h of recovery, 6 of 19 genes were downregulated significantly up to 3.02-fold (p<0.01) (Figure 4f), including Vit_08s0007g00130, Vit_16s0022g00510 and Vit_11s0016g04080. After 5 h of recovery, only two genes among them were downregulated significantly (p<0.01) (Figure 4g), and the other four genes recovered from their downregulated states. However, 3 out of 19 genes, Vit_04s0008g01590, Vit_16s0098g01060 and Vit_19s0085g01050, which expressed highly at 40 °C for 6 h, still maintained high-level expression after 2 h and 5 h of recovery, ranging from 4.49- to 8.49-fold (p<0.01). Therefore, our results indicate that genes in module 17 have different gene functions, and their mechanisms during heat shock and transient states may be complex.
The expression of two putative uncharacterized genes, Vit_07s0185g00040 (ranging from 1.12- to 4.72-fold) and Vit_02s0025g04060 (ranging from 0.47- to 5.66-fold), was also detected during heat shock and recovery. Based on the GGCN analysis, no homologous alignment or annotation information is available about their sequences, domains or gene expression in NCBI (http://www.ncbi.nlm.nih.gov/cdd) or in CRIBI Genomics, University of Padua (http://genomes.cribi.unipd.it/).
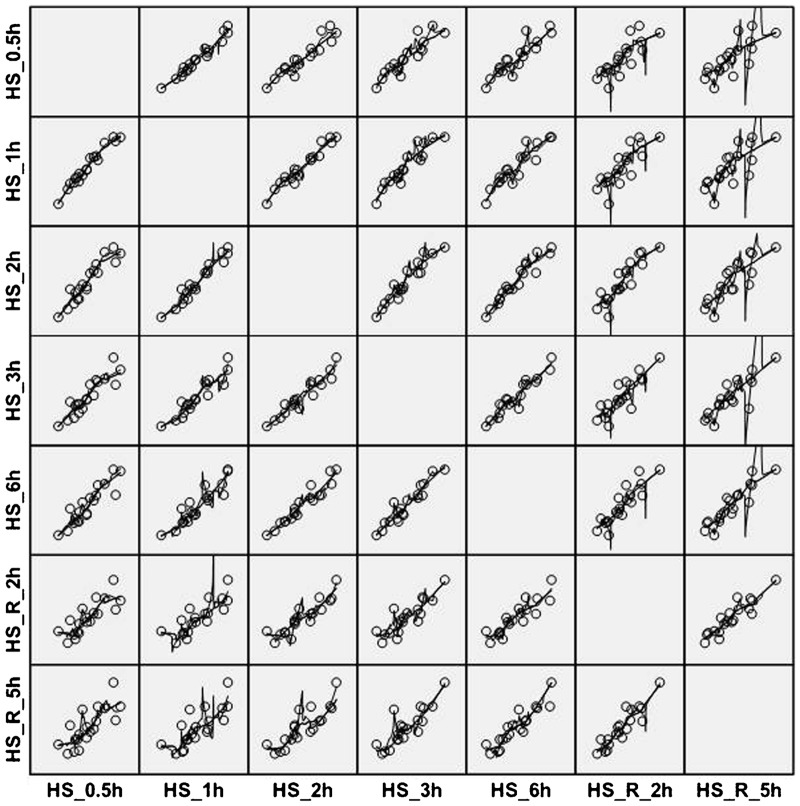
Expression values in response to heat shock and recovery between each two time points were plotted together for the 19 genes in module 17 using the SPSS program28 and treated with LOESS26 (Figure 5). The best goodness-of-fit values were those at adjacent time points. Moreover, most R2 between the dependent and independent variables ‘gene expression value’ and ‘treatment time point’ were close to 1.0 at adjacent time points36 (Table 6), which indicated a strong linear relationship between compared variables. The goodness-of-fit analysis indicated that under the same tempospatial conditions, as a whole network, these genes display a clear co-expression relationship.
Figure 5.
The goodness of fit test of 19 gene expression values in module 17 between each two time points treated with heat shock and subsequent recovery. The fit lines were added by using LOESS in the matrix scatterplot. ‘HS’ represents heat shock treatment. ‘HS_R’ represents recovery after heat shock treatment.
Table 6. ‘Goodness-of-fit’ test of 19 gene expression values in module 17 between each ‘two time points’ treated with heat shock and recovery.
| R2 | HS_0.5 h | HS_1 h | HS_2 h | HS_3 h | HS_6 h | HS_R_2 h | HS_R_5 h |
|---|---|---|---|---|---|---|---|
| HS_0.5 h | 0.961 | 0.880 | 0.825 | 0.829 | 0.659 | 0.591 | |
| HS_1 h | 0.961 | 0.944 | 0.882 | 0.849 | 0.679 | 0.597 | |
| HS_2 h | 0.880 | 0.944 | 0.916 | 0.925 | 0.809 | 0.725 | |
| HS_3 h | 0.825 | 0.882 | 0.916 | 0.905 | 0.754 | 0.727 | |
| HS_6 h | 0.829 | 0.849 | 0.925 | 0.905 | 0.799 | 0.838 | |
| HS_R_2 h | 0.659 | 0.679 | 0.809 | 0.754 | 0.799 | 0.835 | |
| HS_R_5 h | 0.591 | 0.597 | 0.725 | 0.727 | 0.838 | 0.835 |
R2 represents the coefficient of determination between the dependent and independent variables ‘gene expression value’ and ‘treatment time point’ in the linear regression. ‘HS’ represents heat shock treatment. ‘HS_R’ represents recovery after heat shock treatment.
The PCC of gene expression values were significantly greater than 0.78 (Supplementary Table 3). Similarly, during the different time points of heat shock and the recovery process, most PCC values were also greater than 0.78, which indicate that most genes significantly co-express (Supplementary Table 3). Therefore, gene co-expression ‘in response to heat’ represented by module 17 was validated experimentally by qRT-PCR and by PCC analysis of gene expression given that most genes were upregulated together very significantly (p<0.01), and most PCC values were greater than the PCC cutoff value, 0.78, which was used to screen significant co-expression correlation from a large-scale expression data set.
Among the 29 genes in module 17 that corresponded to ‘responses to heat stress’, 10 genes showed no response to heat shock, which could suggest that these genes may co-express in other tempospatial condition heat stress environments or in response to other environment stresses, such as ‘response to high light intensity’, ‘response to oxidative stress’ or ‘response to hydrogen peroxide’, because expression of these genes might be regulated depending on time, space and environmental conditions.37 This process may include many levels, such as chromatin structure, transcription, transcript stability or localization, and translation. The homologous gene comparison for ‘response to heat’ matched quite well between module 17 grapevine genes and those involved in the heat stress response in A. thaliana (Table 5).
Expression patterns of genes in module 17 after low temperature treatment
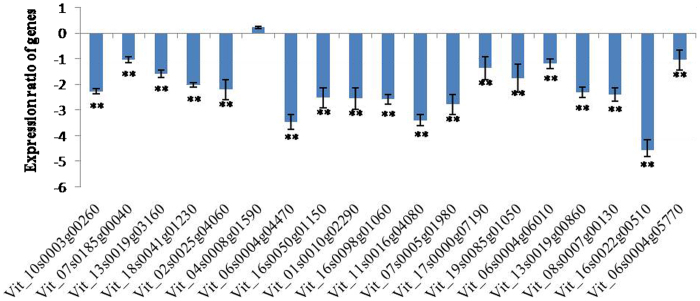
In contrast to the upregulation of these genes, most of the 19 genes were down regulated in response to low temperature (4 °C) treatment (Figure 6), ranging from 1.05- to 4.55-fold (Figure 6). To further test the co-expression relationship between these genes, the PCC of 19 gene expression values were calculated. Supplementary Table 4 shows that 45.91% of them were greater than 0.78; thus, the co-expression relationship of these genes was not very obvious if inferring from PCC values, compared with those after heat shock treatment.
Figure 6.
Gene expression patterns in module 17 after treatment with low temperature at 4 °C for 1 h. The value on the Y-axis is −ΔΔCt. Expression ratio of genes was considered significant if **p<0.01.
Discussion
Plant growth, development and adaptation to the environment are complex, yet highly coordinated, processes. One way to understand these complex processes is to establish gene co-expression networks from which we can predict putative functions of genes in the network because genes sharing a module in a co-expression network are likely involved in similar biological processes.3,7
In this study, we constructed a GGCN at the genome-wide level with publically available microarray data using the efficient heuristic algorithm Qcut, which is based on the optimization of a modularity function (Q), and combined spectral graph partitioning and local search to optimize Q.17 Moreover, nodes were densely linked with each other in a sub-network module, but they were sparse or had no connections between the subnetwork modules. The gene-to-gene PCC derived from gene expression data in Gene Expression Omnibus allowed us to portion these co-expressing genes into network modules in various experimental conditions. The goodness of fit, coefficient of determination and PCC statistical tests of module 17 have confirmed that genes in the same module show co-expression relationships under the same tempo-spatial conditions, which may be associated with the same biological function, one of the important features of a co-expression network.38,39 The homologous gene comparison of ‘response to heat’ between module 17 in grapevine and A. thaliana also demonstrated that partitioning genes into modules from the co-expression network was reliable.
HSPs and chaperones are crucial components of the heat shock regulatory network in plants40 and take a crucial role in response to multiple environmental insults.41,42 These HSPs are also involved in response to cold43 and non-thermal stress treatments, such as salinity,44 drought,45,46 high light stress,47 oxidative stress48 and heavy metal stress.49 Therefore, the biological functions represented by module 17, a module that responds to environmental stresses, may be tested in multiple stresses in the future.
The reliability and biological correlation of the network were further verified by experimentation. The same set of genes in module 17 of the co-expression network exhibited two co-expression patterns, one upregulation (to heat shock treatment) and one downregulation (to cold treatment). The differential response patterns between heat shock and low temperature experimental treatments suggest that other regulatory factors may be involved, which require additional investigation. These covarying patterns could also suggests the complexity of cellular transcriptional activities.14
The co-expression network and partitions into different modules may also help to identify new genes that may putatively be involved in certain biological processes.3 In this research, two putative uncharacterized genes without any gene function information, gene annotation, expression sequence tag(EST), transcriptome data or protein domain prediction were detected in response to heat shock. These genes are worthy of further investigation.
Overall, the study provided a new insight into the module properties of grapevine gene functions, which facilitated the module research of gene functions and the discovery of new genes.
Acknowledgments
This research was supported by the Chinese National Natural Science Foundation Project #31201607.
The authors declare no conflict of interest.
References
- Ruan J, Dean AK, Zhang W. A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC Syst Biol 2010; 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazhnik P, Fuente A, Mendes P. Gene networks: how to put the function in genomics. Trends Biotechnol 2002; 20: 467–472. [DOI] [PubMed] [Google Scholar]
- Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 2007; 48: 381–390. [DOI] [PubMed] [Google Scholar]
- Alm EArkin AP. Biological networks. Curr Opin Struct Biol 2003; 13: 193–202. [DOI] [PubMed] [Google Scholar]
- Luo F, Yang Y, Zhong J et al. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinformatics 2007; 8: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Perez J, Hernandez B, Sunter G, Sponsel VM. Systematic construction and analysis of co-expression networks for identification of functional modules and cis-regulatory elements. In: Proceedings of the 9th International Workshop on Data Mining in Bioinformatics, in Conjuction with Sigkdd’10; 25–28 July 2010; Washington, DC, USA. 2010, pp. 15–24; Arlington, Virginia, USA: ACM press. [Google Scholar]
- Mao L, van Hemert JL, Dash S, Dickerson JA. Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 2009; 10: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yin L, Xue H. Co-expression analysis identifies CRC and AP1 the regulator of Arabidopsis fatty acid biosynthesis. J Integr Plant Biol 2012; 54: 486–499. [DOI] [PubMed] [Google Scholar]
- Heyndrickx KS, Vandepoele K. Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol 2012; 3: 884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler JB, Ficklin SP, Luo F et al. Conserved non-coding regulatory signatures in Arabidopsis co-expressed gene modules. PloS ONE 2012; 7: e45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yin Y, Ma Q et al. Genome-scale identification of cell-wall related genes in Arabidopsis based on co-expression network analysis. BMC Plant Biol 2012; 12: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu S, Zuo K et al. Identification of gene modules associated with drought response in rice by network-based Analysis. PloS ONE 2012; 7: e33748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashutosh P, Prashant M, Mohd PK et al. Co-expression of Arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant Biotechnol J 2014; 12: 69–80. [DOI] [PubMed]
- Stuart JM, Segal E, Koller D et al. A gene-coexpression network for global discovery of conserved genetic modules. Science 2003; 302: 249–255. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007; 449: 463–467. [DOI] [PubMed] [Google Scholar]
- Wang M, Vannozzi A, Wang G et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hort Res 2014; 1: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Zhang W. Identifying network communities with a high resolution. Phys Rev E 2008; 77: 016104. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005; 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K et al. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 2007; 35: D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yuan H, Tadesse MG et al. Integration of multiple data sources for identifying functional modules using Bayesian network. In: Proceedings of IEEE International Workshop on Genomic Signal Processing and Statistics; 2–4 December; Washington, DC, USA. 2012, pp. 13–17; Piscataway, New Jersey, USA: IEEE press. [Google Scholar]
- Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol 2007; 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum 1962; 15: 473–497. [Google Scholar]
- Zhang J, Du X, Wang Q et al. Expression of pathogenesis related genes in response to salicylic acid, methyl jasmonate and 1-aminocyclopropane-1-carboxylic acid in Malus hupehensis (Pamp.) Rehd. BMC Res Notes 2010; 3: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Yuan S, Guo H et al. Evolution and divergence in the coding and promoter regions of the Populus gene family encoding xyloglucan endotransglycosylase/hydrolases. Tree Genet Genomes 2012; 8: 177–194. [Google Scholar]
- Jacoby WG. LOESS: a nonparametric, graphical tool for depicting relationships between variables. Elect Stud 2000; 19: 577–613. [Google Scholar]
- Hagquist C, Stenbeck M. Goodness of fit in regression analysis—R2 and G2 reconsidered. Qual Quant 1998; 32: 229–245. [Google Scholar]
- Norušis MJ. IBM SPSS Statistics 19 Advanced Statistical Procedures Companion. Upper Saddle River, NJ: Pearson Education, Prentice Hall Press, 2012. [Google Scholar]
- Timperio AM, Egidi MG, Zolla L. Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J Proteomics 2008; 71: 391–411. [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U et al. Complexity of the heat stress response in plants. Curr Opin Plant Biol 2007; 10: 310–316. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Bajad S, Shuman J et al. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 2008; 283: 9269–9275. [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S et al. Important roles of drought-and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 2002; 29: 417–426. [DOI] [PubMed] [Google Scholar]
- Liu GT, Wang JF, Cramer G et al. Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol 2012; 12: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E, Huang P, Aron R et al. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol 2006; 36: 1–21. [DOI] [PubMed] [Google Scholar]
- Panikulangara TJ, Eggers-Schumacher G, Wunderlich M et al. Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol 2004; 136: 3148–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Persson S, Mehta T et al. Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol 2006; 142: 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie M, Gascuel O, Lefort V et al. Computational discovery of regulatory elements in a continuous expression space. Genome Biol 2012; 13: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocco DJ, Kohane IS, Butte AJ. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics 2004; 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Yan X, Huang Y, Han J, Zhou XJ. Mining coherent dense subgraphs across massive biological networks for functional discovery. Bioinformatics 2005; 21: i213–i221. [DOI] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P. How do plants feel the heat? Trends Biochem Sci 2012; 37: 118–125. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 2004; 9: 244–252. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Welch W. Role of the major heat shock proteins as molecular chaperones. Annul Rev Cell Biol 1993; 9: 601–634. [DOI] [PubMed] [Google Scholar]
- Janská A, Maršík P, Zelenková S, Ovesná J. Cold stress and acclimation—what is important for metabolic adjustment? Plant Biol 2010; 12: 395–405. [DOI] [PubMed] [Google Scholar]
- Kosová K, Vítámvás P, Urban MO, Prášil IT. Plant proteome responses to salinity stress–comparison of glycophytes and halophytes. Funct Plant Biol 2013; 40: 775–786. [DOI] [PubMed] [Google Scholar]
- Grigorova B, Vaseva I, Demirevska K, Feller U. Combined drought and heat stress in wheat: changes in some heat shock proteins. Biol Plantarum 2011; 55: 105–111. [Google Scholar]
- Xoconostle-Cazares B, Ramirez-Ortega FA, Flores-Elenes L, Ruiz-Medrano R. Drought tolerance in crop plants. Am J Plant Physiol 2010; 5: 241–256. [Google Scholar]
- Carvalho LC, Vilela BJ, Mullineaux PM, Amâncio S. Comparative transcriptomic profiling of Vitis vinifera under high light using a custom-made array and the Affymetrix GeneChip. Mol Plant 2011; 4: 1038–1051. [DOI] [PubMed] [Google Scholar]
- Sokolowska I, Woods AG, Wagner J et al. Mass spectrometry for proteomics-based investigation of oxidative stress and heat shock proteins. In: Oxidative Stress: Diagnostics, Prevention, Therapy. Washington, DC: ACS, 2011: 369–411. [Google Scholar]
- Hossain Z, Komatsu S. Contribution of proteomic studies towards understanding plant heavy metal stress response. Front Plant Sci 2012; 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.