Abstract
Background
The gonadotropin-induced resumption of oocyte meiosis in preovulatory follicles is preceded by expression of epidermal growth factor (EGF)-like peptides, amphiregulin (AREG) and epiregulin (EREG), in mural granulosa and cumulus cells. Both the gonadotropins and the EGF-like peptides possess the capacity to stimulate resumption of oocyte meiosis in vitro via activation of a broad signaling network in cumulus cells. To better understand the rapid genomic actions of gonadotropins (FSH) and EGF-like peptides, we analyzed transcriptomes of cumulus cells at 3 h after their stimulation.
Methods
We hybridized aRNA from cumulus cells to a pig oligonucleotide microarray and compared the transcriptomes of FSH- and AREG/EREG-stimulated cumulus cells with untreated control cells and vice versa. The identified over- and underexpressed genes were subjected to functional genomic analysis according to their molecular and cellular functions. The expression pattern of 50 selected genes with a known or potential function in ovarian development was verified by real-time qRT-PCR.
Results
Both FSH and AREG/EREG increased the expression of genes associated with regulation of cell proliferation, cell migration, blood coagulation and extracellular matrix remodeling. FSH alone induced the expression of genes involved in inflammatory response and in the response to reactive oxygen species. Moreover, FSH stimulated the expression of genes closely related to some ovulatory events either exclusively or significantly more than AREG/EREG (AREG, ADAMTS1, HAS2, TNFAIP6, PLAUR, PLAT, and HSD17B7). In contrast to AREG/EREG, FSH also increased the expression of genes coding for key transcription factors (CEBPB, FOS, ID1/3, and NR5A2), which may contribute to the differing expression profiles of FSH- and AREG/EREG-treated cumulus cells.
Conclusions
The impact of FSH on cumulus cell gene transcription was higher than the impact of EGF-like factors in terms of the number of cell functions affected as well as the number of over- and underexpressed genes. Both FSH and EGF-like factors overexpressed genes involved in the post-ovulatory switch in steroidogenesis and tissue remodelling. However, FSH was remarkably more efficient in the up-regulation of several specific genes essential for ovulation of matured oocytes and also genes that been reported to play an important role in maturation of cumulus-enclosed oocytes in vitro.
Electronic supplementary material
The online version of this article (doi:10.1186/s12958-015-0112-2) contains supplementary material, which is available to authorized users.
Keywords: FSH, Growth factors, Cumulus cell, Transcriptome
Background
The development of mammalian female germ cells requires close contact and metabolic cooperation with the somatic granulosa cells. In the antral follicles, the granulosa cells differentiate into two phenotypically distinct populations: the mural granulosa cells, lining the follicle wall and the cumulus cells, surrounding the oocyte in several layers. Both populations of granulosa cells fulfil different roles during follicle development: the mural granulosa cells are predominantly involved in the perception of signals from outside follicle, the production of steroid hormones and follicular rupture; the cumulus cells provide nutrients and regulatory molecules for oocyte growth, final maturation and ovulation. In turn, the range of function of cumulus cells, including steroidogenesis, gene expression, extracellular matrix formation and metabolism, are modified by factors secreted by the oocyte [1].
During the growth phase, the oocyte is arrested in the first meiotic prophase, but it gains full meiotic competence in several steps [2]. However, these meiotically competent oocytes still remain in the dictyate stage if they are retained within follicles. They only resume meiosis following the preovulatory surge of luteinizing hormone (LH). The resumption of meiosis is regulated inside oocytes by a post-translation mechanism and does not depend on the activation of genes in the oocyte itself [3]. Nevertheless, the LH-induced meiotic resumption is accompanied by a dramatic change in gene expression profiles in mural granulosa and cumulus cells. The changes in the cumulus/granulosa cell transcriptome are under the control of a broad signaling network activated in follicular cells by the LH surge. This network includes major cellular protein kinases including protein kinase A (PKA), phosphoinositide-3-kinase/v-akt murine thymoma viral oncogene homolog (PI3K/AKT), mitogen-activated kinase 3/1 (MAPK3/1) and MAPK14 [4–6] and downstream transcription factors including CREB, AP1, NRIP1, NR5A2 and CEBPB [7–11].
The resumption of meiosis is accompanied by an expansion of cumulus cells, a process that enables the detachment of the oocyte-cumulus complex from the follicle wall and its ovulation to the oviduct. In response to the preovulatory surge of LH, the cumulus cells start the synthesis of a large amount of hyaluronic acid (HA)-enriched extracellular matrix that is deposited into the extracellular spaces and causes the process of expansion [12]. The production of HA is controlled by hyaluronan synthase 2 (HAS2), which is highly expressed in cumulus cells shortly after the preovulatory surge of LH [13]. The retention and organization of HA in the extracellular matrix is mediated by HA-binding proteins, which include versican [14], tumor necrosis factor α-stimulated gene 6 [15], pentraxin 3 [16] and serum-derived members of the inter-α-trypsin inhibitor (IαI) family [17, 18]. The experiment with mice null for the Ptgs2 and Ptger2 gene revealed that the local production of prostaglandin E2 is also essential for the normal expansion of mouse cumulus [19, 20].
Mechanisms of meiotic resumption and cumulus expansion have been studied on a whole follicle culture that is often used in mice and rats or on a simplified model based on a culture of isolated COCs. In the latter case, LH and hCG are inefficient at triggering these events due to the absence of functional LH-receptors on cumulus cells [21, 22]. Functional LH receptors, however, can develop over the course of in vitro culture, under the influence of FSH [22, 23]. For this reason, both the resumption of meiosis and cumulus expansion is routinely stimulated by the addition of FSH, or a combination of FSH and hCG.
The mechanism of LH signal transduction in the preovulatory follicle has been intensively studied over the last decade. It was found in the model of whole follicle cultures in mice and rats that LH binds to its receptors on mural granulosa cells and stimulates the expression of epidermal growth factor (EGF)-like peptides (amphiregulin-AREG, epiregulin-EREG, betacellulin-BTC) [24, 25]. These peptides then act directly on mural granulosa as well as cumulus cells, bind to the EGF-receptor and consequently stimulate the resumption of meiosis and expansion of the cumulus. In addition, a regulatory loop between the mural granulosa and the cumulus cells also ensures the production of AREG and EREG in the cumulus cell compartment [26]. The EGF-like peptides are also produced in the cumulus cells of in vitro-cultured COCs upon stimulation with FSH and trigger the resumption of meiosis [27].
This knowledge offers the possibility of stimulating the meiotic resumption and cumulus expansion of cultured COCs by purified EGF-like peptides. We have recently shown that AREG and EREG stimulate the expansion of cumulus cells, maturation of the oocyte and acquisition of oocyte developmental competence in vitro in pigs [28]. However, we have also pointed out the lower efficiency of EGF-like peptides, compared to FSH, in inducing the expression of expansion-related genes and their inability to activate PKA in cultured COCs [28]. A detailed study of signaling pathways activated by FSH and EGF-like factors in pig cumulus cells confirmed that FSH-stimulated, but not AREG-stimulated resumption of meiosis depends on PKA activity [6]. In addition, this study documented that both modes of stimulation result in the activation of the EGF-receptor tyrosine kinase and MAPK3/1 pathways in cumulus cells, which is essential for both meiotic resumption and cumulus expansion [6]. It is not known how the differences between FSH and AREG/EREG in the activation of signaling pathways affect the gene expression profiles of treated COCs.
We explored the use of microarray hybridization technology to identify rapid global changes in the gene expression profile of cumulus cells stimulated with FSH or EGF-like peptides (AREG/EREG). Both these treatments have been shown to stimulate oocyte meiotic resumption, cumulus cell expansion and to modify cumulus cell steroidogenesis, but it is not completely clear what other molecular and cellular processes in cumulus cells are affected by FSH and EGF-like peptides and what is their relationship to the acquisition of oocyte developmental competence. The current IVM techniques yield oocytes with low developmental competence as compared to their counterparts matured in vivo. The molecular mechanisms underlying this insufficiency are largely unknown. Nevertheless, recent data indicate that the way, by which the maturation of oocytes is stimulated, is crucial for determining the oocyte developmental competence [29–33]. The aim of this study was to assess the differences in impact of FSH and EGF-like peptides on gene expression in cumulus cell with special emphasis on the genes with known relationship to regulation of reproductive function in mammals.
Methods
The study was conducted in the laboratories of IAPG in Libechov in 2012–2015.
Collection of cumulus-oocyte complexes
Ovaries were obtained from premature crossbred gilts (Landrace and Large White), 6–8 months old and 90–120 kg in weight, slaughtered at a local abattoir for commercial purposes. The ovaries were excised and transported to the laboratory in a thermo-flask at 38 °C. The contents of medium-size antral follicles about 3–5 mm in diameter were aspirated with a syringe connected to 20 G needle, pooled in a test-tube and allowed to sediment for 10 min. The sediment was washed twice with PBS, placed in a Petri dish and the COCs were collected by a pipette. Only COCs surrounded by compact multi-layered cumulus were selected for experiments.
Culture of cumulus-oocyte complexes in vitro
The COCs were cultured in M-199 medium (Gibco, Life Technologies, Rockville, USA) supplemented with 0.91 mM sodium pyruvate, 0.57 mM cysteine, 5.5 mM Hepes, antibiotics and fetal calf serum (5 %) (all Sigma, Prague, Czech Republic). Groups of 25–30 COCs were cultured in four-well dishes (Nunclon, Roskilde, Denmark) in 0.5 ml of culture medium at 38.5 °C in a humidified atmosphere of 5 % CO2 for 3 h. The maturation of COCs was stimulated by the addition of human recombinant FSH (100 ng/ml; Gonal F; Merck Serono Europe, London, GB) or recombinant human AREG (100 ng/ml, Sigma) and EREG (100 ng/ml; R&D Systems, Minneapolis, MN, USA) to the culture medium. These concentrations proved to activate various signaling pathways in cumulus cells, stimulate cumulus expansion and oocyte maturation in our previous studies [6, 28]. At the end of the culture period, about 150 COCs were transferred to a Petri dish with 2 ml of PBS and the cumulus cells were mechanically stripped of oocytes by pipetting. The oocytes were removed and PBS with a suspension of the separated cumulus cells was transferred to a microtube and briefly centrifuged in a microcentrifuge. The supernatant was discarded and the pellet of cumulus cells was lysed in 300 μl of RTL buffer (Qiagen, Hilden, Germany) and stored at −80 °C. In each experiment, groups of control, FSH- or AREG/EREG-stimulated COCs were cultured for 42 h as a biological control for the microarray. These COCs were assessed for cumulus expansion and oocyte maturation by methods described in our previous papers [6, 28]. The cumulus expansion index was 0; 2.98 ± 0.05; and 2.74 ± 0.04 and the maturation rate of oocytes to metaphase II was 17.5 ± 1.4; 93.2 ± 3.8; 73.6 ± 2.9 for control, FSH and AREG/EREG group, respectively.
Microarray
Experimental design of microarray experiment. Three different samples (FSH treated, AREG/EREG treated and control untreated cumulus cells), each represented by 3 independently prepared biological replicates, were hybridized to 9 microarrays (Pigoligoarray, www.pigoligoarray.org) as follows (Fig. 1):
Fig. 1.

Design of hybridization scheme of microarray experiment. Each arrow represents one microarray. Samples of cumulus cells stimulated with AREG/EREG, FSH or control cells were labeled with AlexaFluor 555 (depicted in green) or AlexaFluor 647 (depicted in red)
1st and 3rd biological replicate: Control AlexaFluor 647+ AREG/EREG AlexaFluor 555; AREG/EREG AlexaFluor 647 + FSH AlexaFluor 555; FSH AlexaFluor 647 + Control AlexaFluor 555. 2nd biological replicate (dye swap): Control AlexaFluor 555 + AREG /EREG AlexaFluor 647; AREG/EREG AlexaFluor 555 + FSH AlexaFluor 647; FSH AlexaFluor 555 + Control AlexaFluor 647.
Total RNA isolation. Total RNA from all nine samples was isolated using an RNeasy PLUS mini kit (Qiagen) according to the manufacturer’s instructions. RNA integrity was verified using an RNA 6000 Nano LabChip kit with an Agilent 2100 Bioanalyzer. Only RNA samples with a resulting RNA integrity number (RIN) between 8.2 and 9.1 were used for the amplification of aRNA.
aRNA amplification and labeling. aRNA was prepared with one-round amplification using an AminoAllyl MessageAmp II Kit (Ambion, Austin, Texas, USA). The in vitro transcription time was 14 h. Five microgram of aRNA were conjugated with either Alexa Fluor 555 or 647 dye (Invitrogen, Carlsbad, California, USA). Labeled aRNAs were purified using a PicoPure RNA isolation kit (Arcturus) according to the manufacturer’s instructions; 2 μg of each labeled aRNA (Alexa Fluor 555- and Alexa Fluor 647-conjugated aRNA samples according to the experimental design) were mixed together and stored at −80 °C.
Hydration and crosslink of microarrays. Microarray slides were rehydrated for 10 s above a water bath at 50 °C, dried on a heat block for 5 s at 65 °C and cooled for 1 min at room temperature. This procedure was repeated 4 times. UV cross-linking was performed by exposing rehydrated slides to 180 mJ of UV radiation. After crosslinking, the slides were washed in 0.1 % SDS with constant gentle mixing for 5 min followed by rinsing in ddH2O and 3 min of incubation in 100 % ethanol with constant gentle mixing, all at room temperature. Slides were dried by centrifugation for 4 min at 200 g. Just before use, the microarray slides were pre-hybridized at 42 °C for 1 h in pre-hybridization buffer (5 × SSC, 0.1 % SDS, 1 % BSA) using a Tecan HS400Pro hybridization station (TECAN, Austria).
Probe fragmentation and hybridization. Pre-mixed labeled aRNA probes were fragmented using Fragmentation buffer (Ambion Austin, Texas, USA) according to the manufacturer’s instructions. After fragmentation, probes were denatured for 5 min at 90 °C, mixed with 130 μl of SlideHyb Glass Array Hybridization Buffer # 1 (Ambion Austin, Texas, USA) preheated to 68 °C and hybridized for 15 h at 42 °C in Tecan HS400Pro to pre-hybridized microarray slides. After hybridization, slides were washed and dried automatically in the Tecan HS400Pro instrument and stored in the dark until scanned the same day.
Scanning and image analysis. Slides were scanned using a High-Resolution Microarray Scanner (Agilent) at 5 μm resolution with the 20-bit dynamic range setting. Agilent Feature Extraction Software (v10.7) was used for image analysis.
Microarray data analysis. Microarray data background correction, normalization and statistical inference of changes in gene expression were done using the Bioconductor package Limma in the R statistical environment. Briefly, spot and background median signals were imported into the R data-frame and normex background correction was applied. After print-tip loess within-array normalization, A-quantile normalization was used to make the signal intensities more comparable across arrays. A linear model was fitted, with coefficients estimated for all 3 possible comparisons (FSH vs Control, AREG/EREG vs Control and FSH vs AREG/EREG) and for a possible dye-effect. The empirical Bayes method to moderate the standard errors of the estimated changes in expression was applied as implemented in Limma. P-values were adjusted according to the Benjamini and Hochberg method to control for false discovery rate. Only genes with adjusted p-value <0.01 were considered significantly changed.
The functional role of the differentially expressed genes was assessed for gene ontology using the freely available tool Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/).
Quantitative real-time PCR
To validate the microarray data, 50 candidate genes were selected for further analysis by real-time PCR. For this, sequence specific primers (Additional file 1: Table S1) were designed using Beacon Designer (Premier Biosoft, CA, USA). The total RNA was isolated from cumulus cells of 30 COCs cultured for 3 h using an RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Real-time PCR was carried out in a RotorGene 3000 cycler (Corbett Research, Sydney, Australia) using a One-Step RT-PCR Kit (Qiagen) with gene-specific primers. The 20 μl total reaction volume contained QIAGEN OneStep RT-PCR Buffer (1×), dNTP Mix (400 μM final concentration of each), reverse and forward primers (both 400 nM final concentration), SybrGreenI (0.4 μl of 1:1000 stock solution, Molecular Probes, Eugene, OR, USA), RNasine inhibitor (5 IU, Promega, Madison, WI, USA), QIAGEN OneStep RT-PCR Enzyme Mix (0.8 μl), and template RNA (2 μl). The reaction conditions were as follows: reverse transcription at 50 °C for 30 min, pre-denaturation at 95 °C for 15 min, followed by various numbers of PCR cycles, each of which consisted of denaturation at 95 °C for 30 s, annealing at temperature specific for each pair of primers (shown in Additional file 1: Table S1) for 20 s, extension at 72 °C for 20 s, and a final extension step at 72 °C for 5 min. The specificity of PCR product was verified by melting analysis. The relative concentrations of the templates in the different samples were determined using comparative analysis software (Corbett Research). The results for individual target genes were normalized according to the relative concentration of the internal standard, HPRT.
Results
Microarray analysis
We identified 2981 transcripts as overexpressed and 3582 as underexpressed in cumulus cells stimulated with FSH for 3 h compared to control unstimulated cells. A comparison of AREG/EREG-stimulated cells with control cells revealed 1491 transcripts as overexpressed and 1783 transcripts as underexpressed. Altogether, 2899 transcripts were overexpressed and 2581 transcripts underexpressed in the AREG/EREG group compared to the FSH group. The complete microarray data have been deposited in GEO and are accessible via the link http://www.ncbi.nlm.nih.gov/geo/ (GEO Accession: GSE64858). From the identified significantly changed transcripts (p < 0.01), only those transcripts with a log2 fold-change above 1.5 were used in downstream analyses.
The Venn diagrams were constructed to identify the exclusively over- and underexpressed genes after stimulation with AREG/EREG or FSH. As shown in Fig. 2a, we found that 27 genes were exclusively expressed in COCs cultured with EGF-like factors, the expression levels of 55 genes were increased by both AREG/EREG and FSH and 130 transcripts were found to be overexpressed in COCs cultured with FSH alone. A comparison of AREG/EREG-stimulated cells with FSH-stimulated cells revealed 10 overexpressed transcripts. The culture with AREG/EREG decreased the expression levels of 12 genes, 35 transcripts were underexpressed in both AREG/EREG-stimulated and FSH-stimulated COCs, and the expression of 118 genes was only decreased by FSH. A comparison of AREG/EREG-stimulated cells with FSH-stimulated cells revealed 68 underexpressed genes (Fig. 2b).
Fig. 2.
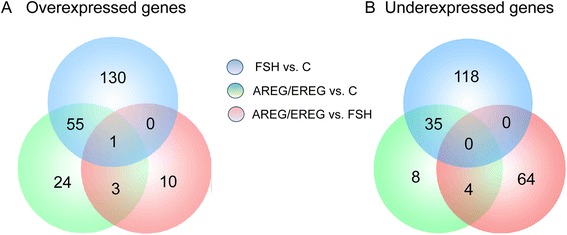
Venn diagrams summarizing microarray data analysis. a Venn diagram showing numbers of overexpressed and (b) underexpressed genes in cumulus cells after stimulation of COCs with FSH or AREG/EREG. Only genes with log2 fold-change above 1.5 were considered for this analysis
The analysis of the data revealed that several genes with a well-recognized function in the regulation of oocyte meiotic resumption and cumulus expansion were only overexpressed by FSH, among them genes associated with signal transduction (AREG), extracellular matrix synthesis and organization (ADAMTS1, HAS2, TNFAIP6, PLAT, and PLAUR) and the regulation of transcription (CEBPB, CREM, FOXO3, and NR4A1). Both AREG/EREG and FSH induced the expression of CYP11A1 and PGR involved in regulation of progesterone synthesis and signaling. Surprisingly, culture with FSH decreased the expression of genes involved in signal transduction (IRS1 and GRB14) and the regulation of transcription (PPARG and SMAD2). Moreover, other genes involved in transcriptional regulation (FOS, FOXO1, JUN, and PNRC1) were underexpressed under both culture conditions. A complete list of exclusively and commonly over/underexpressed genes after FSH and AREG/EREG stimulation is shown in Additional file 2: Table S2.
Gene ontology
The functional role of the differentially expressed genes was assessed for gene ontology using the freely available tool Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/). To increase the predictive ability, only terms with p values ≤0.05 were considered. The selected enriched biological processes are shown in Fig. 3. In general, treatment with FSH affected the expression of approx. double the number of genes with log2 > 1.5 than treatment with AREG/EREG. Both FSH and AREG/EREG stimulated the expression of genes associated with the regulation of cell proliferation and cell migration. Interestingly, both culture conditions increased the expression of genes involved in blood coagulation and extracellular matrix remodeling (F3, FBLN5, TFPI2, and SERPINE1). In addition to AREG/EREG, FSH induced the expression of tissue plasminogen activator (PLAT) and urokinase plasminogen activator receptor (PLAUR). FSH also upregulated a wide variety of genes associated with inflammatory response (ANXA1, BMPR1B, CCL4L1, CEBPB, F3TAC1, F11R, LTA4H, OLR1, PRDX2, TGFB1, and TNFAIP6) and with the response to reactive oxygen species (CYP11A1, CRYAB, MT3, OLR1, PRDX2, SDC1, and SERPINE1). As expected, the cultures with FSH increased the expression of genes involved in gonad development and ovulation (ADAMTS1, BMPR1B, FGF9, FOXO3, PGR, and SDC1). The genes involved in the cell cycle, DNA metabolism, response to stress, cell proliferation and mitotic cell cycle checkpoint were underexpressed under both culture conditions. In the list of transcripts underexpressed in COCs cultured with AREG/EREG, the analysis revealed a group of genes (ADM, FOXO1, FOS, GCLC, and TXNIP) associated with the response to hormonal stimuli (Fig. 3).
Fig. 3.
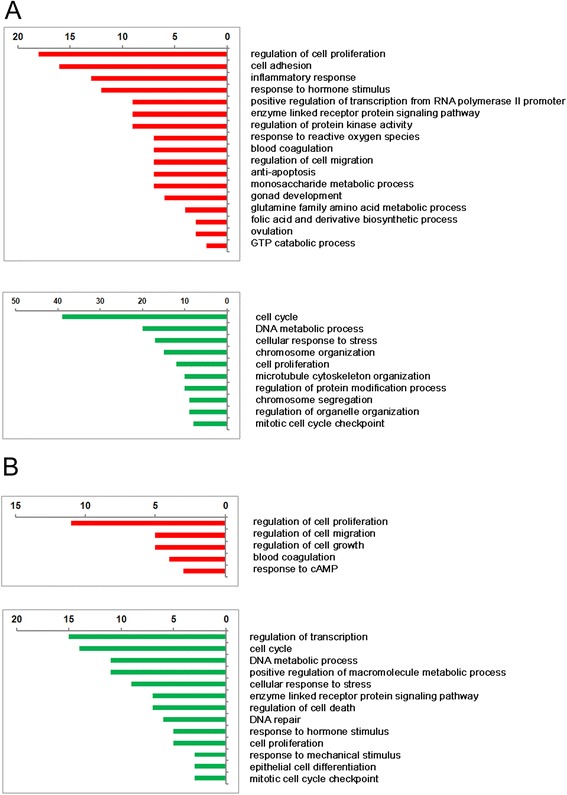
Functional classification and numbers of genes over- and underexpressed in cumulus cells after stimulation with FSH (a) or AREG/EREG (b). Only genes with log2 fold-change above 1.5 were considered for this analysis. The numbers of genes affected by the treatments are shown at the top of the panels. The gene ontology analysis was performed using DAVID Bioinformatic Resources (http://david.abcc.ncifcrf.gov/)
Real-time RT-PCR validation
To confirm the microarray data analysis, we quantified the expression levels of 50 selected genes by quantitative real-time PCR (qRT-PCR). The candidate genes selected were those that had previously been identified as being involved in the physiology of reproduction. Table 1 shows the results of validation for the FSH vs. control and AREG/EREG vs. control microarray. The validation confirmed a significant increase in expression for 21 transcripts (78 %) selected from the list of overexpressed genes originating from the FSH vs. control microarray. The list of confirmed transcripts includes factors essential for cumulus expansion (HAS2 and TNFAIP6), follicle tissue remodeling (ADAMTS1), gonadotropin-dependent EGFR signaling (AREG), polyamine metabolism (ODC1 and SMOX), plasminogen metabolism (PLAT, PLAUR, and SERPINE1) and transcription factors (CEBPB and PGR). However, only 4 transcripts (33 %; FST, PPARG, PTGES and RGS3) selected from the list of underexpressed genes were found to be significantly decreased, as measured by qRT-PCR. Similar results were obtained with the AREG/EREG vs. control microarray. The expression pattern was confirmed for 9 of 12 selected genes overexpressed in the microarray (75 %) and only 3 genes (27 %; JUN, PNRC1, and RGS3) selected from underexpressed ones were confirmed by qRT-PCR. With the AREG/EREG vs. FSH array, qRT-PCR confirmed the microarray results for all transcripts selected from the lists of overexpressed (4) and underexpressed (13) genes (Figs. 4 and 5). Interestingly, qRT-PCR revealed significantly different expression patterns of key transcription factors (FOXO3, ID1, ID3, NFIL3, NR5A2, and PGR) between FSH- and AREG/EREG-stimulated cells. Additionally, the expression levels of CTSL, PGR, SBSN, SDC4, SMOX and TNFAIP6 were increased dramatically in cells cultured with FSH in comparison with AREG/EREG-treated cells.
Table 1.
Verification of selected differentially expressed genes by real time RT-PCR
| Probe name | Gene symbol | Gene description | Fold change in gene expression values determined by RT-PCR (Mean ± SEM) | Significance vs. non-stimulated control | Array results (Fold change) |
|---|---|---|---|---|---|
| FSH vs. Control - overexpressed | |||||
| 31605:45360_15417313:f | ABCC5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 | 0.83 ± 0.12 | ns. | 4.79 |
| 9436:30168_CL9Contig1:f | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 3.72 ± 0.36 | *** | 6.95 |
| 13062:45360_CL23Contig1:f | ANXA1 | Annexun A1 | 0.93 ± 0.10 | ns. | 3.15 |
| 1117:45360_CL10Contig1:r | ANXA2 | Annexin A2 | 0.98 ± 0.07 | ns. | 4.02 |
| 14127:9388_21549088:r | AREG | Amphiregulin | 5.04 ± 0.46 | *** | 4.06 |
| 18133:7423_CL1Contig2:r | CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | 9.68 ± 1.58 | *** | 2.89 |
| 6532:35657_CL1Contig1:r | CYP11A1 | Cytochrome P450, family 11, subfamily A, polypeptide 1 | 11.92 ± 0.90 | *** | 16.20 |
| TC226972:f | FOXO3 | Forkhead box O3 | 1.97 ± 0.21 | ** | 4.22 |
| TC235800:f | HAS2 | Hyaluronan synthase 2 | 47.63 ± 5.11 | *** | 6.59 |
| 9355:35407_CL1Contig1:f | HSD17B7 | Hydroxysteroid (17-beta) dehydrogenase 7 | 3.35 ± 0.30 | *** | 3.87 |
| 5795:24780_CL1Contig1:r | MARCKSL1 | MARCKS-like 1 | 4.44 ± 0.53 | *** | 2.85 |
| 1337:12192_CL1Contig2:f | ODC1 | Ornithine decarboxylase 1 | 6.22 ± 0.85 | ** | 5.26 |
| 15979:340_CL1Contig1:f | PBX1 | Pre-B-cell leukemia homeobox 1 | 0.70 ± 0.10 | * | 4.24 |
| 31453:2453_233792:f | PGR | Progesterone receptor | 23.99 ± 5.10 | ** | 15.57 |
| 8538:3878_CL2Contig1:f | PLAT | Plasminogen activator, tissue | 14.16 ± 2.36 | *** | 5.48 |
| 4143:1539_CL1Contig1:r | PLAUR | Plasminogen activator, urokinase receptor | 2.56 ± 0.20 | *** | 4.00 |
| 33691:1766_29276590:r | PRDX2 | Peroxiredoxin 2 | 6.30 ± 0.44 | *** | 2.88 |
| 13439:18092_CL1Contig1:r | RBKS | Ribokinase | 0.51 ± 0.07 | *** | 7.32 |
| 3373:854_CL1Contig1:f | RGS2 | Regulator of G-protein signaling 2 | 3.47 ± 0.77 | * | 3.13 |
| 33911:29637_CL1Contig1:r | SBSN | Suprabasin | 20.37 ± 1.56 | *** | 21.54 |
| TC204140:f | SDC1 | Syndecan 1 | 2.55 ± 0.57 | * | 13.44 |
| 2269:10415_CL1Contig1:f | SDC4 | Syndecan 4 | 13.79 ± 1.71 | *** | 6.29 |
| TC238171:f | SERPINE1 | Serpin peptidase inhibitor, clade E, member 1 | 3.05 ± 0.50 | * | 14.47 |
| 31918:32605_CL1Contig2:f | SMOX | Spermine oxidase | 33.26 ± 7.94 | ** | 6.29 |
| 5977:12455_CL1Contig1:r | TNFAIP6 | Tumor necrosis factor alpha-induced protein 6 | 2517 ± 105 | *** | 30.54 |
| 19103:6332_CL1Contig1:f | TNFRSF12A | Tumor necrosis factor receptor superfamily, member 12A | 0.52 ± 0.17 | * | 3.04 |
| 9402:4028_CL1Contig1:r | UAP1 | UDP-N-acetylglucosamine pyrophosphorylase 1 | 8.01 ± 1.11 | *** | 5.71 |
| FSH vs. Control - underexpressed | |||||
| 707:4250_CL1Contig1:f | FOS | Proto-oncogene protein c-fos | 8.56 ± 2.43 | * | 0.15 |
| 1893:12937_CL1Contig1:f | FOXO1 | Forkhead box O1 | 0.90 ± 0.09 | ns. | 0.29 |
| 6206:5117_CL1Contig1:f | FST | Follistatin | 0.21 ± 0.02 | *** | 0.10 |
| 15012:1275_CL1Contig1:f | HS3ST1 | Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | 0.80 ± 0.12 | ns. | 0.32 |
| TC210305:f | IRS1 | Insulin receptor substrate 1 | 0.98 ± 0.25 | ns. | 0.14 |
| NM_213880.1 | JUN | Jun proto-oncogene | 1.03 ± 0.14 | ns. | 0.06 |
| 2480:21101_CL1Contig1:f | PCNA | Proliferating cell nuclear antigen | 1.16 ± 0.12 | ns. | 0.29 |
| 18375:27730_CL1Contig1:r | PNRC1 | Proline-rich nuclear receptor coactivator 1 | 0.94 ± 0.15 | ns. | 0.21 |
| 14891:209_CL4Contig1:f | PPARG | Peroxisome proliferator-activated receptor gamma | 0.45 ± 0.04 | *** | 0.32 |
| 1456:37319_CL1Contig1:f | PTGES | Prostaglandin E synthase | 0.70 ± 0.10 | ** | 0.26 |
| 12586:12773_CL1Contig1:f | RGS3 | Regulator of G-protein signaling 3 | 0.78 ± 0.10 | * | 0.17 |
| TC229705:f | TRIB2 | Tribbles pseudokinase 2 | 0.84 ± 0.10 | ns. | 0.33 |
| AREG/EREG vs. Control - overexpressed | |||||
| 13062:45360_CL23Contig1:f | ANXA1 | Annexin A1 | 0.68 ± 0.11 | * | 3.68 |
| 1117:45360_CL10Contig1:r | ANXA2 | Annexin A2 | 2.51 ± 0.41 | ** | 4.41 |
| 45:916_CL3Contig1:f | CTGF (CCN2) | Connective tissue growth factor | 2.20 ± 0.25 | *** | 6.19 |
| 6532:35657_CL1Contig1:r | CYP11A1 | Cytochrome P450, family 11, subfamily A, polypeptide 1 | 2.68 ± 0.44 | ** | 5.82 |
| 14983:2713_CL1Contig1:f | EIF4E | Eukaryotic translation initiation factor 4E | 2.56 ± 0.72 | ns. | 4.14 |
| 977:13547_CL1Contig2:r | ITPKA | Inositol-trisphosphate 3-kinase A | 5.41 ± 1.22 | * | 3.36 |
| 5795:24780_CL1Contig1:r | MARCKSL1 | MARCKS-like 1 | 4.96 ± 0.72 | *** | 3.41 |
| 31453:2453_233792:f | PGR | Progesterone receptor | 4.01 ± 0.58 | *** | 5.25 |
| 33911:29637_CL1Contig1:r | SBSN | Suprabasin | 4.33 ± 0.40 | *** | 4.34 |
| TC204140:f | SDC1 | Syndecan 1 | 1.01 ± 0.01 | ns. | 5.88 |
| TC238171:f | SERPINE1 | Serpin peptidase inhibitor, clade E, member 1 | 6.73 ± 0.65 | *** | 33.82 |
| 19103:6332_CL1Contig1:f | TNFRSF12A | Tumor necrosis factor receptor superfamily, member 12A | 4.34 ± 0.53 | ** | 7.70 |
| AREG/EREG vs. Control - underexpressed | |||||
| 707:4250_CL1Contig1:f | FOS | Proto-oncogene protein c-fos | 3.29 ± 0.57 | ** | 0.09 |
| 1893:12937_CL1Contig1:f | FOXO1 | Forkhead box O1 | 1.06 ± 0.25 | ns. | 0.33 |
| 6206:5117_CL1Contig1:f | FST | Follistatin | 1.01 ± 0.07 | ns. | 0.25 |
| 9260:5151_CL1Contig1:f | GADD45A | Growth arrest and DNA-damage-inducible, alpha | 1.20 ± 0.32 | ns. | 0.35 |
| 2134:17017_CL1Contig1:f | ID1 | Inhibitor of DNA binding 1protein | 1.24 ± 0.16 | ns. | 0.31 |
| NM_213880.1 | JUN | Jun proto-oncogene | 0.54 ± 0.02 | *** | 0.04 |
| 30832:5168_40390252:f | NR5A2 | Nuclear receptor subfamily 5, group A, member 2 | 1.06 ± 0.08 | ns. | 0.31 |
| 18375:27730_CL1Contig1:r | PNRC1 | Proline-rich nuclear receptor coactivator 1 | 0.65 ± 0.16 | * | 0.32 |
| 3373:854_CL1Contig1:f | RGS2 | Regulator of G-protein signaling 2 | 1.29 ± 0.09 | ** | 0.34 |
| 12586:12773_CL1Contig1:f | RGS3 | Regulator of G-protein signaling 3 | 0.36 ± 0.03 | *** | 0.13 |
| TC229705:f | TRIB2 | Tribbles pseudokinase 2 | 0.94 ± 0.11 | ns. | 0.77 |
ns. not significant. *P < 0.05; ** P < 0.01; *** P < 0.001
Fig. 4.
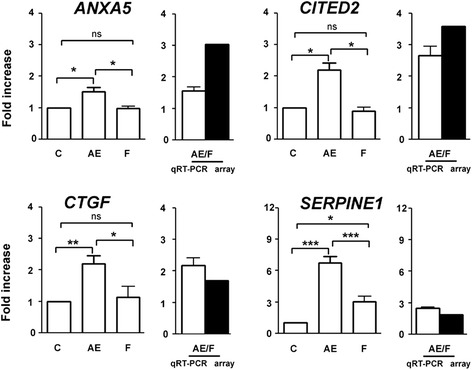
Quantitative real-time PCR validation of genes found to be overexpressed in AREG/EREG vs. FSH microarray. The relative abundance of specific gene mRNA is expressed in arbitrary units as fold increases in the specific gene /HPRT ratio over the level found in the control group at the time 0 h. C control, AE AREG/EREG, F FSH. P < 0.05 for single asterisk; P < 0.01 for double asterisk; P < 0.001 for triple asterisk
Fig. 5.
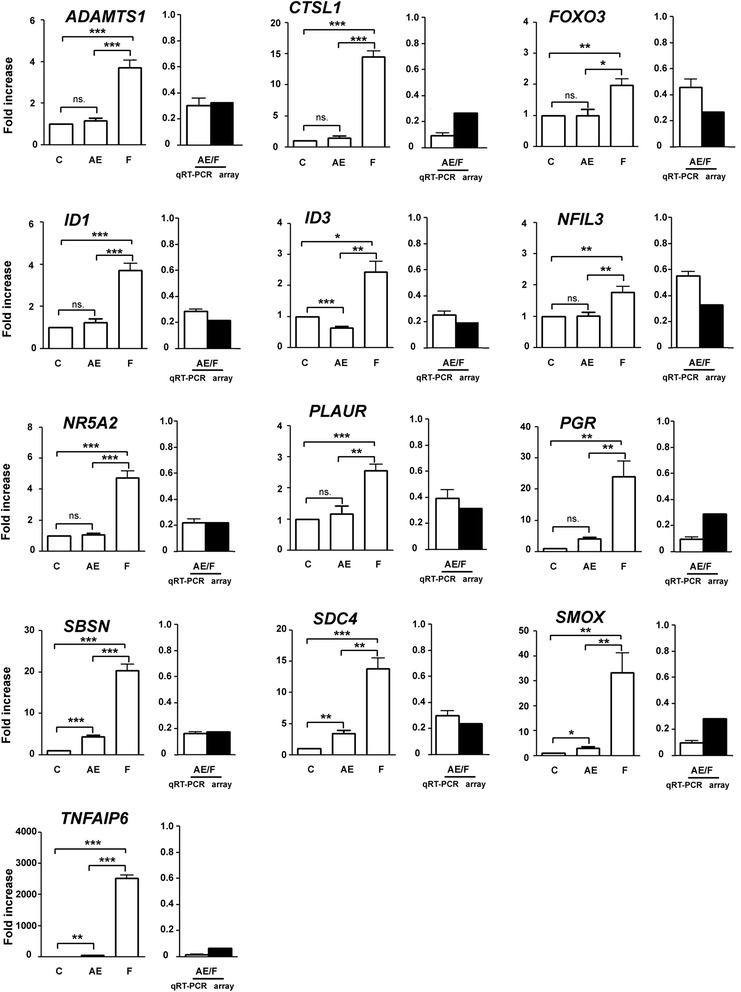
Quantitative real-time PCR validation of genes found to be underexpressed in AREG/EREG vs. FSH microarray. The relative abundance of specific gene mRNA is expressed in arbitrary units as fold increases in the specific gene /HPRT ratio over the level found in the control group at the time 0 h. C control, AE AREG/EREG, F FSH. P < 0.05 for single asterisk; P < 0.01 for double asterisk; P < 0.001 for triple asterisk
Transcription factors. The results obtained from the AREG/EREG vs. FSH microarray led us to suggest the hypothesis that different expression profiles of AREG/EREG- and FSH-stimulated cells might be caused by alterations in the expression of genes coding the key transcription factors. Therefore, the expression levels of 9 transcription factors were measured by qRT-PCR at various time points during the culture (Fig. 6). In general, FSH was found to affect transcription factor expression more efficiently than AREG/EREG. Figure 6 shows that FSH significantly stimulated the expression of CEBPB, FOS, FOXO3, ID1, ID3 and NR5A2. Except for FOS and ID1, the patterns indicate the accumulation of mRNAs reaching its maximum at 4 and 2 h of culture, respectively. The amount of FOS mRNA quickly and dramatically increased within the first hour of culture and then the concentration progressively fell in the subsequent time intervals over the whole culture period. Surprisingly, the expression of JUN, a partner transcription factor of FOS, was neither affected by FSH- nor by AREG/EREG-stimulation. The expression of NFIL3 was also unaffected by any type of stimulation.
Fig. 6.
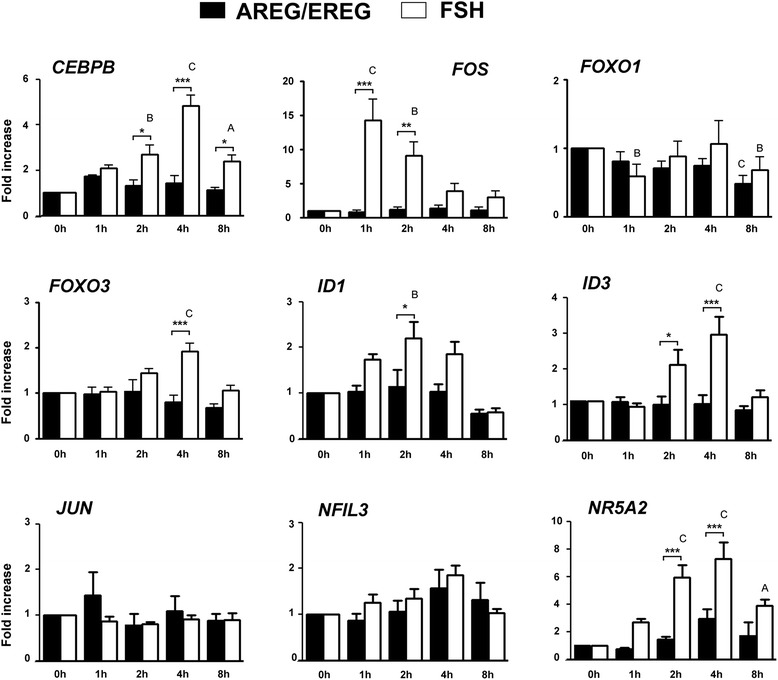
Expression of transcription factors in COCs cultured with AREG/EREG or FSH for various lengths of time. The relative abundance of specific gene mRNA is expressed in arbitrary units as fold increases in the specific gene /HPRT ratio over the level found in the control group of COCs at the time 0 h
Discussion
Design of the study
We have shown previously that an exposure of COCs to FSH as brief as 3 h, followed by culture in hormone-free medium, is sufficient to initiate the synthesis of hyaluronic acid, cumulus expansion and the resumption of pig oocyte meiosis [34]. The rapid expression of the genes critical for meiotic resumption and cumulus expansion (HAS2, PTGS2, TNFAIP6, AREG, and EREG) was confirmed in our previous studies [6, 28, 35]; the expression of some regulatory genes inclusive of AREG became downregulated by 4 h after FSH addition [28]. With the aim of detecting the primary changes in cumulus cell transcriptomes evoked by FSH or AREG/EREG, we compared non-treated controls and treated groups 3 h after the onset of culture. The data reported in this paper demonstrate that the early alterations in the gene expression profile of cumulus cells do not exclusively pertain to genes involved in the regulation of reproductive functions such as cumulus cell extracellular matrix synthesis and organization, steroidogenesis, ovulation and luteinization, but they cover a broad spectrum of cellular functions. Next, the presented data document that even though both FSH and the EGF-like factors can stimulate events associated with ovulation and final oocyte maturation, essential differences exist in their impact on the modification of the gene expression profile of the cumulus cells.
ECM formation
We found that FSH increased the expression of a variety of genes involved in extracellular matrix formation and remodeling, such as the cumulus expansion-related genes HAS2 and TNFAIP6. HAS2 is responsible for the formation of hyaluronan, the main component of the expanded cumulus. Additionally, the expression of HAS2 in human cumulus cells positively correlates with developmental competence of the oocyte [36, 37]. TNFAIP6 is required for the formation of covalent bonds between hyaluronan and the heavy chains of the inter-α-trypsin inhibitor family proteins [38]. Female mice lacking Tnfaip6 demonstrated severe subfertility and impaired cumulus expansion [39]. Furthermore, FSH promoted the expression of A disintegrin and metalloproteinase with thrombospondin motifs 1 (ADAMTS1). In mice, ADAMTS1 is required for the structural remodeling of ovarian follicle and cumulus expansion resulting in ovulation and successful fertilization [40]. In accordance with these findings, Adamts1 null mice demonstrated impaired female (but not male) fertility caused by morphological abnormalities in the uterus and ovaries [41]. FHS also increased the expression of carbohydrate sulfotransferase 7 (CHST7) which may regulate hyaluronan binding to its receptor CD44 by chondroitin sulfonation of CD44 [42].
Both FSH and AREG/EREG enhanced the expression of syndecan 4 (SDC4), a heparan sulfate proteoglycan that provides a mechanical link between the extracellular matrix and cytoskeleton [43]. The higher SDC4 expression in human cumulus cells may predict normal embryo development and pregnancy [44]. Although both culture conditions increased the expression of syndecan 1 (SDC1) as assessed by microarray (SDC1), qRT-PCR revealed that SDC1 expression is unaffected by AREG/EREG stimulation. However, the expression of heparan sulfate glucosamine 3-O-sulfotransferase 1 (HS3ST1) was increased in COC cultured with FSH, indicating the involvement of heparan sulfate and heparan sulfate-binding proteoglycans in extracellular matrix remodeling or signal transduction in response to gonadotropins or EGF-like peptides. Furthermore, AREG/EREG (but not FSH) enhanced the expression of connective tissue growth factor (CTGF), a non-structural matricellular protein that interacts with heparan-sulfate-containing proteoglycans, including syndecans [45]. CTGF has been intensively investigated for its role in the growth and differentiation of granulosa cells and in the formation of the corpus luteum [46]. Ovarian and uterine Ctgf conditional knockout mice exhibit severe subfertility due to multiple reproductive defects including disrupted follicle development and decreased ovulation rates, despite normal cumulus expansion [47]. Interestingly, the phenotype of Ctgf null mice is similar to the phenotype of Smad4- or activin-deficient mice, suggesting that CTGF may be a mediator of SMAD4 and activin signaling [47]. In porcine ovaries, the CTGF mRNA level of granulosa cells increases as a function of follicular development to a maximum in small antral follicles but CTGF mRNA is underexpressed in large antral follicles [48]. These data are in accordance with other studies focused on the role of CTGF in ovarian follicle growth and development [46, 49].
Transcription factors
Both FSH and AREG/EREG treatment affected the expression levels of various genes involved in transcription regulation. FSH promoted the expression of CEBPB and CEBPD, the members of a family of basic-leucine zipper transcription factors that interact with the CCAAT box motifs present in gene promoters. CEBPA and CEBPB have been previously found to be essential for ovulation and luteinization in mice [50]. Double-mutant females lacking both Cebpa and Cebpb genes in granulosa cells failed to ovulate and were completely infertile [50]. Mice null for CEBPB were subfertile and CEBPB seems to play an important role in the AREG-induced expression of Tnfaip6 and Ptgs2 in mouse COCs [11]. In pigs, CEBPB mRNA significantly increased after ovulation in the corpus hemorrhagicum [51]. Furthermore, the CEBPB-dependent transcription of the key genes Areg, Ereg and Ptgs2 is inhibited by the interaction of CEBPB with the NFIL3 transcription factor on the gene promoters [52]. Interestingly, NFIL3 expression was found to be unaffected by culture with AREG/EREG in our study, but it was only increased by FSH.
Quantitative RT-PCR analysis also revealed that FSH, but not AREG/EREG, increased the expression of NR5A2, a transcription factor essential for female fertility. NR5A2 transcripts were highly abundant in granulosa cells and distinctly absent in theca interna and ovarian stroma [53]. Conditional knockout of Nr5a2 in granulosa cells results in infertility, null mice are not able to ovulate due to the failure of preovulatory follicle tissue remodeling caused by abnormally low mRNA levels for proteases implicated in the ovulatory process [10]. NR5A2 seems to be essential for progesterone production [54] and the transcriptional regulation of a variety of genes important for steroidogenesis, such as Star, Cyp11a1, Fdx1, and Hsd3b1 [55–57]. In accordance with such findings, NR5A2 was found to be required for the formation and maintenance of the corpus luteum, promotion of decidualization and for placental formation in mice [57]. However, granulosa-specific targeted disruption of the Nr5a2 gene yielded contradictory findings about a possible role of this transcription factor in cumulus expansion [10, 54].
Surprisingly, FOS was rapidly but transiently induced by FSH within 1 h after addition to culture media. These data are consistent with previously published results in rats [58, 59]. In pigs, FOS is present in preovulatory follicles but lower in the corpus luteum than in antral follicles [60], indicating possible downregulation by LH signaling. Interestingly, the FSH vs. control microarray revealed that BATF3, an inhibitor of the AP1 complex [61, 62], was overexpressed in response to FSH and the transcript was assessed as underexpressed in the AREG/EREG and FSH microarray. However, the role of AP1/FOS during folliculogenesis and ovulation is poorly understood, and it may be involved in many cellular processes, e. g. in the regulation of steroidogenesis [63] or granulosa cell proliferation [64].
In contrast to AREG/EREG, FSH induced the expression of the ID1 and ID3 transcription factors. In ovine follicles, the ID1 and ID3 proteins are present in granulosa cells, particularly in the peri-oocytic region [65], indicating the involvement of oocyte-derived factors such as GDF9 and BMP15. Indeed, GFP9 and GMP15 were found to stimulate ID1 expression in ovine granulosa cells, and both ID1 and ID3 mRNA levels were increased by BMP6 and decreased by activin A [65]. In porcine granulosa cells, FSH promoted the expression levels of ID2 and ID3 [46]. We also found that AREG/EREG (but not FSH) enhanced the expression of Cbp/P300-interacting transactivator 2 (CITED2). This transcription factor is involved in gonad development [66] and negatively regulates the expression of genes coding matrix metalloproteinases in chondrocytes [67].
In agreement with the microarray results, qRT-PCR confirmed the downregulation of PPARG mRNA in COCs cultured in the presence of FSH. In mice, the level of Pparg mRNA is enhanced in a progesterone-dependent manner in the granulosa cells of PMSG-primed mice treated with hCG [68]. Granulosa cell-specific deletion of Pparg resulted in a significant impairment of follicle rupture and subfertility [68]. However, other Pparg null mice didn’t exhibit any problems in their ovulation process and were subfertile due to abnormal implantation [69].
Polyamine metabolism
FSH also stimulated the expression of ODC1 and SMOX, enzymes involved in polyamine metabolism. Xenopus oocytes lacking ODC1 activity exhibit high levels of reactive oxygen species and high caspase 3 activity [70]. Female mice overexpressing Sat1 mRNA, whose product is a rate-limiting enzyme in polyamine catabolism, were found to be infertile due to ovarian hypofunction and hypoplastic uteri [71]. The ovaries of these mice contained the primary and small secondary follicles, but larger developing follicles and corpora lutea were absent, indicating disruption of the late phases of folliculogenesis and ovulation. SMOX oxidizes spermine to produce spermidine, 3-aminopropanal and hydrogen peroxide [72]. Exogenous hydrogen peroxide on its own effectively induced cumulus expansion in mouse COCs [73]. Moreover, broad-range scavengers of reactive oxygen species (ROS) inhibited LH-induced cumulus expansion, activation of EGFR and MAPK3/1 and expression of cumulus expansion-related genes (Has2, Ptgs2, Tnfaip6) [73]. Antioxidants may even reduce the ovulation rate in mice [73, 74] and rabbits [75]. Interestingly, AREG/EREG induced the expression of neutrophil cytosol factor 2 (NCF2) which is required for the activation of NADPH oxidase and superoxide production [76].
ROS
The generation of ROS in rat follicles induced by LH was previously described to be coupled with mitochondrial steroidogenesis via Star/Cyp11A1 [77]. To protect against oxidative damage and subsequent apoptosis, the increase in steroidogenic capacity is correlated with increased activity of the antioxidants [78]. Both FSH and AREG/EREG promoted expression of glutaredoxin 1 (GLRX), which reduces low-molecular-weight disulfides and proteins [79]. Moreover GLRX was shown to reduce mixed disulfides between various signaling proteins and glutathione, regenerating their activities and affecting signal transduction and cellular response [79]. FSH induced the expression of a variety of genes involved in the response to oxidative damage (EPHX3, GSTM3, KLHDC10, PRDX2, MT2A, and MT3). Glutathione S-transferase M3 (GSTM3) has well-established roles in the detoxification and clearance of a variety of electrophilic compounds, including xenobiotics as well as substances generated by ROS damage to intracellular molecules [80], but also acts as a tumor suppressor [81]. Peroxiredoxin 2 (PRDX2) is an antioxidant enzyme reducing hydrogen peroxide and alkyl hydroperoxides by using reducing agents such as thioredoxin [82]. Prdx2 was previously found to be expressed in the ovaries of eCG-primed rats in response to hCG administration, localized in the granulosa and theca cells of preovulatory follicles in the ovaries of eCG-primed rats [74]. PRDX2 is proposed to be part of a control mechanism for the balance between the production and elimination of hydrogen peroxide during the periovulatory period [74]. In mouse granulosa cells, suppression of the Prdx2 gene using siRNA inhibits cell proliferation, promotes apoptosis and increases the production of endogenous hydrogen peroxide [83]. Furthermore, the expression of PRDX2 in the cumulus cells of women positively correlates with embryo quality [84].
Coagulation factors
Surprisingly, both FSH and AREG/EREG induced the expression of genes involved in “blood coagulation”, namely plasminogen activator inhibitor 1 (SERPINE1), tissue factor (F3), and tissue factor pathway inhibitor 2 (TFPI2). Moreover, FSH itself increased the expression levels of urokinase plasminogen activator surface receptor (PLAUR), tissue-type plasminogen activator (PLAT), junctional adhesion molecule A (F11R), CD9 antigen (CD9). PLAT, SERPINE1 and PLAUR are components of the plasminogen activation system, which regulates the conversion of plasminogen to plasmin. Numerous studies have reported the expression of these genes in antral follicles after exposure to a gonadotropin surge [85–87], but the role of plasmin during the ovulation process is still unclear. Plasmin is assumed to play a role in collagen breakdown and stigma development [88]. On the other hand, plasminogen-deficient females exhibited normal ovulation efficiency compared to plasminogen wild-type mice [89] and the reduced fertility of plasminogen knockout females may be caused by poor general health [90]. Some studies indicate that plasmin activity is involved in cumulus expansion and/or extracellular matrix remodeling [91], and progesterone production by the corpus luteum [92]. In pigs, plasmin detached sperm bound to zona pellucida and thus reduced the risk of polyspermy [93].
Conclusions
The stimulation of in vitro cultured COCs by both FSH and EGF-like factors caused a rapid and extensive rearrangement of gene expression patterns in cumulus cells. These early changes in the cumulus cell transcriptome not only pertained to events associated with the final maturation of oocytes and expansion of the cumulus cells, but they covered a broad spectrum of general cellular functions including signal transduction, regulation of the cell cycle, growth and proliferation, cytoskeleton and microtubule organization, metabolism, apoptosis, response to stress and last but not least the regulation of transcription. As expected, the impact of FSH on cumulus cell gene transcription was higher than the impact of EGF-like factors in terms of the number of cell functions affected as well as the number of over- and underexpressed genes. As for the regulation of ovarian functions, both FSH and EGF-like factors overexpressed genes involved in the post-ovulatory switch in steroidogenesis (CYP11A1 and PGR) and tissue remodeling (F3, SERPINE1, and TIMP1). However, FSH was remarkably more efficient in the up-regulation of several specific genes that are considered to be essential for the successful ovulation of matured oocytes (ADAMTS1, HAS2, HSD17B7, PLAT, PLAUR, and TNFAIP6) and that have also been reported to play an essential role in the maturation of cumulus-enclosed oocytes in vitro (ADAMTS1, HAS2, and TNFAIP6).
We have shown in our previous work that EGF-like peptides do not mimic all effects of FSH on cultured pig COCs, nevertheless they provided oocytes with the developmental competence comparable or even higher than those stimulated with gonadotropins [28]. These data were confirmed in further studies with pig and bovine COCs [32, 33, 94]. Moreover, a synergistic effect of AREG and oocyte-derived paracrine factors (GDF9, BMP15) and cAMP on promotion of developmental competence of in vitro cultured oocytes was recently reported [32, 33]. The molecular background of this feature was identified as a promotion of EGFR signaling system in the somatic cell compartment, which was critical for successful maturation of less competent COCs isolated from small size follicles [33]. Taken together with the data of the present study, we may conclude that in a simplified model of in vitro culture of COCs isolated from growing follicles, both cAMP-PKA signaling induced by gonadotropins and action of oocyte-derived paracrine signaling are required for proper function of the EGFR activated by EGF-like peptides. The application of this knowledge may increase efficiency of the current IVM/IVF systems for use in animal breeding, biomedicine and for the treatment of infertility.
Acknowledgments
This work was supported by Grant Agency of the Czech Republic (grant P502/11/0593), National Agency for Agricultural Research (grant QJ 1510138), Institute of Animal Physiology and Genetics (Institutional Research Concept RVO 67985904 and Internal Grant 14/14). The authors would like to thank Mr. B. J. Watson-Jones for reading the manuscript and language corrections.
Abbreviations
- ADAMTS1
A disintegrin and metalloproteinase with trombospondin motif 1
- ADM
Adrenomedullin
- AKT
v-akt murine thymoma viral oncogene homolog
- ANXA1
Annexin A1
- ANXA2
Annexin A2
- ANXA5
Annexin A5
- AP1
activator protein 1
- AREG
Amphiregulin
- BATF3
Basic leucine zipper transcriptional factor ATF-like 3
- BMP6
Bone morphogenetic protein 6
- BMP15
Bone morphogenetic protein 15
- BMPR1B
Bone morphogenetic protein receptor type-1B
- BTC
Betacellulin
- CCL4L1
Chemokine (C-C motif) ligand 4-like 1
- CEBPA
CCAAT/enhancer binding protein alpha
- CEBPB
CCAAT/enhancer binding protein beta
- CD9
CD 9 antigen
- CEBPD
CCAAT/enhancer binding protein delta
- CHST7
Carbohydrate sulfotransferase 7
- CITED2
Cbp/P300-interacting transactivator 2
- COC
Cumulus-oocyte complex
- CREB
cAMP response element binding protein
- CREM
cAMP response element modulator
- CRYAB
Crystallin, alpha- B
- CTSL
Cathepsin L
- CTGF
Connective tissue growth factor
- CYP11A1
Cytochrome P450, family 11, subfamily A, polypeptide 1
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- EPHX3
Epoxide hydrolase 3
- EREG
Epiregulin
- F3
Coagulation factor 3
- F11R
F11 receptor
- FBLN5
Fibulin-5
- FDX1
Ferredoxin 1
- FGF9
Fibroblast growth factor 9
- FOS
Proto-oncogene protein c-fos
- FOXO1
Forkhead box protein O1
- FOXO3
Forkhead box protein O1
- FST
Follistatin
- FSH
Follicle stimulating hormone
- GCLC
Glutamate cysteine ligase catalytic subunit
- GDF9
Growth differentiation factor 9
- GLRX
Glutaredoxin 1
- GRB14
Growth factor receptor-bound protein 14
- GRB14
Growth factor receptor-bound protein 14
- GSTM3
Glutathione S-transferase Mu 3
- GV
Germinal vesicle
- GVBD
Germinal vesicle breakdown
- HA
hyaluronic acid
- HAS2
Hyaluronan synthase 2
- HS3ST1
heparan sulfate glucosamine 3-O-sulfotransferase 1
- HSD17B7
Hydoxysteroid (17-beta) dehydrogenase 7
- HSD3B1
Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1
- ID1/3
Inhibitor of DNA-binding protein 1/3
- IRS1
Insulin receptor substrate 1
- JUN
Jun proto-oncogene
- KLHDC10
Kelch domain-containing protein 10
- LH
Luteinizing hormone
- LTA4H
Leukotriene A-4 hydrolase
- MII
metaphase II
- MAPK3/1
Mitogen-activated protein kinase 3/1
- MT2A
Metallothionein 2A
- MT3
Metallothionein 3
- NCF2
Neutrophil cytosol factor 2
- NFIL3
Nuclear factor interleukin-3-regulated
- NR4A1
Nuclear receptor subfamily 4, group A, member 1
- NR5A2
Nuclear receptor subfamily 5, group A, member 2
- NRIP1
Nuclear receptor-interacting protein 1
- ODC1
Ornithine decarboxylase 1
- OLR1
Oxidized low-density lipoprotein receptor 1
- PGR
progesterone receptor
- PI3K
phosphoinositide-3-kinase
- PKA
Protein kinase A
- PLAT
plasminogen activator, tissue
- PLAUR
Plasminogen activator, urokinase receptor
- PNRC1
Proline-rich nuclear receptor coactivator 1
- PPARG
Peroxisome proliferator-activated receptor gamma
- PRDX2
Peroxiredoxin-2
- PTGS2
Prostaglandin-endoperoxide synthase 2
- PTGER
Prostaglandin E receptor 2
- PTGES
Prostaglandin E synthase
- RGS3
Regulator of G-protein signaling 3
- SAT1
Spermidine/spermine N1-acetyltransferase 1
- SBSN
Suprabasin
- SDC1
Syndecan 1
- SDC4
Syndecan 4
- SERPINE1
Serpin peptidase inhibitor, clade E, member 1
- SMAD2
SMAD family member 2
- SMAD4
SMAD family member 4
- SMOX
Spermine oxidase
- SNAP
S-nitroso-N-acetyl-penicillamine
- TAC1
Tachykinin precursor 1
- TFPI2
Tissue factor pathway inhibitor 2
- TGFB1
Transforming growth factor beta-1
- TNFAIP6
Tumor necrosis factor alpha-induced protein 6
- TXNIP
Thioredoxin-interacting protein
Additional files
List of primers used for microarray data analysis verification. List and data of primers used in qRT-PCR. F, forward primer; R, reverse primer. (XLSX 18 kb)
List of over- and underexpressed genes in cumulus cells stimulated with FSH or AREG/EREG depicted in Venn diagrams. A complete list of genes depicted in Venn diagrams (Fig. 2). (XLSX 47 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MB designed the experiments, analyzed the microarray data, conducted RT-PCR, and wrote the manuscript. LN performed RT-PCR, analyzed the microarray data and wrote the manuscript. KV designed the experiments, performed the microarray, analyzed the microarray data and wrote the manuscript. PV designed the experiments, performed the microarray, analyzed the microarray data and wrote the manuscript. RP conceived the study, isolated and cultured the COCs, prepared samples for mRNA isolation and wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008;14:159–77. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- 2.Eppig JJ, O’Brien MJ. Comparison of preimplantation developmental competence after mouse oocyte growth and development in vitro and in vivo. Theriogenology. 1998;49:415–22. doi: 10.1016/S0093-691X(97)00413-5. [DOI] [PubMed] [Google Scholar]
- 3.Crozet N, Kanka J, MotliK J, Fulka J. Nucleolar fine structure and RNA synthesis in bovine oocytes from antral follicles. Gamete Res. 1986;14:65–73. doi: 10.1002/mrd.1120140108. [DOI] [Google Scholar]
- 4.Wayne CM, Fan HY, Cheng X, Richards JS. Follicle-stimulating hormone induces multiple signaling cascades: evidence that activation of Rous sarcoma oncogene, RAS, and the epidermal growth factor receptor are critical for granulosa cell differentiation. Mol Endocrinol. 2007;21:1940–57. doi: 10.1210/me.2007-0020. [DOI] [PubMed] [Google Scholar]
- 5.Conti M, Hsieh M, Musa Zamah A, Oh JS. Novel signaling mechanisms in the ovary during oocyte maturation and ovulation. Mol Cell Endocrinol. 2012;356:65–73. doi: 10.1016/j.mce.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prochazka R, Blaha M, Nemcova L. Signaling pathways regulating FSH- and amphiregulin-induced meiotic resumption and cumulus cell expansion in the pig. Reproduction. 2012;144:535–46. doi: 10.1530/REP-12-0191. [DOI] [PubMed] [Google Scholar]
- 7.Ning G, Ouyang H, Wang S, Chen X, Xu B, Yang J, et al. 3',5'-cyclic adenosine monophosphate response element binding protein overexpressed cytochrome P450 lanosterol 14alpha-demethylase expression involved in follicle-stimulating hormone-induced mouse oocyte maturation. Mol Endocrinol. 2008;22:1682–94. doi: 10.1210/me.2007-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards JS, Pangas SA. The ovary: basic biology and clinical implications. J Clin Invest. 2010;120:963–72. doi: 10.1172/JCI41350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tullet JM, Pocock V, Steel JH, White R, Milligan S, Parker MG. Multiple signaling defects in the absence of RIP140 impair both cumulus expansion and follicle rupture. Endocrinology. 2005;146:4127–37. doi: 10.1210/en.2005-0348. [DOI] [PubMed] [Google Scholar]
- 10.Duggavathi R, Volle DH, Mataki C, Antal MC, Messaddeq N, Auwerx J, et al. Liver receptor homolog 1 is essential for ovulation. Genes Dev. 2008;22:1871–6. doi: 10.1101/gad.472008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, et al. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324:938–41. doi: 10.1126/science.1171396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eppig JJ. FSH stimulates hyaluronic acid synthesis by oocyte-cumulus cell complexes from mouse preovulatory follicles. Nature. 1979;281:483–4. doi: 10.1038/281483a0. [DOI] [PubMed] [Google Scholar]
- 13.Fülöp C, Salustri A, Hascall VC. Coding sequence of a hyaluronan synthase homologue expressed during expansion of the mouse cumulus-oocyte complex. Arch Biochem Biophys. 1997;337:261–6. doi: 10.1006/abbi.1996.9793. [DOI] [PubMed] [Google Scholar]
- 14.Russell DL, Doyle KM, Ochsner SA, Sandy JD, Richards JS. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J Biol Chem. 2003;278:42330–9. doi: 10.1074/jbc.M300519200. [DOI] [PubMed] [Google Scholar]
- 15.Fülöp C, Kamath RV, Li Y, Otto JM, Salustri A, Olsen BR, et al. Coding sequence, exon-intron structure and chromosomal localization of murine TNF-stimulated gene 6 that is specifically expressed by expanding cumulus cell-oocyte complexes. Gene. 1997;202:95–102. doi: 10.1016/S0378-1119(97)00459-9. [DOI] [PubMed] [Google Scholar]
- 16.Salustri A, Garlanda C, Hirsch E, De Acetis M, Maccagno A, Bottazzi B, et al. PTX3 plays a key role in the organization of the cumulus oophorus extracellular matrix and in in vivo fertilization. Development. 2004;131:1577–86. doi: 10.1242/dev.01056. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Mao SJ, McLean LR, Powers RW, Larsen WJ. Proteins of the inter-alpha-trypsin inhibitor family stabilize the cumulus extracellular matrix through their direct binding with hyaluronic acid. J Biol Chem. 1994;269:28282–7. [PubMed] [Google Scholar]
- 18.Nagyova E, Camaioni A, Prochazka R, Salustri A. Covalent transfer of heavy chains of inter-alpha-trypsin inhibitor family proteins to hyaluronan in in vivo and in vitro expanded porcine oocyte-cumulus complexes. Biol Reprod. 2004;71:1838–43. doi: 10.1095/biolreprod.104.029595. [DOI] [PubMed] [Google Scholar]
- 19.Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, et al. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell. 1997;91:197–208. doi: 10.1016/S0092-8674(00)80402-X. [DOI] [PubMed] [Google Scholar]
- 20.Hizaki H, Segi E, Sugimoto Y, Hirose M, Saji T, Ushikubi F, et al. Abortive expansion of the cumulus and impaired fertility in mice lacking the prostaglandin E receptor subtype EP(2) Proc Natl Acad Sci U S A. 1999;96:10501–6. doi: 10.1073/pnas.96.18.10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng XR, Hsueh AJ, LaPolt PS, Bjersing L, Ny T. Localization of luteinizing hormone receptor messenger ribonucleic acid expression in ovarian cell types during follicle development and ovulation. Endocrinology. 1991;129:3200–7. doi: 10.1210/endo-129-6-3200. [DOI] [PubMed] [Google Scholar]
- 22.Prochazka R, Nemcova L, Nagyova E, Scsukova S, Mlynarcikova A. Development of functional LH receptors on pig cumulus-oocyte complexes cultured in vitro by a novel two-step culture system. Mol Reprod Dev. 2009;76:751–61. doi: 10.1002/mrd.21039. [DOI] [PubMed] [Google Scholar]
- 23.Hattori MA, Takesue K, Nishida N, Kato Y, Fujihara N. Inhibitory effect of retinoic acid on the development of immature porcine granulosa cells to mature cells. J Mol Endocrinol. 2000;25:53–61. doi: 10.1677/jme.0.0250053. [DOI] [PubMed] [Google Scholar]
- 24.Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303:682–4. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- 25.Ashkenazi H, Cao X, Motola S, Popliker M, Conti M, Tsafriri A. Epidermal growth factor family members: endogenous mediators of the ovulatory response. Endocrinology. 2005;146:77–84. doi: 10.1210/en.2004-0588. [DOI] [PubMed] [Google Scholar]
- 26.Shimada M, Hernandez-Gonzalez I, Gonzalez-Robayna I, Richards JS. Paracrine and autocrine regulation of epidermal growth factor-like factors in cumulus oocyte complexes and granulosa cells: key roles for prostaglandin synthase 2 and progesterone receptor. Mol Endocrinol. 2006;20:1352–65. doi: 10.1210/me.2005-0504. [DOI] [PubMed] [Google Scholar]
- 27.Downs SM, Chen J. EGF-like peptides mediate FSH-induced maturation of cumulus cell-enclosed mouse oocytes. Mol Reprod Dev. 2008;75:105–14. doi: 10.1002/mrd.20781. [DOI] [PubMed] [Google Scholar]
- 28.Prochazka R, Petlach M, Nagyova E, Nemcova L. Effect of epidermal growth factor-like peptides on pig cumulus cell expansion, oocyte maturation, and acquisition of developmental competence in vitro: comparison with gonadotropins. Reproduction. 2011;141:425–35. doi: 10.1530/REP-10-0418. [DOI] [PubMed] [Google Scholar]
- 29.Albuz FK, Sasseville M, Lane M, Armstrong DT, Thompson JG, Gilchrist RB. Simulated physiological oocyte maturation (SPOM): a novel in vitro maturation system that substantially improves embryo yield and pregnancy outcomes. Hum Reprod. 2010;25:2999–3011. doi: 10.1093/humrep/deq246. [DOI] [PubMed] [Google Scholar]
- 30.Kawashima I, Okazaki T, Noma N, Nishibori M, Yamashita Y, Shimada M. Sequential exposure of porcine cumulus cells to FSH and/or LH is critical for appropriate expression of steroidogenic and ovulation-related genes that impact oocyte maturation in vivo and in vitro. Reproduction. 2008;136:9–21. doi: 10.1530/REP-08-0074. [DOI] [PubMed] [Google Scholar]
- 31.Richani D, Ritter LJ, Thompson JG, Gilchrist RB. Mode of oocyte maturation affects EGF-like peptide function and oocyte competence. Mol Hum Reprod. 2013;19:500–9. doi: 10.1093/molehr/gat028. [DOI] [PubMed] [Google Scholar]
- 32.Sugimura S, Ritter LJ, Sutton-McDowall ML, Mottershead DG, Thompson JG, Gilchrist RB. Amphiregulin co-operates with bone morphogenetic protein 15 to increase bovine oocyte developmental competence: effects on gap junction-mediated metabolite supply. Mol Hum Reprod. 2014;20:499–513. doi: 10.1093/molehr/gau013. [DOI] [PubMed] [Google Scholar]
- 33.Sugimura S, Ritter LJ, Rose RD, Thompson JG, Smitz J, Mottershead DG, et al. Promotion of EGF receptor signaling improves the quality of low developmental competence oocytes. Dev Biol. 2015;403:139–49. doi: 10.1016/j.ydbio.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Prochazka R, Kalab P, Nagyova E. Epidermal growth factor-receptor tyrosine kinase activity regulates expansion of porcine oocyte-cumulus cell-complexes in vitro. Biol Reprod. 2003;68:797–803. doi: 10.1095/biolreprod.102.005520. [DOI] [PubMed] [Google Scholar]
- 35.Nemcova L, Nagyova E, Petlach M, Tomanek M, Prochazka R. Molecular mechanisms of insulin-like growth factor 1 promoted synthesis and retention of hyaluronic acid in porcine oocyte-cumulus complexes. Biol Reprod. 2007;76:1016–24. doi: 10.1095/biolreprod.106.057927. [DOI] [PubMed] [Google Scholar]
- 36.McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, et al. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19:2869–74. doi: 10.1093/humrep/deh535. [DOI] [PubMed] [Google Scholar]
- 37.Cillo F, Brevini TA, Antonini S, Paffoni A, Ragni G, Gandolfi F. Association between human oocyte developmental competence and expression levels of some cumulus genes. Reproduction. 2007;134:645–50. doi: 10.1530/REP-07-0182. [DOI] [PubMed] [Google Scholar]
- 38.Rugg MS, Willis AC, Mukhopadhyay D, Hascall VC, Fries E, Fülöp C, et al. Characterization of complexes formed between TSG-6 and inter-alpha-inhibitor that act as intermediates in the covalent transfer of heavy chains onto hyaluronan. J Biol Chem. 2005;280:25674–86. doi: 10.1074/jbc.M501332200. [DOI] [PubMed] [Google Scholar]
- 39.Fülöp C, Szántó S, Mukhopadhyay D, Bárdos T, Kamath RV, Rugg MS, et al. Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development. 2003;130:2253–61. doi: 10.1242/dev.00422. [DOI] [PubMed] [Google Scholar]
- 40.Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, et al. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biol Reprod. 2010;83:549–57. doi: 10.1095/biolreprod.110.084434. [DOI] [PubMed] [Google Scholar]
- 41.Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, et al. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest. 2000;105:1345–52. doi: 10.1172/JCI8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruffell B, Poon GF, Lee SS, Brown KL, Tjew SL, Cooper J, et al. Differential use of chondroitin sulfate to regulate hyaluronan binding by receptor CD44 in Inflammatory and Interleukin 4-activated Macrophages. J Biol Chem. 2011;286:19179–790. doi: 10.1074/jbc.M110.200790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beauvais DM, Rapraeger AC. Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol. 2004;2:3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 2011;26:1035–51. doi: 10.1093/humrep/der036. [DOI] [PubMed] [Google Scholar]
- 45.Luft FC. CCN2, the connective tissue growth factor. J Mol Med Berl. 2008;86:1–3. doi: 10.1007/s00109-007-0287-x. [DOI] [PubMed] [Google Scholar]
- 46.Winterhager E, Gellhaus A. The role of the CCN family of proteins in female reproduction. Cell Mol Life Sci. 2014;71:2299–311. doi: 10.1007/s00018-014-1556-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagashima T, Kim J, Li Q, Lydon JP, DeMayo FJ, Lyons KM, et al. Connective tissue growth factor is required for normal follicle development and ovulation. Mol Endocrinol. 2011;25:1740–59. doi: 10.1210/me.2011-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wandji SA, Gadsby JE, Barber JA, Hammond JM. Messenger ribonucleic acids for MAC25 and connective tissue growth factor (CTGF) are inversely regulated during folliculogenesis and early luteogenesis. Endocrinology. 2000;141:2648–57. doi: 10.1210/endo.141.7.7576. [DOI] [PubMed] [Google Scholar]
- 49.Kawamura K, Cheng Y, Suzuki N, Deguchi M, Sato Y, Takae S, et al. Hippo signaling disruption and Akt stimulation of ovarian follicles for infertility treatment. Proc Natl Acad Sci U S A. 2013;110:17474–9. doi: 10.1073/pnas.1312830110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan HY, Liu Z, Johnson PF, Richards JS. CCAAT/enhancer-binding proteins (C/EBP)-α and -β are essential for ovulation, luteinization, and the expression of key target genes. Mol Endocrinol. 2011;25:253–68. doi: 10.1210/me.2010-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillio-Meina C, Hui YY, LaVoie HA. Expression of CCAAT/enhancer binding proteins alpha and beta in the porcine ovary and regulation in primary cultures of granulosa cells. Biol Reprod. 2005;72:1194–204. doi: 10.1095/biolreprod.104.035246. [DOI] [PubMed] [Google Scholar]
- 52.Li F, Liu J, Jo M, Curry TE., Jr A role for nuclear factor interleukin-3 (NFIL3), a critical transcriptional repressor, in down-regulation of periovulatory gene expression. Mol Endocrinol. 2011;25:445–59. doi: 10.1210/me.2010-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinshelwood MM, Repa JJ, Shelton JM, Richardson JA, Mangelsdorf DJ, Mendelson CR. Expression of LRH-1 and SF-1 in the mouse ovary: localization in different cell types correlates with differing function. Mol Cell Endocrinol. 2003;207:39–45. doi: 10.1016/S0303-7207(03)00257-0. [DOI] [PubMed] [Google Scholar]
- 54.Bertolin K, Gossen J, Schoonjans K, Murphy BD. The orphan nuclear receptor Nr5a2 is essential for luteinization in the female mouse ovary. Endocrinology. 2014;155:1931–43. doi: 10.1210/en.2013-1765. [DOI] [PubMed] [Google Scholar]
- 55.Gerrits H, Paradé MC, Koonen-Reemst AM, Bakker NE, Timmer-Hellings L, Sollewijn Gelpke MD, et al. Reversible infertility in a liver receptor homologue-1 (LRH-1)-knockdown mouse model. Reprod Fertil Dev. 2014;26:293–306. doi: 10.1071/RD12131. [DOI] [PubMed] [Google Scholar]
- 56.Imamichi Y, Mizutani T, Ju Y, Matsumura T, Kawabe S, Kanno M, et al. Transcriptional regulation of human ferredoxin 1 in ovarian granulosa cells. Mol Cell Endocrinol. 2013;370:1–10. doi: 10.1016/j.mce.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C, Large MJ, Duggavathi R, DeMayo FJ, Lydon JP, Schoonjans K, et al. Liver receptor homolog-1 is essential for pregnancy. Nature Med. 2013;19:1061–6. doi: 10.1038/nm.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pennybacker M, Herman B. Follicle-stimulating hormone increases c-fos mRNA levels in rat granulosa cells via a protein kinase C-dependent mechanism. Mol Cell Endocrinol. 1991;80:11–20. doi: 10.1016/0303-7207(91)90138-I. [DOI] [PubMed] [Google Scholar]
- 59.Delidow BC, Lynch JP, White BA, Peluso JJ. Regulation of proto-oncogene expression and deoxyribonucleic acid synthesis in granulosa cells of perifused immature rat ovaries. Biol Reprod. 1992;47:428–35. doi: 10.1095/biolreprod47.3.428. [DOI] [PubMed] [Google Scholar]
- 60.Rusovici R, LaVoie HA. Expression and distribution of AP-1 transcription factors in the porcine ovary. Biol Reprod. 2003;69:64–74. doi: 10.1095/biolreprod.102.013995. [DOI] [PubMed] [Google Scholar]
- 61.Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–63. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- 62.Williams KL, Nanda I, Lyons GE, Kuo CT, Schmid M, Leiden JM, et al. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur J Immunol. 2001;31:1620–7. doi: 10.1002/1521-4141(200105)31:5<1620::AID-IMMU1620>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 63.Beshay VE, Havelock JC, Sirianni R, Ye P, Suzuki T, Rainey WE, et al. The mechanism for protein kinase C inhibition of androgen production and 17alpha-hydroxylase expression in a theca cell tumor model. J Clin Endocrinol Metab. 2007;92:4802–9. doi: 10.1210/jc.2007-1394. [DOI] [PubMed] [Google Scholar]
- 64.Delidow BC, White BA, Peluso JJ. Gonadotropin induction of c-fos and c-myc expression and deoxyribonucleic acid synthesis in rat granulosa cells. Endocrinology. 1990;126:2302–6. doi: 10.1210/endo-126-5-2302. [DOI] [PubMed] [Google Scholar]
- 65.Hogg K, Etherington SL, Young JM, McNeilly AS, Duncan WC. Inhibitor of differentiation (Id) genes are expressed in the steroidogenic cells of the ovine ovary and are differentially regulated by members of the transforming growth factor-beta family. Endocrinology. 2010;151:1247–56. doi: 10.1210/en.2009-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Combes AN, Spiller CM, Harley VR, Sinclair AH, Dunwoodie SL, Wilhelm D, et al. Gonadal defects in Cited2-mutant mice indicate a role for SF1 in both testis and ovary differentiation. Int J Dev Biol. 2010;54:683–9. doi: 10.1387/ijdb.092920ac. [DOI] [PubMed] [Google Scholar]
- 67.Sun HB. CITED2 mechanoregulation of matrix metalloproteinases. Ann N Y Acad Sci. 2010;1192:429–36. doi: 10.1111/j.1749-6632.2009.05305.x. [DOI] [PubMed] [Google Scholar]
- 68.Kim J, Sato M, Li Q, Lydon JP, Demayo FJ, Bagchi IC, et al. Peroxisome proliferator-activated receptor gamma is a target of progesterone regulation in the preovulatory follicles and controls ovulation in mice. Mol Cell Biol. 2008;28:1770–82. doi: 10.1128/MCB.01556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cui Y, Miyoshi K, Claudio E, Siebenlist UK, Gonzalez FJ, Flaws J, et al. Loss of the peroxisome proliferation-activated receptor gamma (PPARgamma) does not affect mammary development and propensity for tumor formation but leads to reduced fertility. J Biol Chem. 2002;277:17830–5. doi: 10.1074/jbc.M200186200. [DOI] [PubMed] [Google Scholar]
- 70.Zhou Y, Ma C, Karmouch J, Katbi HA, Liu XJ. Antiapoptotic role for ornithine decarboxylase during oocyte maturation. Mol Cell Biol. 2009;29:1786–95. doi: 10.1128/MCB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pietilä M, Alhonen L, Halmekytö M, Kanter P, Jänne J, Porter CW. Activation of polyamine catabolism profoundly alters tissue polyamine pools and affects hair growth and female fertility in transgenic mice overexpressing spermidine/spermine N1-acetyltransferase. J Biol Chem. 1997;272:18746–51. doi: 10.1074/jbc.272.30.18746. [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, Casero RA. Mammalian polyamine catabolism: a therapeutic target, a pathological problem, or both? J Biochem. 2006;139:17–25. doi: 10.1093/jb/mvj021. [DOI] [PubMed] [Google Scholar]
- 73.Shkolnik K, Tadmor A, Ben-Dor S, Nevo N, Galiani D, Dekel N. Reactive oxygen species are indispensable in ovulation. Proc Natl Acad Sci U S A. 2011;108:1462–7. doi: 10.1073/pnas.1017213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park JI, Jeon HJ, Jung NK, Jang YJ, Kim JS, Seo YW, et al. Periovulatory expression of hydrogen peroxide-induced sulfiredoxin and peroxiredoxin 2 in the rat ovary: gonadotropin regulation and potential modification. Endocrinology. 2012;153:5512–21. doi: 10.1210/en.2012-1414. [DOI] [PubMed] [Google Scholar]
- 75.Miyazaki T, Sueoka K, Dharmarajan AM, Atlas SJ, Bulkley GB, Wallach EE. Effect of inhibition of oxygen free radical on ovulation and progesterone production by the in-vitro perfused rabbit ovary. J Reprod Fertil. 1991;91:207–12. doi: 10.1530/jrf.0.0910207. [DOI] [PubMed] [Google Scholar]
- 76.Okamura N, Babior BM, Mayo LA, Peveri P, Smith RM, Curnutte JT. The p67-phox cytosolic peptide of the respiratory burst oxidase from human neutrophils. Functional aspects. J Clin Invest. 1990;85:1583–7. doi: 10.1172/JCI114608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yacobi K, Tsafriri A, Gross A. Luteinizing hormone-induced caspase activation in rat preovulatory follicles is coupled to mitochondrial steroidogenesis. Endocrinology. 2007;148:1717–26. doi: 10.1210/en.2006-1533. [DOI] [PubMed] [Google Scholar]
- 78.Hanukoglu I. Antioxidant protective mechanisms against reactive oxygen species (ROS) generated by mitochondrial P450 systems in steroidogenic cells. Drug Metab Rev. 2006;38:171–96. doi: 10.1080/03602530600570040. [DOI] [PubMed] [Google Scholar]
- 79.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–17. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 80.Qi L, Zou ZQ, Wang LY, Gao S, Fan YC, Long B, et al. Methylation of the glutathione-S-transferase M3 gene promoter is associated with oxidative stress in acute-on-chronic hepatitis B liver failure. Tohoku J Exp Med. 2012;228:43–51. doi: 10.1620/tjem.228.43. [DOI] [PubMed] [Google Scholar]
- 81.Sun Y, Wang Y, Yin Y, Chen X, Sun Z. GSTM3 reverses the resistance of hepatoma cells to radiation by regulating the expression of cell cycle/apoptosis-related molecules. Oncol Lett. 2014;8:1435–40. doi: 10.3892/ol.2014.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 83.Yang S, Luo A, Hao X, Lai Z, Ding T, Ma X, et al. Peroxiredoxin 2 inhibits granulosa cell apoptosis during follicle atresia through the NFKB pathway in mice. Biol Reprod. 2011;84:1182–9. doi: 10.1095/biolreprod.110.087569. [DOI] [PubMed] [Google Scholar]
- 84.Lee MS, Liu CH, Lee TH, Wu HM, Huang CC, Huang LS, et al. Association of creatin kinase B and peroxiredoxin 2 expression with age and embryo quality in cumulus cells. J Assist Reprod Genet. 2010;27:629–39. doi: 10.1007/s10815-010-9459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li M, Karakji EG, Xing R, Fryer JN, Carnegie JA, Rabbani SA, et al. Expression of urokinase-type plasminogen activator and its receptor during ovarian follicular development. Endocrinology. 1997;138:2790–9. doi: 10.1210/endo.138.7.5221. [DOI] [PubMed] [Google Scholar]
- 86.Cao M, Buratini J, Jr, Lussier JG, Carrière PD, Price CA. Expression of protease nexin-1 and plasminogen activators during follicular growth and the periovulatory period in cattle. Reproduction. 2006;13:125–37. doi: 10.1530/rep.1.00849. [DOI] [PubMed] [Google Scholar]
- 87.Li Q, Jimenez-Krassel F, Kobayashi Y, Ireland JJ, Smith GW. Effect of intrafollicular indomethacin injection on gonadotropin surge-induced expression of select extracellular matrix degrading enzymes and their inhibitors in bovine preovulatory follicles. Reproduction. 2006;131:533–43. doi: 10.1530/rep.1.00926. [DOI] [PubMed] [Google Scholar]
- 88.Murdoch WJ. Proteolytic and cellular death mechanisms in ovulatory ovarian rupture. Biol Signals Recept. 2000;9:102–14. doi: 10.1159/000014629. [DOI] [PubMed] [Google Scholar]
- 89.Ny A, Leonardsson G, Hägglund AC, Hägglöf P, Ploplis VA, Carmeliet P, et al. Ovulation in plasminogen-deficient mice. Endocrinology. 1999;140:5030–5. doi: 10.1210/endo.140.11.7113. [DOI] [PubMed] [Google Scholar]
- 90.Ploplis VA, Carmeliet P, Vazirzadeh S, Van Vlaenderen I, Moons L, Plow EF, et al. Effects of disruption of the plasminogen gene on thrombosis, growth, and health in mice. Circulation. 1995;92:2585–93. doi: 10.1161/01.CIR.92.9.2585. [DOI] [PubMed] [Google Scholar]
- 91.Liu YX, NY T. Sarkar D, Loskutoff D, Hsueh AJ. Identification and regulation of tissue plasminogen activator activity in rat cumulus-oocyte complexes. Endocrinology. 1986;119:1578–87. doi: 10.1210/endo-119-1-236. [DOI] [PubMed] [Google Scholar]
- 92.Wahlberg P, Bodén I, Paulsson J, Lund LR, Liu K, Ny T. Functional corpora lutea are formed in matrix metalloproteinase inhibitor-treated plasminogen-deficient mice. Endocrinology. 2007;148:1226–34. doi: 10.1210/en.2006-0669. [DOI] [PubMed] [Google Scholar]
- 93.Coy P, Jiménez-Movilla M, García-Vázquez FA, Mondéjar I, Grullón L, Romar R. Oocytes use the plasminogen-plasmin system to remove supernumerary spermatozoa. Hum Reprod. 2012;27:1985–93. doi: 10.1093/humrep/des146. [DOI] [PubMed] [Google Scholar]
- 94.Ritter LJ, Sugimura S, Gilchrist RB. Oocyte induction of EGF responsiveness in somatic cells is associated with the acquisition of porcine oocyte developmental competence. Endocrinology. 2015;156:2299–22312. doi: 10.1210/en.2014-1884. [DOI] [PubMed] [Google Scholar]


