Abstract
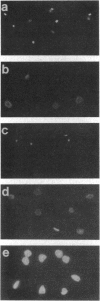
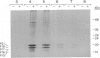
Kinetoplast DNA (kDNA), the mitochondrial DNA of trypanosomes, is a highly condensed disc-shaped network of catenated DNA circles consisting of maxicircles, the equivalent of conventional mitochondrial DNA, and several thousand smaller circular DNAs termed minicircles. Upon cell lysis, kDNA expands, giving rise to a two-dimensional network of catenated circles with an overall diameter close to that of the whole cell. To identify proteins associated with the condensed form of kDNA in the cell, proteins were reversibly crosslinked to kDNA in whole cells of Crithidia fasciculata by formaldehyde treatment. Crosslinked networks were purified and found to retain a condensed structure which becomes fully expanded upon proteinase K treatment or reversal of the crosslinks by heating at 65 degrees C. Five low molecular weight proteins released from the kDNA by heat treatment were purified by polyacrylamide gel electrophoresis and their amino-terminal sequences were determined. PCR amplification and sequence analysis of cDNA sequences between these amino-terminal sequences and the miniexon (spliced leader) sequence present at the 5' end of all C. fasciculata mRNAs predicts the presence of 9-amino acid presequences with features characteristic of mitochondrial presequences on three of the proteins. Two of these proteins are lysine-rich basic proteins. These findings suggest that basic proteins may play a role in the condensation of kDNA in the kinetoplast and that these proteins are imported into the kinetoplast by a mechanism involving a cleavable presequence.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borst P., Hoeijmakers J. H. Kinetoplast DNA. Plasmid. 1979 Jan;2(1):20–40. doi: 10.1016/0147-619x(79)90003-9. [DOI] [PubMed] [Google Scholar]
- Caron F., Jacq C., Rouvière-Yaniv J. Characterization of a histone-like protein extracted from yeast mitochondria. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4265–4269. doi: 10.1073/pnas.76.9.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J Biol Chem. 1992 Feb 15;267(5):3368–3374. [PubMed] [Google Scholar]
- Englund P. T. The replication of kinetoplast DNA networks in Crithidia fasciculata. Cell. 1978 May;14(1):157–168. doi: 10.1016/0092-8674(78)90310-0. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992 Feb 15;267(5):3358–3367. [PubMed] [Google Scholar]
- Hartl F. U., Pfanner N., Nicholson D. W., Neupert W. Mitochondrial protein import. Biochim Biophys Acta. 1989 Jan 18;988(1):1–45. doi: 10.1016/0304-4157(89)90002-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Muhich M. L., Hughes D. E., Simpson A. M., Simpson L. The monogenetic kinetoplastid protozoan, Crithidia fasciculata, contains a transcriptionally active, multicopy mini-exon sequence. Nucleic Acids Res. 1987 Apr 10;15(7):3141–3153. doi: 10.1093/nar/15.7.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntambi J. M., Marini J. C., Bangs J. D., Hajduk S. L., Jimenez H. E., Kitchin P. A., Klein V. A., Ryan K. A., Englund P. T. Presence of a bent helix in fragments of kinetoplast DNA minicircles from several trypanosomatid species. Mol Biochem Parasitol. 1984 Jul;12(3):273–286. doi: 10.1016/0166-6851(84)90084-7. [DOI] [PubMed] [Google Scholar]
- Ray D. S. Conserved sequence blocks in kinetoplast minicircles from diverse species of trypanosomes. Mol Cell Biol. 1989 Mar;9(3):1365–1367. doi: 10.1128/mcb.9.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. R., Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991 Aug 22;352(6337):731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- Schouten J. P. Hybridization selection of covalent nucleic acid-protein complexes. 2. Cross-linking of proteins to specific Escherichia coli mRNAs and DNA sequences by formaldehyde treatment of intact cells. J Biol Chem. 1985 Aug 15;260(17):9929–9935. [PubMed] [Google Scholar]
- Silver L. E., Torri A. F., Hajduk S. L. Organized packaging of kinetoplast DNA networks. Cell. 1986 Nov 21;47(4):537–543. doi: 10.1016/0092-8674(86)90618-5. [DOI] [PubMed] [Google Scholar]
- Simpson L. Kinetoplast DNA in trypanosomid flagellates. Int Rev Cytol. 1986;99:119–179. doi: 10.1016/s0074-7696(08)61426-6. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Larsen P. L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988 Jun 17;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]