Abstract
We developed 21,499 genome-wide insertion–deletion (InDel) markers (2- to 54-bp in silico fragment length polymorphism) by comparing the genomic sequences of four (desi, kabuli and wild C. reticulatum) chickpea [Cicer arietinum (L.)] accessions. InDel markers showing 2- to 6-bp fragment length polymorphism among accessions were abundant (76.8%) in the chickpea genome. The physically mapped 7,643 and 13,856 markers on eight chromosomes and unanchored scaffolds, respectively, were structurally and functionally annotated. The 4,506 coding (23% large-effect frameshift mutations) and regulatory InDel markers were identified from 3,228 genes (representing 11.7% of total 27,571 desi genes), suggesting their functional relevance for trait association/genetic mapping. High amplification (97%) and intra-specific polymorphic (60–83%) potential and wider genetic diversity (15–89%) were detected by genome-wide 6,254 InDel markers among desi, kabuli and wild accessions using even a simpler cost-effective agarose gel-based assay. This signifies added advantages of this user-friendly genetic marker system for manifold large-scale genotyping applications in laboratories with limited infrastructure and resources. Utilizing 6,254 InDel markers-based high-density (inter-marker distance: 0.212 cM) inter-specific genetic linkage map (ICC 4958 × ICC 17160) of chickpea as a reference, three major genomic regions harboring six flowering and maturity time robust QTLs (16.4–27.5% phenotypic variation explained, 8.1–11.5 logarithm of odds) were identified. Integration of genetic and physical maps at these target QTL intervals mapped on three chromosomes delineated five InDel markers-containing candidate genes tightly linked to the QTLs governing flowering and maturity time in chickpea. Taken together, our study demonstrated the practical utility of developing and high-throughput genotyping of such beneficial InDel markers at a genome-wide scale to expedite genomics-assisted breeding applications in chickpea.
Keywords: chickpea, flowering time, InDel, maturity time, QTL
1. Introduction
Chickpea [Cicer arietinum (L.)] is a self-pollinated annual diploid legume with a genome size of ∼740 Mb.1–3 Globally, chickpea is the third most important food legume complementing cereals and considered a vital human dietary source of protein abundant in essential amino acids.2,3 In the present era of climatic variability, resource scarcity and fast-growing population, it is necessary to enhance productivity as well as sustainability of agriculture for circumventing the global food insecurity. The prime objective of chickpea genomics-assisted breeding was thus inclined towards developing high-yielding durable stress tolerant (climate resilient) chickpea cultivars to meet the dietary demand of increasing population for attaining food security. To achieve these goals, an effective molecular dissection of these complex quantitative stress tolerance and yield-contributing traits by identification (map-based cloning) of potential trait-associated genes/quantitative trait loci (QTLs) through genetic/association mapping and their deployment in marker-assisted genetic enhancement of chickpea is essential. Significant progress in this regard has been made primarily through development and high-throughput genotyping of numerous genome/gene-derived simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers in diverse natural and mapping populations to expedite the process of genetic and association mapping in chickpea.4–12 Consequently, hitherto very limited number of markers tightly linked to genes/QTLs governing traits of agronomic importance has been identified to be exploited for marker-assisted genetic improvement of chickpea.13 Several studies have documented the narrow genetic base causing low intra-specific marker polymorphism as the major hindrance for chickpea genetic improvement.2,14 Henceforth, it is imperative to develop and use numerous genome-wide informative sequence-based genetic markers in generating high-density linkage maps and for high-resolution genetic and association mapping in order to delineate functionally relevant trait-regulatory genes/QTLs for genomics-assisted crop improvement of chickpea.
Insertion-deletions (InDels) are one of the major sources of structural variation found widely distributed across the genomes of diverse plant species, including Arabidopsis, rice, wheat and tomato.15,16 InDels generally arise due to certain cellular mechanisms, including movement of transposable elements, replication slippage and unequal crossing-over within the genome.17 InDel is an important phenomenon, which can exert a deleterious as well as beneficial effect on particular loci within the genome.18,19 These InDels are valuable complement to other sequence-based genetic markers like SSRs and SNPs and well recognized as an effective marker system for genetic analysis in crop plants primarily due to myriad desirable inherent genetic attributes such as multi-allelic and co-dominant inheritance and wide genomic distribution. Besides, the InDel markers are easily detectable at a genome-wide scale (gene level) in silico with low cost, labor and time via comparison of freely accessible genomic (transcriptomic) sequence resources of different available genotypes through computational genomics tools. In addition to the user-friendly mining approach, these markers can be preferably selected on the basis of their predicted fragment length variations and further validated/genotyped in germplasm lines and mapping populations using a simple cost-effective agarose gel-based assay. This makes InDels an ideal marker system of choice for diverse genomics-assisted breeding applications in crop plants.15–17,20–23
Much recently, the completion of genome sequencing of desi (cv. ICC 4958) and kabuli (cv. CDC Frontier) cultivars of chickpea has accelerated the process of genome resequencing of diverse desi, kabuli and wild accessions.2,14 It would be thus interesting to develop informative InDel markers in silico at a genome-wide scale by comparing these publicly accessible genomic (gene) sequences of different chickpea accessions with sub-optimal use of resources. Furthermore, the large-scale validation and genotyping of these developed InDel markers in natural germplasm lines and mapping populations with higher marker genetic polymorphic potential, could be utilized for construction of high-resolution genetic linkage maps as well as molecular mapping and genetic association analysis in order to identify trait-regulating genes/QTLs for marker-assisted genetic enhancement of chickpea.
In light of the above, the present study has made an effort to develop large-scale genome-wide InDel markers and to determine their genomic constitution in the chickpea genome. The potential of these markers was assessed to detect intra-specific polymorphism and genetic diversity among desi, kabuli and wild accessions, and construct high-density inter-specific genetic linkage map of chickpea. As a proof of concept, this InDel markers-based high-resolution genetic linkage map was utilized as a reference to identify and map the major genomic regions harboring robust QTLs governing flowering and maturity time, with an aim to accelerate genomics-assisted breeding in chickpea.
2. Materials and methods
2.1. Discovery of genome-wide InDel markers in chickpea
The high-quality FASTQ sequence reads generated from whole-genome resequencing of four chickpea accessions (desi cv. ICC 4958 and ICC 4951, kabuli cv. ICC 12968 and wild Cicer reticulatum cv. ICC 17160) were acquired.14,24 The mapping of sequence reads of three chickpea accessions onto the chromosome pseudomolecules of reference desi (ICC 4958) genome and detection of high-quality InDels among accessions were performed following Jain et al.20 A highly stringent criteria with a minimum read depth of 10 and a minimum sequence variant frequency (polymorphism call rate) of >85% as well as the presence of at least one InDel within a 50-bp window were considered relevant for screening and detection of high-quality (minimal false-positive) InDels among four chickpea accessions. About 200–300 bp high-quality genomic sequences of ICC 4958 flanking the either side of identified InDels were retrieved. The forward and reverse primers targeting these InDels-carrying sequences were designed using the generic primer interface of BatchPrimer3 (http://probes.pw.usda.gov/batchprimer3) to develop genome-wide InDel markers. The genomic distribution of InDel markers on chromosome pseudomolecules of desi chickpea genome was determined and visualized using Circos visualization tool.11 To determine the precise distribution of InDel markers in the diverse intergenic as well as coding and non-coding sequence components of genes/genomes (chromosomes/pseudomolecules and unanchored scaffolds), the physical positions (bp) of InDel markers were integrated/correlated with the GFF file containing genome annotation of desi chickpea (CGAP v1.014) using the customized Perl scripts. The putative function of InDel markers-carrying genes was predicted according to desi genome annotation14 and PFAM database v27.0 (http://pfam.sanger.ac.uk).
2.2. Experimental validation, polymorphism and genetic diversity potential of InDel markers
To assess the amplification and polymorphic potential of developed InDel markers, the InDel markers showing ≥10 bp in silico fragment length polymorphism among accessions were PCR amplified using the genomic DNA of 24 chickpea accessions. This included four accessions (ICC 4958, ICC 4951, ICC 12968 and ICC 17160) from which the InDel markers were originally identified, and 20 additional desi (9) and kabuli (11) accessions (Supplementary Table S1). The standard PCR constituents and touchdown thermal cycling profiling for PCR amplification and 2.5% agarose gel to resolve the amplified PCR products were utilized as per Kujur et al.10 For experimental validation of InDel markers revealing 2–9 bp in silico fragment length polymorphism among accessions, these markers were PCR amplified using the genomic DNA of 24 aforesaid chickpea accessions. The amplified PCR products were purified and sequenced in both forward and reverse directions twice on a capillary-based automated DNA sequencer (Applied Biosystems, ABI 3730xl DNA Analyzer, Vernon Hills, IL, USA) using the BigDye Terminator v3.1 sequencing kit. The high-quality consensus sequences obtained for each InDel marker were aligned and compared among accessions used. The genotyping data of validated InDel markers were utilized to estimate the average polymorphic alleles per marker, percent polymorphism and polymorphism information content (PIC) among chickpea accessions employing the PowerMarker v3.51.25 The genetic diversity and phylogenetic relationships among accessions were determined using the genotyping data of validated polymorphic genome/gene-derived InDel markers. The Nei and Li similarity coefficient-based neighbor joining (NJ) method (with 1,000 bootstrap replicates) of PowerMarker v3.51 was employed for clustering analysis and construction of unrooted phylogenetic tree among accessions.
2.3. Genetic linkage map construction
The InDel markers showing polymorphism between ICC 4958 and ICC 17160 were PCR amplified and genotyped using 190 individuals and parental accessions derived from an F5 inter-specific mapping population (ICC 4958 × ICC 17160), following the aforementioned methods of agarose gel- and PCR amplicons resequencing-based assays. The genotyping data were analyzed employing the χ2-test (P < 0.05) to assess their goodness-of-fit to the expected Mendelian 1:1 segregation ratio. The MAPMAKER/EXP 3.026 was used to estimate linkage analysis among the InDel markers, basing upon which different linkage groups (LGs) were classified. For further validation of linkages among markers, the genotyping data of markers grouped by MAPMAKER were analyzed through JoinMap 4.1 (http://www.kyazma.nl/index.php/mc.JoinMap) at higher logarithm of odds (LOD) threshold (4.0–8.0) with Kosambi mapping function. The InDel markers were incorporated into defined LGs (designated LG1 to LG8) based on their centiMorgan (cM) genetic distances and corresponding marker physical positions (bp) on the chromosomes. An inter-specific genetic linkage map was finally constructed and visualized using MapChart v2.2.10
2.4. QTL mapping
The 190 individuals along with parental accessions of an inter-specific mapping population (ICC 4958 × ICC 17160) were grown in the field for two consecutive years during crop-growing season at two diverse geographical locations of India. Additionally, greenhouse trial was performed to measure the flowering and maturity time response of mapping individuals under both long- and short-day conditions at 22 ± 2°C. The days to 50% flowering (DF) and maturity (DM) time of each mapping individual (10–12 representative plants from each individual) was measured by counting the number of days from sowing (first irrigation) to the stage when 50% of their plants have begun to flower and 90% of pods have matured and turned yellow, respectively. The homogeneity, frequency distribution, coefficient of variation (CV) and broad-sense heritability (H2) of DF and DM traits in mapping population were determined following Bajaj et al.27
For QTL mapping, the genotyping data of InDel markers physically/genetically mapped on eight LGs/chromosomes and field phenotypic data (DF and DM) of 190 mapping individuals and parental accessions were correlated using composite interval mapping (CIM) function of MapQTL 6.28 The LOD threshold score of more than 4.0 at 1,000 permutations was considered significant (P < 0.05) to identify and map the major QTLs on LGs governing DF and DM traits in chickpea. At significant LOD, the positional genetic effect and phenotypic variation explained (PVE) by QTLs were measured. QTL Network v2.0 was used to determine the additive effect of marker loci underlying the QTLs. At 1.5-LOD support intervals (95% bootstrap CI), the confidence intervals (CI) around each significant major QTL peak was estimated.
3. Results and discussion
3.1. Genomic constitution of InDel markers in chickpea genome
The comparison of resequencing data of three chickpea accessions ICC 4951 (cultivated desi), ICC 12968 (cultivated kabuli) and ICC 17160 (wild C. reticulatum) with reference genome of desi accession (ICC 4958) developed a total of 21,499 InDel markers (Fig. 1, Supplementary Table S2). These developed markers with diverse potential of detecting in silico InDel polymorphism in all possible-pair combination of four different chickpea accessions were physically mapped on the eight chromosomes and unanchored scaffolds of desi chickpea genome (Fig. 2A–D). A maximum of 16 839 (78.3%) markers showing in silico InDel-based fragment length polymorphism between ICC 4958 and ICC 17160, followed by 4699 (21.8%) markers between ICC 4958 and ICC 12968 and 3252 (15.1%) markers between ICC 4958 and ICC 4951, and minimum of 1173 (5.4%) markers between ICC 4951 and ICC 17160 were identified (Fig. 2A–C). Interestingly, markers designed from 694 (3.2%) InDels were found to be commonly polymorphic among all four chickpea accessions used. The observed InDel marker-based genetic polymorphism overall reflect the close phylogenetic relatedness of desi rather than kabuli with wild accessions and evolutionary divergence of cultivated accessions from a common wild progenitor C. reticulatum as well as existence of a domestication-led bottleneck in cultivated desi and kabuli chickpea.2,14,29,30 Remarkably, in silico fragment length polymorphism detected by markers based on their size (bp) of InDels varied from 2 to 54 bp with an average of 6 bp (Supplementary Table S3). More than 76.8% (16,510) InDel markers showed 2 to 6 bp in silico fragment length polymorphism, whereas remaining 23.2% (4,989) markers had 7 to 54 bp fragment length polymorphism (Supplementary Figure S1). This trend of proportionate distribution of two different kinds of fragment length polymorphisms (2 to 6 bp: 75.3 to 87.5% and 7 to 54 bp: 12.5 to 24.7%) showing InDel markers detected in each possible-pair combination of four chickpea accessions used, remained almost similar (Supplementary Figure S1). A relatively higher frequency of markers with smaller size InDels (2 to 6 bp) identified in chickpea could be due to the use of NGS-based short sequence reads (<200 bp) and/or user-specific computational genomics tools/algorithms for detection of InDels among chickpea accessions at a genome-wide scale.33
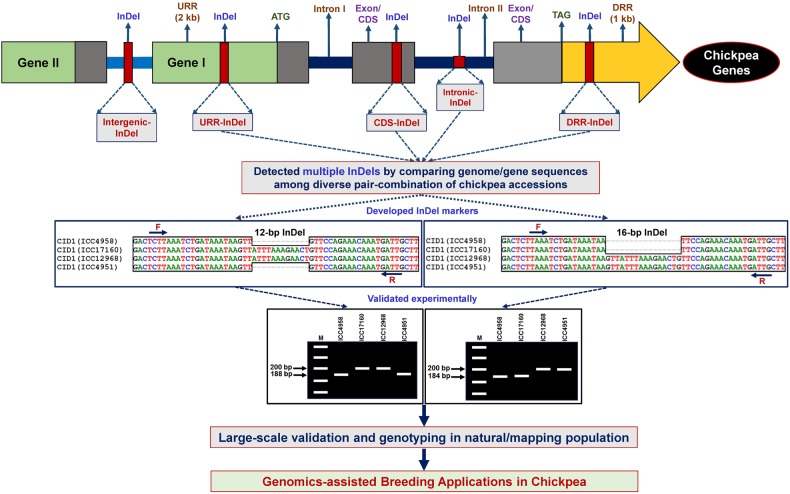
Figure 1.
Schematic illustrating the major steps followed for successful development, validation and genotyping of InDel markers derived from diverse intergenic and gene (coding and non-coding) sequence components of chickpea genome for their genomics-assisted breeding applications. The forward (F) and reverse (R) primers designed from the sequences flanking the target InDels were developed as InDel markers and amplified in a diverse set of desi, kabuli and wild natural chickpea accessions and individuals of a mapping population. This figure is available in black and white in print and in colour at DNA Research online.
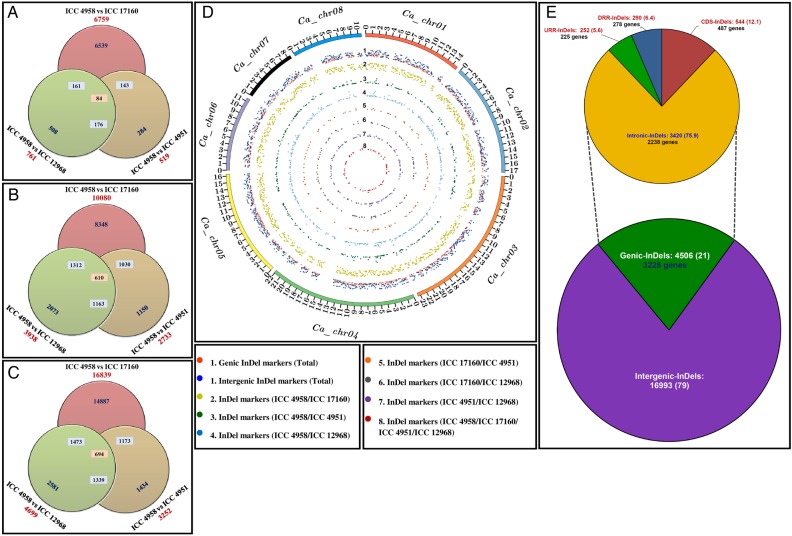
Figure 2.
Genomic constitution and genome-wide distribution pattern of 21,499 InDel markers developed by comparing the genome sequences of four different desi (ICC 4958 and ICC 4951), kabuli (ICC 12968) and wild (ICC 17160) chickpea accessions. InDel markers identified from diverse possible-pair combination (indicated by individual circles) of four accessions that were physically mapped on eight chromosomes (A) and unanchored scaffolds (B) as well as sum of the whole chickpea genome (C) are depicted by Venn diagrams. (D) The frequency and relative distribution of 7,643 InDel markers physically mapped on eight chromosomes of desi chickpea genome are illustrated by a Circos circular ideogram. The outermost circles signify the different physical size (Mb) of eight chromosomes coded with multiple colors as per the pseudomolecule size documented in desi chickpea genome.14 (E) Relative frequency of 21,499 InDel markers identified from the intergenic and various coding and non-coding (introns and regulatory regions) sequence components of 3,228 genes annotated from desi genome. Parenthesis indicates the proportion (%) of InDel markers. The CDS (coding sequences), URR (upstream regulatory region) and DRR (downstream regulatory region) of protein-coding genes were defined according to the available gene annotation of desi chickpea genome.14 This figure is available in black and white in print and in colour at DNA Research online.
Of the designed 21,499 InDel markers, 7,643 and 13,856 markers were physically mapped on eight chromosomes (Fig. 2D) and unanchored scaffolds of desi chickpea genome (Supplementary Table S1). Highest and lowest number of InDel markers were mapped on chromosomes 3 (1,468 markers with a mean map density: 15.9 kb) and 7 (488 markers with a mean map density: 17.3 kb), respectively. Interestingly, almost a similar level of map density of InDel markers was observed in all eight chickpea chromosomes, while it ranged from 15.4 kb for chromosome 6 to 18.8 kb in chromosome 8 with an average of 16.3 kb (Supplementary Table S1). Notably, only 120 (23.1%) of 520 Mb draft scaffold sequence (covering ∼70% of 740 Mb estimated genome size of desi chickpea) has been anchored onto eight chromosome pseudomolecules of chickpea genome so far.14 The probability of physical mapping of InDel markers derived from the unanchored scaffolds onto chromosomes can be enriched by a subsequent increase in anchoring proportion of these scaffolds across the eight chickpea chromosomes. The detailed structural annotation of 21,499 InDel markers exhibited the presence of 16,993 (79%) and 4,506 (21%) markers in the intergenic regions and 3,228 protein-coding genes, respectively (Fig. 2E). Notably, we observed the presence of InDel markers in the 3,228 genes representing 11.7% of the total genes (27,571) annotated from desi chickpea genome. This is almost comparable with the proportionate occurrence (9–10%) of InDels-carrying genes documented in many crop plants, including sorghum and Arabidopsis.23 The gene-derived InDel markers included a maximum of 3,420 (75.9%) markers from the intronic sequence components of 2,238 genes, followed by 544 (12.1%) markers from the CDS of 487 genes, 290 (6.4%) markers from the downstream regulatory regions (DRRs) of 278 genes and minimum of 252 (5.6%) markers from the upstream regulatory regions (URRs) of 225 genes (Fig. 2E). Among the coding sequence variants, 87% of InDel markers affected initiation and stop codons as well as frameshift mutations. Interestingly, in both the genes and intergenic regions of desi chickpea genome, we observed a significantly higher relative distribution of InDel markers revealing 2 to 6 bp (75.7 to 77.1%) than 7 to 54 bp (22.9 to 24.3%) in silico fragment length polymorphism among four chickpea accessions (Supplementary Figure S1). The functional annotation of 4,506 InDel markers-containing 3,228 genes revealed their maximum correspondence with genes encoding growth, development and metabolism-related proteins (34%), followed by transcription factors (14%) and signal transduction (10%) and disease resistance-related proteins (6%), and minimum to transposons/retrotransposons (1%). In contrast to previous reports (60–70%) in many crop plants,23,31 a lower abundance of transposons/retrotransposons-associated InDel markers was observed in chickpea genome.
Collectively, the structurally and functionally annotated InDel markers derived from the diverse sequence components of the genome/genes are specifically responsible for frameshift mutations due to disruption of ORFs (open-reading frames) in the coding regions. These probably have large impact on transcriptional gene regulation (expression) leading to alteration of gene functions in chickpea. While comparing the average frequency between coding-InDel polymorphism and non-synonymous SNP substitutions detected among four chickpea accessions, we observed a higher frequency of non-synonymous mutations (506 SNPs/Mb) than that of InDels (252 InDels/Mb) causing frameshift mutations in the coding regions of chickpea genes.
3.2. Amplification and polymorphic potential of InDel markers
To determine the amplification and polymorphic potential of designed InDel markers, 6,580, including 3,042 and 3,538 InDel markers revealing 10–54 and 2–9 bp in silico fragment length polymorphism among four chickpea accessions, were selected for experimental validation using the agarose gel- and PCR amplicons resequencing-based assays, respectively. The overall criteria used to select InDel markers from the total 21,499 designed markers for our experimental validation study is depicted in Supplementary Figure S2. All the 6,580 selected markers were validated primarily by gel- and sequencing-based assays using the genomic DNA of four chickpea accessions (ICC 4958, ICC 4951, ICC 12968 and ICC 17160), from which the InDel markers were originally detected. This included 694 markers showing in silico fragment length polymorphism commonly among all four chickpea accessions. Notably, 6,482 of 6,580 markers produced single reproducible PCR amplicons in 2.5% agarose gel with a mean amplification success rate of 98.5% (Fig. 3A). Of these, 6,254 (96.5%) amplified markers showing in silico polymorphism at least between two combination of four chickpea accessions were validated experimentally using both agarose gel- and PCR amplicons resequencing-based assays (Fig. 3A, Supplementary Figure S3). The validation and genotyping of InDel markers particularly by PCR amplicons resequencing-based assay confirmed the presence of expected InDels, which further corresponded well with their in silico fragment length polymorphism detected among four chickpea accessions (Supplementary Figure S3).
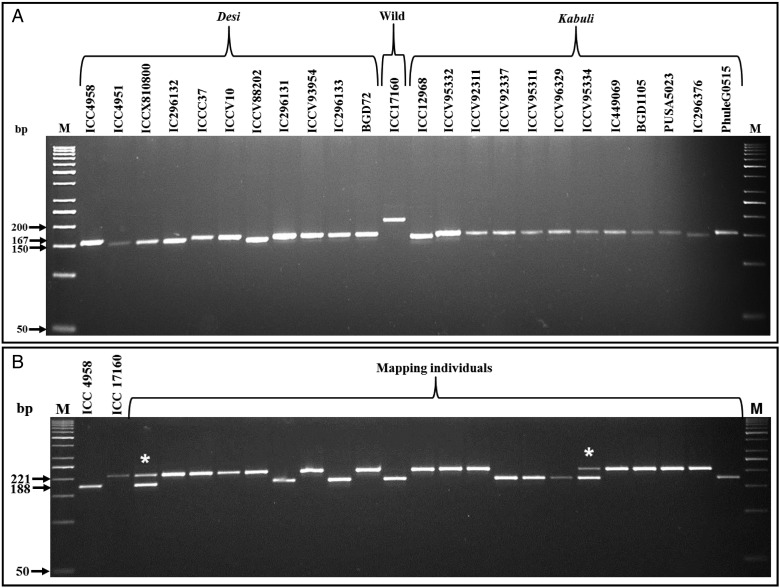
Figure 3.
(A) Allelic polymorphism detected among 24 desi, kabuli and wild chickpea accessions along with four control accessions (ICC 4958, ICC 4951, ICC 12968 and ICC 17160; from which sequences the InDel markers were originally identified) using a representative intergenic InDel marker (CID_C_1862732) in gel-based assay. A maximum number of three polymorphic alleles was amplified among accessions. The detailed information regarding accessions are provided in the Supplementary Table S1. (B) Allelic segregation pattern of a selected intronic InDel marker (CID_C_11555849) in a representative set of 22 individuals and two parental accessions of an inter-specific mapping population (ICC 4958 × ICC 17160) scanned using agarose gel-based assay. Asterisk indicates the heterozygous alleles amplified by this InDel marker in two selected mapping individuals. The amplified fragment sizes (bp) of the polymorphic alleles are indicated. M: 50 bp DNA ladder size standard. The identities of two InDel markers with their detailed information are mentioned in the Supplementary Table S3.
The 6,254 informative polymorphic InDel markers were further genotyped in a diverse set of 24 desi, kabuli and wild accessions to assess their potential for detecting polymorphism among these chickpea accessions (Fig. 3A, Supplementary Figure S3). These markers overall produced 15,623 alleles (with an average PIC of 0.71) among accessions. The number of alleles detected by these markers varied from 1 to 3 with an average of 2.5 alleles per marker. Six thousand forty-eight (96.7%, mean PIC: 0.65) of 6,254 markers showed polymorphism between cultivated and wild chickpea accessions. Five thousand one hundred ninty-two (83%, mean PIC: 0.60) of 6,254 markers were found to be polymorphic between desi and kabuli. Notably, 4,315 (69%) markers revealed polymorphism among 11 desi accessions (1 to 3 alleles with mean PIC: 0.57), whereas 3,746 (59.9%) markers exhibited polymorphism among 12 kabuli accessions (1 to 2 alleles with mean PIC: 0.51).
The inter-specific (96.7%) and also intra-specific (60–83%) polymorphic potential detected by the InDel markers among desi, kabuli and wild chickpea accessions is much higher than that estimated especially with random genome-wide SSR and SNP markers (∼35%) as well as in silico polymorphic SSR and SNP markers (50–60%).5,7,32,33 The informative InDel markers with high intra-specific polymorphic potential developed by us at a genome-wide scale could serve as a valuable resource for their immense use in genomics-assisted breeding applications of chickpea. The aforementioned marker validation outcomes specifically inferred that 3,042 validated InDel markers revealing 10–54 bp in silico fragment length polymorphism among desi, kabuli and wild accessions have efficiency to be resolved and genotyped even by a simpler cost-effective agarose gel-based assay using numerous chickpea accessions.
3.3. Genetic diversity potential of InDel markers
The evaluation of genetic diversity level based on pair-wise similarity among 24 desi, kabuli and wild chickpea accessions using 6,254 genome/gene-derived polymorphic InDel markers revealed a wide range of distance coefficient from 0.15 to 0.89 with an average of 0.57. The genetic distance among 23 cultivated desi and kabuli accessions ranged from 0.20 to 0.81 with an average of 0.51. The level of genetic diversity (15 to 89%) measured among the accessions using the InDel markers is comparatively much higher than that estimated previously with the microsatellite and SNP markers.5,7,10,11,34 A higher efficiency of InDel markers in assaying genetic diversity implies their significance in establishing distinctness among cultivated (desi and kabuli) and wild accessions and thus could be deployed in genomics-assisted varietal improvement program in chickpea. The genetic relationship among 24 desi, kabuli and wild chickpea accessions was depicted by an unrooted phylogenetic tree (Supplementary Figure S4). The genome/gene-derived InDel markers clearly discriminated all these accessions from each other and clustered into two major desi and kabuli groups. One wild C. reticulatum accession ICC 17160 being divergent from cultivated accessions; however, showing close evolutionary relationships with desi chickpea was clustered within a desi group. This overall reflects the correspondence of clustering patterns observed among these accessions with the known cultivar-specific origination, pedigree relationships and parentage.5,7,10,11,34
3.4. Generation of a high-density InDel marker-based inter-specific chickpea genetic linkage map
For constructing a high-resolution inter-specific genetic linkage map, 6,219 InDel markers revealing polymorphism between parental accession ICC 4958 and ICC 17160 were genotyped among 190 individuals of a F5 mapping population (ICC 4958 × ICC 17160). The co-dominant inheritance pattern of the InDel markers was evident from their efficiency to discriminate homozygous mapping individuals of either of the parental accessions from heterozygous individuals (Fig. 3B). This suggests the broader practical applicability of developed genome-wide InDel markers for multiple large-scale rapid genotyping applications, including their suitability in construction of high-density genetic linkage maps and molecular mapping of high-resolution QTLs. By performing linkage analysis using the 6,219 marker genotyping data, we mapped 6,197 InDel markers across eight LGs of an inter-specific chickpea genetic map (Table 1, Fig. 4A). Highest number of markers were mapped on CaLG04 (1,123 markers), followed by CaLG03 (1,011 markers) and lowest on CaLG07 (457 markers). The generated eight LGs-based genetic maps spanned a total map length of 1,311.9 cM, with a mean inter-marker distance of 0.212 cM (Table 1). CaLG03 and CaLG07 had longest and shortest map length spanning 199.4 and 135.3 cM, respectively. Most saturated genetic map was constructed in case of CaLG04 (mean inter-marker distance: 0.162 cM), followed by CaLG06 (0.175 cM) and least saturated map was generated for CaLG01 (0.253 cM) (Table 1). An InDel marker-based inter-specific genetic linkage map (ICC 4958 × ICC 17160) generated in our study had greater map density (inter-marker distance: 0.212 cM) and thus highly saturated when compared with that reported so far in multiple intra- and inter-specific mapping population-derived genetic maps (0.50–8.01 cM) of chickpea.5,7,8–10,12,27,35
Table 1.
Characteristics of an inter-specific genetic linkage map constructed using a 190 F5 chickpea mapping population (ICC 4958 × ICC 17160)
| Linkage groups (LGs) | InDel markers mapped | Map length covered (cM) | Mean inter-marker distance (cM) |
|---|---|---|---|
| CaLG01 | 651 | 164.8 | 0.253 |
| CaLG02 | 697 | 171.9 | 0.247 |
| CaLG03 | 1,011 | 199.4 | 0.197 |
| CaLG04 | 1,123 | 181.9 | 0.162 |
| CaLG05 | 808 | 167.1 | 0.207 |
| CaLG06 | 805 | 140.9 | 0.175 |
| CaLG07 | 457 | 135.3 | 0.296 |
| CaLG08 | 645 | 150.6 | 0.233 |
| Total | 6,197 | 1,311.9 | 0.212 |
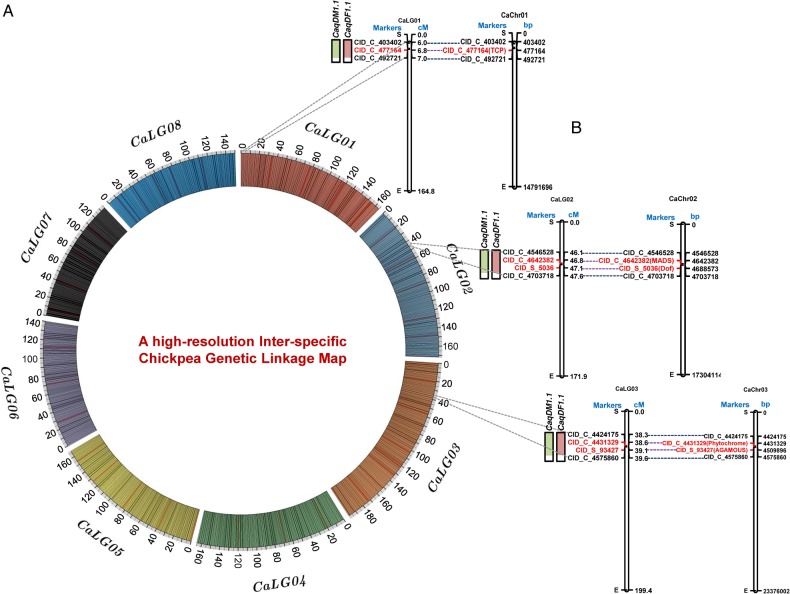
Figure 4.
(A) A high-resolution inter-specific genetic map (ICC 4958 × ICC 17160) constructed by integrating 6,197 InDel markers on eight LGs of chickpea, are depicted in a Circos circular ideogram. The outermost circles represent the different genetic map length (cM) (spanning 20 cM uniform genetic distance intervals between bins) of eight LGs coded with multiple colors. (B) The integration of genetic and physical maps delineated five InDel markers-containing candidate genes at three major genomic regions harboring six robust DF and DM QTLs mapped on three desi chromosomes 1, 2 and 3. The InDel markers-carrying genes showing strong linkage with DF and DM QTLs selected as potential candidate for flowering and maturity time regulation in chickpea are highlighted with red color. The genetic (cM)/physical (bp) distance and identities of the InDel markers mapped on the LGs/chromosomes are indicated on the right and left side of the LGs/chromosomes, respectively. The detail information regarding InDel markers and QTLs are mentioned in Supplementary Table S3 and Table 2. This figure is available in black and white in print and in colour at DNA Research online.
3.5. Molecular mapping of QTLs governing flowering and maturity time in chickpea
A significant phenotypic difference of DF (25.7–111.0 with 81% H2) and DM (86.4–151.3 with 85% H2) traits in 190 mapping individuals (ICC 4958 × ICC 17160) and two parental accessions across 2 years was observed based on ANOVA (Supplementary Table S4). ANOVA outcomes inferred existence of a highly significant difference (P < 0.001) of DF and DM traits among the mapping individuals although significant environmental (years) effects observed on these two traits in both seasons/years. A bi-directional transgressive segregation, including normal frequency distribution of DF and DM traits in mapping individuals and parental accessions across 2 years, was obtained (Supplementary Figure S5A and B). These results collectively indicate that the complex quantitative genetic inheritance pattern of DF and DM traits in our developed inter-specific mapping population (ICC 4958 × ICC 17160) are regulated by multiple genes and thus have significance to be utilized for QTL mapping.
The 2 years multi-location field phenotyping data of DF and DM traits and genotyping information of 6,197 InDel markers genetically mapped on eight LGs were integrated for molecular mapping of QTLs. This analysis identified three major genomic regions harboring three of each significant (LOD: 8.1–11.5) DF (CaqDF1.1, CaqDF2.1 and CaqDF3.1) and DM (CaqDM1.1, CaqDM2.1 and CaqDM3.1) QTLs that were mapped across three LGs (CaLG01, CaLG02 and CaLG03) (Table 2, Fig. 4B). The individual major QTL explained 17.5–27.5% and 16.4–26.9% phenotypic variations (R2) for DF and DM traits, respectively. The PVE measured for all three of each robust DF and DM QTLs in combination was 68 and 65%, respectively. The identified six DF and DM QTLs being validated and exhibited consistent PVE (>10% R2) at higher LOD (8.1–11.5) across two seasons (years) were considered as robust QTLs. Three major genomic regions (1.036, 1.555 and 1.292 cM marker genetic intervals on CaLG01, CaLG02 and CaLG03, respectively) underlying these six robust DF and DM QTLs spanned with 45 InDel markers, were mapped on three different genomic regions with similar marker intervals across three LGs (Fig. 4B). The molecular mapping and clustering of multiple DF and DM QTLs especially on a single major genomic region of three LGs infers complex genetic inheritance pattern of these quantitative traits in chickpea. All these six DF and DM QTLs exhibited positive additive gene effects (3.9–6.4) for early flowering and maturity time with major allelic contributions from ICC 4958 (Table 2). This could be due to strong positive correlation (Pearson's correlation coefficient, r: 91%, P < 0.0001) between DF and DM traits observed in an inter-specific mapping population (Supplementary Figure S5C). The five InDel markers tightly linked to six major DF and DM robust QTLs identified by both single marker- and CIM-based QTL analysis (Table 2) could have potential to be deployed in marker-assisted genetic enhancement for developing early flowering and maturing chickpea cultivars.
Table 2.
Molecular mapping of significant QTLs governing DF and DM traits on chickpea chromosomes
| QTLs | LGs/chromosomes | Marker intervals with genetic positions (cM) | QTL physical intervals (kb) | Markers tightly linked with QTLs | 2012 |
2013 |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| LOD | PVE (R2%) | A | LOD | PVE (R2%) | A | |||||
| CaqDF1.1 | CaLG(Chr01) | 1.036 cM [CID_C_403402 (5.997) to CID_C_492721 (7.033)] | 89.3 | CID_C_477164 (TCP TF) | 9.8 | 21.8 | 5.8 | 10.7 | 23.6 | 6.1 |
| CaqDF2.1 | CaLG(Chr02) | 1.555 [CID_C_4546528 (46.061) to CID_C_4703718 (47.616)] | 157.2 | CID_C_4642382 (MADS TF) CID_S_5036 (DOF Zinc Finger TF) |
8.5 | 17.5 | 3.9 | 9.7 | 20.4 | 4.9 |
| CaqDF3.1 | CaLG(Chr03) | 1.292 [CID_C_4424175 (38.314) to CID_C_4575860 (39.606)] | 151.7 | CID_C_4431329 (Phytochrome F-box) CID_S_93427 (AGAMOUS) |
10.3 | 24.7 | 4.6 | 11.5 | 27.5 | 5.6 |
| CaqDM1.1 | CaLG(Chr01) | 1.036 cM [CID_C_403402 (5.997) to CID_C_492721 (7.033)] | 89.3 | CID_C_477164 (TCP TF) | 9.5 | 20.5 | 4.7 | 10.1 | 22.7 | 6.4 |
| CaqDM2.1 | CaLG(Chr02) | 1.555 [CID_C_4546528 (46.061) to CID_C_4703718 (47.616)] | 157.2 | CID_C_4642382 (MADS TF) CID_S_5036 (DOF Zinc Finger TF) |
8.1 | 16.4 | 4.1 | 9.4 | 19.6 | 4.5 |
| CaqDM3.1 | CaLG(Chr03) | 1.292 [CID_C_4424175 (38.314) to CID_C_4575860 (39.606)] | 151.7 | CID_C_4431329 (Phytochrome F-box) CID_S_93427 (AGAMOUS) |
9.8 | 23.5 | 4.2 | 11.2 | 26.9 | 5.1 |
CaqDF1.1 (Cicer arietinum QTL for days to 50% flowering time on chromosome 1 number 1) and CaqDM1.1 (Cicer arietinum QTL for days to maturity time on chromosome 1 number 1). PVE: percentage of phenotypic variation explained by QTLs. A: Additive effect; positive additive effect infers alleles from ICC 4958 with early flowering and maturity trait values. Details regarding CID_C and CID_S markers are mentioned in the Supplementary Table S3. TF: transcription factor.
To ascertain the novelty of our identified QTLs, the major genomic regions harboring six robust DF and DM QTLs were compared with that documented in earlier QTL mapping studies utilizing diverse inter- and intra-specific chickpea mapping populations.4,6,13,36–40 We could not find any correspondence of these identified DF and DM QTLs with known QTLs reported previously based on their congruent genetic or physical positions on chickpea LGs/chromosomes. This indicates that DF and DM trait-associated QTLs identified by us are novel and possibly exhibit population-specific genomic distribution.
3.6. Delineation of candidate gene-based InDel markers regulating flowering and maturity time in chickpea
The integration of genetic linkage map information of InDel markers spanning six robust DF and DM QTLs with that of physical maps of desi chickpea genome demarcated these three major QTL intervals into 89.3 (chromosome 1)–157.2 (chromosome 2) kb physical genomic regions on chromosomes 1, 2 and 3 (Table 2, Fig. 4B). The structural and functional annotation of these target genomic physical intervals underlying DF and DM QTLs on three desi chromosomes identified multiple candidate protein-coding desi chickpea genes. Interestingly, five InDel markers-containing genes among these, at three target major QTL intervals exhibited tight linkage with six DF and DM-regulating QTLs based on our high-resolution QTL mapping (Table 2, Fig. 4B). One InDel marker (CID_C_477164) in the CDS of a TCP [Teosinte branched 1 (tb1) Cycloidea, Proliferating cell factor (PCF)] transcription factor (TF) gene localized at CaqDF1.1 and CaqDM1.1 QTL region was associated strongly (R2: 25.8–28.6% at 9.8–10.7 LOD) with DF and DM traits (Table 2, Fig. 4B). Another two InDel markers (CID_C_4642382 and CID_S_5036) in the intron and CDS of MADS [MCM (Minichromosome maintenance) AGAMOUS DEFICIENS SRF (human serum response factor)] and DOF (DNA-binding One zinc Finger)-TF genes, respectively, annotated at CaqDF2.1 and CaqDM2.1 QTL region revealed strong association (R2: 17.5–20.4% at 8.5–9.7 LOD) with DF and DM traits. Additionally, two InDel markers (CID_C_4431329 and CID_S_93427) in the DRR and introns of Phytochrome F-box and AGAMOUS genes, respectively annotated at CaqDF3.1 and CaqDM3.1 QTL interval had strong association (R2: 28.7–30.2% at 10.3–11.5 LOD) with DF and DM traits (Table 2, Fig. 4B). These InDel markers-carrying genes are known to be involved in transcriptional regulation of diverse known flowering time pathways in crop plants, including legumes.41–44 The MADS, TCP and DOF transcription factors, and Phytochrome F-box and AGAMOUS genes have significant role in regulating the ABC model of flower development by defining the genetic mechanism underlying the floral organ identity and floral architecture as well as through controlling signal perception during floral transitions from vegetative to reproductive phase specifically in Arabidopsis and other crop plants.45–48 In this context, the InDel markers-associated QTLs and five potential candidate genes regulating flowering and maturity time delineated in the present study, thus have functional relevance to understand the genetic basis of complex flowering and maturity time traits in chickpea.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org
Funding
The authors gratefully acknowledge the financial support for this study provided by a research grant (102/IFD/SAN/2161/2013-14) from the Department of Biotechnology (DBT), Government of India. Funding to pay the Open Access publication charges for this article was provided by the National Institute of Plant Genome Research (NIPGR).
Supplementary Material
Acknowledgements
S.D. acknowledges the DBT for Junior Research Fellowship award. The authors thank the Editor and reviewer for critically evaluating the manuscript and providing constructive comments.
References
- 1.Kumar J., Choudhary A.K., Solanki R.K., Pratap A.. 2011, Towards marker-assisted selection in pulses: a review, Plant Breed., 130, 297–313. [Google Scholar]
- 2.Varshney R.K., Song C., Saxena R.K. et al. 2013, Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement, Nat. Biotechnol., 31, 240–6. [DOI] [PubMed] [Google Scholar]
- 3.Varshney R.K., Mohan S.M., Gaur P.M. et al. 2013, Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics, Biotechnol. Adv., 31, 1120–34. [DOI] [PubMed] [Google Scholar]
- 4.Cobos M.J., Rubio J., Fernández-Romero M.D. et al. 2007, Genetic analysis of seed size, yield and days to flowering in a chickpea recombinant inbred line population derived from a kabuli × desi cross, Ann. Appl. Biol., 151, 33–42. [Google Scholar]
- 5.Nayak S.N., Zhu H., Varghese N. et al. 2010, Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome, Theor. Appl. Genet., 120, 1415–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gowda S.J.M., Radhika P., Mhase L.B., Jamadagni B.M., Gupta V.S., Kadro N.Y.. 2011, Mapping of QTLs governing agronomic and field traits in chickpea, J. Appl. Genet., 52, 9–21. [DOI] [PubMed] [Google Scholar]
- 7.Gujaria N., Kumar A., Dauthal P. et al. 2011, Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.), Theor. Appl. Genet., 122, 1577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiremath P.J., Farmer A., Cannon S.B. et al. 2011, Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa, Plant Biotechnol. J., 9, 922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaur R., Aza S., Jeena G. et al. 2012, High-throughput SNP discovery and genotyping for constructing a saturated linkage map of chickpea (Cicer arietinum L.), DNA Res., 19, 357–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kujur A., Bajaj D., Saxena M.S. et al. 2013, Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping, DNA Res., 20, 355–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kujur A., Bajaj D., Upadhyaya H.D. et al. 2015, Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea, Front. Plant Sci., 6, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxena M.S., Bajaj D., Das S. et al. 2014, An integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea, DNA Res., 21, 695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varshney R.K., Thudi M., Nayak S.N. et al. 2014, Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.), Theor. Appl. Genet., 127, 445–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain M., Misra G., Patel R.K. et al. 2013, A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.), Plant J., 74, 715–29. [DOI] [PubMed] [Google Scholar]
- 15.Yang J., Wang Y., Shen H., Yang W.. 2014, In silico identification and experimental validation of insertion-deletion polymorphisms in tomato genome, DNA Res., 21, 429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y., Cui X., Li R. et al. 2015, Development of genome-wide insertion/deletion markers in rice based on graphic pipeline platform, J. Integr. Plant. Biol., doi:10.1111/jipb.12354. [DOI] [PubMed] [Google Scholar]
- 17.Moghaddam M.S., Song Q., Mamidi S.. 2014, Developing market class specific InDel markers from next generation sequence data in Phaseolus vulgaris L., Front. Plant Sci., 5, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson C.E., Edamura K.N., Cleary J.D.. 2005, Repeat instability: mechanisms of dynamic mutations, Nat. Rev. Genet., 6, 729–42. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Diaz M., Kunkel T.A.. 2006, Mechanism of a genetic glissando: structural biology of InDel mutations, Trends Biochem. Sci., 31, 206–14. [DOI] [PubMed] [Google Scholar]
- 20.Jain M., Moharana K.C., Shankar R., Kumari R., Garg R.. 2014, Genome-wide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance, Plant Biotechnol. J., 12, 253–64. [DOI] [PubMed] [Google Scholar]
- 21.Li Y.H., Liu B., Reif J.C. et al. 2014, Development of insertion and deletion markers based on biparental resequencing for fine mapping seed weight in soybean, Plant Genome, 7, 1–8. [Google Scholar]
- 22.Wang Y., Lu J., Chen S. et al. 2014, Exploration of presence/absence variation and corresponding polymorphic markers in soybean genome, J. Integr. Plant. Biol., 56, 1009–19. [DOI] [PubMed] [Google Scholar]
- 23.Shen X., Liu Z., Mocoeur A.R.J., Xia Y., Jing H.C.. 2015, PAV markers in Sorghum bicolour: genome pattern, affected genes and pathways, and genetic linkage map construction, Theor. Appl. Genet., 128, 623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misra G., Priya P., Bandhiwal N. et al. 2014, The chickpea genomic web resource: visualization and analysis of the desi-type Cicer arietinum nuclear genome for comparative exploration of legumes, BMC Plant Biol., 14, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu K., Muse S.V.. 2005, PowerMarker: an integrated analysis environment for genetic marker analysis, Bioinformatics, 21, 2128–9. [DOI] [PubMed] [Google Scholar]
- 26.Lander E.S., Green P., Abrahamson J. et al. 1987, MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations, Genomics, 1, 174–81. [DOI] [PubMed] [Google Scholar]
- 27.Bajaj D., Upadhyaya H.D., Khan Y. et al. 2015, A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea, Sci. Rep., 5, 9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Ooijen J.W. 2009, MapQTL 6: Software for the mapping of quantitative trait loci in experimental populations of diploid species, B.V. Kyazma: Wageningen, Netherlands. [Google Scholar]
- 29.Abbo S., Berger J., Turner N.C.. 2003, Evolution of cultivated chickpea: four bottlenecks limit diversity and constrain adaptation, Funct. Plant. Biol., 30, 1081–7. [DOI] [PubMed] [Google Scholar]
- 30.Burger J.C., Champan M.A., Burke J.M.. 2008, Molecular insights into the evolution of crop plants, Am. J. Bot., 95, 113–22. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L.M., Luo H., Liu Z.Q. et al. 2014, Genome-wide patterns of large-size presence/absence variants in sorghum, J. Integr. Plant. Biol., 56, 24–37. [DOI] [PubMed] [Google Scholar]
- 32.Hiremath P.J., Kumar A., Penmetsa R.V. et al. 2012, Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes, Plant Biotechnol. J., 10, 716–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agarwal G., Jhanwar S., Priya P. et al. 2012, Comparative analysis of kabuli chickpea transcriptome with desi and wild chickpea provides a rich resource for development of functional markers, PLoS ONE, 7, e52443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bharadwaj C., Srivastava R., Chauhan S.K. et al. 2011, Molecular diversity and phylogeny in geographical collection of chickpea (Cicer sp.) accessions, J. Genet., 90, e94–100. [PubMed] [Google Scholar]
- 35.Deokar A.A., Ramsay L., Sharpe A.G. et al. 2014, Genome-wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly, BMC Genomics, 15, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho S.H., Kumar J., Shultz J.L., Anupama K., Tefera F., Muehlbauer F.J.. 2002, Mapping genes for double podding and other morphological traits in chickpea, Euphytica, 128, 285–92. [Google Scholar]
- 37.Anbessa Y., Warkentin T., Vandenberg A., Ball R.. 2006, Inheritance of time to flowering in chickpea in a short-season temperate environments, J. Hered., 97, 55–61. [DOI] [PubMed] [Google Scholar]
- 38.Cobos M.J., Winter P., Kharrat M. et al. 2009, Genetic analysis of agronomic traits in a wide cross of chickpea, Field Crops Res., 111, 130–6. [Google Scholar]
- 39.Aryamanesh N., Nelson M.N., Yan G., Clarke H.J., Siddique K.H.M.. 2010, Mapping a major gene for growth habit and QTLs for Ascochyta blight resistance and flowering time in a population between chickpea and Cicer reticulatum, Euphytica, 173, 307–19. [Google Scholar]
- 40.Jamalabadi J.G., Saidi A., Karami E., Kharkesh M., Talebi R.. 2013, Molecular mapping and characterization of genes governing time to flowering, seed weight, and plant height in an intraspecific genetic linkage map of chickpea (Cicer arietinum), Biochem. Genet., 51, 387–97. [DOI] [PubMed] [Google Scholar]
- 41.Laurie R.E., Diwadkar P., Jaudal M. et al. 2011, The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time, Plant Physiol., 156, 2207–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andres F., Coupland G.. 2012, The genetic basis of flowering responses to seasonal cues, Nat. Rev. Genet., 13, 627–39. [DOI] [PubMed] [Google Scholar]
- 43.Zhai H., Lu S., Liang S. et al. 2014, GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean, PLoS ONE, 9, e89030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weller J.L., Martínez R.O.. 2015, Genetic control of flowering time in legumes, Front. Plant Sci., 6, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smaczniak C., Immink R.G., Muiño J.M. et al. 2012, Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development, Proc. Natl. Acad. Sci. USA, 109, 1560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noguero M., Atif R.M., Ochatt S., Thompson R.D.. 2013, The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants, Plant Sci., 209, 32–45. [DOI] [PubMed] [Google Scholar]
- 47.Dreni L., Kater M.M.. 2014, MADS reloaded: evolution of the AGAMOUS subfamily genes, New Phytol., 201, 717–32. [DOI] [PubMed] [Google Scholar]
- 48.Lopez J.A., Sun Y., Blair P.B., Mukhtar M.S.. 2015, TCP three-way handshake: linking developmental processes with plant immunity, Trends Plant Sci., 20, 238–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.