Abstract
Curcumin (diferuloylmethane), a polyphenolic compound, is a component of Curcuma longa, commonly known as turmeric. It is a well-known anti-inflammatory, anti-oxidative, and anti-lipidemic agent and has recently been shown to modulate several diseases via epigenetic regulation. Many recent studies have demonstrated the role of epigenetic inactivation of pivotal genes that regulate human pathologies, such as neurocognitive disorders, inflammation, obesity, and cancers. Epigenetic changes involve changes in DNA methylation, histone modifications, or altered microRNA expression patterns which are known to be interconnected and play a key role in tumor progression and failure of conventional chemotherapy. The majority of epigenetic changes are influenced by lifestyle and diets. In this regard, dietary phytochemicals as dietary supplements have emerged as a promising source that are able to reverse these epigenetic alterations, to actively regulate gene expression and molecular targets that are known to promote tumorigenesis, and also to prevent age-related diseases through epigenetic modifications. There have been several studies which reported the role of curcumin as an epigenetic regulator in neurological disorders, inflammation, and in diabetes apart from cancers. The epigenetic regulatory roles of curcumin include (1) inhibition of DNA methyltransferases (DNMTs), which has been well defined from the recent studies on its function as a DNA hypomethylating agent; (2) regulation of histone modifications via regulation of histone acetyltransferases (HATs) and histone deacetylases (HDACs); and (3) regulation of micro RNAs (miRNA). This review summarizes the current knowledge on the effect of curcumin in the treatment and/or prevention of inflammation, neurodegenerative diseases, and cancers by regulating histone deacetylases, histone acetyltransferases, and DNA methyltransferases.
Keywords: Curcumin, Epigenetics, DNA methylation, Histone modification, Cancer, Nrf2
Introduction
Curcumin, commonly known as turmeric, is extracted from the rhizomes of the Curcuma longa plant. It has been the most commonly used spice in India for ages and is regularly used in Ayurveda and traditional Chinese medicine. Curcumin has many medicinal properties that have been studied widely by researchers all over the world [1, 2]. Curcumin constitutes 80 % of the curcuminoid complex, with the remainder constituted by demethoxycurcumin (17 %) and bisdemethoxycurcumin (3 %) [3]. Curcumin is a promising medicinal agent, as it regulates several key molecular signaling pathways that modulate survival, pathways governing anti-oxidative properties (e.g., nuclear factor E2-related factor 2, Nrf2), and inflammation pathways (e.g., nuclear factor kappa B, NF-κB) [4]. Apart from its role in regulating Nrf2 in different types of cancers, curcumin modulates Nrf2 expression in many different types of human pathologies, including neurocognitive disorders, kidney disorders, and diabetes. Curcumin is a well-known anti-inflammatory agent. It regulates the anti-inflammatory response by downregulating the enzymatic activities of cyclooxygenase (COX-2) and inducible nitric oxide synthase (iNOS) by suppressing the transcription factor NF-κB, which in turn suppresses tumorigenesis [5, 6]. Curcumin also downregulates the expression levels of NF-κB-regulated gene products, such as tumor necrosis factor (INF), 5-lipoxygenase (5-LOX), interleukins (IL-1, IL-6, IL-8), adhesion molecules, C-reactive protein (CRP), and chemokine receptor type 4 (CXCR-4) [7, 8].
Treating mouse liver and small intestine with curcumin, our group performed a global gene expression study and identified curcumin-regulated Nrf2-dependent genes. Our findings reveal the induction and suppression of several genes related to apoptosis and cell cycle control, cell adhesion, kinases, and phosphatases, along with transcription factors. The results showed many phase II detoxification/antioxidant enzyme genes regulated by Nrf2 among these identified genes, demonstrating the potential roles of Nrf2 and curcumin in chemo-prevention [9]. A recent study in rats identified curcumin as a neuroprotectant against hemin-induced damage in primary cultures of cerebellar granule neurons (CGNs), wherein the protective effect of curcumin was attenuated by the inhibition of the heme oxygenase system or glutathione (GSH) synthesis by tin mesoporphyrin and buthionine sulfoximine, respectively, in hemin-induced toxicity. Furthermore, after 24-h incubation with curcumin, glutathione reductase, glutathione S-transferase, and superoxide dismutase activities were increased by 1.4-, 2.3-, and 5.2-fold, respectively, suggesting that Nrf2 and an antioxidant response may play important roles in the protective effect of this antioxidant against hemin-induced neuronal death [10]. Curcumin is also being researched for its beneficial effects in diabetes: curcumin attenuated glucose intolerance without affecting high-fat diet (HFD)-induced body weight gain. Curcumin treatment reversed the levels of total or nuclear Nrf2 contents and its downstream target heme oxygenase-1, which were reduced by HFD feeding; this was seen as a result of induction in the nuclear translocation of Nrf2 by curcumin [11]. A study conducted to determine the molecular mechanisms involved in the putative anti-oxidative effects of curcumin against experimental stroke showed that curcumin protects neurons against ischemic injury. This neuroprotective effect involves the Akt/Nrf2 pathway, which is consistent with the fact that Nrf2 participates in the neuroprotective effects of curcumin against oxidative damage [12]. The role of Nrf2 in reno-protection has been reported in several studies. In a study of ischemia-reperfusion injury, Nrf2−/− mice showed significantly worse renal function, vascular permeability, and survival compared to wild-type mice. The streptozotocin (STZ)-induced diabetic nephropathy model revealed that Nrf2−/− mice developed severe renal injury with greater oxidative DNA damage than wild-type mice. Nrf2−/− mice showed higher renal damage and interstitial fibrosis by cyclosporin A treatment [13–15]. It was also reported that curcumin (100 mg/kg) administration significantly decreased infiltration of renal macrophages and renal production of pro-inflammatory cytokines, such as TNF-α and IL-1β, along with NF-κB inhibition in STZ diabetic animals [16]. A study showing attenuation of arsenic-induced hepatotoxicity and oxidative injuries upon curcumin treatment revealed that curcumin treatment relieved arsenic-induced elevation of serum alanine amino transferase (ALT) and aspartate aminotransferase (AST) activities, augmentation of hepatic malondialdehyde (MDA), and the reduction of blood and hepatic GSH levels via Nrf2 activation [17].
Epigenetic Regulation by Curcumin
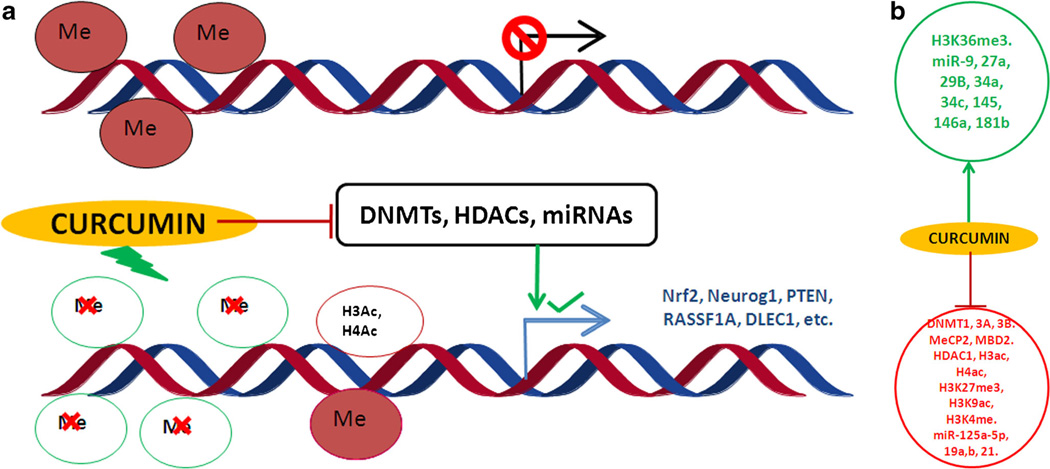
The studies discussed earlier have focused on the genetic modulation of various molecular aspects. However, epigenetic regulation—which includes changes in DNA methylation, histone modifications, and alterations in microRNA (miRNA) expression levels without any changes in the DNA sequence—constitutes an important mechanism by which dietary components, such as curcumin, can selectively activate or inactivate gene expression (Fig. 1a). DNA methylation and histone modifications are crucial epigenetic modifications of the genome that are involved in regulating many cellular processes, such as embryonic development, transcription, and chromatin structure and chromosome stability. DNA methylation represses transcription directly by inhibiting the binding of specific transcription factors and indirectly by recruiting methyl-CpG-binding proteins [18]. DNA methylation of several important genes has been reported in a wide variety of diseases, including several types of cancers, including those of the prostate, colon, and lung, where one or more genes are repressed due to hypermethylation [19]. DNA methylation also plays important roles in several other disorders, such as diabetes, neurocognitive diseases, autoimmune disorders, and inflammatory disorders [20]. While both DNA methylation and histone modification are involved in establishing patterns of gene repression during development, DNA methylation leads to stable long-term repression, whereas histone methylation causes local formation of heterochromatin that is readily reversible. Hence, understanding the cross-talk between DNA methylation and histone modification is paramount in understanding normal development, somatic cell reprogramming, and tumorigenesis [21]. There are different types of histone modifications, including methylation, acetylation, phosphorylation, ubiquitylation, and sumoylation, which regulate transcription, repair, replication, or condensation [22]. The regulatory role of each histone modification is governed by the sites that are methylated or acetylated. The histone acetyltransferases (HATs) and histone deacetylases (HDACs) are two families of enzymes and have opposing actions that regulate the acetylation of lysines, which is a highly dynamic process. There are two major classes of HATs: type-A and type-B, and four classes of HDACs: HDACs 1, 2, 3, and 4 [23]. In this review, we will examine the roles of curcumin in DNA methylation and different histone modifications in various diseases and will summarize the current knowledge on the effects of curcumin in the treatment and/or prevention of inflammation, neurodegenerative diseases, and cancers through its regulation on histone HDACs, HATs, and DNA methyltransferases.
Fig. 1.
a The effect of epigenetic changes on transcription of genes upon curcumin treatment, b Induced and suppressed histone-modifying enzymes, DNA methyl transferases, and microRNAs
DNA Methylation and Curcumin
Methylation of cytosine residues in 5′-cytosine–guanosine (CpG) in the promoter regions of genes is an epigenetic mechanism that controls gene transcription, genome stability, and genetic imprinting. Methylation is regulated by a group of enzymes called DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) in the presence of S-adenosyl-methionine (SAM); SAM serves as a methyl donor for the methylation of cytosine residues at the C-5 position to yield 5-methylcytosine. Several studies have demonstrated that the transcriptional silencing of several tumor suppressor genes (TSGs) is due to the aberrant hypermethylation of promoter CpG islands (>55 % CG content), leading to a variety of solid and blood cancers [24]. Treatment with hypomethylating agents both in vitro and in vivo is an effective way of restoring gene expression and normal patterns of differentiation and apoptosis in malignant cells. Bioactive components from dietary supplements, such as the potent hypomethylating agent curcumin [25, 26, 27••], may represent attractive agents for cancer prevention or treatment. The importance of methylation and its reversal by curcumin in different pathologies is reviewed.
Reversal of DNA Methylation by Curcumin in Different Disorders
As mentioned previously, DNA methylation of several important genes can be observed in a wide variety of diseases, including cancers of the prostate, colon, and lung, where one or more genes are repressed due to hypermethylation. DNA methylation is also known to play an important role in several other disorders, such as diabetes, neurocognitive diseases, autoimmune disorders, and inflammatory disorders.
Cancer epigenetics is characterized by heritable patterns of specific DNA methylation and disrupted cellular pathways mediated by DNA methyltransferases [28]. DNA methylation serves as an excellent target in the treatment of acute myeloid leukemia (AML) as it has been shown that inactivation of genes due to DNA methylation plays a major role in AML development. However, the mechanism by which curcumin elicits its DNA hypomethylation to reactivate silenced tumor suppressor genes and to present a potential treatment option for AML remains unclear. A study reported that curcumin downregulated DNMT1 expression in AML cell lines, both in vitro and in vivo, and in primary AML cells ex vivo. In addition, curcumin reduced the expression of the positive regulators of DNMT1, p65 and Sp1, which correlates with reductions in the binding of these transcription factors to the DNMT1 promoter in AML cell lines, demonstrating the promise of curcumin as a potential treatment for AML [29]. More recently, dimethoxycurcumin, a synthetic structural analog of curcumin, along with other epigenetic modifiers [e.g., 5-azacytidine (5AZA) and decitabine], was shown to augment the effect of DNMT inhibitors on DNA methylation to reactivate the silenced genes in leukemia cells [30]. In addition to its regulatory role in blood cancers, curcumin is a potent epigenetic regulatory agent in many solid tumors. As a potent DNMT inhibitor, curcumin at a concentration of 5 µM reverses CpG methylation at the promoter region of Neurog1, a cancer methylation marker in LNCaP human prostate cancer cells; at 2.5 µM, curcumin reverses the hypermethylation of Nrf2 in transgenic adenocarcinoma of mice prostate cells (TRAMP C1 cells) [26, 27••]. A study conducted in three colorectal cancer cell lines, HCT116, HT29, and RKO, showed non-specific global hypomethylation with 5-aza-CdR treatment. In contrast, curcumin treatment resulted in methylation changes at selected, partially methylated loci instead of fully methylated CpG sites [31]. A recent study showed that curcumin induced a decrease in the AgNOR (argyrophilic nucleolar) protein, whose expression is elevated in malignant cells compared to normal cells and reflects the rapidity rate of cancer cell proliferation. This change in expression may be mediated by the global DNA hypermethylation that is observed following low-concentration curcumin treatment [32]. Curcumin treatment also significantly inhibits cell proliferation and increases the apoptosis rate by up-regulating phosphatase tensin homolog (PTEN), whose expression is associated with decreased DNA methylation. In addition, curcumin treatment causes the suppression of DNA methyltransferase 3b (DNMT3b) both in vivo and in vitro [33]. In addition to pure curcumin, the novel curcumin analogs EF31 and UBS109 also show demethylating effects. In one study, MiaPaCa-2 and PANC-1 cells were treated with a vehicle control, curcumin, EF31, or UBS109. The analogs resulted in significantly higher inhibition of proliferation and cytosine methylation compared to curcumin. Demethylation was associated with the re-activation of silenced p 16, SPARC, E-cadherin, HSP-90, and NF-κB, leading to the downregulation of DNMT-1 expression [34]. A study using U251 and U81 glioblastoma cells also showed that treatment with 30 µM curcumin for 4 days decreased the promoter hypermethylation of the receptor activator of NF-κB (RANK), resulting in RANK gene expression and activation [35]. Furthermore, in a different study, curcumin treatment in MCF-7 cells enhanced the mRNA and protein levels of Ras-association domain family protein 1A (RASSF1A) and decreased its promoter methylation. This study also demonstrated that curcumin downregulated the DNA methylation activity of nuclear extracts and also downregulated the mRNA and protein levels of DNMT1 in MCF-7 cells. These effects may be associated with the disruption of binding between the NF-κB/Sp1 complex and the promoter region of DNMT1 induced by curcumin [36].
In addition to regulating cancer, curcumin also regulates inflammatory disorders through epigenetic modulation. Neprilysin (NEP, EP24.11), a zinc-dependent metallopeptidase, is expressed at relatively low levels in the brain due to its promoter hypermethylation [37]. NEP is emerging as a potent inhibitor of AKT/protein kinase B. One study showed that curcumin treatment induced the reactivation of the NEP gene via CpG demethylation. In curcumin-treated N2a/APPswe cells, the upregulation of NEP expression was also concomitant with AKT inhibition and the subsequent suppression of NF-κB and its downstream proinflammatory targets, including COX-2 and iNOS [38], Furthermore, the Fanconi anemia (FA) complementation group F (FANCF) gene plays an important role in the FA pathway and is frequently inactivated due to promoter hypermethylation in specific types of cancer [39]. Curcumin treatment produces a 5-fold increase in FANCF gene expression due to the demethylation of 12 of the 15 CpGs present in the promoter region [40]. In a study in non-small cell lung cancer cells comparing the effects of three different curcuminoids in demethylating the promoter region of Wnt inhibitory factor (WIF-1), bisdemethoxycurcumin possessed a strong demethylation function in vitro compared to the other two curcuminoids in restoring WIF-1 expression in A549 cells [41]. A recent study conducted in our laboratory demonstrated that curcumin decreased the protein expression levels of DNA methyltransferases (DNMT1, 3A, and 3B) and induced the demethylation of Deleted in lung and esophageal cancer 1 (DLEC1) in human colorectal adenocarcinoma HT-29 cells. The results also showed that curcumin treatment decreased colony formation; however, the effect of curcumin was attenuated in DLEC1 knockdown cells (Guo et al., manuscript submitted). A study using MCF7 and MDA MB231 breast cancer cells showed that curcumin treatment decreased the expression levels of DNMT1, 3A, and 3B and increased p21WAF1 expression [42].
Histone Modifications and Curcumin
Epigenetic alterations might occur as early events in carcinogenesis and might precede genetic alterations during oncogenic transformation [43]. The activation or repression of a gene is not determined by histone acetylation or methylation alone. Rather, it is determined by the modification of different residues. However, promoter DNA methylation generally suppresses gene expression. A histone octamer consisting of an H3/H4 tetramer and two H2A/H2B dimers wraps DNA, forming the nucleosome. Generally, histone modifications play a key role in maintaining the highly folded chromatin structure, which is closely linked to gene expression [44–46]. Histone acetylation in chromatin is a very widely studied histone modification. The different groups of histone-modifying enzymes that play crucial roles in histone acetylation are (1) HATs that add acetyl groups to lysine residues, (2) HDACs or lysine deacetylases (KDACs) that remove acetyl groups from lysine residues, (3) histone methyltransferases (HMTs) that add methyl groups to arginine or lysine residues, and (4) histone demethylases (HDMs) that remove methyl groups from arginine or lysine residues, in turn regulating the conformation of chromatin structure to facilitate or hinder the association of DNA repair proteins or transcription factors to chromatin [47, 48]. Collectively, the aberrant enrichment of HAT and HDAC activities may trigger carcinogenesis [49]. The different residues that are modified could play active roles in either activation or repression. For instance, trimethylation of lysine 4 on histone H3 (H3K4me3) results in transcriptional gene activation, whereas trimethylation of lysine 9 on histone H3 (H3K9me3) and trimethylation of lysine 27 on histone H3 (H3K27me3) at gene promoters repress transcription [47]. However, acetylation increases chromatin accessibility and is generally associated with transcriptional activation, as it neutralizes the DNA– histone interactions, resulting in a relaxed, open chromatin conformation that facilitates access of transcriptional activators to their cognate recognition elements and initiates/ enhances transcription.
Regulation of Histone Modifications by Curcumin
Curcumin, a HAT inhibitor, significantly reduces H3ac levels in the IL-6 promoter, along with IL-6 mRNA expression and IL-6 protein secretion by rheumatoid arthritis synovial fibroblasts RASFs. Thus, hyperacetylation of histone H3 in the IL-6 promoter induces the increase in IL-6 production by RASFs and thereby participates in rheumatoid arthritis (RA) pathogenesis [50]. One study showed that the anti-inflammatory activity of curcumin is regulated by epigenetically inhibiting the expression of Triggering receptor expressed on myeloid cells 1 (TREM-1), a potent amplifier of Toll-like receptor (TLR)-initiated inflammatory responses. The inhibition was observed both in vitro in primary bone marrow-derived macrophages and in vivo in the lungs of mice with sepsis. Curcumin inhibited p300 activity in the TREM-1 promoter region, leading to the hypoacetylation of lysine residues in histones 3 and 4 (H3Kac and H4Kac) [51]. In another study, the effect of curcumin on histone acetylation and proinflammatory cytokine secretion under high-glucose conditions in human monocytes was studied using human monocytic (THP-1) cells. The results showed that curcumin treatment not only significantly reduced HAT activity and the level of p300, a co-activator of NFκB and acetylated CBP/p300 gene expression, but also induced HDAC2 expression. The results indicate that curcumin decreases high glucose-induced cytokine production in monocytes via epigenetic changes involving NF-κB [52]. Histone acetylation and HDAC regulation have been implicated as playing important roles in Alzheimer’s disease (AD) [53–55]. In one study, HDAC alterations to histone acetylation at the promoter regions of AD-related genes, such as Presenilin 1 (PS 1) and beta-site amyloid precursor protein cleaving enzyme 1 (BACE1), were studied in neuroblastoma N2a cells. To establish the cellular models of AD, the cells were transfected with Swedish mutated human amyloid precursor protein (APP) (N2a/APPswe) and wild-type APP (N2a/APPwt). Curcumin, a natural selective inhibitor of p300 in HATs, significantly suppressed PS1 and BACE1 expression by inhibiting H3 acetylation at their promoter regions in N2a/APPswe cells [56]. When the combinatorial effects of SAHA and curcumin were investigated as a protective treatment against amyloid-beta neurotoxicity in vitro, the combination provided comprehensive protection against Aβ25–35-induced neuronal damage in PC12 cells via HDAC regulation [57]. The epigenetic regulatory role of curcumin was recently studied in the management of neuropathic pain in a chronic construction injury model in rats. Chromatin immunoprecipitation (ChIP) analysis revealed the dose-dependent reduction in the recruitment of p300/CBP (CREB binding protein) and acetyl-histone H3/acetyl-histone H4 to the promoters of the BDNF and COX-2 genes upon curcumin administration, indicating the therapeutic role of curcumin in neuropathic pain management [58]. In addition to this effect, it has previously been reported that curcumin plays a key role as a p300/CBP inhibitor of histone acetyl-transferase [59, 60]. Lipopolysaccharide (LPS)-induced cardiac hypertrophy is known to accentuate p300 transcriptional activity, and LPS-mediated cardiac hypertrophy can be reversed by blocking p300-HAT activity upon treatment with curcumin at a dose of 100 µg/kg of body weight in mice [61]. A recent study revealed that diabetic rats treated with curcumin had significantly decreased blood urea nitrogen and creatinine and increased albumin; these variables are associated with the development of diabetic nephropathy These results demonstrated that curcumin induced post-translational modifications of histone H3, expression of HSP-27, and MAP kinase p38 in diabetic kidneys [62]. Studies on the anti-cancer potential of curcumin in medulloblastoma reported that curcumin treatment of medulloblastoma cells induced apoptosis and cell cycle arrest at the G2/M phase. Curcumin also reduced histone deacetylase (HDAC) 4 expression and activity while increasing tubulin acetylation, indicating its potential therapeutic role in medulloblastoma [63]. Neurogl, a gene that is epigenetically regulated and is considered an important biomarker in prostate cancer, is upregulated upon treatment with curcumin in LNCaP cells by increasing HDAC1, 4, 5, and 8 expression while decreasing HDAC3 expression. Curcumin also decreases the enrichment of H3K27me3 at both the Neurogl promoter region and the global level [26], In addition, DLEC1 expression is regulated by HDAC inhibition in human colorectal adenocarcinoma HT-29 cells upon curcumin treatment. Finally, curcumin inhibits subtypes of HDACs (i.e., HDACs 4, 5, 6, and 8) (under review, Guo et al. Biochemical Pharmacology).
Regulatory Role of Curcumin in miRNA Expression
MicroRNAs (miRNAs) are small non-coding RNA molecules ranging from 17 to 25 nucleotides in length that are endogenously expressed; they induce target mRNA degradation or repress mRNA translation by imperfect binding to their 3′-untranslated region. The RNA polymerase II, Drosha, transcribes miRNA gene into a primary transcript (pri-miRNA) in the nucleus, where the microprocessing complex, DGCR8, processes the hairpin stem-loop structure into a precursor miRNA (pre-miRNA). The 70-nucleotide-long pre-miRNA is then exported into the cytoplasm, where it undergoes a second processing by Dicer, where one strand of the hairpin is incorporated into the ribonucleoprotein complex called the miRNA-induced silencing complex (miRNA-RISC) [64, 65]. Studies have shown that miRNA control the self-renewal and differentiation of embryonic stem cells (ESCs), and the aberrant expression and/or functions of miRNAs have been implicated in tumorigenesis [66]. A significant proportion of miRNA genes (20–40 %) are located close to CpG islands, and these miRNAs can be inactivated by epigenetic mechanisms [67]. Some of the signaling pathways that are regulated by miRNAs include the vascular endothelial growth factor (VEGF), NF-κB, protein kinase B, and mitogen-activated protein kinase (MAPK) pathways, all of which are known to be targets of curcumin [68]. miRNAs represent very promising therapeutic targets for cancer treatment because their aberrant expression has been linked to cancer stem cell dysregulation and, thus, oncogenesis [69]. The function of histone lysine N-methyltransferase (EZH2) in pancreatic cancer was studied in pancreatic cancer cells using difluorinated curcumin (CDF), a novel analog of curcumin. The results revealed decreased EZH2 expression and increased expression of a panel of tumor-suppressor microRNAs (e.g., let-7a, b, c, and d; miR-26a; miR-101; miR-146a; and miR-200b and c), which are most commonly lost in pancreatic cancer [70]. In addition, studies have shown that curcumin treatment alters miRNA expression in the PxBC-3 human pancreatic cancer cell line. For example, in one study, curcumin upregulated miRNA-22 and downregulated miRNA-199a expression. The upregulation of miRNA-22 expression by either curcumin or transfection with miRNA-22 mimetics in the PxBC-3 pancreatic cancer cell line suppressed the expression of its target genes, SP1 transcription factor (SP1) and estrogen receptor 1 (ESR1). Conversely, inhibiting miRNA-22 with antisense enhanced SP1 and ESR1 expression [71]. Another study showed that treatment with curcumin and its analog, CDF, upregulated miR-200 expression and downregulated miR-21 expression, and the downregulation of miR-21 resulted in PTEN induction [72]. miR-203 is often hypermethylated in bladder cancers; curcumin induces the hypomethylation of the miR-203 promoter and subsequently upregulates miR-203 expression. As a result of increased miR-203 expression, the miR-203 target genes Akt2 and Src are downregulated, which leads to a decrease in proliferation and an increase in bladder cancer cell apoptosis [73]. A global miRNA expression profiling study performed in human ARPE-19 cells treated with curcumin showed that curcumin alters the expression of H2O2-modulated miRNAs that are known to be regulators of antioxidant defense and the renin–angiotensin system. These results were confirmed using RT-PCR and western blot. Compared with controls, cells treated with curcumin alone downregulated 20 miRNAs and upregulated nine miRNAs. While exposure to H2O2 downregulated 18 miRNAs and up-regulated 29 miRNAs, cells that had been pre-treated with curcumin and were later exposed to H2O2 showed a significant reduction in the H2O2-induced expression of 17 miRNAs [74]. In an engrafting melanoma model developed by injecting murine B78H1 cells in the flanks of C57BL/6 mice, oral curcumin treatment was administered through a 4 % curcumin diet. The results showed that curcumin intake altered miRNA expression, with mmu-miR-205-5p being expressed over 100-fold higher than controls [75].
Conclusion
The aberrant activation or inactivation of harmful and beneficial genes by epigenetic alterations has gained much focus in recent years. Epigenetic regulation, which includes changes in DNA methylation, and histone modifications and alterations in microRNA (miRNA) expression without any changes in the DNA sequence, constitutes an important mechanism by which dietary components, such as curcumin, can selectively activate or inactivate gene expression (Fig. 1b). The role of curcumin in regulating different disorders has been long studied. Contemporary research shows that curcumin can epigenetically regulate the expression of important genes by reversing DNA methylation and altering histone modifications and by targeting several miRNAs that play a key role in diseases (Table 1). Several studies have shown the potential of curcumin in regulating various intracellular pathways, thereby attenuating certain diseases, including cancers, diabetic nephropathy, and neurocognitive disorders (e.g., Alzheimer’s disease). Additionally, its potency can also be attributed to the regulation of epigenetic mechanisms, as has been shown recently. Furthermore, several studies have shown that curcumin is a potent DNMT and HDAC inhibitor and can regulate miRNA expression in several disorders. Therefore, it is imperative to identify more epigenetic targets of curcumin for the treatment of other disorders.
Table 1.
Examples of the effect of curcumin on DNA methylation, histone modifications, and miRNA activity
| Dietary agent | Molecular mechanism | Validated target(s) | Experimental model | Concentration/do se | Treatment duration |
References | |
|---|---|---|---|---|---|---|---|
| DNA methylation | Curcumin | ↓ Methylation in promoter region | Nrf2 | TRAMPC1 mouse prostate cells | 2.5 µM | 5 days | [27••] |
| Curcumin | ↓ Methylation in promoter region | Neurogl | LNCaP human prostate cancer cells | 5 µM | 7 days | [26] | |
| Curcumin | ↓ DNMT3B | PTEN | HSC primary cells (RAT) | 20 µM | 2 days | [33] | |
| EF31 (curcumin analogs) | ↓ DNMT1 | pi6, SPARC, and E-cadherin |
MiaPaCa-2 and PANC-1 pancreatic cancer cells |
750 nM and 1.25 µM |
2 days | [34] | |
| UBS109 | ↓ DNMT1 | 250 nM | 2 days | [34] | |||
| Curcumin | ↓ Methylation in promoter region | RANK | U87 and U251 glioblastoma cells | 30 µM | 4 days | [35] | |
| Curcumin | ↓ DNMT1, 3A, 3B, ↓ MeCP2, ↓MBD2 |
p21WAFl | MCF 7 and MDA MB 231 cells | 10 µM | 4 days | [42] | |
| Curcumin | ↓ DNMT1 | p15 (INK4B) | K562, THP1, Kasumi and MV4-11 cells, primary AML cells |
10 and 20 µM | 1 and 3 days | [29] | |
| Curcumin | ↓ Methylation in promoter region | FANCF | SiHa cell line | 20 µM | 4 days | [40] | |
| Curcumin | ↓ Methylation in promoter region | RASSF1A | MCF 7 cells | 10 and 20 µM | 3 days | [36] | |
| Histone modifications | Curcumin | ↓ Total HDAC activity, ↓ H3K27me3 | Neurogl | LNCaP human prostate cancer cells | 5 µM | 7 days | [26] |
| Dimethoxy curcumin | ↓H3K36me3 | p15andCDHl | CEM human leukemia cells | 1 µM | 3 days | [30] | |
| Curcumin | ↓ HDAC1 | p300 | Raji cells (human Burkitt’s lymphoma) | 12.5 µmol/1 | 6-48 h | [76] | |
| Curcumin | ↓ H3ac and H4ac | PARP–caspase 3 | A172 (human glioblastoma cell lines) | 4 µg/ml | 4 days | [77] | |
| Curcumin | P300/CBP HAT inhibition | BDNF and COX-2 | Chronic construction injury rat model (CCI) |
40 and 60 mg/kg | 7 days | [58] | |
| Curcumin | ↓ H3K4me, ↓ H3ac and H4ac | TREM-1 | BMDM (macrophages from WT mice) | 10 µmol/1 | 1–4 h | [51] | |
| Curcumin | ↓ HDAC 1 | p21WAFl | MCF 7 and MDA MB 231 cells | 10 µM | 4 days | [42] | |
| Curcumin | ↓ H3K9Ac | NKG2D | Jurkat and HUT78 cells | 4 µM | 1 day | [78] | |
| Curcumin | ↓ H3ac | IL-6 | RA synovial fibroblasts | 20 µM | 8h | [50] | |
| Curcumin | ↓ H3ac | PS1,BACE1 | N2a cells neuroblastoma cells | 10 µM | 2 days | [56] | |
| microRNAs | Curcumin | ↑ miR-29B | PTEN (by decreasing DNMT3B) |
HSC-T6 cells | 20 µM | 2 days | [33] |
| CDF (difluorinated curcumin) | ↑ miR-34a and miR-34c | Notch-1 | SW620 colon cancer cell line; archival formalin-fixed paraffin-embedded tissues from normal colonic mucosa and colon tumors |
100 µM | 3 days | [79] | |
| Curcumin | ↓ miR-125a-5p | Tumor protein 53 (TP53) |
HONE 1 and HK1 nasopharyngeal carcinoma cell lines |
16 and 36 µM | 3 days | [80••] | |
| Curcumin | ↓ miR-19 a and ↓ miR-19B | PTEN, p-AKT, P-MDM2, p53 |
MCF7 | 1 µM | 4 days | [81] | |
| CDF (difluorinated curcumin) | ↑ miR-146a | EGFR | Human pancreatic cancer cell lines | 200-500 nM | 4 days | [82] | |
| Curcumin | ↑ miR-181b | CXCL1 and CXCL2 | MDA-MB-231 | 25 µM | 1 day | [83] | |
| Curcumin | ↑ miR-9 | Akt, FOXO1 | SKOV3 ovarian cancer cell lines | 10-60 µM | 3 days | [84] | |
| Curcuminoids | ↑ miR-27a | ZBTB10 | SW-480 and HT-29 | 2.5-10 µg/ml | 1 day | ||
| EF24 (curcumin analog) | | miR-21 | PTEN and PDCD4 | DU145 and B16 murine melanoma cells |
5 µM | 1 day | [85] | |
| Curcumin | ↑ miR-145 | I1-6 and sIL-6R | Head and neck cancer cells | 10 or 20 µmol/l | 1 day | [86] |
Acknowledgments
We thank all members of Kong laboratory for their help in the preparation of this review. This work was supported by R01AT007065 from the National Center for Complementary and Alternative Medicines (NCCAM) and the Office of Dietary Supplements (ODS).
Footnotes
Compliance with Ethics Guidelines.
Conflict of Interest Sarandeep S. S. Boyanapalli and Ah-Ng. Tony Kong declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently have been highlighted as:
•• Of major importance
- 1.Aggarwal BB, Sung B. Pharmacological basis for the role of curcumin in chronic diseases: an age-old spice with modern targets. Trends Pharmacol Sci. 2009;30(2):85–94. doi: 10.1016/j.tips.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Ammon HP, Wahl MA. Pharmacology of Curcuma longa. Planta Med. 1991;57(1):1–7. doi: 10.1055/s-2006-960004. [DOI] [PubMed] [Google Scholar]
- 3.Lao CD, Ruffin MT, Normolle D, Heath DD, Murray SI, Bailey JM, et al. Dose escalation of a curcuminoid formulation. BMC Complement Altern Med. 2006;6:10. doi: 10.1186/1472-6882-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65(11):1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surh YJ, Chun KS, Cha HH, Han SS, Keum YS, Park KK, et al. Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: down-regulation of COX-2 and iNOS through suppression of NF-kappa B activation. Mutat Res. 2001;480-481:243–268. doi: 10.1016/s0027-5107(01)00183-x. [DOI] [PubMed] [Google Scholar]
- 6.Jobin C, Bradham CA, Russo MP, Juma B, Narula AS, Brenner DA, et al. Curcumin blocks cytokine-mediated NF-kappa B activation and proinflammatory gene expression by inhibiting inhibitory factor I-kappa B kinase activity. J Immunol. 1999;163(6):3474–3483. [PubMed] [Google Scholar]
- 7.Skommer J, Wlodkowic D, Pelkonen J. Gene-expression profiling during curcumin-induced apoptosis reveals downregulation of CXCR4. Exp Hematol. 2007;35(1):84–95. doi: 10.1016/j.exphem.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Rao CV. Regulation of COX and LOX by curcumin. Adv Exp Med Biol. 2007;595:213–226. doi: 10.1007/978-0-387-46401-5_9. [DOI] [PubMed] [Google Scholar]
- 9.Shen G, Xu C, Hu R, Jain MR, Gopalkrishnan A, Nair S, et al. Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol Cancer Ther. 2006;5(1):39–51. doi: 10.1158/1535-7163.MCT-05-0293. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Reyes S, Guzman-Beltran S, Medina-Campos ON, Pedraza-Chaverri J. Curcumin pretreatment induces Nrf2 and an antioxidant response and prevents hemin-induced toxicity in primary cultures of cerebellar granule neurons of rats. Oxidative Med Cell Longev. 2013;2013:801418. doi: 10.1155/2013/801418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He HJ, Wang GY, Gao Y, Ling WH, Yu ZW, Jin TR. Curcumin attenuates Nrf2 signaling defect, oxidative stress in muscle and glucose intolerance in high fat diet-fed mice. World J Diabetes. 2012;3(5):94–104. doi: 10.4239/wjd.v3.i5.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Li Q, Wang X, Yu S, Li L, Wu X, et al. Neuroprotection by curcumin in ischemic brain injury involves the Akt/Nrf2 pathway. PLoS One. 2013;8(3):e59843. doi: 10.1371/journal.pone.0059843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M, Grigoryev DN, Crow MT, Haas M, Yamamoto M, Reddy SP, et al. Transcription factor Nrf2 is protective during ischemic and nephrotoxic acute kidney injury in mice. Kidney Int. 2009;76(3):277–285. doi: 10.1038/ki.2009.157. [DOI] [PubMed] [Google Scholar]
- 14.Shin DH, Park HM, Jung KA, Choi HG, Kim JA, Kim DD, et al. The NRF2-heme oxygenase-1 system modulates cyclosporin A-induced epithelial-mesenchymal transition and renal fibrosis. Free Radic Biol Med. 2010;48(8):1051–1063. doi: 10.1016/j.freeradbiomed.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang T, Huang Z, Lin Y, Zhang Z, Fang D, Zhang DD. The protective role of Nrf2 in streptozotocin-induced diabetic nephropathy. Diabetes. 2010;59(4):850–860. doi: 10.2337/db09-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soetikno V, Sari FR, Veeraveedu PT, Thandavarayan RA, Harima M, Sukumaran V, et al. Curcumin ameliorates macrophage infiltration by inhibiting NF-kappaB activation and proinflammatory cytokines in streptozotocin induced-diabetic nephropathy. Nutr Metab. 2011;8(1):35. doi: 10.1186/1743-7075-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao S, Duan X, Wang X, Dong D, Liu D, Li X, et al. Curcumin attenuates arsenic-induced hepatic injuries and oxidative stress in experimental mice through activation of Nrf2 pathway, promotion of arsenic methylation and urinary excretion. Food Chem Toxicol. 2013;59:739–747. doi: 10.1016/j.fct.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Robertson KD. DNA methylation and chromatin—unraveling the tangled web. Oncogene. 2002;21(35):5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 19.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19(11):7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 21.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009;19(3):706–709. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 26.Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, et al. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurogl by curcumin in prostate LNCaP cells. AAPS J. 2011;13(4):606–614. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khor TO, Huang Y, Wu TY, Shu L, Lee J, Kong AN. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol. 2011;82(9):1073–1078. doi: 10.1016/j.bcp.2011.07.065.Demethylation of Nrf2 was found to be associated with the re-expression of Nrf2 and one of its downstream target gene, NQO-1, one of the major anti-oxidative stress enzymes, both at the mRNA and protein levels. Taken together, our current study suggests that CUR can elicit its prostate cancer chemopreventive effect, potentially at least in part, through epigenetic modification of the Nrf2 gene with its subsequent induction of the Nrf2-mediated anti-oxidative stress cellular defense pathway.
- 28.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Peng Y, Wu LC, Xie Z, Deng Y, Hughes T, et al. Curcumin down-regulates DNA methyltransferase 1 and plays an anti-leukemic role in acute myeloid leukemia. PLoS One. 2013;8(2):e55934. doi: 10.1371/journal.pone.0055934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan HE, Carlson S, Abdallah I, Buttolph T, Glass KC, Fandy TE. Curcumin and dimethoxycurcumin induced epigenetic changes in leukemia cells. Pharm Res. 2014 doi: 10.1007/s11095-014-1502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Link A, Balaguer F, Shen Y, Lozano JJ, Leung HC, Boland CR, et al. Curcumin modulates DNA methylation in colorectal cancer cells. PLoS One. 2013;8(2):e57709. doi: 10.1371/journal.pone.0057709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewinska A, Adamczyk J, Pajak J, Stoklosa S, Kubis B, Pastuszek P, et al. Curcumin-mediated decrease in the expression of nucleolar organizer regions in cervical cancer (HeLa) cells. Mutat Res Genet Toxicol Environ Mutagen. 2014;771:43–52. doi: 10.1016/j.mrgentox.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Zheng J, Wu C, Lin Z, Guo Y, Shi L, Dong P, et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation—a novel mechanism suppressing liver fibrosis. FEBS J. 2014;281(1):88–103. doi: 10.1111/febs.12574. [DOI] [PubMed] [Google Scholar]
- 34.Nagaraju GP, Zhu S, Wen J, Farris AB, Adsay VN, Diaz R, et al. Novel synthetic curcumin analogues EF31 and UBS109 are potent DNA hypomethylating agents in pancreatic cancer. Cancer Lett. 2013;341(2):195–203. doi: 10.1016/j.canlet.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Wu B, Yao X, Nie X, Xu R. Epigenetic reactivation of RANK in glioblastoma cells by curcumin: involvement of STAT3 inhibition. DNA Cell Biol. 2013;32(6):292–297. doi: 10.1089/dna.2013.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du L, Xie Z, Wu LC, Chiu M, Lin J, Chan KK, et al. Reactivation of RASSF1A in breast cancer cells by curcumin. Nutr Cancer. 2012;64(8):1228–1235. doi: 10.1080/01635581.2012.717682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen KL, Wang SS, Yang YY, Yuan RY, Chen RM, Hu CJ. The epigenetic effects of amyloid-beta(1-40) on global DNA and neprilysin genes in murine cerebral endothelial cells. Biochem Biophys Res Commun. 2009;378(1):57–61. doi: 10.1016/j.bbrc.2008.10.173. [DOI] [PubMed] [Google Scholar]
- 38.Deng Y, Lu X, Wang L, Li T, Ding Y, Cao H, et al. Curcumin inhibits the AKT/NF-kappaB signaling via CpG demethylation of the promoter and restoration of NEP in the N2a cell line. AAPS J. 2014;16(4):649–657. doi: 10.1208/s12248-014-9605-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9(5):568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 40.Parashar G, Parashar NC, Capalash N. Curcumin causes promoter hypomethylation and increased expression of FANCF gene in SiHa cell line. Mol Cell Biochem. 2012;365(1–2):29–35. doi: 10.1007/s11010-012-1240-z. [DOI] [PubMed] [Google Scholar]
- 41.Liu YL, Yang HP, Gong L, Tang CL, Wang HJ. Hypomethylation effects of curcumin, demethoxycurcumin and bisdemethoxycurcumin on WIF-1 promoter in non-small cell lung cancer cell lines. Mol Med Rep. 2011;4(4):675–679. doi: 10.3892/mmr.2011.473. [DOI] [PubMed] [Google Scholar]
- 42.Mirza S, Sharma G, Parshad R, Gupta SD, Pandya P, Ralhan R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J Breast Cancer. 2013;16(1):23–31. doi: 10.4048/jbc.2013.16.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nephew KP, Huang TH. Epigenetic gene silencing in cancer initiation and progression. Cancer Lett. 2003;190(2):125–133. doi: 10.1016/s0304-3835(02)00511-6. [DOI] [PubMed] [Google Scholar]
- 44.Berlowitz L, Pallotta D. Acetylation of nuclear protein in the heterochromatin and euchromatin of mealy bugs. Exp Cell Res. 1972;71(1):45–48. doi: 10.1016/0014-4827(72)90261-3. [DOI] [PubMed] [Google Scholar]
- 45.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 46.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128(4):651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010;245(3):378–393. doi: 10.1016/j.taap.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang K, Dent SY. Histone modifying enzymes and cancer: going beyond histones. J Cell Biochem. 2005;96(6):1137–1148. doi: 10.1002/jcb.20615. [DOI] [PubMed] [Google Scholar]
- 49.Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin Exp Metastasis. 2008;25(2):183–189. doi: 10.1007/s10585-007-9131-5. [DOI] [PubMed] [Google Scholar]
- 50.Wada TT, Araki Y, Sato K, Aizaki Y, Yokota K, Kim YT, et al. Aberrant histone acetylation contributes to elevated interleukin-6 production in rheumatoid arthritis synovial fibroblasts. Biochem Biophys Res Commun. 2014;444(4):682–686. doi: 10.1016/j.bbrc.2014.01.195. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Z, Syed MA, Panchal D, Rogers D, Joo M, Sadikot RT. Curcumin mediated epigenetic modulation inhibits TREM-1 expression in response to lipopolysaccharide. Int J Biochem Cell Biol. 2012;44(11):2032–2043. doi: 10.1016/j.biocel.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 52.Yun JM, Jialal I, Devaraj S. Epigenetic regulation of high glucose-induced proinflammatory cytokine production in monocytes by curcumin. J Nutr Biochem. 2011;22(5):450–458. doi: 10.1016/j.jnutbio.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Epigenetics MG. A role for epigenetics in cognition. Science. 2010;329(5987):27. doi: 10.1126/science.329.5987.27. [DOI] [PubMed] [Google Scholar]
- 54.Xu K, Dai XL, Huang HC, Jiang ZF. Targeting HDACs: a promising therapy for Alzheimer’s disease. Oxidative Med Cell Longev. 2011;2011:143269. doi: 10.1155/2011/143269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35(4):870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu X, Deng Y, Yu D, Cao H, Wang L, Liu L, et al. Histone acetyl-transferase p300 mediates histone acetylation of PS1 and BACE1 in a cellular model of Alzheimer’s disease. PLoS One. 2014;9(7):el03067. doi: 10.1371/journal.pone.0103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meng J, Li Y, Camarillo C, Yao Y, Zhang Y, Xu C, et al. The antitumor histone deacetylase inhibitor SAHA and the natural flavonoid curcumin exhibit synergistic neuroprotection against amyloid-beta toxicity. PLoS One. 2014;9(1):e85570. doi: 10.1371/journal.pone.0085570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu X, Li Q, Chang R, Yang D, Song Z, Guo Q, et al. Curcumin alleviates neuropathic pain by inhibiting p300/CBP histone acetyl-transferase activity-regulated expression of BDNF and cox-2 in a rat model. PLoS One. 2014;9(3):e91303. doi: 10.1371/journal.pone.0091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morimoto T, Sunagawa Y, Kawamura T, Takaya T, Wada H, Nagasawa A, et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008;118(3):868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puglia C, Frasca G, Musumeci T, Rizza L, Puglisi G, Bonina F, et al. Curcumin loaded NLC induces histone hypoacetylation in the CNS after intraperitoneal administration in mice. Eur J Pharm Biopharm. 2012;81(2):288–293. doi: 10.1016/j.ejpb.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 61.Chowdhury R, Nimmanapalli R, Graham T, Reddy G. Curcumin attenuation of lipopolysaccharide induced cardiac hypertrophy in rodents. ISRN Inflamm. 2013;2013:539305. doi: 10.1155/2013/539305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tikoo K, Meena RL, Kabra DG, Gaikwad AB. Change in post-translational modifications of histone H3, heat-shock protein-27 and MAP kinase p38 expression by curcumin in streptozotocin-induced type I diabetic nephropathy. Br J Pharmacol. 2008;153(6):1225–1231. doi: 10.1038/sj.bjp.0707666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SJ, Krauthauser C, Maduskuie V, Fawcett PT, Olson JM, Rajasekaran SA. Curcumin-induced HDAC inhibition and attenuation of medulloblastoma growth in vitro and in vivo. BMC Cancer. 2011;11:144. doi: 10.1186/1471-2407-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 65.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 67.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sarkar FH, Li Y. Cell signaling pathways altered by natural chemo-preventive agents. Mutat Res. 2004;555(1-2):53–64. doi: 10.1016/j.mrfmmm.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 69.Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and sternness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bao B, Ali S, Banerjee S, Wang Z, Logna F, Azmi AS, et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72(1):335–345. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7(3):464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 72.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, et al. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70(9):3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Saini S, Arora S, Majid S, Shahryari V, Chen Y, Deng G, et al. Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev Res. 2011;4(10):1698–1709. doi: 10.1158/1940-6207.CAPR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Howell JC, Chun E, Farrell AN, Hur EY, Caroti CM, Iuvone PM, et al. Global microRNA expression profiling: curcumin (diferuloylmethane) alters oxidative stress-responsive microRNAs in human ARPE-19 cells. Mol Vis. 2013;19:544–560. [PMC free article] [PubMed] [Google Scholar]
- 75.Dahmke IN, Backes C, Rudzitis-Auth J, Laschke MW, Leidinger P, Menger MD, et al. Curcumin intake affects miRNA signature in murine melanoma with mmu-miR-205-5p most significantly altered. PLoS One. 2013;8(12):e81122. doi: 10.1371/journal.pone.0081122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y, Shu W, Chen W, Wu Q, Liu H, Cui G. Curcumin, both histone deacetylase and p300/CBP-specific inhibitor, represses the activity of nuclear factor kappa B and Notch 1 in Raji cells. Basic Clin Pharmacol Toxicol. 2007;101(6):427–433. doi: 10.1111/j.1742-7843.2007.00142.x. [DOI] [PubMed] [Google Scholar]
- 77.Kang SK, Cha SH, Jeon HG. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006;15(2):165–174. doi: 10.1089/scd.2006.15.165. [DOI] [PubMed] [Google Scholar]
- 78.Fernandez-Sanchez A, Baragano Raneros A, Carvajal Palao R, Sanz AB, Ortiz A, Ortega F, et al. DNA demethylation and histone H3K9 acetylation determine the active transcription of the NKG2D gene in human CD8+ T and NK cells. Epigenetics. 2013;8(1):66–78. doi: 10.4161/epi.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58. doi: 10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gao W, Chan JY, Wong TS. Curcumin exerts inhibitory effects on undifferentiated nasopharyngeal carcinoma by inhibiting the expression of miR-125a-5p. Clin Sci. 2014;127(9):571–579. doi: 10.1042/CS20140010.Primary nasopharyngeal carcinoma (NPC) exhibited a significantly higher expression level of miR-125a-5p than healthy controls. miR-125a-5p inhibited the expression of tumour protein 53 (TP53), and curcumin treatment upregulated the expression of TP53. Taken together, these results indicate that curcumin exerted inhibitory effects on NPC by inhibiting the expression of miR-125a-5p and, subsequently, enhancing the expression of’TP53.
- 81.Li X, Xie W, Xie C, Huang C, Zhu J, Liang Z, et al. Curcumin modulates miR-19/PTEN/AKT/p53 axis to suppress bisphenol A- induced MCF-7 breast cancer cell proliferation. Phytother Res. 2014;28(10):1553–1560. doi: 10.1002/ptr.5167. [DOI] [PubMed] [Google Scholar]
- 82.Ali S, Ahmad A, Aboukameel A, Ahmed A, Bao B, Banerjee S, et al. Deregulation of miR-146a expression in a mouse model of pancreatic cancer affecting EGFR signaling. Cancer Lett. 2014;351(1):134–142. doi: 10.1016/j.canlet.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kronski E, Fiori ME, Barbieri O, Astigiano S, Mirisola V, Killian PH, et al. miR1 81b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL1 and −2. Mol Oncol. 2014;8(3):581–595. doi: 10.1016/j.molonc.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao SF, Zhang X, Zhang XJ, Shi XQ, Yu ZJ, Kan QC. Induction of micro RNA-9 mediates cytotoxicity of curcumin against SKOV3 ovarian cancer cells. Asian Pac J Cancer Prev. 2014;15(8):3363–3368. doi: 10.7314/apjcp.2014.15.8.3363. [DOI] [PubMed] [Google Scholar]
- 85.Yang CH, Yue J, Sims M, Pfeffer LM. The curcumin analog EF24 targets NF-kappaB and miRNA-21, and has potent anticancer activity in vitro and in vivo. PLoS One. 2013;8(8):e71130. doi: 10.1371/journal.pone.0071130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu CC, Tsai LL, Wang ML, Yu CH, Lo WL, Chang YC, et al. miR145 targets the SOX9/ADAM17 axis to inhibit tumor-initiating cells and IL-6-mediated paracrine effects in head and neck cancer. Cancer Res. 2013;73(11):342S40. doi: 10.1158/0008-5472.CAN-12-3840. [DOI] [PubMed] [Google Scholar]