Abstract
Toll-like receptors (TLRs) play a crucial role in innate- and adaptive immunity. The TLR pathways were shown to play key functional roles in experimental acute and chronic kidney injury, including the allo-immune response after experimental renal transplantation. Data about the precise impact of TLRs and their negative regulators on human renal transplant outcomes however are limited and contradictory. We studied twelve non-synonymous single nucleotide polymorphisms (SNPs) of which eleven in TLR1-8 and one in SIGIRR in a final cohort comprising 1116 matching donors and recipients. TLR3 p.Leu412Phe and SIGIRR p.Gln312Arg significantly deviated from Hardy-Weinberg equilibrium and were excluded. The frequency distribution of the minor alleles of the remaining 10 TLR variants were compared between patients with end-stage renal disease (recipients) and controls (kidney donors) in a case-control study. Secondly, the associations between the minor allele frequency of the TLR variants and delayed graft function, biopsy-proven acute rejection and death-censored graft failure after transplantation were investigated with Cox regression. Carrier frequencies of the minor alleles of TLR1 p.His305Leu (OR = 4.79, 95% CI = 2.35–9.75, P = 0.0002), TLR1 p.Asn248Ser (OR = 1.26, 95% CI = 1.07–1.47, P = 0.04) and TLR8 p.Met1Val (OR = 1.37, 95% CI = 1.14–1.64, P = 0.008) were significantly higher in patients with ESRD, with little specificity for the underlying renal disease entity (adjusted for age, gender and donor-recipient relatedness). The minor allele frequency of none of the TLR variants significantly associated with the surrogate and definite outcomes, even when multivariable models were created that could account for TLR gene redundancy. In conclusion, genetic variants in TLR genes were associated with the prevalence of ESRD but not renal transplant outcomes. Therefore, our data suggests that specific TLR signaling routes might play a role in the final common pathway of primary renal injury. A role for TLR signaling in the context of renal transplantation is probably limited.
Introduction
Toll-like receptors (TLRs) are pattern recognition receptors (PRR), which can be activated by both pathogen-associated molecular patterns (PAMPs) and endogenous ligands called damage-associated molecular patterns (DAMPs) leading to the induction of an inflammatory response [1, 2]. Single Ig IL-1-related receptor (SIGIRR) is one of the negative regulators of the TLR signalling pathway and is involved in reducing inflammation upon TLR activation to prevent excessive inflammation [3, 4]. TLRs play a part in both the innate and the subsequent adaptive immunity and are of special interest in renal diseases; TLRs are expressed on murine and human leukocytes and renal endothelial and epithelial cells, including podocytes [1, 2]. TLRs are crucial in the antibacterial defence mechanisms during renal infection, however this immune response is detrimental during a sterile inflammatory response including acute and chronic kidney injury and the allo-immune response after transplantation [2, 5]. In renal transplant patients, TLR4 is the most frequently studied TLR family member that is activated by DAMPs that are released during an episode of renal injury and in particular during ischemia-reperfusion injury after long-term cold storage of the transplants. One of the most well-known DAMPs recognized by TLR4 is High-mobility group protein B1 (HMGB1), which is highly expressed in renal transplants of deceased but not living donors after surgery [6]. Different studies have shown that if the donor or recipient inherits or possess a TLR4 loss-of-function single nucleotide polymorphisms (SNPs) such as p.Asp299Gly allele A/G and p.Thr399Ile allele C/T, recipients were less likely to experience delayed graft function (DGF) or acute rejection (AR) [7, 8]. Importantly, recipients with these particular TLR4 variants experienced more episodes of infections [7] highlighting a possible double-edge sword for TLRs in the context of transplantation. Unfortunately, there are conflicting data on the role of TLR4 and other TLR signalling sequence variants on renal outcome in renal transplant recipients [7–18]. This might be explained by the variety in the patient databases that have been used. In addition, studies vary in their definition of study endpoints or studies use only one single endpoint [7–18]. Large cohorts that are adequately powered to investigate the impact of especially multiple SNPs are needed since these pattern recognition receptors are known to be redundant. The aim of the current study is therefore to investigate the impact of SNPs in genes that are involved in TLR pathways on outcomes in the context of renal transplantation.
Material and Methods
Study population
Samples were included from a study cohort as described before [19,20]. Between March 1993 and February 2008, 1271 matching donor and recipient peripheral blood mononuclear cells (PBMCs) were obtained from patients who underwent kidney transplantation at the University Medical Center Groningen, The Netherlands. The exclusion criteria were: cases of re-transplantation, combined kidney/pancreas or kidney/liver transplantation, technical problems during surgery, the unavailability of DNA and loss to follow-up. The institutional ethical review board of the University Medical Center Groningen approved the study (METc 2014/077). Written informed consent was obtained from all patients. None of the living transplant donors were from a vulnerable population and all living donors provided written informed consent. In case of deceased donation, the donors provided informed consent when they registered their donation status and by law, no additional consent was needed. The study was conducted according to the principles of the declaration of Helsinki. The final statistical analyses were performed on 1116 individuals (2232 samples), corresponding to 92% of the donor and recipient pairs after exclusion of patients with primary non-functioning grafts (see below).
DNA isolation, quality control and SNP selection criteria
DNA samples were analyzed for absorbance at 260 nm with NanoDrop spectrophotometer (ND–1000, NanoDrop Technologies) and DNA concentration was calculated by the NanoDrop nucleic acid application module. As a measure of DNA purity 260/280 and 260/230 absorbance ratios were assessed. Where samples failed to meet the minimum DNA concentration and purity recommended for Illumina genotyping, repeated isolation attempts were made. In this study, 12 non-synonymous SNPs in TLR and SIGIRR genes obtained from NCBI (inclusion criterion: minor allele frequency >1%) were analyzed for their association with the various renal outcomes as defined below. Genotyping of the selected SNP was performed using the Illumina VeraCode GoldenGate Assay kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Genotype clustering and calling was performed using Beadstudio Software (Illumina). Of the chosen SNPs, rs4986790 (TLR4 p.Asp299Gly) and rs4986791 (TLR4 p.Thr399Ile) are in linkage disequilibrium (LD) (r2 = 1, SNP Annotation and Proxy Search, Broad institute).
Case-control study for end-stage renal disease
We performed a case-control study to investigate the association between the TLR SNPs and the prevalence of end-stage renal disease by comparing the recipients as cases with the donors as their respective controls. Cases were separately evaluated for the association of the allele frequencies of the variants in specific groups of renal diseases, namely congenital nephropathy (N = 216), immunecomplex-mediated glomerulonephritis (N = 299), infective pyelonephritis (N = 126) and renovascular disease (N = 102) with the full group of controls (donors, N = 1116). We took into account that donors and recipients are related in case of living related renal transplantation.
Study endpoints for longitudinal analyses after transplantation
The endpoints used in longitudinal analyses were: delayed graft function (DGF), defined as the requirement for dialysis within the first week after transplantation due to the need for additional renal replacement therapy (patients with subsequent non-functioning and loss of their allograft, referred to as primary non-function (PNF, N = 60) [19], were excluded from analyses), time to the first episode of biopsy-proven acute rejection (BPAR) and death-censored graft failure (defined as the need for dialysis or re-transplantation, patients with PNF were excluded). Data on rejection type (antibody- or T cell-mediated rejection) was unavailable due to the lack of a standardized method for the determination of donor-specific antibodies over time.
Statistical analyses
Statistical analyses were performed using the R platform for statistical computing version 3.1.1. (www.r-project.org) and PLINK version 1.07 for Mac OS X 10.10.4 (S. Purcell, http://pngu.mgh.harvard.edu/purcell/plink) [21]. We followed the protocol for statistical analyses in genetic studies as described by Clarke et al. [22]. Two-sided P-values <0.05 were considered statistically significant after Bonferroni correction for multiple comparisons. Minor allele frequencies were calculated as the sum of the minor alleles divided by 2 times the total number of patients. Hardy-Weinberg equilibria for the variants in the donors (as a healthy control group) were calculated and when SNPs were in disequilibrium (after Bonferroni correction), they were excluded from further analyses. In the case-control study, odds ratios and corresponding 95% confidence intervals were calculated with additive genetic logistic regression models correcting for age and gender. Donor-recipient relatedness in case of living related renal transplantation was taken into consideration by applying the DFAM algorithm in PLINK. Because we had a large database with a fixed amount of patients before genotyping started, type II error percentages (100%—power) for the significantly associated variants were calculated post-hoc according to the statistical methods as described by Skol et al. [23]. The prevalence of end-stage renal disease in The Netherlands was estimated at 0.1% based on the number of patient on renal replacement therapy at time of analysis. The association between single TLR variants and delayed graft function was calculated with univariable logistic regression models and the odds ratios with corresponding 95% confidence intervals were presented. The association between single TLR variants and biopsy-proven acute rejection and death-censored graft failure was calculated with univariable Cox regression models and the hazard ratios with corresponding 95% confidence intervals were presented. Corresponding P-values were calculated with log rank tests. In the regression models, all P-values underwent Bonferroni correction for multiple comparisons. For the 3 endpoints, we next constructed multivariable models per endpoint that included parameters that are known to influence these endpoints (e.g. cold ischemia time for delayed graft function) and compared these clinical models to models that additionally included all TLR variants in both donors and recipients in order to account for gene redundancy. We used the Akaike information criterion (AIC) to compare the relative goodness-of-fit of the models for their association with the endpoints. Lower AIC values are indicative of a better goodness-of-fit of the model. In this way we wanted to investigate whether including all TLR variants in one multivariable model as a method to account for redundancy between TLRs genes, might provide additional information besides parameters that have been known to influence the outcomes. In this case, the role of all TLR variants as a group rather than the possible role for a single variant is investigated.
Results
Study characteristics and distribution of the TLR gene variants
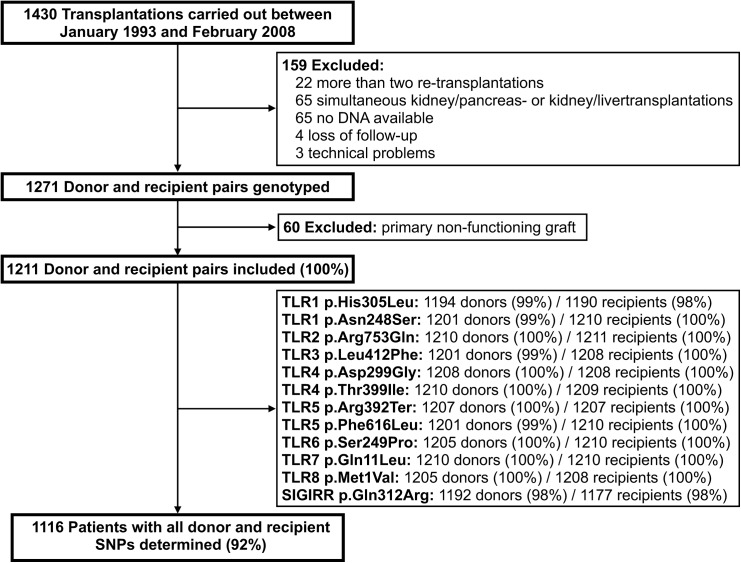
Fig 1 shows the flowchart of the in- and excluded patients and samples. The characteristics of cohort are presented in Table 1. There were no significant differences in donor, recipient and transplant characteristics between the included and excluded patient cohorts, which supports our statement that inclusion was not systematically biased. The genotypic distributions of the SNPs in donors and recipients are displayed in Table 2. Of the total of 1211 patients included in the study, 1116 (92%) had all donor and recipient SNPs determined. After Bonferroni correction for multiple testing, TLR3 p.Leu412Phe (P = 0.046) and SIGIRR p.Gln312Arg (P < 0.0001) appeared in Hardy-Weinberg disequilibrium in controls (donors) and these SNPs were therefore excluded from further analysis.
Fig 1. Flowchart of the in- and excluded samples.
Table 1. Characteristics of the study group, subdivided by included and excluded patients.
| Variable | TotalN = 1271 | IncludedN = 1116 | ExcludedN = 155 | P-value 1 |
|---|---|---|---|---|
| Donor characteristics | ||||
| Age (mean years ± SD) | 44 ± 14 | 45 ± 14 | 44 ± 16 | 1 |
| Male N (%) | 645 (51%) | 554 (50%) | 91 (59%) | 0.7 |
| Donor type N (%) | ||||
| Living donor | 282 (22%) | 249 (22%) | 33 (21%) | 1 |
| Deceased donor (DBD + DCD) | 989 (78%) | 867 (78%) | 122 (79%) | |
| Recipient characteristics | ||||
| Age (mean years ± SD) | 48 ± 13 | 48 ± 13 | 48 ± 13 | 1 |
| Male N (%) | 739 (58%) | 655 (59%) | 84 (54%) | 1 |
| Initial immunosuppression N (%) | ||||
| Corticosteroids | 1201 (95%) | 1053 (94%) | 148 (95%) | 1 |
| Calcineurin inhibitor (CsA or TAC) | 1182 (93%) | 1039 (93%) | 143 (92%) | 1 |
| Proliferation inhibitor (MPA or AZA) | 979 (77%) | 863 (77%) | 116 (75%) | 1 |
| mTOR inhibitor | 38 (3%) | 31 (3%) | 7 (5%) | 1 |
| Induction therapy N (%) | ||||
| Anti-thymocyte globulin | 103 (8%) | 90 (8%) | 13 (8%) | 1 |
| Anti-CD3 moab | 19 (2%) | 15 (1%) | 4 (3%) | 1 |
| Interleukin–2 receptor antagonist | 199 (16%) | 171 (15%) | 28 (18%) | 1 |
| First transplant N (%) | 1143 (90%) | 1001 (90%) | 142 (92%) | 1 |
| Transplant characteristics | ||||
| Cold ischemia time (mean hours ± SD) | ||||
| Living donors | 2.7 ± 1.9 | 2.7 ± 2.0 | 2.6 ± 0.7 | 1 |
| Deceased donors | 20.7 ± 6.5 | 20.6 ± 6.4 | 21.6 ± 6.8 | 1 |
| HLA no. of 0 mismatches N (%) 2 | 241 (23%) | 213 (23%) | 28 (22%) | 1 |
DBD = deceased brain death, DCD = deceased cardiac death, SD = standard deviation, CsA = cyclosporine A, TAC = tacrolimus, MPA = mycophenolic acid, AZA = azathioprine, mTOR = mammalian target of rapamycin, moab = monoclonal antibody, HLA = human leukocyte antigen. HLA = Human leukocyte antigen.
1Bonferroni corrected for multiple testing
2Data for N = 221 were missing; N = 195 (20%) in the included patients, N = 26 in the excluded (17%), P = 0.9.
Table 2. Allele frequency distributions and possible phenotypical consequences of the single nucleotide polymorphisms in TLR-related genes.
| Gene | Chr | HGVS name (rs number) | Phenotype (refs) | A/a | 1000Genomes library | Donor | Recipient | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A (%) | A/a (%) | a/a (%) | MAF (%) | A/A (%) | A/a (%) | a/a (%) | MAF (%) | A/A (%) | A/a (%) | a/a (%) | MAF (%) | |||||
| TLR1 | 4 | p.His305Leu (rs3923647) | GOF [24,25] LOF [26] | T/a | 95.4 | 4.2 | 0.4 | 2.5 | 95.1 | 4.7 | 0.2 | 2.6 | 84.3 | 11.6 | 4.1 | 9.5 |
| TLR1 | 4 | p.Asn248Ser (rs4833095) | LOF [25–27] | C/t | 53.7 | 36.6 | 9.7 | 28.0 | 54.3 | 38.1 | 7.6 | 25.9 | 50.3 | 39.0 | 10.7 | 30.0 |
| TLR2 | 4 | p.Arg753Gln (rs5743708) | LOF [28–30] | G/a | 95.2 | 4.8 | 0 | 2.4 | 89.7 | 10.1 | 0.2 | 5.1 | 80.8 | 19.1 | 0.1 | 9.6 |
| TLR3 | 4 | p.Leu412Phe (rs3775291) | LOF [31,32] GOF [33] | C/t | 44.7 | 45.7 | 9.5 | 32.4 | 49.0 | 44.6 | 6.4 | 28.7 | 52.3 | 41.2 | 6.5 | 27.1 |
| TLR4 | 9 | p.Asp299Gly (rs4986790) | LOF [34,35] | A/g | 89.3 | 10.1 | 0.6 | 5.7 | 89.3 | 10.6 | 0.2 | 5.4 | 88.7 | 11.0 | 0.2 | 5.8 |
| TLR4 | 9 | p.Thr399Ile (rs4986791) | LOF [34,35] | C/t | 89.1 | 10.3 | 0.1 | 5.3 | 89.1 | 10.9 | 0.1 | 5.5 | 88.3 | 11.5 | 0.2 | 6.0 |
| TLR5 | 1 | p.Arg392Ter (rs5744168) | LOF [36–38] | G/a | 88.1 | 11.7 | 0.2 | 6.1 | 86.5 | 13.4 | 0.1 | 7.1 | 84.2 | 15.2 | 0.6 | 8.2 |
| TLR5 | 1 | p.Phe616Leu (rs5744174) | GOF [39–41] | A/g | 34.8 | 48.5 | 16.7 | 41.0 | 30.6 | 51.0 | 18.4 | 42.6 | 32.4 | 49.8 | 17.8 | 44.6 |
| TLR6 | 4 | p.Ser249Pro (rs5743810) | GOF [24] | G/a | 35.8 | 46.7 | 17.5 | 40.9 | 35.3 | 48.0 | 16.7 | 41.0 | 39.5 | 44.0 | 16.5 | 38.6 |
| TLR7 | X | p.Gln11Leu (rs17900) | LOF [42,43] | A/t | 71.7 | 23.3 | 5.0 | 16.7 | 68.7 | 20.1 | 11.2 | 21.1 | 71.3 | 15.9 | 12.8 | 21.7 |
| TLR8 | X | p.Met1Val(rs3764880) | GOF [44,45] | A/g | 66.4 | 15.0 | 18.6 | 26.1 | 68.9 | 18.6 | 12.5 | 22.6 | 63.7 | 16.7 | 19.6 | 27.9 |
| SIGIRR | 11 | p.Gln312Arg (rs3210908) | ND | C/t | 60.8 | 34.2 | 5.0 | 22.1 | 51.2 | 47.0 | 1.8 | 25.4 | 38.3 | 49.2 | 12.5 | 37.1 |
Donor and recipient genotype are displayed as dominant (A/A), heterozygous (A/a) or recessive (a/a). Chr = chromosome, HGVS = Human Genome Variation Society LOF = loss of function, GOF = gain of function, ND = not determined, MAF = minor allele frequency.
Frequency distribution of the variant alleles comparing patients with end-stage renal disease and healthy controls
We wanted to find out whether there was a difference in the carrier distribution of the minor allele alleles between patients with end-stage renal disease (recipients) and healthy controls (donors) in a case-control approach. After correction for multiple comparisons, age, gender and donor-recipient relatedness, the carrier frequency of the minor allele for TLR1 p.His305Leu (OR = 4.79, 95% CI = 2.35–9.75, P = 0.0002), TLR1 p.Asn248Ser (OR = 1.26, 95% CI = 1.07–1.47, P = 0.04) and TLR8 p.Met1Val (OR = 1.37, 95% CI = 1.14–1.64, P = 0.008) was significantly higher in patients with end-stage renal disease (Table 3). In Table 4, we separated the patients by cause of end-stage renal disease in order to investigate whether the carrier frequency of the minor alleles for the TLR variants showed specificity for certain underlying disease entities. The minor allele for TLR1 p.His305Leu showed the strongest association with each of the underlying renal disease group tested: congenital nephropathy (OR = 4.94, 95% CI = 2.26–10.84, P = 0.0002), immunecomplex-mediated glomerulonephritis (OR = 4.91, 95% CI = 2.31–10.41, P = 0.0001), infective pyelonephritis (OR = 6.71, 95% CI = 3.10–14.53, P < 0.0001) and renovascular disease (OR = 4.37, 95% CI = 1.83–10.43, P = 0.003). TLR1 p.Asn248Ser and TLR8 p.Met1Val showed some specificity for immunecomplex-mediated glomerulonephritis (respectively OR = 1.33, 95% CI = 1.05–1.67, P = 0.05 and OR = 1.52, 95% CI = 1.18–1.97, P = 0.004) and renovascular disease (respectively OR = 1.53, 95% CI = 1.10–2.11, P = 0.03 and OR = 1.86, 95% CI = 1.25–2.77, P = 0.002), however the estimated type II error ranged between 9–28%.
Table 3. Case-control study for the association between TLR single nucleotide polymorphsims and end-stage renal disease.
| Gene | HGVS name | MAF cases (%) | MAF controls (%) | OR1 | 95% CI 1 | P-value 2 | Type II error when P < 0.05 3 |
|---|---|---|---|---|---|---|---|
| TLR1 | p.His305Leu | 9.5 | 2.6 | 4.79 | 2.35–9.75 | 0.0002 | 0% |
| TLR1 | p.Asn248Ser | 30.0 | 25.9 | 1.26 | 1.07–1.47 | 0.04 | 6% |
| TLR2 | p.Arg753Gln | 9.6 | 5.1 | 0.73 | 0.22–2.43 | 1 | |
| TLR4 | p.Asp299Gly | 5.8 | 5.4 | 1.27 | 0.52–3.12 | 1 | |
| TLR4 | p.Thr399Ile | 6.0 | 5.5 | 1.77 | 0.57–5.51 | 1 | |
| TLR5 | p.Arg392Ter | 8.2 | 7.1 | 2.93 | 1.03–8.33 | 0.4 | |
| TLR5 | p.Phe616Leu | 44.6 | 42.6 | 0.92 | 0.82–1.04 | 1 | |
| TLR6 | p.Ser249Pro | 38.6 | 41.0 | 0.92 | 0.81–1.04 | 1 | |
| TLR7 | p.Gln11Leu | 21.7 | 21.1 | 1.05 | 0.85–1.30 | 1 | |
| TLR8 | p.Met1Val | 27.9 | 22.6 | 1.37 | 1.14–1.64 | 0.008 | 1% |
1Per allele odds ratios (OR) and 95% confidence intervals (CI) based on additive genetic logistic regression models adjusted for age and gender, taking case-control relatedness into consideration (DFAM algorithm). HGVS = Human Genome Variation Society.
2 P-values are Bonferroni corrected.
3Estimates of the type II errors (100%–power) were calculated according to the methods by Skol et al. [23] with the data as mentioned in the table and an end-stage renal disease prevalence of 0.1% (estimate in The Netherlands).
Table 4. Case-control study for the association between TLR single nucleotide polymorphisms and end-stage renal disease per underlying disease category.
| Gene | HGVS name | Renal disease | N cases | MAF cases(%) | MAF controls(%) | OR 1 | 95% CI 1 | P-value 2 | Type II error when P < 0.05 3 |
|---|---|---|---|---|---|---|---|---|---|
| TLR1 | p.His305Leu | Congenital | 216 | 10.4 | 2.6 | 4.94 | 2.26–10.84 | 0.0002 | 0% |
| Glomerulonephritis | 299 | 8.4 | 2.6 | 4.91 | 2.31–10.41 | 0.0001 | 0% | ||
| Pyelonephritis | 126 | 11.5 | 2.6 | 6.71 | 3.10–14.53 | <0.0001 | 0% | ||
| Renovascular | 102 | 9.8 | 2.6 | 4.37 | 1.83–10.43 | 0.003 | 0% | ||
| TLR1 | p.Asn248Ser | Congenital | 216 | 25.9 | 25.9 | 1.02 | 0.76–1.37 | 1 | |
| Glomerulonephritis | 299 | 32.6 | 25.9 | 1.33 | 1.05–1.67 | 0.05 | 16% | ||
| Pyelonephritis | 126 | 27.0 | 25.9 | 1.27 | 0.94–1.71 | 0.4 | |||
| Renovascular | 102 | 33.3 | 25.9 | 1.53 | 1.10–2.11 | 0.03 | 28% | ||
| TLR8 | p.Met1Val | Congenital | 216 | 24.6 | 22.6 | 1.18 | 0.86–1.64 | 0.9 | |
| Glomerulonephritis | 299 | 29.4 | 22.6 | 1.52 | 1.18–1.97 | 0.004 | 9% | ||
| Pyelonephritis | 126 | 28.1 | 22.6 | 1.24 | 0.85–1.81 | 0.8 | |||
| Renovascular | 102 | 33.6 | 22.6 | 1.86 | 1.25–2.77 | 0.002 | 17% |
1Per allele odds ratios (OR) and 95% confidence intervals (CI) based on additive genetic logistic regression models adjusted for age and gender, taking case-control relatedness into consideration (DFAM algorithm). HGVS = Human Genome Variation Society.
2 P-values are Bonferroni corrected.
3 Post hoc estimates of the type II errors (100%–power) were calculated according to the methods by Skol et al. [23] with the data as mentioned in the table and an end-stage renal disease prevalence of 0.1% (estimate in The Netherlands).
Role of TLR sequence variants in relation to delayed graft function
Delayed graft function (DGF) occurred in 328 of 1116 patients (29%). DGF was observed in 37% (317/867) of recipients receiving a graft from a deceased donor and 4% (11/249) in recipients of a living donor. We analysed the association of TLR SNPs with DGF in all included patients and separately in recipients of a deceased donor, since TLR activation is more prominently observed in the context of deceased donation [6]. In univariable analyses, where all donor and recipient SNPs were tested separately, none of the TLR SNPs associated with DGF after Bonferroni correction (Table 5). As expected, cold ischemia time (P < 0.0001), donor age (P = 0.0008) and recipient age (P = 0.0005) were significantly associated with the occurrence of DGF. The model that included these 3 parameters had an Akaike information criterion (AIC) of 1272.5. Since TLRs are highly redundant, we also tested the performance of a logistic regression model that included all TLR variants in one multivariable model. Adding all donor and recipient SNPs to the crude multivariable model of cold ischemia time, donor age and recipient age again resulted in a higher AIC (1289.4), showing no additional value for variants in TLR genes in explaining the occurrence of DGF. Similar results were obtained when only recipients of a deceased donor were analysed (S1 Table). We therefore conclude that variants in TLR genes are not associated with the occurrence of delayed graft function after transplantation.
Table 5. Association of TLR single nucleotide polymorphism with delayed graft function in univariable logistic regression analysis.
| Gene | HGVS name | Allele combination 1 | Donor | Recipient | ||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value 2 | OR | 95% CI | P-value 2 | |||
| TLR1 | p.His305Leu | T/a | 0.69 | 0.34–1.30 | 1 | 1.12 | 0.75–1.67 | 1 |
| a/a | 2.37 | 0.09–60.01 | 1 | 0.55 | 0.23–1.13 | 1 | ||
| TLR1 | p.Asn248Ser | C/t | 0.86 | 0.65–1.13 | 1 | 0.85 | 0.64–1.12 | 1 |
| t/t | 0.65 | 0.37–1.11 | 1 | 1.64 | 1.07–2.50 | 0.4 | ||
| TLR2 | p.Arg753Gln | G/a | 0.71 | 0.44–1.11 | 1 | 0.74 | 0.52–1.04 | 1 |
| a/a | - | - | - | - | - | - | ||
| TLR4 | p.Asp299Gly | A/g | 1.30 | 0.86–1.94 | 1 | 0.82 | 0.53–1.24 | 1 |
| g/g | 2.48 | 0.10–62.79 | 1 | 1.18 | 0.05–12.33 | 1 | ||
| TLR4 | p.Thr399Ile | C/t | 1.28 | 0.85–1.90 | 1 | 0.85 | 0.56–1.28 | 1 |
| t/t | - | - | - | 1.18 | 0.05–12.37 | 1 | ||
| TLR5 | p.Arg392Ter | G/a | 1.35 | 0.94–1.93 | 1 | 1.08 | 0.75–1.54 | 1 |
| a/a | - | - | - | 1.46 | 0.30–6.00 | 1 | ||
| TLR5 | p.Phe616Leu | A/g | 0.92 | 0.69–1.24 | 1 | 0.78 | 0.59–1.05 | 1 |
| g/g | 1.15 | 0.80–1.67 | 1 | 0.88 | 0.60–1.28 | 1 | ||
| TLR6 | p.Ser249Pro | G/a | 1.00 | 0.75–1.33 | 1 | 0.91 | 0.68–1.20 | 1 |
| a/a | 0.97 | 0.66–1.41 | 1 | 1.01 | 0.69–1.46 | 1 | ||
| TLR7 | p.Gln11Leu | A/t | 0.88 | 0.63–1.23 | 1 | 0.83 | 0.57–1.19 | 1 |
| t/t | 1.25 | 0.83–1.85 | 1 | 0.74 | 0.48–1.09 | 1 | ||
| TLR8 | p.Met1Val | A/g | 0.96 | 0.68–1.34 | 1 | 1.09 | 0.77–1.55 | 1 |
| g/g | 1.04 | 0.70–1.53 | 1 | 1.12 | 0.80–1.54 | 1 | ||
OR = odds ratio (1per allele combination as compared to the homozygous dominant allele combination)
CI = confidence interval, HGVS = Human Genome Variation Society. The results represent univariable crude models, i.e. no other independent variables were included.
2 P-values are Bonferroni corrected.
Role of TLR sequence variants in relation to acute rejection
The median time of freedom-of-rejection was 51 months (interquartile range 1–105 months). The overall cumulative incidence of biopsy-proven acute rejection (BPAR) after renal transplantation was 34% (378/1116). In univariable analyses, where all donor and recipient SNPs were tested separately, none of the SNPs in the TLR genes associated with BPAR after Bonferroni correction (Table 6). In our cohort, most of the variation in the occurrence of BPAR could be explained by a preceding period of DGF (P = 0.01), recipient age (P < 0.0001) and the number of HLA mismatches (P < 0.0001), which resulted in an AIC of 4125.6. When we added all donor and recipient TLR variants to the model that included DGF, recipient age and number of HLA mismatches, again a higher AIC was calculated (AIC = 4142.0), indicating no additional explanatory value by the TLR variants for the cumulative incidence of BPAR.
Table 6. Association of TLR single nucleotide polymorphism with biopsy-proven acute rejection.
| Gene | HGVS name | Allele combination 1 | Donor | Recipient | ||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value 2 | HR | 95% CI | P-value 2 | |||
| TLR1 | p.His305Leu | T/a | 0.68 | 0.39–1.18 | 1 | 1.60 | 1.20–2.12 | 0.1 |
| a/a | 1.86 | 0.26–13.21 | 1.02 | 0.60–1.74 | ||||
| TLR1 | p.Asn248Ser | C/t | 0.97 | 0.78–1.20 | 1 | 1.09 | 0.88–1.35 | 1 |
| t/t | 1.37 | 0.95–1.98 | 1.04 | 0.72–1.48 | ||||
| TLR2 | p.Arg753Gln | G/a | 0.84 | 0.59–1.20 | 1 | 1.32 | 1.04–1.68 | 0.9 |
| a/a | - | - | 2.92 | 0.41–20.84 | ||||
| TLR4 | p.Asp299Gly | A/g | 0.85 | 0.60–1.20 | 1 | 0.90 | 0.65–1.25 | 1 |
| g/g | - | - | 1.05 | 0.15–7.49 | ||||
| TLR4 | p.Thr399Ile | C/t | 0.91 | 0.65–1.27 | 1 | 0.94 | 0.68–1.30 | 1 |
| t/t | - | - | 1.06 | 0.15–7.53 | ||||
| TLR5 | p.Arg392Ter | G/a | 1.00 | 0.75–1.34 | 0.3 | 0.80 | 0.59–1.09 | 1 |
| a/a | 10.52 | 1.47–75.19 | 1.04 | 0.33–3.24 | ||||
| TLR5 | p.Phe616Leu | A/g | 1.08 | 0.85–1.36 | 1 | 0.92 | 0.74–1.16 | 1 |
| g/g | 1.03 | 0.76–1.38 | 0.97 | 0.72–1.30 | ||||
| TLR6 | p.Ser249Pro | G/a | 0.89 | 0.72–1.12 | 1 | 0.73 | 0.59–0.92 | 0.1 |
| a/a | 0.86 | 0.64–1.16 | 1.06 | 0.81–1.40 | ||||
| TLR7 | p.Gln11Leu | A/t | 0.99 | 0.76–1.28 | 1 | 0.91 | 0.68–1.22 | 1 |
| t/t | 1.25 | 0.92–1.28 | 1.17 | 0.88–1.56 | ||||
| TLR8 | p.Met1Val | A/g | 1.25 | 0.98–1.61 | 1 | 1.07 | 0.81–1.41 | 1 |
| g/g | 0.96 | 0.70–1.32 | 1.20 | 0.93–1.54 | ||||
HR = hazard ratio (1per allele combination as compared to the homozygous dominant allele combination)
CI = confidence interval, HGVS = Human Genome Variation Society. The results represent univariable crude models, i.e. no other independent variables were included.
2 P-values are calculated by log rank tests after Bonferroni correction for multiple comparisons.
Role of TLR sequence variants in relation to graft failure
Median overall graft survival was 5.5 years (interquartile range 2.9–8.9 years). The overall cumulative incidence of death-censored graft failure was 191/1116 (17%) of which 124/215 (66%, 11% of total) failed due to rejection. In line with our negative findings concerning the lack of association of the TLR variants with the surrogate endpoints DGF and BPAR, none of the variants associated with the cumulative incidence of death-censored graft failure (Table 7). In our cohort, patients that underwent an episode of BPAR had worse death-censored graft survival (P < 0.0001, AIC = 2504.3). Again, a multivariable model that included all donor and recipient TLR variants plus the occurrence of an episode of BPAR did not improve the goodness-of-fit of the model (AIC = 2530.2), therefore indicating no additional value for the TLR variants in explaining the development of death-censored graft failure after transplantation.
Table 7. Association of TLR single nucleotide polymorphism with death-censored graft survival.
| Gene | HGVS name | Allele combination 1 | Donor | Recipient | ||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value 2 | HR | 95% CI | P-value 2 | |||
| TLR1 | p.His305Leu | T/a | 1.02 | 0.52–2.00 | 1 | 1.01 | 0.65–1.56 | 1 |
| a/a | - | - | 0.56 | 0.23–1.37 | ||||
| TLR1 | p.Asn248Ser | C/t | 1.13 | 0.84–1.52 | 1 | 1.11 | 0.82–1.50 | 1 |
| t/t | 0.93 | 0.51–1.69 | 1.44 | 0.92–2.24 | ||||
| TLR2 | p.Arg753Gln | G/a | 1.13 | 0.72–1.77 | 1 | 1.29 | 0.93–1.80 | 1 |
| a/a | - | - | - | - | ||||
| TLR4 | p.Asp299Gly | A/g | 1.15 | 0.74–1.80 | 1 | 0.80 | 0.50–1.29 | 1 |
| g/g | - | - | - | - | ||||
| TLR4 | p.Thr399Ile | C/t | 1.17 | 0.76–1.81 | 1 | 0.74 | 0.45–1.20 | 1 |
| t/t | - | - | - | - | ||||
| TLR5 | p.Arg392Ter | G/a | 1.31 | 0.90–1.90 | 1 | 0.84 | 0.55–1.27 | 1 |
| a/a | - | - | 0.77 | 0.11–5.52 | ||||
| TLR5 | p.Phe616Leu | A/g | 1.03 | 0.75–1.43 | 1 | 1.20 | 0.87–1.64 | 1 |
| g/g | 0.82 | 0.53–1.28 | 0.70 | 0.44–1.13 | ||||
| TLR6 | p.Ser249Pro | G/a | 1.19 | 0.87–1.64 | 1 | 0.85 | 0.62–1.17 | 1 |
| a/a | 0.96 | 0.62–1.48 | 1.08 | 0.73–1.61 | ||||
| TLR7 | p.Gln11Leu | A/t | 0.83 | 0.57–1.19 | 1 | 0.89 | 0.59–1.35 | 1 |
| t/t | 0.96 | 0.61–1.50 | 1.41 | 0.96–2.06 | ||||
| TLR8 | p.Met1Val | A/g | 0.99 | 0.68–1.44 | 1 | 1.05 | 0.71–1.56 | 1 |
| g/g | 1.20 | 0.80–1.80 | 1.28 | 0.91–1.82 | ||||
HR = hazard ratio (1per allele combination as compared to the homozygous dominant allele combination)
CI = confidence interval, HGVS = Human Genome Variation Society. The results represent univariable crude models, i.e. no other independent variables were included.
2 P-values are calculated by log rank tests after Bonferroni correction for multiple comparisons.
Discussion
TLR-signaling and control by their negative regulators is of special interest in renal diseases because of their expression pattern in murine and human kidneys and the role they play in experimental models of acute and chronic renal injury [46–55]. Contribution of other TLRs (besides the TLR2-4-6 axis) on progression to pre-transplant ESRD and renal outcome after transplantation is a relatively unexplored field. In this large cohort of renal transplant recipients, we observed that 1) TLR1 p.His305Leu, TLR1 p.Asn248Ser and TLR8 p.Met1Val significantly associated with the prevalence of end-stage renal disease, and 2) SNPs in TLR genes do not explain the occurrence of delayed graft function, biopsy-proven acute rejection and subsequently death-censored graft failure after transplantation.
Thus far, relatively few studies have investigated the contribution of non-synonymous polymorphisms in TLR-related genes and their association with renal diseases of the native kidneys and the development of chronic kidney disease (CKD) [12,56–62]. Table 8 summarizes the studies that we could identify from the literature in which the same TLR variants were analyzed as in our study. Lee et al. described a higher frequency of the minor allele for TLR1 p.Asn248Ser in pediatric patients with IgA nephropathy as compared to healthy controls [56], which might be compared to the higher frequency of the minor allele for this variant in patients with immunecomplex-mediated glomerulonephritis in our cohort. Unfortunately, in this study TLR1 p.His305Leu, the variant that showed a robust and very relevant association with end-stage renal disease in our cohort, was not investigated. TLR4 p.Asp299Gly and p.Thr399Ile are by far the most studied TLR variants in the literature [12,58–62]. Only the study by Akil et al. found a significant association between the minor allele frequency of any of the two TLR4 variants and the prevalence of chronic kidney disease [60]. The vast majority of studies however did not find an association, which is in line with our study and because p.Asp299Gly and p.Thr399Ile are in high linkage disequilibrium, one would expect a similar effect for both variants in this case. Interestingly, two [60,61] of three studies [60–62] found an association of the minor allele frequency for the TLR4 variant p.Asp299Gly with pyelonephritis (both with and without secondary chronic kidney disease), which could unfortunately not be investigated in such detail in our study. In line with our study, Cheng et al. did not find an association of the minor allele of TLR5 variant p.Arg392Ter with native kidney diseases [57]. The residual TLR variants, which includes TLR8 p.Met1Val that showed an association with end-stage renal disease in our cohort, have not been described before. Due to the lack of functional TLR pathway testing in patients carrying these TLR SNPs associated with end-stage renal disease, we consider our study hypothesis-generating.
Table 8. Association of the TLR gene polymorphisms with renal outcomes as described in the literature.
| Gene | HGVS name | Author, year | Reference | Country | Native / Transplant | Disease | Controls | Effect of the minor allele |
|---|---|---|---|---|---|---|---|---|
| TLR1 | p.His305Leu | - | - | - | - | - | - | - |
| TLR1 | p.Asn248Ser | Lee, 2011 | [56] | Korean | Native | GN | Healthy | Increased risk |
| Cheng, 2013 | [57] | Taiwan | Native | PN | no PN | No effect | ||
| TLR2 | p.Arg753Gln | Mutlubas, 2009 | [58] | Turkey | Native | CKD | Healthy | Increased risk |
| Soylu, 2010 | [59] | Turkey | Native | GN | no GN | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | DGF | no DGF | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | AR | no AR | No effect | ||
| Mutlubas, 2009 | [58] | Turkey | Transplant | GF | no GF | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | GF | no GF | No effect | ||
| TLR4 | p.Asp299Gly | Nogueira, 2007 | [12] | Brazil | Native | CKD | Healthy | No effect |
| Mutlubas, 2009 | [58] | Turkey | Native | CKD | Healthy | No effect | ||
| Akil, 2012 | [60] | Turkey | Native | CKD | no CKD | Increased risk | ||
| Bayram, 2013 | [62] | Turkey | Native | CKD | no CKD | No effect | ||
| Soylu, 2010 | [59] | Turkey | Native | GN | no GN | No effect | ||
| Karoly, 2007 | [61] | Hungary | Native | PN | no PN | Increased risk | ||
| Akil, 2012 | [60] | Turkey | Native | PN | no PN | Increased risk | ||
| Bayram, 2013 | [62] | Turkey | Native | PN | no PN | No effect | ||
| Nogueira, 2007 | [12] | Brazil | Transplant | DGF 2 | no DGF 2 | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | DGF 2 | no DGF 2 | No effect | ||
| Ducloux, 2005 | [7] | France | Transplant | AR 2 | no AR 2 | Decreased risk | ||
| Palmer, 2006 | [8] | United States | Transplant | AR 1 | no AR 1 | Decreased risk | ||
| Fekete, 2006 | [63] | Hungary | Transplant | AR 2 | no AR 2 | Decreased risk | ||
| Nogueira, 2007 | [12] | Brazil | Transplant | AR 2 | no AR 2 | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | AR 2 | no AR 2 | No effect | ||
| Krichen, 2013 | [18] | Tunesia | Transplant | AR 2 | no AR 2 | No effect | ||
| Nogueira, 2007 | [12] | Brazil | Transplant | PN 2 | no PN 2 | No effect | ||
| Ducloux, 2005 | [7] | France | Transplant | GF 2 | no GF 2 | No effect | ||
| Mutlubas, 2009 | [58] | Turkey | Transplant | GF 2 | no GF 2 | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | GF 2 | no GF 2 | No effect | ||
| TLR4 | p.Thr399Ile | Nogueira, 2007 | [12] | Brazil | Native | CKD | Healthy | No effect |
| Mutlubas, 2009 | [58] | Turkey | Native | CKD | Healthy | No effect | ||
| Bayram, 2013 | [62] | Turkey | Native | CKD | no CKD | No effect | ||
| Soylu, 2010 | [59] | Turkey | Native | GN | no GN | No effect | ||
| Nogueira, 2007 | [12] | Brazil | Transplant | DGF | no DGF 2 | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | DGF | no DGF 2 | No effect | ||
| Ducloux, 2005 | [7] | France | Transplant | AR 2 | no AR 2 | Decreased risk | ||
| Palmer, 2006 | [8] | United States | Transplant | AR 1 | no AR 1 | Decreased risk | ||
| Nogueira, 2007 | [12] | Brazil | Transplant | AR 2 | no AR 2 | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | AR 2 | no AR 2 | No effect | ||
| Nogueira, 2007 | [12] | Brazil | Transplant | PN 2 | no PN 2 | No effect | ||
| Ducloux, 2005 | [7] | France | Transplant | GF 2 | no GF 2 | No effect | ||
| Mutlubas, 2009 | [58] | Turkey | Transplant | GF 2 | no GF 2 | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | GF 2 | no GF 2 | No effect | ||
| TLR5 | p.Arg392Ter | Cheng, 2013 | [57] | Taiwan | Native | PN | no PN | No effect |
| Krüger, 2010 | [13] | Germany | Transplant | DGF | no DGF | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | AR | no AR | No effect | ||
| Krüger, 2010 | [13] | Germany | Transplant | GF | no GF | No effect | ||
| TLR5 | p.Phe616Leu | - | - | - | - | - | - | - |
| TLR6 | p.Ser249Pro | - | - | - | - | - | - | - |
| TLR7 | p.Gln11Leu | - | - | - | - | - | - | - |
| TLR8 | p.Met1Val | - | - | - | - | - | - | - |
1SNP in donors
2SNP in recipients.
Native = renal diseases of the native kidneys, transplant = renal diseases after transplantation, GN = glomerulonephritis, Healthy = healthy controls, PN = pyelonephritis, CKD = chronic kidney disease, DGF = delayed graft function, AR = acute rejection, GF = graft failure.
Chronic renal allograft failure on the other hand is known to develop from both immune and non-immune damage to the graft. In our study we were unable to show an association of any of the TLR SNPs in either donor or recipient with surrogate and definite outcomes. Table 8 provides a list of the same TLR SNPs and their association with transplant outcomes as described in the literature [7,8,12,13,18,58,63]. Comparable to our study, Krüger et al. [13] and Mutlubas et al. [58] did not identify an association of TLR2 p.Arg753Gln with renal transplant outcomes. In the context of renal transplantation, TLR4 p.Asp299Gly and p.Thr399Ile are also among the most studied TLR SNPs [7,8,12,13,18,58,63]. Contrary to the well-known role for TLR4 in experimental ischemia-reperfusion injury, none of the studies (including the current) found an association with either of the two TLR4 SNPs with the development of delayed graft function [12,13]. Delayed graft function is a heterogeneous and arbitrary outcome measure, that includes the effect of donation type, donor/recipient age, ischemia times (cold and warm) but also allo-immune phenomena, which makes a direct comparison with experimentally controlled ischemia-reperfusion injury difficult and this difference should be taken into account when interpreting genetic analyses. A potential role for TLR4 SNPs in the context of acute rejection generated conflicting results with half of the studies describing a protective effect [7,8,63] and half of the studies describing no effect at all [12,13,18]. Our study, which is the largest investigating both TLR4 SNPs to date, is in line with the latter studies. Interestingly, none of the studies (including the current) describes an effect on the cumulative incidence of graft failure [7,13,58]. One could wonder why a loss-of-function polymorphism in TLR4 leads to a lower incidence of acute rejection, but not graft failure. If there is a role for TLR4 polymorphisms in renal transplantation, it is probably a minor one that is overshadowed by other immune and non-immune phenomena that take place in the graft. Finally, Krüger et al. [13], in line with our study, did not find an association of TLR5 p.Arg392Ter with renal transplant outcomes. Even though experimental renal transplantation studies have consistently shown that TLR engagement can break allograft tolerance while inhibition of for instance TLR2 and -4 signalling improves allograft acceptance [48,53,64–66], in human renal transplantation the consequences of non-synonymous SNPs in TLR genes appear to be not as devastating for transplant outcome.
In conclusion, TLR gene polymorphisms are enriched in patients with end-stage renal disease and may contribute to the final common pathway of renal injury whereas after renal transplantation, this effect for the TLR gene polymorphisms was not observed. This difference in effect size by the TLR gene polymorphisms highlights that the development of chronic kidney disease of the native kidneys and chronic kidney disease in the context of renal transplantation might be explained by different risk factors.
Supporting Information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Dutch Kidney Foundation (www.nierstichting.nl), grant no. C06.6023. The funder had no role in study design, data collection, analysis, preparation of the manuscript or decision to publish. European Union FP6 program me grant no. EU_FP-6 037697. The funder had no role in study design, data collection, analysis, preparation of the manuscript or decision to publish.
References
- 1. Gluba A, Banach M, Hannam S, Mikhailidis DP, Sakowicz A, Rysz J. The role of Toll-like receptors in renal diseases. Nat Rev Nephrol. 2010;6(4):224–35. 10.1038/nrneph.2010.16 [DOI] [PubMed] [Google Scholar]
- 2. Leemans JC, Kors L, Anders H-J, Florquin S. Pattern recognition receptors and the inflammasome in kidney disease. Nat Rev Nephrol. 2014;10(7):398–414. 10.1038/nrneph.2014.91 [DOI] [PubMed] [Google Scholar]
- 3. Liew FY, Xu D, Brint EK, O’Neill LAJ. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5(6):446–58. [DOI] [PubMed] [Google Scholar]
- 4. Kawai T, Akira S. TLR signaling. Seminars in Immunology. 2007. p. 24–32. [DOI] [PubMed] [Google Scholar]
- 5. Sivick KE, Mobley HLT. Waging war against uropathogenic Escherichia coli: Winning back the urinary tract. Infection and Immunity. 2010. p. 568–85. 10.1128/IAI.01000-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kruger B, Krick S, Dhillon N, Lerner SM, Ames S, Bromberg JS, et al. Donor Toll-like receptor 4 contributes to ischemia and reperfusion injury following human kidney transplantation. Proc Natl Acad Sci U S A. 2009;106(9):3390–5. 10.1073/pnas.0810169106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ducloux D, Deschamps M, Yannaraki M, Ferrand C, Bamoulid J, Saas P, et al. Relevance of Toll-like receptor–4 polymorphisms in renal transplantation. Kidney Int. 2005;67(6):2454–61. [DOI] [PubMed] [Google Scholar]
- 8. Palmer SM, Burch LH, Mir S, Smith SR, Kuo PC, Herczyk WF, et al. Donor polymorphisms in Toll-like receptor–4 influence the development of rejection after renal transplantation. Clin Transplant. 2006;20(1):30–6. [DOI] [PubMed] [Google Scholar]
- 9. Cervera C, Lozano F, Saval N, Gimferrer I, Ibañez A, Suárez B, et al. The influence of innate immunity gene receptors polymorphisms in renal transplant infections. Transplantation. 2007;83(11):1493–500. [DOI] [PubMed] [Google Scholar]
- 10. Fekete A, Viklicky O, Hubacek JA, Rusai K, Erdei G, Treszl A, et al. Association between heat shock protein 70s and toll-like receptor polymorphisms with long-term renal allograft survival. Transpl Int. 2006;19(3):190–6. [DOI] [PubMed] [Google Scholar]
- 11. Mutlubas F, Mir S, Berdeli A, Ozkayin N, Sozeri B. Association Between Toll-like Receptors 4 and 2 Gene Polymorphisms With Chronic Allograft Nephropathy in Turkish Children. Transpl Proc. 2009;41(5):1589–93. [DOI] [PubMed] [Google Scholar]
- 12. Nogueira E, Ozaki KS, Macusso GD, Quarim RF, Câmara NOS, Pacheco-Silva A. Incidence of Donor and Recipient Toll-Like Receptor–4 Polymorphisms in Kidney Transplantation. Transplant Proc. 2007;39(2):412–4. [DOI] [PubMed] [Google Scholar]
- 13. Krüger B, Banas MC, Walberer A, Böger CA, Farkas S, Hoffmann U, et al. A comprehensive genotype-phenotype interaction of different Toll-like receptor variations in a renal transplant cohort. Clin Sci (Lond). 2010;119(12):535–44. [DOI] [PubMed] [Google Scholar]
- 14. Israni A, Leduc R, Holmes J, Jacobson PA, Lamba V, Guan W, et al. Single-nucleotide polymorphisms, acute rejection, and severity of tubulitis in kidney transplantation, accounting for center-to-center variation. Transplantation. 2010;90(12):1401–8. 10.1097/TP.0b013e3182000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Srivastava P, Singh A, Kesarwani P, Jaiswal PK, Singh V, Mittal RD. Association studies of Toll-like receptor gene polymorphisms with allograft survival in renal transplant recipients of North India. Clin Transpl. 2012;26(4):581–8. [DOI] [PubMed] [Google Scholar]
- 16. Eikmans M, de Canck I, van der Pol P, Baan CC, Haasnoot GW, Mallat MJK, et al. The Functional Polymorphism Ala258Ser in the Innate Receptor Gene Ficolin–2 in the Donor Predicts Improved Renal Transplant Outcome. Transplantation Journal. 2012. p. 478–85. [DOI] [PubMed] [Google Scholar]
- 17. Kim TH, Jeong K-H, Kim SK, Lee SH, Ihm CG, Lee TW, et al. TLR9 gene polymorphism (rs187084, rs352140): association with acute rejection and estimated glomerular filtration rate in renal transplant recipients. Int J Immunogenet. 2013. December;40(6):502–8. 10.1111/iji.12069 [DOI] [PubMed] [Google Scholar]
- 18. Krichen H, Gorgi Y, Dhaouadi T, Mecheri Y, Sfar I, Bardi R, et al. Toll-like receptor 4 and CD14 gene polymorphisms in Tunisian kidney transplantation. Transplant Proc. 2013;45(10):3472–7. 10.1016/j.transproceed.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 19. Damman J, Daha MR, Leuvenink HG, Van Goor H, Hillebrands JL, Dijk MC Van, et al. Association of complement C3 gene variants with renal transplant outcome of deceased cardiac dead donor kidneys. Am J Transplant. 2012;12(3):660–8. 10.1111/j.1600-6143.2011.03880.x [DOI] [PubMed] [Google Scholar]
- 20. Reznichenko A, Snieder H, van den Born J, de Borst MH, Damman J, van Dijk MCRF, et al. CUBN as a novel locus for end-stage renal disease: Insights from renal transplantation. PLoS One. 2012;7(5). 10.1371/journal.pone.0036512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M a R, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6(2):121–33. 10.1038/nprot.2010.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006. February;38(2):209–13. [DOI] [PubMed] [Google Scholar]
- 24. Randhawa AK, Shey MS, Keyser A, Peixoto B, Wells RD, de Kock M, et al. Association of human TLR1 and TLR6 deficiency with altered immune responses to bcg vaccination in south african infants. PLoS Pathog. 2011;7(8). 10.1371/journal.ppat.1002174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shey MS, Randhawa AK, Bowmaker M, Smith E, Scriba TJ, de Kock M, et al. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin–6 secretion. Genes Immun. 2010;11(7):561–72. 10.1038/gene.2010.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Omueti KO, Mazur DJ, Thompson KS, Lyle EA, Tapping RI. The polymorphism P315L of human toll-like receptor 1 impairs innate immune sensing of microbial cell wall components. J Immunol. 2007;178(10):6387–94. [DOI] [PubMed] [Google Scholar]
- 27. Johnson CM, Lyle EA, Omueti KO, Stepensky VA, Yegin O, Alpsoy E, et al. Cutting edge: A common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol. 2007;178(12):7520–4. [DOI] [PubMed] [Google Scholar]
- 28. Xiong Y, Song C, Snyder GA, Sundberg EJ, Medvedev AE. R753Q polymorphism inhibits toll-like receptor (TLR) 2 tyrosine phosphorylation, dimerization with TLR6, and recruitment of myeloid differentiation primary response protein 88. J Biol Chem. 2012;287(45):38327–37. 10.1074/jbc.M112.375493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schröder NWJ, Diterich I, Zinke A, Eckert J, Draing C, von Baehr V, et al. Heterozygous Arg753Gln polymorphism of human TLR–2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J Immunol. 2005;175(4):2534–40. [DOI] [PubMed] [Google Scholar]
- 30. Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun. 2000;68(11):6398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ranjith-Kumar CT, Miller W, Sun J, Xiong J, Santos J, Yarbrough I, et al. Effects of single nucleotide polymorphisms on Toll-like receptor 3 activity and expression in cultured cells. J Biol Chem. 2007;282(24):17696–705. [DOI] [PubMed] [Google Scholar]
- 32. Gorbea C, Makar KA, Pauschinger M, Pratt G, Bersola JLF, Varela J, et al. A role for toll-like receptor 3 variants in host susceptibility to enteroviral myocarditis and dilated cardiomyopathy. J Biol Chem. 2010;285(30):23208–23. 10.1074/jbc.M109.047464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sironi M, Biasin M, Cagliani R, Forni D, De Luca M, Saulle I, et al. A common polymorphism in TLR3 confers natural resistance to HIV–1 infection. J Immunol. American Association of Immunologists; 2012. January 15;188(2):818–23. [DOI] [PubMed] [Google Scholar]
- 34. Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25(2):187–91. [DOI] [PubMed] [Google Scholar]
- 35. Kiechl S, Lorenz E, Reindl M, Wiedermann CJ, Oberhollenzer F, Bonora E, et al. Toll-like receptor 4 polymorphisms and atherogenesis. N Engl J Med. 2002;347(3):185–92. [DOI] [PubMed] [Google Scholar]
- 36. Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires’ disease. J Exp Med. 2003;198(10):1563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hawn TR, Wu H, Grossman JM, Hahn BH, Tsao BP, Aderem A. A stop codon polymorphism of Toll-like receptor 5 is associated with resistance to systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2005;102(30):10593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. West TE, Chantratita N, Chierakul W, Limmathurotsakul D, Wuthiekanun V, Myers ND, et al. Impaired TLR5 functionality is associated with survival in melioidosis. J Immunol. 2013;190(7):3373–9. 10.4049/jimmunol.1202974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klimosch SN, Försti A, Eckert J, Knezevic J, Bevier M, von Schönfels W, et al. Functional TLR5 genetic variants affect human colorectal cancer survival. Cancer Res. 2013;73(24):7232–42. 10.1158/0008-5472.CAN-13-1746 [DOI] [PubMed] [Google Scholar]
- 40. Sheridan J, Mack DR, Amre DK, Israel DM, Cherkasov A, Li H, et al. A non-synonymous coding variant (L616F) in the TLR5 gene is potentially associated with Crohn’s disease and influences responses to bacterial flagellin. PLoS One. Public Library of Science; 2013. January 11;8(4):e61326 10.1371/journal.pone.0061326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dhiman N, Ovsyannikova IG, Vierkant RA, Ryan JE, Shane Pankratz V, Jacobson RM, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: Preliminary results. Vaccine. 2008;26(14):1731–6. 10.1016/j.vaccine.2008.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oh D-Y, Baumann K, Hamouda O, Eckert JK, Neumann K, Kücherer C, et al. A frequent functional toll-like receptor 7 polymorphism is associated with accelerated HIV–1 disease progression. AIDS. 2009;23(3):297–307. 10.1097/QAD.0b013e32831fb540 [DOI] [PubMed] [Google Scholar]
- 43. Askar E, Ramadori G, Mihm S. Toll-like receptor 7 rs179008/Gln11Leu gene variants in chronic hepatitis C virus infection. J Med Virol. 2010. November;82(11):1859–68. 10.1002/jmv.21893 [DOI] [PubMed] [Google Scholar]
- 44. Wang C-H, Eng H-L, Lin K-H, Liu H-C, Chang C-H, Lin T-M. Functional Polymorphisms of TLR8 Are Associated With Hepatitis C Virus Infection. Immunology. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oh D-Y, Taube S, Hamouda O, Kücherer C, Poggensee G, Jessen H, et al. A functional toll-like receptor 8 variant is associated with HIV disease restriction. J Infect Dis. 2008;198(5):701–9. 10.1086/590431 [DOI] [PubMed] [Google Scholar]
- 46. Leemans JC, Stokman G, Claessen N, Rouschop KM, Teske GJD, Kirschning CJ, et al. Renal-associated TLR2 mediates ischemia/reperfusion injury in the kidney. J Clin Invest. 2005. October 1;115(10):2894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu H, Chen G, Wyburn KR, Yin J, Bertolino P, Eris JM, et al. TLR4 activation mediates kidney ischemia/reperfusion injury. J Clin Invest. 2007;117(10):2847–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Farrar CA, Keogh B, McCormack W, O’Shaughnessy A, Parker A, Reilly M, et al. Inhibition of TLR2 promotes graft function in a murine model of renal transplant ischemia-reperfusion injury. The FASEB Journal. 2012. p. 799–807. 10.1096/fj.11-195396 [DOI] [PubMed] [Google Scholar]
- 49. Shigeoka AA, Holscher TD, King AJ, Hall FW, Kiosses WB, Tobias PS, et al. TLR2 is constitutively expressed within the kidney and participates in ischemic renal injury through both MyD88-dependent and -independent pathways. J Immunol. 2007;178(10):6252–8. [DOI] [PubMed] [Google Scholar]
- 50. Pulskens WP, Rampanelli E, Teske GJ, Butter LM, Claessen N, Luirink IK, et al. TLR4 promotes fibrosis but attenuates tubular damage in progressive renal injury. J Am Soc Nephrol. 2010;21(8):1299–308. 10.1681/ASN.2009070722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pulskens WP, Teske GJ, Butter LM, Roelofs JJ, van der Poll T, Florquin S, et al. Toll-like receptor–4 coordinates the innate immune response of the kidney to renal ischemia/reperfusion injury. PLoS One. 2008;3(10). 10.1371/journal.pone.0003596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leemans JC, Butter LM, Pulskens WPC, Teske GJD, Claessen N, van der Poll T, et al. The role of toll-like receptor 2 in inflammation and fibrosis during progressive renal injury. PLoS One. 2009;4(5). 10.1371/journal.pone.0005704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noris M, Cassis P, Azzollini N, Cavinato R, Cugini D, Casiraghi F, et al. The Toll-IL-1R member Tir8/SIGIRR negatively regulates adaptive immunity against kidney grafts. J Immunol. 2009;183(7):4249–60. 10.4049/jimmunol.0803549 [DOI] [PubMed] [Google Scholar]
- 54. Skuginna V, Lech M, Allam R, Ryu M, Clauss S, Susanti HE, et al. Toll-like receptor signaling and sigirr in renal fibrosis upon unilateral ureteral obstruction. PLoS One. 2011;6(4). 10.1371/journal.pone.0019204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lech M, Avila-Ferrufino A, Allam R, Segerer S, Khandoga A, Krombach F, et al. Resident dendritic cells prevent postischemic acute renal failure by help of single Ig IL–1 receptor-related protein. J Immunol. 2009;183(6):4109–18. 10.4049/jimmunol.0900118 [DOI] [PubMed] [Google Scholar]
- 56. Lee JS, Park H-K, Suh J-S, Hahn W-H, Kang SW, Park HJ, et al. Toll-like receptor 1 gene polymorphisms in childhood IgA nephropathy: a case-control study in the Korean population. Int J Immunogenet. 2011;38(2):133–8. 10.1111/j.1744-313X.2010.00978.x [DOI] [PubMed] [Google Scholar]
- 57. Cheng C-H, Lee Y-S, Chang C-J, Lin T-Y. Genetic polymorphisms in Toll-like receptors among pediatric patients with renal parenchymal infections of different clinical severities. PLoS One. 2013;8(3):e58687 10.1371/journal.pone.0058687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mutlubas F, Mir S, Berdeli A, Ozkayin N, Sozeri B. Association Between Toll-like Receptors 4 and 2 Gene Polymorphisms With Chronic Allograft Nephropathy in Turkish Children. Transplant Proc. 2009;41(5):1589–93. 10.1016/j.transproceed.2009.02.079 [DOI] [PubMed] [Google Scholar]
- 59. Soylu A, Kizildaǧ S, Kavukçu S, Cingöz S, Türkmen M, Demir BK, et al. TLR–2 Arg753Gln, TLR–4 Asp299Gly, and TLR–4 Thr399Ile polymorphisms in Henoch Schonlein purpura with and without renal involvement. Rheumatol Int. 2010;30(5):667–70. 10.1007/s00296-009-1052-y [DOI] [PubMed] [Google Scholar]
- 60. Akil I, Ozkinay F, Onay H, Canda E, Gumuser G, Kavukcu S. Assessment of toll-like receptor–4 gene polymorphism on pyelonephritis and renal scar. Int J Immunogenet. 2012;39(4):303–7. 10.1111/j.1744-313X.2012.01090.x [DOI] [PubMed] [Google Scholar]
- 61. Karoly E, Fekete A, Banki NF, Szebeni B, Vannay A, Szabo AJ, et al. Heat shock protein 72 (HSPA1B) gene polymorphism and toll-like receptor (TLR) 4 mutation are associated with increased risk of urinary tract infection in children. Pediatr Res. 2007;61(3):371–4. [DOI] [PubMed] [Google Scholar]
- 62. Bayram MT, Soylu A, Ateş H, Kizildaǧ S, Kavukçu S. TLR–4 polymorphisms and leukocyte TLR–4 expression in febrile UTI and renal scarring. Pediatr Nephrol. 2013;28(9):1827–35. 10.1007/s00467-013-2478-8 [DOI] [PubMed] [Google Scholar]
- 63. Fekete A, Viklický O, Hubácek JA, Rusai K, Erdei G, Treszl A, et al. Association between heat shock protein 70s and toll-like receptor polymorphisms with long-term renal allograft survival. Transpl Int. 2006;19(3):190–6. [DOI] [PubMed] [Google Scholar]
- 64. Wang S, Schmaderer C, Kiss E, Schmidt C, Bonrouhi M, Porubsky S, et al. Recipient Toll-like receptors contribute to chronic graft dysfunction by both MyD88- and TRIF-dependent signaling. Dis Model Mech. 2010. January 1;3(1–2):92–103. 10.1242/dmm.003533 [DOI] [PubMed] [Google Scholar]
- 65. Wu J-F, Chen C-H, Ni Y-H, Lin Y-T, Chen H-L, Hsu H-Y, et al. Toll-like receptor and hepatitis B virus clearance in chronic infected patients: a long-term prospective cohort study in Taiwan. J Infect Dis. 2012;206(5):662–8. 10.1093/infdis/jis420 [DOI] [PubMed] [Google Scholar]
- 66. Chen L, Wang T, Zhou P, Ma L, Yin D, Shen J, et al. TLR engagement prevents transplantation tolerance. Am J Transplant. 2006;6(10):2282–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.