Abstract
Large scale antibody responses in Plasmodium vivax malaria remains unexplored in the endemic setting. Protein microarray analysis of asexual-stage P. vivax was used to identify antigens recognized in sera from residents of hypoendemic Peruvian Amazon. Over 24 months, of 106 participants, 91 had two symptomatic P. vivax malaria episodes, 11 had three episodes, 3 had four episodes, and 1 had five episodes. Plasmodium vivax relapse was distinguished from reinfection by a merozoite surface protein-3α restriction fragment length polymorphism polymerase chain reaction (MSP3α PCR-RFLP) assay. Notably, P. vivax reinfection subjects did not have higher reactivity to the entire set of recognized P. vivax blood-stage antigens than relapse subjects, regardless of the number of malaria episodes. The most highly recognized P. vivax proteins were MSP 4, 7, 8, and 10 (PVX_003775, PVX_082650, PVX_097625, and PVX_114145); sexual-stage antigen s16 (PVX_000930); early transcribed membrane protein (PVX_090230); tryptophan-rich antigen (Pv-fam-a) (PVX_092995); apical merozoite antigen 1 (PVX_092275); and proteins of unknown function (PVX_081830, PVX_117680, PVX_118705, PVX_121935, PVX_097730, PVX_110935, PVX_115450, and PVX_082475). Genes encoding reactive proteins exhibited a significant enrichment of non-synonymous nucleotide variation, an observation suggesting immune selection. These data identify candidates for seroepidemiological tools to support malaria elimination efforts in P. vivax-endemic regions.
Introduction
Plasmodium vivax malaria affects human populations in Asia, Latin America, and parts of Africa.1,2 Plasmodium vivax malaria is conventionally considered to be less of a public health concern than the more lethal malaria parasite P. falciparum, which dominates malaria-related mortality predominantly in sub-Saharan Africa.3 The biology of P. vivax fundamental differs from P. falciparum particularly with regard to how this species maintains itself in human populations, particularly the ability to relapse from the dormant liver forms, the hypnozoite, and its wider tolerance of temperature conditions enabling sporogonic development.3 For these reasons, and because the parasite cannot be propagated in vitro, studies of this form of malaria are neglected. In an era when malaria eradication has been put on the global health agenda,4 an exigent need has emerged to understand detailed aspects of P. vivax, including parasite antigen-specific human immune responses and to develop rapid tools to assess transmission on a population level.
Current estimates of the global prevalence of P. vivax infection are based on identifying potential regions of transmission using a geo-referenced parasite-prevalence rate combined with biological masking to indicate malaria-compatible transmission zones.5 Quantifying the global burden of P. vivax malaria is complicated by the biology of the parasite: P. vivax relapse from dormant hypnozoite liver stage over time and space—often asymptomatically—which allows for human migration to disperse infection within an endemic region or reintroduce the parasite into areas where malaria may have been eliminated. The potential range of P. vivax transmission is vast, the biology of the parasite complicates incidence and prevalence estimates, and the public health impact of P. vivax remains. Regional elimination and global eradication require accurate and population-deployable tools to estimate parasite prevalence and malarial disease incidence.1,2,4,6
Because determining the presence of malaria parasitemia or exposure is time intensive, often insensitive, and expensive, interest has grown in using serological tools to monitor infection status and transmission dynamics—so-called seroepidemiology. This approach has been particularly useful for malaria7,8 and additionally for vector-borne diseases such as lymphatic filariasis in which mass drug administration campaigns have been carried out.9–14 Antigens for antibody detection have included lysates of in vitro grown P. falciparum schizonts8,15–20 and recombinant proteins based on vaccine candidates (circumsporozoite protein, merozoite surface protein 1 [MSP1], and apical membrane antigen-1 [AMA-1]) have been commonly used. By analogy, for seroepidemiological studies of P. vivax, recombinant PvMSP1 and PvAMA-1 have been used to determine seroprevalence of antibodies to P. vivax in populations of Vanuatu, Solomon Islands.7 More recently, a proof-of-principle array study using 152 predicted asexual-stage proteins was reported in which sera from Korean P. vivax malaria patients were analyzed for anti-P. vivax antibodies using a wheat germ expression system.21 Recent work from Papua New Guinea—where all four major Plasmodium spp. circulate at intense level—has led to the development of focused protein microarrays22 composed of both P. falciparum and P. vivax recombinant proteins. Because P. vivax samples from humans have low parasitemia, and only in limited quantities from non-human primates, seroepidemiology studies using P. vivax asexual-stage parasite lysates has only infrequently been done, hence recombinant proteins are essential for studying exposure to P. vivax in endemic populations.
To understand the P. vivax antigens predominantly recognized by the antibody response of naturally infected humans in a low-transmission region, we analyzed the serological reactivity of patients with P. vivax malaria using custom-made protein microarrays composed of asexual blood-stage antigens predicted by transcriptional profiling23,24 and heterologously produced by a prokaryotic cell-free expression system.25–27 With the validation of molecular tools to distinguish P. vivax relapses and reinfections that are common in our Amazonian village study population,28,29 we compared the level of IgG responses between consecutive episodes of symptomatic P. vivax malaria, in particular, to determine whether naturally acquired antibody responses were boosted by subsequent infection. More generally, we intended to systematically and comprehensively identify P. vivax antigens of potential use for seroepidemiological studies and possibly for identifying vaccine candidates.
Materials and Methods
Ethics statement.
This study was approved by the following institutional review boards: Ethical Committee of Universidad Peruana Cayetano Heredia, Lima, Peru; Ethical Committee of Asociación Benéfica PRISMA, Lima, Peru; the Directorate of Health of Loreto-Peru; the Humans Research Protection Program of the University of California at San Diego, La Jolla, CA; and the Institutional Review Board of the Johns Hopkins Bloomberg School of Public Health, Baltimore, MD. All participants provided written informed consent and were coded through all study participation. Specimens were also coded before protein microarray analysis.
Study sites.
The field activities of this study were carried out from May 2005 to July 2008 in northeastern Peru, in villages near to the city of Iquitos in the province of Maynas, the capital of the Amazon Department of Loreto (map in Ref. 28). Considering the region's geographical isolation from the rest of Peru, health services within the surrounding areas of Iquitos are relatively good and accessible. Three health posts provide medical services to the study villages described as follows. Santo Tomas health post, located 16 km by road from Iquitos and surrounded by the Nanay River, is a referral health center for three villages: La Unión, 12 de Diciembre, and Santo Tomás, collectively 2,650 inhabitants. San Jose de Lupuna health post, located 10 km from Iquitos is accessible only through the Nanay River and is the referral center for four villages: San Pedro, Santa Rita, Fray Martín, and San Jose de Lupuna, with collectively 1,250 inhabitants. Padrecocha health post, located 6 km from Iquitos is accessible only through the Nanay River and is a referral health post for three villages: San Andrés, Nueva Vida, and Padrecocha, with collectively 1,800 inhabitants.
Blood sampling.
Blood samples were collected from patients diagnosed by conventional light microscopy on site at Peruvian Ministry of Health establishments according to national norms. Patients identified as infected with only P. vivax (microscopy only) were invited to participate in the study. At each enrollment site, venous blood was collected in EDTA Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ) and taken to the field project laboratory where samples were aliquoted and frozen at −20°C. Samples were shipped on dry ice to Universidad Peruana Cayetano Heredia in Lima for molecular diagnostic analysis.
Molecular confirmation of P. vivax infection.
DNA was extracted from 200 μL of thawed, anticoagulated whole blood using the Qiagen Blood Kit (Qiagen, Valencia, CA). The diagnosis of P. vivax infections was confirmed in all patients using a genus and species-specific nested polymerase chain reaction (PCR) assay based on an 18S ribosomal RNA gene fragment specific for the genus Plasmodium, as previously reported.28,30 A 1,200-bp (base pair) fragment allowed for Plasmodium genus-level identification; identification of P. vivax and P. falciparum species was done using a 120-bp fragment specific for P. vivax and a 205-bp fragment specific for P. falciparum; samples containing P. falciparum or mixed infections were not considered for further analysis, P. malariae or P. ovale were not found.
Molecular genotyping.
Genotyping of P. vivax isolates was performed by PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the P. vivax MSP3α (PvMSP3α) gene as previously described.31
Case definition and data analysis.
Relapse was defined as identical PvMSP3α PCR-RFLP patterns when primary and subsequent infection parasites were compared. Reinfection was defined as different genotypes present in primary versus subsequent infection parasites by PvMSP3α PCR-RFLP patterns.28,29,32,33
Protein microarray analysis using a P. vivax asexual blood-stage proteome array.
A proteome array was designed containing 2,233 P. vivax recombinant proteins, representing 1,936 predicted P. vivax asexual blood-stage proteins and comprised as recently described.22 Proteins predicted to be secreted or present on the parasite surface, on the basis of the presence of a secretory signal peptide or transmembrane domains, were annotated as such in PlasmoDB34,35 and preferentially included. Additional selection for expression of P. vivax genes included expression microarray evidence for blood-stage expression.23,24 Putative cytoplasmic proteins, lacking both signal peptides and transmembrane domains, were also selected (isoelectric point [pI] < 7.6 for P. vivax genes). Plasmodium vivax vir genes were excluded because of their variability. Both single and multi-exon genes were selected. Exons of multi-exon genes were cloned and expressed separately. Furthermore, large genes or exons were further divided into overlapping segments to limit amplicon length between 300 and 3,000 nucleotides. Amplicons were labeled with the exon number and the total number of exons, such as “1o2” for exon 1 of a 2-exon gene. Genes that were further divided into segments were labeled s1, s2, and so forth.
The array was fabricated as previously described.36 In brief, target sequences were amplified genes from P. vivax genomic DNA (Sal I strain [MRA-552, MR4]) and cloned into the PXT7 plasmid with a T7 transcription terminator and encoded 5′ polyhistidine (HIS) and 3′ hemagglutinin (HA) epitopes. Recombinant proteins were expressed by an Escherichia coli cell-free in vitro transcription and translation system (Rapid Translation System 100 High Yield [RTS 100 HY] kits from 5 PRIME, Gaithersburg, MD) according to the manufacturer's instructions. Proteome arrays were printed as previously described, with each recombinant protein spotted once in each array.36 Each array contained 256 negative control spots made with an in vitro transcription–translation master mix without plasmid DNA. Once printed, recombinant protein expression was verified using anti-polyhistidine (clone His-1; Sigma-Aldrich, St. Louis, MO) and anti-hemagglutinin (clone 3F10; Roche, Indianapolis, IN) monoclonal antibodies, as previously described.36 All signal intensities were corrected for spot-specific background. A recombinant protein was deemed to be present on the slide if its spot's mean fluorescence intensity was higher than the mean of the “no DNA” control spots plus two standard deviations.
Overall, 1,470 different P. vivax recombinant proteins (66%) were expressed with both His and HA tags, confirming that the majority of recombinant proteins were fully expressed. These represented 1,328 P. vivax-predicted asexual blood-stage proteins. Furthermore 1,949 P. vivax recombinant proteins (87%) were expressed with at least one tag suggesting that these recombinant proteins were at least partially expressed.22
Microarray data analysis.
A reactive protein was defined as a protein that had a signal intensity values > 2-fold the mean of the “no DNA” intensity values for the same sample (i.e., column on the matrix) in at least 10% of the samples in the Pv-infected group, Pv reinfected group or the Pv relapse group. The background-corrected signal intensity values for reactive antigens were log 2 transformed and normalized by subtracting the median of the “no DNA” spots in the log space. t tests were used to identify differentially reactive antigens. The Benjamini–Hochberg (BH) method was used for multiple test correction, which estimates the False Discovery Rate (FDR) and provide corrected P values. Statistical analysis and figure generation was performed in the R environment (www.r-project.org). Paired Student's t test was used to compare differences in total antibody responses between relapsed and reinfected subjects.
Analysis of genome-wide P. vivax single nucleotide polymorphisms.
The genes encoding were compared with the four globally represented and annotated P. vivax genomes (parasites from Brazil, India, Mauritania, and North Korea), available at: https://www.broadinstitute.org/annotation/genome/plasmodium_vivax/GenomeDescriptions.html (accessed May 5, 2015).
Results
Number of P. vivax malaria episodes.
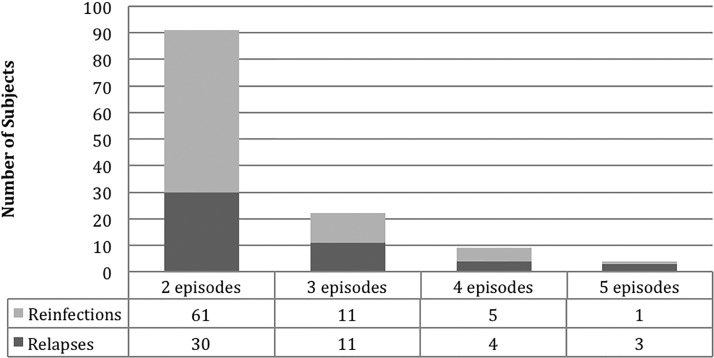
In the study period, 106 participants were enrolled who had 1–5 episodes of symptomatic light microscope- and PCR-confirmed P. vivax malaria (Table 1, Figure 1 ). The median age was 24 years, main age was 27 years, and 48% of the subjects were women. Occupation and demographics of the study participants were typical of the region (Table 2).
Table 1.
Characteristics of participants and recurrent episodes (relapses plus reinfections)
| Number | Mean age (years) [min–max] | Gender (%) | Median age (years) | Mean time to relapse/reinfection (months) [(min–max] | Mean lifetime malaria episodes | |
|---|---|---|---|---|---|---|
| Participants | 106 | 27 [3–83] | Female (48) | 24 | 8.5 [1–24.2] | 4.8 |
| Recurrences | 126 | 8.5 [1–24.2] | ||||
| Relapses | 48 | 26 [3–83] | Female (38) | 18 | 6 [1.2–20.4] | 4.3 |
| Reinfections | 78 | 28 [5–76] | Female (49) | 25 | 10 [1–24.2] | 5.2 |
max = maximum; min = minimum.
Figure 1.
Number of Plasmodium vivax relapses and reinfections in patient population.
Table 2.
Occupation/demographic characteristics of Plasmodium vivax malaria subjects
| Occupation | Relapses (%) | Reinfections (%) | Total (%) |
|---|---|---|---|
| Student | 23 (48) | 25 (32) | 48 (38) |
| Agriculture | 7 (15) | 12 (15) | 19 (15) |
| Homemaker | 6 (13) | 20 (26) | 26 (21) |
| Crafting | 0 (0) | 2 (3) | 2 (2) |
| Transportation | 1 (2) | 3 (4) | 4 (3) |
| Commerce | 1 (2) | 1 (1) | 2 (2) |
| Construction | 0 (0) | 8 (10) | 8 (6) |
| Unemployed | 4 (8) | 4 (5) | 8 (6) |
| Fisherman | 4 (8) | 2 (3) | 6 (5) |
| Logging | 0 (0) | 1 (1) | 1 (1) |
| < 5 years old | 2 (4) | 0 (0) | 2 (2) |
| Total | 48 (100) | 78 (100) | 126 (100) |
All infected participants were given standard antimalarial treatment following guidelines from the Peruvian Ministry of Health (chloroquine for 3 days (10 mg/kg on days 1 and 2 and 5 mg/kg on day 3), plus primaquine (0.5 mg/kg/day for 7 days)). Of the 106 subjects, 91 had two episodes, 11 had three episodes, 3 had four episodes, and 1 had five episodes. Using PvMSP3α genotyping, of the recurrent infection episodes, 48 had identical PvMSP3α PCR-RFLP patterns (hereafter referred to as “genotype”) compared with a previous infection leading to classification as relapse; 78 had different genotypes and were classified as reinfection. This classification is limited by the possibility of a subject harboring more than one P. vivax clone and hence overestimates the rate of reinfection compared with relapse. Subjects with P. vivax malaria tended to report more lifetime episodes of malaria than the episodes observed during the study period (Table 1). The average number of self-described malaria lifetime episodes for all participants was 4.8; for relapsing subjects, 4.3; and for reinfection subjects, 5.2.
Descriptive analysis of serological reactivity to P. vivax antigens using a protein microarray.
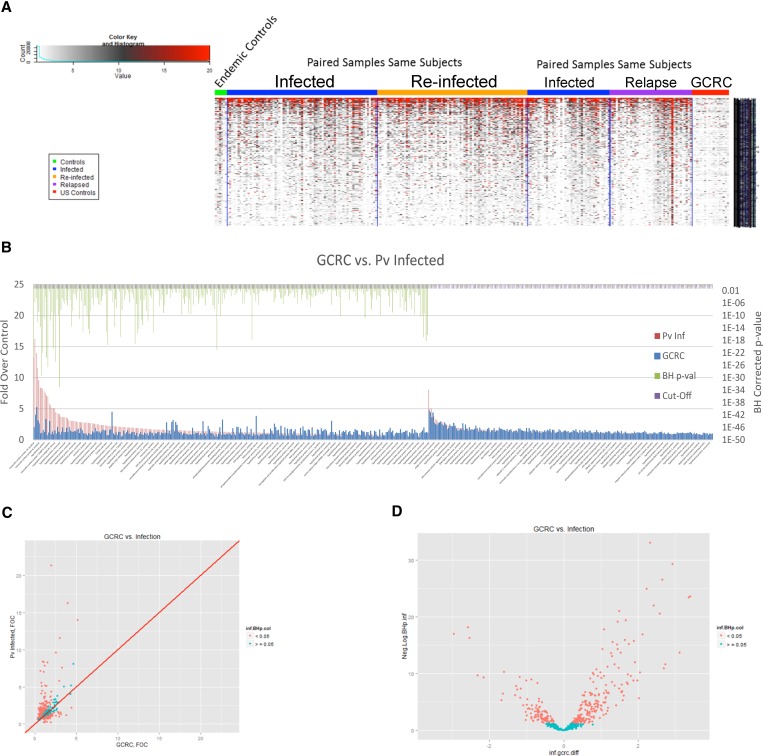
Endemic controls were not used for this determination because they have substantial reactivity against P. vivax antigens and therefore are not an appropriate control for the discovery of markers of exposure as shown in the heat map of the reactive antigens (Figure 2 ). Therefore, differentially reactive antigens in the text indicates results in relation to the negative control group with no history of malaria (N = 18, U.S. residents with no history of malaria). Of the 2,233 P. vivax protein spots (derived from 1,470 genes) on the protein microarray (see Supplemental Table 1 for complete list of arrayed recombinant proteins), 304 proteins spots were reactive when comparing P. vivax-infected subjects to a negative control group after BH P value correction (Figure 2).
Figure 2.
Protein microarray analysis comparing serological responses of Plasmodium vivax relapse vs. reinfection. (A) Key to heat map, indicating color associated with signal intensity (left). Displayed at the left of the heat map are results of uninfected controls from the Iquitos region (“endemic controls”); the middle two panels of the heat map show results of paired, sequential samples from P. vivax infection of same subjects comparing total Infected (this category does not distinguish relapse from re-infection) vs. Re-infected; the right two panels of the heat map show results of paired, sequential samples from P. vivax infected-subjects that compare total Infected vs. Relapse (based on shared molecular genotyping—see Methods). (B) Protein microarray analysis of sera from first observed episode of Plasmodium vivax malaria. Recognized proteins are indicated with a green bars above them on the left side of the figure, where the P value is less than 0.05 (corrected for multiple measures by the Benjamini Hochberg test (BH) P value). All P. vivax-infected (relapse plus reinfection) are indicated by (Pv Inf) and negative controls are non-malaria infected individuals from the USA controls (GCRC, General Clinical Research Center). The full list of recognized proteins is in Supplementary Table 1. (C) Scatter plots with the GCRC mean fold over control (FOC) values on the x-axis, the mean P. vivax Infected FOC values on the y-axis and the identity line in red are graphed (Pv antigens with BH P values < 0.05 are colored in salmon and the others in cyan). (D) Volcano plots with the log2 (fold difference) on the x-axis and the −log10(BH P value) on the y-axis (P. vivax antigens with BH P values < 0.05 are colored in salmon and the others in cyan) to illustrate the difference in antibody profiles between the groups.
The most highly recognized differentially reactive P. vivax proteins were MSP 4, 7, 8, and 10 (PVX_003775, PVX_082650, PVX_097625, and PVX_114145); sexual-stage antigen s16 (PVX_000930); early transcribed membrane protein (ETRAMP) (PVX_090230); tryptophan-rich antigen (Pv-fam-a) (PVX_092995); AMA 1 (PVX_092275); and proteins of unknown function (PVX_081830, PVX_117680, PVX_118705, PVX_121935, PVX_097730, PVX_110935, PVX_115450, and PVX_082475). The top 10 differentially reactive proteins and gene for all P. vivax-infected subjects are listed in Table 3. The complete list of differentially reactive proteins is presented in Supplemental Table 2.
Table 3.
Top 10 reactive proteins recognized in Plasmodium vivax malaria patients
| Protein | Exon | No. of exons | No. of tm domains | SignalP positive | Molecular weight (Da) | Isolectric point | Selection criteria | Description |
|---|---|---|---|---|---|---|---|---|
| PVX_114145 | 1 | 1 | 0 | Yes | 52,341 | 4.79 | SE_secreted | MSP10, putative |
| PVX_081830 | 2 | 2 | 0 | No | 56,607 | 4.64 | ME_cytoplasmic | Hypothetical protein |
| PVX_117680 | 1 | 2 | 0 | No | 128,624 | 5.47 | ME_cytoplasmic | Hypothetical protein, conserved |
| PVX_000930 | 1 | 2 | 2 | Yes | 15,068 | 10.46 | ME_secreted | Sexual stage antigen s16, putative |
| PVX_097625 | 1 | 1 | 1 | Yes | 54,742 | 4.92 | SE_secreted | MSP8, putative |
| PVX_118705 | 1 | 1 | 1 | No | 50,808 | 10.51 | SE_secreted | Hypothetical protein, conserved |
| PVX_121935 | 2 | 2 | 2 | Yes | 27,735 | 8.85 | ME_secreted | Hypothetical protein |
| PVX_097730 | 1 | 1 | 0 | Yes | 46,332 | 3.25 | SE_secreted | Hypothetical protein |
| PVX_110935 | 1 | 1 | 0 | Yes | 42,780 | 3.7 | SE_secreted | Hypothetical protein |
| PVX_082650 | 1 | 1 | 0 | Yes | 47,963 | 5.2 | SE_secreted | MSP7 (MSP7), putative |
MSP = merozoite surface protein; tm = transmembrane.
Reactive P. vivax antigens were not found more commonly in reinfection than in relapse and reinfection, and relapse had similar cumulative IgG signals.
Plasmodium vivax parasitemia may reflect one of several clinical states, relapse, which may be symptomatic or, most often in the Peruvian Amazon region, asymptomatic new infection,37 or a continuing previously un- or inadequately treated blood-stage infection. Subjects studied here represent either relapse or reinfection because blood-stage P. vivax is not known to be resistant to chloroquine in the region and all patients were treated with directly observed therapy (DOT) with standard therapy (chloroquine and primaquine).
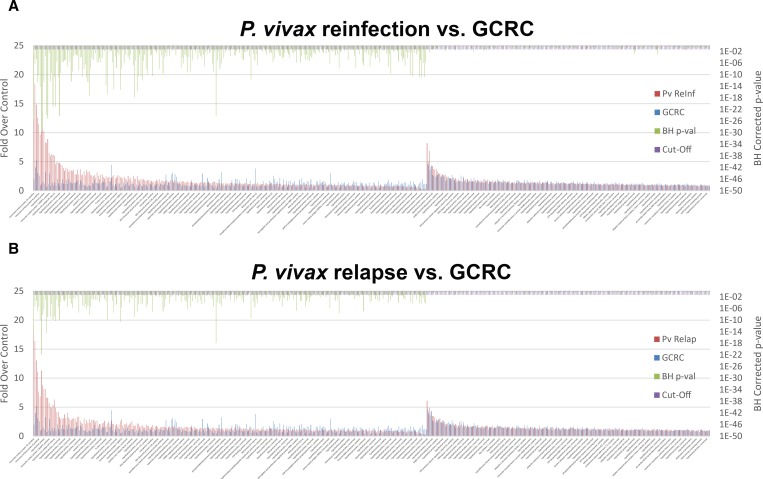
The antigens recognized by P. vivax-infected subjects were not more likely to have higher cumulative signal for subjects that were reinfected than for subjects undergoing a relapse (Figure 3 ). In addition, reinfected subjects did not recognize more of the antigens (285) than did the subjects that relapsed (219). In this study, a new P. vivax infection was only determined for symptomatic subjects.
Figure 3.
Protein microarray analysis comparing serological responses of subjects with Plasmodium vivax (Pv) reinfection to relapse (A and B, respectively). The number of reactive proteins and signal intensity of these proteins (top 10 proteins shown in Table 3) is not significantly different (paired Student's t test) in the reinfection group compared with the relapse group (complete list in Supplemental Table 2). For clarity, proteins that are not recognized are not shown. (C) The sum of intensities (y axis) for each sample (x axis), the median of these sums and a Volcano plot (D) demonstrates antibody titers and number of anti-Pv antigen antibodies.
Operationally, subjects who develop a recurrent infection within 28 days after prior episode were considered as treatment failure or recrudescence and could potentially have been due to tolerance or resistance to treatment. In this study, no subject developed a recurrent episode within 28 days after prior treatment. One subject developed a recurrent episode after 29 days but it had a different PvMSP3α pattern, hence was considered a reinfection for this analysis.
Paired analysis reveals that relapse was not associated with a greater increase in reactivity than reinfection.
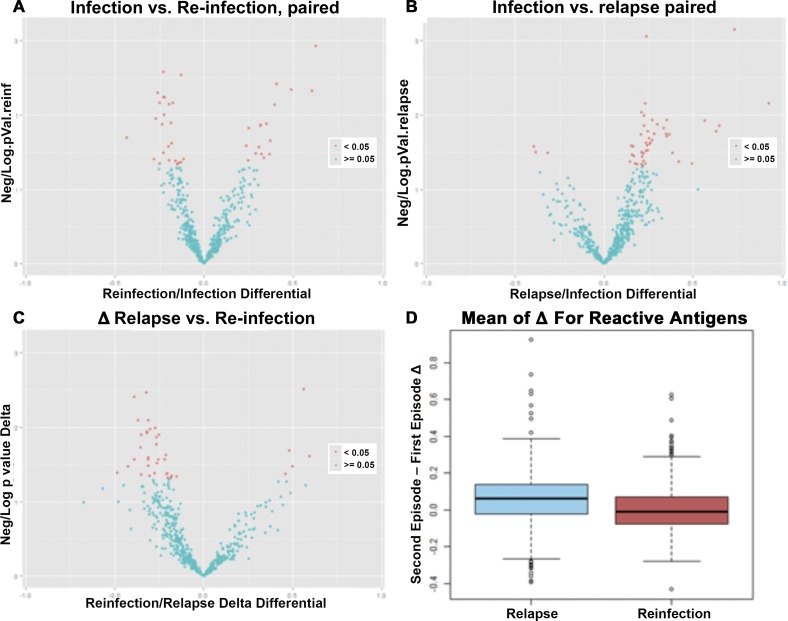
Although looking at the infected, reinfected, and relapse groups allowed us to find markers of exposure, there were multiple confounding factors that complicated comparison of the reinfected and the relapse groups. One confounder is precise knowledge of previous to P. vivax infections before episode 1 in this study, which would be useful in understanding changes in antibody profile after reinfection versus relapse. The current dataset had 113 subjects for whom samples for episodes one and two were available, of which 73 were defined as reinfection and 40 as relapse. Paired analysis looking at the changes elicited by the second episode for relapse subjects did not show increase after episode 2 compared with reinfection subjects; this latter group appeared to have just as many recognized antigens decreasing at episode 2 as increasing (Figure 4 ). Comparing the deltas (Second Episode–First Episode for each subject) observed in the two subject groups, the deltas for the reactive antigens in the relapse group were not greater in scope and magnitude than the deltas in the relapse group (Figure 4).
Figure 4.
Paired protein microarray analysis comparing changes (indicated as delta) in serological response between episodes 1 and 2 of subjects with Plasmodium vivax (Pv) relapse or reinfection, as defined by MSP3α PCR-RFLP. Volcano plots comparing episodes 1 and 2 for reinfection and relapse (A and B, respectively) show log2 of the fold change on the x axis and the –log10 (P value) on the y axis. (C) Comparison of reinfection to relapse using a volcano plot comparing the deltas for each subject and each antigen with the log2 of the fold difference on the x axis and the −log10 (P value) on the y axis. A boxplot of the mean of the deltas is shown. A boxplot of the mean of the deltas (D) illustrates the median and distribution of the mean of the deltas for the relapse group on the left and the reinfection group on the right.
Enrichment of genetic diversity of genes encoding differentially reactive antigens.
The non-synonymous genetic diversity (π) of genes encoding the set of recognized P. vivax antigens was compared with coding sequences found in the P. vivax genome as a whole. This analysis, using globally representative P. vivax strains from Brazil, India, Mauritania, and North Korea, only included annotated genes; vir gene family members, hypothetical genes, were not included. The set of genes encoding the recognized P. vivax antigens as a whole (combining proteins differentially recognized by both relapse and reinfection patients) had a significantly higher level of single nucleotide polymorphism (SNP) diversity than the rest of the coding genes, as assessed by the Mann–Whitney test (W = 249,177, P = 0.0007472). Genes encoding the set of recognized proteins were significantly more likely to have 1 or more non-synonymous SNPs (W = 249,791, P = 0.0006419). In contrast, the level of synonymous SNPs in genes encoding recognized proteins was not significantly different (W = 291,635, P = 0.3948). This analysis suggests that positive immune selection for amino acid polymorphism in these P. vivax antigens has taken place.
Discussion
This study is the first large-scale analysis in which immune responses to relapsed versus reinfection of P. vivax infection in a hypoendemic human population were compared. The first major finding was that there was no observable difference in seroreactivity between relapsed versus reinfection P. vivax infections. The second major finding is that we identified a large set of serodiagnostic antigens that have potential utility for seroepidemiological studies of transmission in P. vivax-endemic areas. Third, genome-scale analysis of the diversity of the serodiagnostic antigens recognized by P. vivax-infected subjects suggests that immune selection has led to antigenic changes in the parasite. These results collectively demonstrate that most of the antigens that could lead to important antibody responses are not being investigated; and immune responses differ between relapse and reinfection, which has important implications for understanding P. vivax epidemiology, transmission, and vaccine development.
Infections by malaria parasites are known to lead to broadly reactive anti-Plasmodium antibody responses.38 How to analyze large-scale antibody responses in key areas of malaria research such as delineating target antigens of protective immunity or how to identify new proteins for seroepidemiological analyses of malaria transmission in endemic regions are important applications of current systems immunology methodologies.27 In this study, sera from symptomatically relapsed or reinfected P. vivax-infected subjects in the low-transmission region of the Peruvian Amazon were used to probe genome-scale P. vivax protein microarrays.
That the cumulative total antibody response was not greater in reinfection compared with relapsing infection has implications for understanding immunological mechanisms and consequences of relapsing P. vivax malaria. Previous studies have suggested that asymptomatic relapse is common in P. vivax malaria,37 despite the more widely recognized phenomenon of relapse leading to symptomatic infection and the seeking of medical care. The present investigation provides evidence that reinfection resulting in clinical symptoms is not associated with an enhanced immune memory-driven antibody response than relapsing symptomatic infection. This observation might be explained in several ways, but raises the possibility that any partially effective immune response to a previous P. vivax strain might have been maximized in terms of driving elevations of antibody response. Alternatively, it is also possible that lower parasitemia in relapse compared with reinfection might be associated with a lower antibody response, but the present data are not consistent with this possibility because the parasitemias were only semiquantitatively assessed (1–4 + system) in this study.
An important limitation of this study is the accurate classification of P. vivax relapse versus reinfection in patients at continued risk for new infections, an issue addressed in other reports when P. vivax-infected subjects leave a malaria-endemic region and experience what can definitively classified as relapse in a malaria non-endemic place.39,40 Our classification of relapse is limited by the possibility that subjects may harbor more than one P. vivax clone. Hence our data conservatively underestimate the rate of relapse, and for this reason the lack of difference in antibody responses between relapse and reinfection on a systems level should remain valid. However, there is still no reliable method to differentiate P. vivax relapse from reinfection in the endemic setting, an important area for future research.
This protein microarray analysis provides immediate tools to apply to seroepidemiology of malaria with regard to surveillance, control, and elimination. Future work should use these tools to establish the population decay kinetics of antibody responses to different specific antigens, thereby allowing estimates of timing of previous infection. Comparison of E. coli to eukaryotic recombinant protein expression systems may be important to improve the performance of such serological assays based on the protein microarray results. Such studies will be important because previous seroepidemiological studies have relied on blood-stage vaccine candidate antigens selected for high titer, immunodominant and sustained antibody responses rather than with regard to seroreactivity in human populations that would diminish over time. The use of parasite lysates for enzyme-linked immunosorbent assay or immunofluorescent microscopy is not applicable to such studies of decay kinetics because of their heterogeneity and low sensitivity/specificity for inferring previous time of infection. The ideal antigen(s) for determining recent versus remote malaria transmission from mosquitos to residents of endemic areas would have the properties of being associated with rapid versus sustained decay kinetics.
The P. vivax antigen yielding the most consistently high-intensity reactivity on the protein array was PvMSP10; other top-ranked antigens included MSP7, MSP8 and several hypothetical proteins, and a putatively sexual-stage antigen s16. Antibodies to PvMSP10 have previously been readily detected in humans with P. vivax malaria41,42 using E. coli-expressed41 and wheat germ in vitro expression system–expressed protein.42 Studies with P. falciparum MSP10 demonstrated a lack of cross-reactivity with other MSPs (PfMSP-1, -4, -5 and -8), consistent with the 95% specificity for P. vivax infection found for antibodies detected against wheat germ–expressed PvMSP10.42 No experimental evidence has emerged to date regarding the viability of any Plasmodium spp. MSP10 as a candidate for an asexual blood-stage-based vaccine.
This large-scale analysis of antibody responses to P. vivax infection has identified a large set of serodiagnostic antigens with potential utility for seroepidemiological studies of transmission in endemic areas. This work sets the stage for testing these antigens in well-characterized human populations in diverse malaria-endemic settings around the world and for planning new approaches to malaria vaccine development aimed at the neglected parasite, P. vivax.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paula Maguina of the University of California for her scientific, logistical, and ethics compliance support that were essential for the completion of this work. We also thank Huw Davies of University of California, Irvine, for kind discussions and Emma Sperling of Antigen Discovery Inc., who helped with assays and data generation.
Footnotes
Financial support: This work was supported by United States Public Health Service grants U19AI089681, D43TW007120, R01AI067727, and K24AI068903 (Joseph M. Vinetz); AI066791, AI095916, and AI089686 (Philip Felgner); and AI075692 (Ruobing Wang and Xiaowu Liang), all from the U.S. National Institutes of Health.
Conflict of interest: The authors have read the journal's policy and declare the following potential conflicts: Philip Felgner has an equity interest in Antigen Discovery, Inc., which is developing products related to the research described in this article. In addition, this author serves on the advisory board of ADI and receives compensation for these services. The terms of this arrangement have been reviewed and approved by the University of California in accordance with its conflict of interest policies. Douglas Molina is an employee of Antigen Discovery, Inc. No other author declares a conflict of interest. This does not alter our adherence to ASTMH/AJTMH journal policies on sharing data and materials.
Authors' addresses: Raul Chuquiyauri and Joseph M. Vinetz, Department of Medicine, Division of Infectious Diseases, University of California San Diego, La Jolla, CA, Laboratorio ICEMR-Amazonia, Laboratorios de Investigación y Desarrollo, Facultad de Ciencias y Filosofía, Universidad Peruana Cayetano Heredia, Lima, Perú, and Instituto de Medicina Tropical Alexander von Humboldt, Universidad Peruana Cayetano Heredia, Lima, Perú, E-mails: raulharo@yahoo.com and jvinetz@ucsd.edu. Douglas M. Molina, Philip Felgner, and Xiaowu Liang, Antigen Discovery Inc., Irvine, CA, E-mails: dmolina@antigendiscovery.com, pfelgner@uci.edu, and xliang@antigendiscovery.com. Eli L. Moss and Daniel E. Neafsey, Malaria Research Group, Broad Institute of MIT and Harvard University, Cambridge, MA, E-mails: emoss@broadinstitute.org and neafsey@broadinstitute.org. Ruobing Wang, Seattle Biomed, Seattle, WA, E-mail: ruobing.wang@seattlebiomed.org. Malcolm J. Gardner, Department of Medicine, Division of Global Public Health, University of California San Diego, La Jolla, CA, E-mail: malcolm.gardner@seattlebiomed.org. Kimberly C. Brouwer and Robert H. Gilman, Department of International Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, E-mails: kbrouwer@ucsd.edu and gilmanbob@gmail.com. Sonia Torres and Alejandro Llanos-Cuentas, Instituto de Medicina Tropical Alexander von Humboldt, Universidad Peruana Cayetano Heredia, Lima, Perú, E-mails: soniatadimi@yahoo.com and elmer.llanos@upch.pe.
References
- 1.White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. Malaria. Lancet. 2014;383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 2.Cotter C, Sturrock HJ, Hsiang MS, Liu J, Phillips AA, Hwang J, Gueye CS, Fullman N, Gosling RD, Feachem RG. The changing epidemiology of malaria elimination: new strategies for new challenges. Lancet. 2013;382:900–911. doi: 10.1016/S0140-6736(13)60310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009;9:555–566. doi: 10.1016/S1473-3099(09)70177-X. [DOI] [PubMed] [Google Scholar]
- 4.Alonso PL, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, Collins F, Doumbo OK, Greenwood B, Hall BF, Levine MM, Mendis K, Newman RD, Plowe CV, Rodriguez MH, Sinden R, Slutsker L, Tanner M. A research agenda to underpin malaria eradication. PLoS Med. 2011;8:e1000406. doi: 10.1371/journal.pmed.1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gething PW, Elyazar IR, Moyes CL, Smith DL, Battle KE, Guerra CA, Patil AP, Tatem AJ, Howes RE, Myers MF, George DB, Horby P, Wertheim HF, Price RN, Mueller I, Baird JK, Hay SI. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis. 2012;6:e1814. doi: 10.1371/journal.pntd.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feachem RG, Phillips AA, Hwang J, Cotter C, Wielgosz B, Greenwood BM, Sabot O, Rodriguez MH, Abeyasinghe RR, Ghebreyesus TA, Snow RW. Shrinking the malaria map: progress and prospects. Lancet. 2010;376:1566–1578. doi: 10.1016/S0140-6736(10)61270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook J, Reid H, Iavro J, Kuwahata M, Taleo G, Clements A, McCarthy J, Vallely A, Drakeley C. Using serological measures to monitor changes in malaria transmission in Vanuatu. Malar J. 2010;9:169. doi: 10.1186/1475-2875-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tongol-Rivera P, Kano S, Miguel E, Tongol P, Suzuki M. Application of seroepidemiology in identification of local foci in a malarious community in Palawan, The Philippines. Am J Trop Med Hyg. 1993;49:608–612. doi: 10.4269/ajtmh.1993.49.608. [DOI] [PubMed] [Google Scholar]
- 9.Hamlin KL, Moss DM, Priest JW, Roberts J, Kubofcik J, Gass K, Streit TG, Nutman TB, Eberhard ML, Lammie PJ. Longitudinal monitoring of the development of antifilarial antibodies and acquisition of Wuchereria bancrofti in a highly endemic area of Haiti. PLoS Negl Trop Dis. 2012;6:e1941. doi: 10.1371/journal.pntd.0001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joseph H, Maiava F, Naseri T, Silva U, Lammie P, Melrose W. Epidemiological assessment of continuing transmission of lymphatic filariasis in Samoa. Ann Trop Med Parasitol. 2011;105:567–578. doi: 10.1179/2047773211Y.0000000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noordin R, Muhi J, Md Idris Z, Arifin N, Kiyu A. Duration of detection of anti-BmR1 IgG4 antibodies after mass-drug administration (MDA) in Sarawak, Malaysia. Trop Biomed. 2012;29:191–196. [PubMed] [Google Scholar]
- 12.Nuti M, Ferrari JD, Au AC. Seroepidemiology of Bancroftian filariasis in the Seychelles Islands. Tropenmed Parasitol. 1982;33:25–27. [PubMed] [Google Scholar]
- 13.Steel C, Kubofcik J, Ottesen EA, Nutman TB. Antibody to the filarial antigen Wb123 reflects reduced transmission and decreased exposure in children born following single mass drug administration (MDA) PLoS Negl Trop Dis. 2012;6:e1940. doi: 10.1371/journal.pntd.0001940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tisch DJ, Bockarie MJ, Dimber Z, Kiniboro B, Tarongka N, Hazlett FE, Kastens W, Alpers MP, Kazura JW. Mass drug administration trial to eliminate lymphatic filariasis in Papua New Guinea: changes in microfilaremia, filarial antigen, and Bm14 antibody after cessation. Am J Trop Med Hyg. 2008;78:289–293. [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold BF, Priest JW, Hamlin KL, Moss DM, Colford JM, Jr, Lammie PJ. Serological measures of malaria transmission in Haiti: comparison of longitudinal and cross-sectional methods. PLoS One. 2014;9:e93684. doi: 10.1371/journal.pone.0093684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook J, Speybroeck N, Sochanta T, Somony H, Sokny M, Claes F, Lemmens K, Theisen M, Soares IS, D'Alessandro U, Coosemans M, Erhart A. Sero-epidemiological evaluation of changes in Plasmodium falciparum and Plasmodium vivax transmission patterns over the rainy season in Cambodia. Malar J. 2012;11:86. doi: 10.1186/1475-2875-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Carvalho ME, Ferreira MU, De Souza MR, Ninomia RT, Matos GF, Camargo LM, Ferreira CS. Malaria seroepidemiology: comparison between indirect fluorescent antibody test and enzyme immunoassay using bloodspot eluates. Mem Inst Oswaldo Cruz. 1992;87:205–208. doi: 10.1590/s0074-02761992000200006. [DOI] [PubMed] [Google Scholar]
- 18.Fowkes FJ, Richards JS, Simpson JA, Beeson JG. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: a systematic review and meta-analysis. PLoS Med. 2010;7:e1000218. doi: 10.1371/journal.pmed.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart L, Gosling R, Griffin J, Gesase S, Campo J, Hashim R, Masika P, Mosha J, Bousema T, Shekalaghe S, Cook J, Corran P, Ghani A, Riley EM, Drakeley C. Rapid assessment of malaria transmission using age-specific sero-conversion rates. PLoS One. 2009;4:e6083. doi: 10.1371/journal.pone.0006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren M, Collins WE, Jeffery GM, Skinner JC. The seroepidemiology of malaria in middle America. V. Antibody responses in an indicator population from an endemic area with attack phase antimalaria activities. Am J Trop Med Hyg. 1983;32:1209–1215. [PubMed] [Google Scholar]
- 21.Lu F, Li J, Wang B, Cheng Y, Kong DH, Cui L, Ha KS, Sattabongkot J, Tsuboi T, Han ET. Profiling the humoral immune responses to Plasmodium vivax infection and identification of candidate immunogenic rhoptry-associated membrane antigen (RAMA) J Proteomics. 2014;102C:66–82. doi: 10.1016/j.jprot.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 22.Finney OC, Danziger SA, Molina DM, Vignali M, Takagi A, Ji M, Stanisic DI, Siba PM, Liang X, Aitchison JD, Mueller I, Gardner MJ, Wang R. Predicting antidisease immunity using proteome arrays and sera from children naturally exposed to malaria. Mol Cell Proteomics. 2014;13:2646–2660. doi: 10.1074/mcp.M113.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, Ginsburg H, Nosten F, Day NP, White NJ, Carlton JM, Preiser PR. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci USA. 2008;105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westenberger SJ, McClean CM, Chattopadhyay R, Dharia NV, Carlton JM, Barnwell JW, Collins WE, Hoffman SL, Zhou Y, Vinetz JM, Winzeler EA. A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis. 2010;4:e653. doi: 10.1371/journal.pntd.0000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molina DM, Finney OC, Arevalo-Herrera M, Herrera S, Felgner PL, Gardner MJ, Liang X, Wang R. Plasmodium vivax pre-erythrocytic-stage antigen discovery: exploiting naturally acquired humoral responses. Am J Trop Med Hyg. 2012;87:460–469. doi: 10.4269/ajtmh.2012.12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, Oloo JA, Blair PL, Aguiar JC, Baldi P, Davies DH, Felgner PL. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, Burk CR, Waisberg M, Jasinskas A, Tan X, Doumbo S, Doumtabe D, Kone Y, Narum DL, Liang X, Doumbo OK, Miller LH, Doolan DL, Baldi P, Felgner PL, Pierce SK. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA. 2010;107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuquiyauri R, Penataro P, Brouwer KC, Fasabi M, Calderon M, Torres S, Gilman RH, Kosek M, Vinetz JM. Microgeographical differences of Plasmodium vivax relapse and re-infection in the Peruvian Amazon. Am J Trop Med Hyg. 2013;89:326–338. doi: 10.4269/ajtmh.13-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosek M, Yori PP, Gilman RH, Calderon M, Zimic M, Chuquiyauri R, Jeri C, Pinedo-Cancino V, Matthias MA, Llanos-Cuentas A, Vinetz JM. High degree of Plasmodium vivax diversity in the Peruvian Amazon demonstrated by tandem repeat polymorphism analysis. Am J Trop Med Hyg. 2012;86:580–586. doi: 10.4269/ajtmh.2012.11-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roshanravan B, Kari E, Gilman RH, Cabrera L, Lee E, Metcalfe J, Calderon M, Lescano AG, Montenegro-James S, Calampa C, Vinetz JM. Endemic malaria in the Peruvian Amazon region of Iquitos. Am J Trop Med Hyg. 2003;69:45–52. [PubMed] [Google Scholar]
- 31.Cui L, Mascorro CN, Fan Q, Rzomp KA, Khuntirat B, Zhou G, Chen H, Yan G, Sattabongkot J. Genetic diversity and multiple infections of Plasmodium vivax malaria in western Thailand. Am J Trop Med Hyg. 2003;68:613–619. doi: 10.4269/ajtmh.2003.68.613. [DOI] [PubMed] [Google Scholar]
- 32.Chuquiyauri R, Paredes M, Penataro P, Torres S, Marin S, Tenorio A, Brouwer KC, Abeles S, Llanos-Cuentas A, Gilman RH, Kosek M, Vinetz JM. Socio-demographics and the development of malaria elimination strategies in the low transmission setting. Acta Trop. 2011;121:292–302. doi: 10.1016/j.actatropica.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui L, Mascorro CN, Fan Q, Rzomp KA, Jhuntirat B, Zhou G, Chen H, Yan G, Sattabongkot J. Genetic diversity and multiple infections of Plasmodium vivax malaria in western Thailand. Am J Trop Med Hyg. 2003;68:613–619. doi: 10.4269/ajtmh.2003.68.613. [DOI] [PubMed] [Google Scholar]
- 34.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr, Treatman C, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 36.Molina D, Finney OC, Aravelo-Herrera M, Herrera S, Felgner PL, Gardner MJ, Liang X, Wang R. Plasmodium vivax pre-erythrocytic-stage antigen discovery: exploiting naturally acquired humoral responses. Am J Trop Med Hyg. 2012;87:460–469. doi: 10.4269/ajtmh.2012.12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van den Eede P, Soto-Calle VE, Delgado C, Gamboa D, Grande T, Rodriguez H, Llanos-Cuentas A, Anne J, D'Alessandro U, Erhart A. Plasmodium vivax sub-patent infections after radical treatment are common in Peruvian patients: results of a 1-year prospective cohort study. PLoS One. 2011;6:e16257. doi: 10.1371/journal.pone.0016257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crompton PD, Moebius J, Portugal S, Waisberg M, Hart G, Garver LS, Miller LH, Barillas C, Pierce SK. Malaria immunity in man and mosquito: insights into unsolved mysteries of a deadly infectious disease. Annu Rev Immunol. 2014;32:157–187. doi: 10.1146/annurev-immunol-032713-120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imwong M, Snounou G, Pukrittayakamee S, Tanomsing N, Kim JR, Nandy A, Guthmann JP, Nosten F, Carlton J, Looareesuwan S, Nair S, Sudimack D, Day NP, Anderson TJ, White NJ. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 40.Chen N, Auliff A, Rieckmann K, Gatton M, Cheng Q. Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J Infect Dis. 2007;195:934–941. doi: 10.1086/512242. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Leal O, Sierra AY, Barrero CA, Moncada C, Martinez P, Cortes J, Lopez Y, Salazar LM, Hoebeke J, Patarroyo MA. Identifying and characterising the Plasmodium falciparum merozoite surface protein 10 Plasmodium vivax homologue. Biochem Biophys Res Commun. 2005;331:1178–1184. doi: 10.1016/j.bbrc.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y, Wang B, Sattabongkot J, Lim CS, Tsuboi T, Han ET. Immunogenicity and antigenicity of Plasmodium vivax merozoite surface protein 10. Parasitol Res. 2014;113:2559–2568. doi: 10.1007/s00436-014-3907-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.