Abstract
A number of approaches for Cas9-mediated transcriptional activation have recently been developed, allowing target genes to be overexpressed from their endogenous genomic loci. However, these approaches have thus far been limited to cell culture, and this technique has not been demonstrated in vivo in any animal. The technique involving the fewest separate components, and therefore the most amenable to in vivo applications, is the dCas9-VPR system, where a nuclease-dead Cas9 is fused to a highly active chimeric activator domain. In this study, we characterize the dCas9-VPR system in Drosophila cells and in vivo. We show that this system can be used in cell culture to upregulate a range of target genes, singly and in multiplex, and that a single guide RNA upstream of the transcription start site can activate high levels of target transcription. We observe marked heterogeneity in guide RNA efficacy for any given gene, and we confirm that transcription is inhibited by guide RNAs binding downstream of the transcription start site. To demonstrate one application of this technique in cells, we used dCas9-VPR to identify target genes for Twist and Snail, two highly conserved transcription factors that cooperate during Drosophila mesoderm development. In addition, we simultaneously activated both Twist and Snail to identify synergistic responses to this physiologically relevant combination. Finally, we show that dCas9-VPR can activate target genes and cause dominant phenotypes in vivo, providing the first demonstration of dCas9 activation in a multicellular animal. Transcriptional activation using dCas9-VPR thus offers a simple and broadly applicable technique for a variety of overexpression studies.
Keywords: CRISPR-Cas9; gene activation; overexpression, gain-of-function
IT has recently become possible to activate transcription of target genes from their native genomic locus using nuclease-dead Cas9 (dCas9) fused to transcriptional activator domains (Mali et al. 2013; Gilbert et al. 2014; Tanenbaum et al. 2014; Zalatan et al. 2014; Chavez et al. 2015; Konermann et al. 2015). Activating genes from their endogenous transcription start site (TSS) offers several benefits that are complementary to traditional overexpression studies based on cloned cDNAs. For example, the dCas9-mediation activation technique is preferable for genes that are difficult to clone, e.g., if they occur in multiple splice isoforms and/or are very large. In addition, there is evidence that dCas9-mediated activation leads to target gene activation at physiologically relevant levels, as opposed to many existing techniques (Chavez et al. 2015). Cas9-mediated activation also has the benefits that it is easily multiplexed and that it is rapidly scalable for genome-wide studies because the target specificity is provided by easy-to-synthesize 20-bp single guide RNAs (sgRNAs) (Gilbert et al. 2014; Chen et al. 2015; Konermann et al. 2015).
The first attempts to activate transcription by fusing dCas9 to activator domains such as VP64 yielded very low levels of overexpression (Gilbert et al. 2013; Maeder et al. 2013; Mali et al. 2013; Perez-Pinera et al. 2013). However, three strategies to substantially increase the effectiveness of dCas9 activators have subsequently been described. In the dCas9-VPR system (Chavez et al. 2015), dCas9 is directly fused to a chimeric activator (composed of the VP64, p65, and Rta domains), based on a systematic screen of 20 candidate activator domains. In a second strategy, termed “SunTag” (Gilbert et al. 2014; Tanenbaum et al. 2014), dCas9 is fused to multiple copies of an epitope tag and is cotransfected with a single-chain antibody fused to the VP64 activator domain, thus recruiting multiple VP64 domains to each molecule of dCas9. The third strategy, which has been developed independently by two groups (Zalatan et al. 2014; Konermann et al. 2015), involves inserting specific RNA hairpin sequences into exposed portions of the sgRNA, and co-expressing proteins that specifically recognize these hairpin sequences and are fused to additional activator domains.
While all of these approaches show promise in cell culture, none has yet been demonstrated in vivo in any multicellular animal. We reasoned that, because the dCas9-VPR system requires a single activator component in addition to the sgRNA, it would be most amenable to stable transgenesis for in vivo studies. dCas9-VPR has been shown to efficiently activate gene expression in yeast, human, mouse, and Drosophila cells, yet previous studies in Drosophila cells have been limited to just two target genes and utilized pools of up to five sgRNAs per gene (Chavez et al. 2015). In this study, we first show that dCas9-VPR functions robustly in Drosophila cells on an array of target genes, both singly and in multiplex. We test a number of sgRNAs per target gene and conclude that a single highly active sgRNA is sufficient to activate transcription and that there is substantial variability in sgRNA effectiveness. We also confirm previous observations that target gene activation levels are inversely proportional to their basal expression levels. We use dCas9-VPR to activate the transcription factors Twist and Snail in cells, both singly and together, and then use RNAseq to identify transcriptional targets of these two conserved factors. Finally, we adapt the dCas9-VPR system for Gal4-UAS activation and show that this approach can activate target genes in vivo at levels sufficient to induce dominant phenotypes. Together, our results demonstrate the ease and utility of the dCas9-VPR system in Drosophila cells and in vivo.
Materials and Methods
Cloning of Cas9 activators and sgRNA
dCas9-VPR has been previously described (Chavez et al. 2015). UAS-driven transgenes were cloned into pWalium20 (Ni et al. 2011) using Gibson cloning (Gibson et al. 2009; Gibson 2011). A Kozak sequence (GCCACC) was added upstream of the start codon, and the ftz intron between the CDS and the 3′ UTR was removed.
Single guides were cloned into pCFD3 (Port et al. 2014) using a BbsI digest, as described in Housden et al. (2014). Double-guides (targeting wg, hnt, cut, and elav) were cloned into pCFD4 (Port et al. 2014) using Gibson cloning, following the author’s protocols. All guide sequences are available in Supporting Information, Table S1. Nuclease efficiency scores were calculated using the algorithm described in Housden et al. (2014), accessed via an online tool (http://www.flyrnai.org/evaluateCrispr/). Briefly, these values are based on an empirical analysis of the cutting efficiency of a library of sgRNAs, based on the position of each nucleotide at each of the 20 positions within the protospacer.
Cell culture and transfection
S2R+ cells were cultured in Schneider’s Drosophila medium (Millipore, Gibco) containing 10% fetal bovine serum and penicillin/streptomycin (at 1000 units/ml and 1000 mcg/ml, respectively). Cells were transfected using Effectene Transfection Reagent (Qiagen) using the manufacturer’s protocol, except that twice the number of recommended suspension cells were seeded per well. For pActin-driven experiments, 50 ng of gRNAs and 150 ng dCas9 were transfected in 24-well plates. For UAS experiments, equal amounts (either 66 or 100 ng) of all components were transfected in 12- or 24-well plates.
Quantitative PCR
Three or four days after transfection, total RNA was collected using TRIZOL (Life Technologies) following the manufacturer’s instructions. Total RNA was purified using an RNeasy MinElute Cleanup Kit (Qiagen), including a 30-min on-column DNase treatment. Equal volumes of total RNA were used as template for first-strand complementary DNA (cDNA) synthesis using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR (qPCR) was conducted using iQ Supermix (Bio-Rad) on a C1000 Thermal Cycler (Bio-Rad), and fold-change was calculated using the 2ΔΔCt method (Livak and Schmittgen 2001), with error propagated using standard methods and with rp49 as a reference gene. The primers used for qPCR are listed in Table S2.
Western blotting
Cells were harvested 3 days after transfection. The following primary antibodies were used: anti-tubulin (Sigma T5168, 1:10,000), anti-Wg (4D4; DSHB, 1:400), anti-Hnt (1G9; DSHB, 1:500), anti-Cas9 (Abcam 191468, 1:500), and anti-FLAG (Sigma F3165, 1:10,000), with 5% BSA as a blocking reagent. HRP-coupled sheep anti-mouse (Amersham NXA931, 1:5000) was used as a secondary antibody, and signal was detected with Pierce ECL or SuperSignal West Pico reagents (Thermo).
RNAseq
S2R+ cells were transfected with Actin:dCas9-VPR along with either a negative control sgRNA that does not target the Drosophila genome (QUAS #1; Table S1) or a pool of five sgRNAs targeting either snail, twist, or a combination of both pools (Table S1). Total RNA was obtained as described above, and RNA integrity was confirmed by Bioanalyzer (Agilent). Between 2.0 and 2.5 M 100-bp single-end reads were generated for each sample using Illumina Hi-Seq at the Columbia Genome Center, following standard protocols for Illumina library preparation and sequencing. Reads were mapped to the Drosophila melanogaster genome (BDGP R5 assembly) using TopHat (Trapnell et al. 2009), and only uniquely mapped reads (between 76.4 and 83.3% of the reads for each sample) were used for further analysis. FPKM and read count values were obtained using Cufflinks (Trapnell et al. 2010) and HTSeq (Anders et al. 2015), respectively. Two biological replicates were sequenced per sample, and duplicate runs were highly correlated (Pearson’s correlation ≥0.99 for all experiments). To eliminate potentially confounding effects of low read counts, we filtered out genes with <1 of 1 M reads recorded for each sample. The “nbionomTest” of the Bioconductor package DESeq (Anders and Huber 2010) was then used to obtain differentially expressed gene lists at a multiple hypothesis testing-adjusted P-value of 0.05.
For each activation experiment, we defined the target genes as the union of the (1) differentially expressed genes in induced sample compared to control sample and (2) genes that are not expressed (0 or very few reads) in control but highly expressed in induced samples or vice versa. These gene lists were used for Gene Ontology (GO) analysis and further comparison with chromatin immunoprecipitation (ChIP) data.
To compare our data with published ChIP data, we downloaded Snail and Twist ChIP data from the Berkeley Drosophila transcription Network Project (MacArthur et al. 2009) (http://bdtnp.lbl.gov/Fly-Net/) and updated the wiggle file genome coordinates to the R5 genome assembly. We then pooled binding-site information of the two replicates and identified genes with the nearest TSS to the binding peak as the putative target gene. Read stacks were generated using the Integrated Genomics Visualizer (Robinson et al. 2011) after pooling the two BAM files for each experiment.
All RNAseq data have been deposited in Gene Expression Omnibus (accession no. GSE71430).
Transgenic flies
Transgenic 10X-UAS:3xFLAG-Cas9-VP64 and VPR constructs and double sgRNA-plasmids in pCFD4 (both described above) were integrated into the attP40 landing site on the second chromosome (Markstein et al. 2008) using standard phiC31 transformation methods.
For activation experiments, flies of the genotype w;UAS:dCas9-VP64/CyO;dpp-Gal4/TM6b,Tb or w;UAS:dCas9-VPR/CyO;dpp-Gal4/TM6b,Tb were crossed to homozygous sgRNA-wg flies (yv;sgRNA-wg). Wing discs from non-Tb larvae (i.e., those containing dpp-Gal4) were costained with an anti-FLAG antibody to differentiate those larvae expressing the activator constructs from their siblings receiving the CyO balancer chromosome and an anti-Wg antibody to test for ectopic Wg expression.
Immunohistochemistry
In vivo experiments were conducted at 27°. Wandering-stage larval wing discs were dissected in PBS, fixed for 25–30 min in 4% paraformaldehyde in PBS, and then stained using standard protocols. Antibodies used were mouse anti-Wingless (4D4; DSHB, 1:100) and rabbit anti-FLAG (Sigma F7425, 1:500). Secondary antibodies coupled to Alexa 488 and 555 (Invitrogen) were used at 1:400, samples were imaged on a Zeiss LSM 780 confocal microscope, and maximum-intensity projections are shown.
Results and Discussion
Human codon-optimized dCas9-VPR works robustly in Drosophila cells
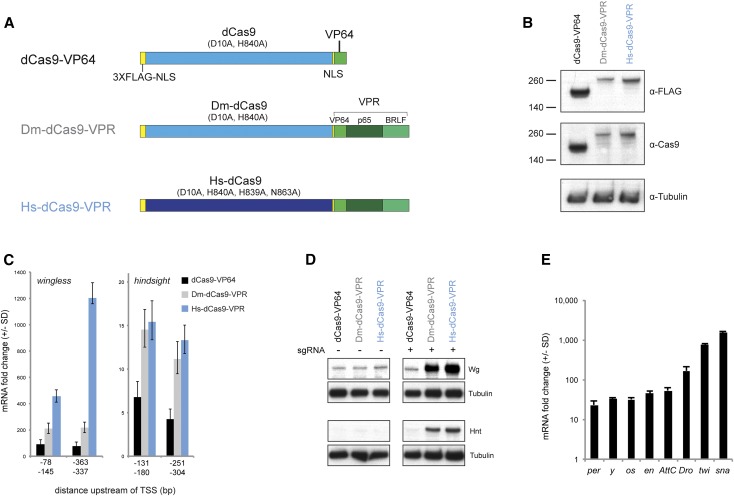
We first compared the activity of the published dCas9-VPR activator, which is human codon-optimized and contains four nuclease-attenuating mutations (D10A, H839A, H840A, and N863A), to a Drosophila codon-optimized dCas9-VPR that contains two of these mutations (D10A and H840A), thought to be sufficient to remove nuclease activity (Mali et al. 2013; Perez-Pinera et al. 2013) (Figure 1A). We cotransfected these constructs, under UAS control, together with a plasmid encoding pActin-Gal4, and pairs of two sgRNAs targeting a window from −400 to −50 upstream of the TSS of two endogenous genes: wingless (wg) and hindsight (hnt, aka pebbled). We confirmed efficient translation of all of the activator constructs via Western blot (Figure 1B), demonstrating that differential activity was not due to activator protein levels.
Figure 1.
dCas9-VPR activates target gene expression in Drosophila S2R+ cells. (A) Schematics of the constructs tested in this study. Dm-dCas9 is codon-optimized for Drosophila, Hs-dCas9 for human. (B) Western blot analysis of dCas9 activators demonstrating that constructs are effectively translated. (C) qPCR analysis of wg and hnt activation. For each gene, two pairs of sgRNAs located upstream of the TSS were tested. Each sgRNA pair was expressed from a single plasmid driving expression from the U6:3 and U6:1 promoters, respectively (see Materials and Methods). (D) Western blot analysis of Wg and Hnt activation. (E) qPCR analysis of eight additional endogenous genes by Hs-dCas9-VPR. In B–D, UAS-driven constructs were cotransfected with pActin-Gal4. In E, Hs-dCas9-VPR was expressed using the Actin promoter.
In all four cases, the published Hs-dCas9-VPR construct substantially outperformed dCas9-VP64 and Dm-dCas9-VPR (Figure 1, C and D). The superior performance of Hs-dCas9-VPR was seen both via qPCR (Figure 1C) and via Western blots against the target genes (Figure 1D). It is unlikely that codon optimization caused this difference, as the two VPR constructs were expressed at equivalent levels (Figure 1B), suggesting that the four nuclease-attenuating mutations may be important for maximal function. We used the Hs-dCas9-VPR construct (hereafter shortened to “dCas9-VPR”) in all subsequent experiments.
To test whether dCas9-VPR can activate a range of target genes, we cotransfected cells with Actin:dCas9-VPR with pools of two to six sgRNAs targeting each of eight additional genes (per, y, os, en, AttC, Dro, twi, and Sna). In all cases, we observed robust activation ranging over two orders of magnitude (Figure 1E). Importantly, we note that two additional genes that we targeted (cut and elav) were not upregulated using either of two pairs of sgRNAs per gene (data not shown). In agreement with previous reports (Chavez et al. 2015; Konermann et al. 2015), we found that the level of activation of a given gene was inversely correlated with its basal expression level (Figure S1). In other words, dCas9 activation is most effective for genes that are expressed at low levels in a given cell type and does not strongly upregulate genes that are already transcriptionally active.
Design principles for sgRNAs
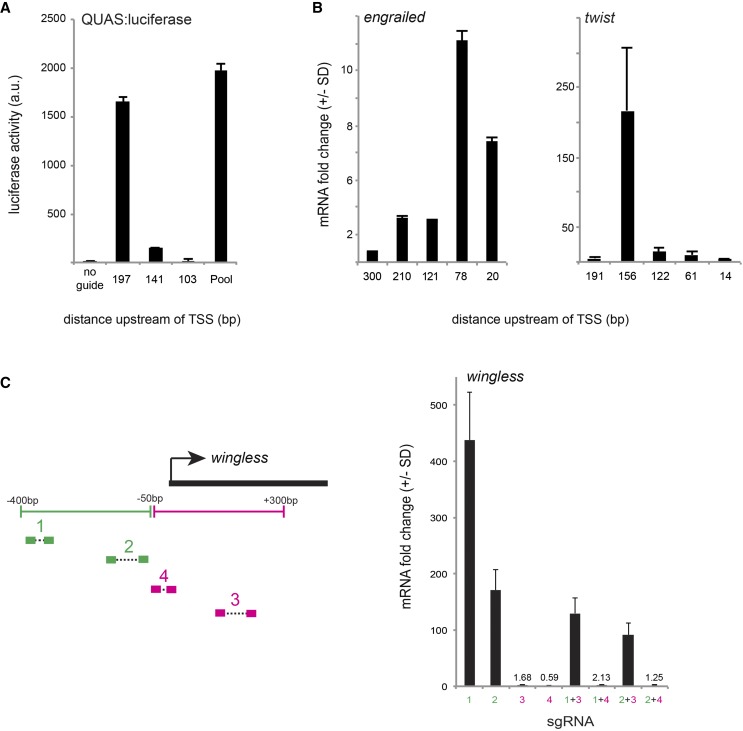
The initial characterization of dCas9-VPR employed pools of up to five sgRNAs per target gene (Chavez et al. 2015). We therefore wanted to know whether such groups of sgRNAs have synergistic effects or whether a single guide within the pool is largely responsible for activation. To address this question, we transfected three guides that target upstream of the TSS of a reporter construct (QUAS:Luciferase), both singly and in combination. The effect of the pooled sgRNAs could be almost completely attributed to the activity of a single highly active sgRNA with an essentially additive effect of the other two minimally active guides (Figure 2A). Next, we tested five nonoverlapping guides targeting immediately upstream of two endogenous genes, twist and engrailed. In both cases, there was marked heterogeneity in guide efficiency, with one guide giving substantially higher activation than any of the others (Figure 2B). Recent studies utilizing alternative Cas9-activator strategies have similarly found that individual sgRNAs vary widely in their ability to activate target gene activity (Tanenbaum et al. 2014; Konermann et al. 2015). These differences in activation are not correlated to the predicted sgRNA nuclease efficiency score, which is based on empirical analysis of cutting efficiency relative to the probability of a given nucleotide at each of the 20 positions within the sgRNA (Figure S2A) (Housden et al. 2014). Neither are these differences due to differential sgRNA-binding capability, as activation levels were uncorrelated with sgRNA GC content (Figure S2B). Furthermore, sgRNA performance was not related to differential bioavailability, as sgRNA concentration was not limiting over the wide range of concentrations tested (Figure S2C). Together, these results suggest that certain single sgRNAs are largely responsible for activation, but we do not currently understand the specific design principals for these particularly effective sgRNAs.
Figure 2.
Effects of individual sgRNA on target gene activation. (A) Three nonoverlapping sgRNAs tiling the region upstream of a QUAS:luciferase reporter construct were transfected either singly or in combination. (B) Five nonoverlapping sgRNAs targeting the upstream region of two endogenous genes, engrailed and twist, differ in their effectiveness. (C) Four pairs of sgRNAs targeting the regions upstream and downstream of the wg TSS were tested singly and in combination. sgRNAs downstream of the TSS do not activate transcription, and their presence can reduce or completely block transcription in the presence of an effective sgRNA.
We next considered the effect of sgRNA placement relative to the TSS. Two previous studies have systematically examined the effect of sgRNA placement relative to the TSS (Gilbert et al. 2014; Konermann et al. 2015). Gilbert et al. (2014) calculated an optimal window of activation range from −400 to −50 bp upstream of the TSS, whereas Konermann et al. (2015) found that a smaller window from −100 to 0 bp upstream of the TSS is optimal. In our experiments, the most active sgRNA was not necessarily within 100 bp of the TSS, and we observed that several sgRNAs within this window were not effective (Figure 2B). Furthermore, our experiments with pairs of sgRNAs targeting wg (Figure 1C) showed that a pair of sgRNAs located −337 and 363 bp upstream of the TSS gave far better activation than a pair at −78 and −145 bp upstream, while a trend in the opposite direction was true for hnt. Together, our results demonstrate that it is important to test a variety of sgRNAs in a window from −400 to 0 upstream of the TSS to maximize activation. We suggest that a good compromise for future studies is to express sgRNAs from the pCFD4 plasmid (Port et al. 2014), which contains sites for co-expression of two separate sgRNAs driven by the U6:3 and U6:1 promoters, respectively.
Many sgRNAs targeting early in the first exon of genes have been generated by a variety of laboratories for the purpose of generating null mutations via Cas9-mediated mutagenesis (reviewed in Housden et al. 2014). We therefore asked whether such existing sgRNA reagents could be useful for Cas9-mediated transcriptional activation. However, previous studies have shown that dCas9–sgRNA complexes targeting in the first exon, downstream of the TSS, can prohibit activation by blocking transcript elongation (Cheng et al. 2013; Qi et al. 2013). To verify this in our system, we examined the activation efficiency of four pairs of sgRNAs targeting a region from −400 bp upstream to 400 bp downstream of the wg TSS, both singly and in combination. sgRNAs targeting downstream of the TSS did not activate transcription, and in fact these sgRNAs reduced or completely blocked the effect of upstream sgRNAs (Figure 2C). We therefore conclude that Cas9-activator studies should avoid using sgRNAs that target downstream of the TSS. These guides, however, may prove useful for future studies using dCas9 for transcriptional repression.
Identification of transcription factor targets using multiplexed Cas9 activation and RNAseq
Cas9-based transcriptional activation has the notable benefit that multiple genes can be simultaneously targeted using a pool of sgRNAs (Zalatan et al. 2014; Chavez et al. 2015; Konermann et al. 2015). We validated the efficacy of multiplexed gene activation in Drosophila cells by cotransfecting Actin:dCas9-VPR with guides targeting three target genes: twist, snail, and engrailed. We observed robust activation of all three genes singly, as pairs, and as a pool of three (Figure S3).
Given the effectiveness of dCas9-VPR, we reasoned that combining Cas9-based activation with RNAseq should provide a conceptually simple approach for identifying transcription factor target genes. We focused on Twist and Snail, two highly conserved transcription factors that function in the Drosophila embryo to specify mesoderm specification and subsequent development (Leptin 1991). Twist is a basic helix-loop-helix activator (Thisse et al. 1988; Murre et al. 1989), and Snail is a zinc-finger transcription factor, classically considered to be a repressor (Boulay et al. 1987; Nieto 2002; Barrallo-Gimeno and Nieto 2005). However, a recent study has suggested that Snail may have additional roles as a transcriptional activator (Rembold et al. 2014). Importantly, the genome-wide targets of both genes have been characterized via independent means, allowing for direct comparison with our data (Sandmann et al. 2007; Zeitlinger et al. 2007; Macarthur et al. 2009).
We transfected S2R+ cells with sgRNAs targeting twist and snail singly and in combination (5 sgRNAs per gene), as well as a nontargeting sgRNA negative control, and then used RNAseq to identify differentially expressed genes. This approach should identify direct and indirect targets of both genes (i.e., genes that are secondarily activated by direct targets) and should identify target genes of both factors individually, as well as those genes that are only activated by both factors acting together.
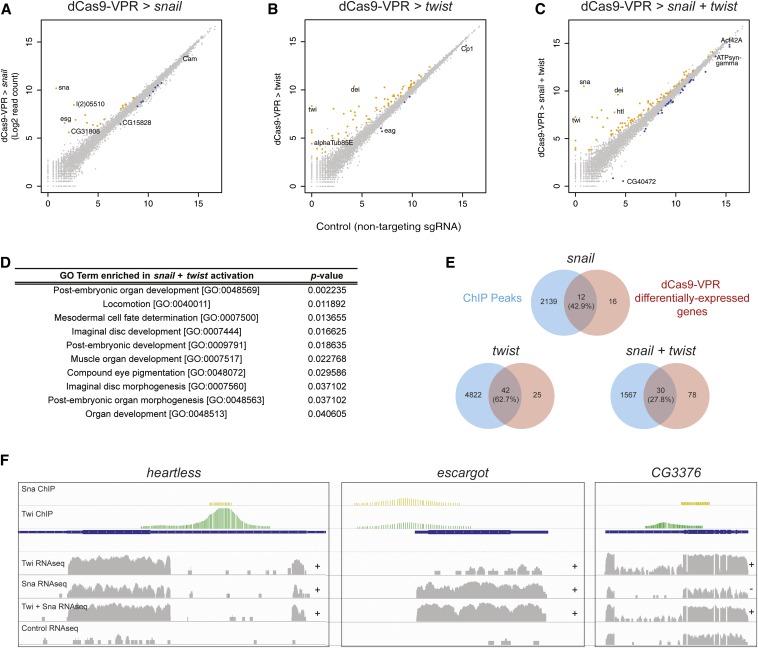
RNAseq confirmed that snail and twist themselves were highly activated by dCas9-VPR, whether targeted singly or together (Figure 3, A–C; Table S3). In each experiment, we also identified a number of additional genes that were significantly differentially expressed (P-value cutoff = 0.05) following overexpression of twist (66 genes), snail (27 genes), or both (106 genes; Figure 3, A–C; Table S3).
Figure 3.
Identification of snail and twist target genes using dCas9-VPR and RNAseq. (A–C) Differential expression analysis following activation of snail (A), twist (B), or both (C). Read counts are plotted on a log2 scale. Colored circles indicate significant difference from control values at P < 0.05. (D) GO term enrichment for snail + twist targets, including several terms associated with mesoderm development. (E) Venn diagrams demonstrating the proportion of differentially expressed genes that also show ChIP peaks for snail, twist, or both. (F) Representative examples showing RNAseq data together with previous ChIP data. The “+” and “−” indicate significant upregulation and downregulation, respectively, relative to control expression levels.
One important caveat is that dCas9-VPR may have off-target activation effects (Kuscu et al. 2014; Wu et al. 2014). Indeed, it has been shown that dCas9 is capable of binding to DNA sequences with up to nine consecutive mismatches in the PAM-distal region (Kuscu et al. 2014). We therefore analyzed each of the predicted off-target binding sites according this rule (Gratz et al. 2014) and asked whether any nearby gene is upregulated in our RNAseq experiments. Among 77 potential off-target sites for the snail or twist sgRNAs, 5 fell near genes that were differentially expressed in our analysis. While 3 of these genes were also near ChIP sites for Twist or Snail, and thus may be genuine targets, 2 are not near ChIP peaks (CG32813 and CG15154) and should be considered off-target effects. In future studies, we strongly recommend using one of the existing online sgRNA design tools to minimize off-target binding sites in the genome (reviewed in Housden et al. 2014).
Gene Ontology enrichment analysis showed that the genes coregulated by Snail and Twist are enriched for terms related to mesoderm and muscle development, as expected (Figure 3D). A subset of these terms was also significantly enriched among targets of Twist alone, including muscle organ development (P = 0.003163), but no terms were following Snail activation alone, consistent with the observation that these factors act synergistically (Rembold et al. 2014). Furthermore, of the genes upregulated by Snail and Twist together, 38 genes (35.8%) were upregulated only upon co-expression of Snail + Twist. Repression of target genes, as opposed to activation, was observed in a substantially higher proportion of Snail-regulated genes than Twist-regulated genes (37.0 compared to 7.6%), consistent with the observation that Snail commonly acts as a repressor (Barrallo-Gimeno and Nieto 2005). However, we also noted that Snail and Twist together led to the down-regulation of 23 genes not repressed by either factor individually (Table S3), suggesting that the presence of Twist may contribute to the repressive activity of Snail, although this effect could be indirect, i.e., mediated by an additional factor that is regulated by Twist and/or Snail.
To begin to differentiate between direct and indirect targets, we calculated the proportion of differentially expressed genes that are adjacent to known ChIP peaks for snail and twist. A highly significant proportion of our predicted target genes were adjacent to ChIP peaks for the relevant factor (P < 0.0001; χ2 test; Figure 3E), suggesting that these are direct Snail and Twist targets. These include known target genes such as heartless (Shishido et al. 1993), inflated (Sandmann et al. 2007), and escargot (Fuse et al. 1996) (Figure 3F and Table S3) and also include new, uncharacterized targets such as CG6330 and CG3376 (Figure 3F). For the target gene CG3376, Snail and Twist had opposite effects on the expression levels, but in combination led to an increase in CG3376 levels (Figure 3F). In contrast, for the majority of target genes identified in this study, we observed that snail and twist, both singly and in combination, promoted target gene activation rather than repression, consistent with recent observations that snail has a dual role as a transcriptional activator (e.g., htl and esg, Figure 3F). The remaining genes, which are not adjacent to ChIP peaks, are likely indirect targets (Table S3).
The number of differentially expressed genes in the present study is far less than the number of observed ChIP peaks (representing between 1.3 and 6.8% of the ChIP peaks; Figure 3E). This difference may be partially due to the difference in cell type (S2R+ cells vs. embryonic tissue) or false positives from ChIP experiments based on cross-linking conditions, but we suggest that this may also reflect the fact that transcription factor occupancy does not necessarily correlate with transcription. Because the approach described here relies on a direct analysis of target gene transcription, it should therefore be less prone to false positives than ChIP studies.
In vivo activation using dCas9-VPR
To date, all studies of Cas9 activators have been conducted in cell culture, and in vivo activation has not yet been demonstrated in any multicellular animal (Gilbert et al. 2014; Tanenbaum et al. 2014; Zalatan et al. 2014; Chavez et al. 2015; Konermann et al. 2015). We therefore tested whether the dCas9-VPR system functions in vivo in Drosophila.
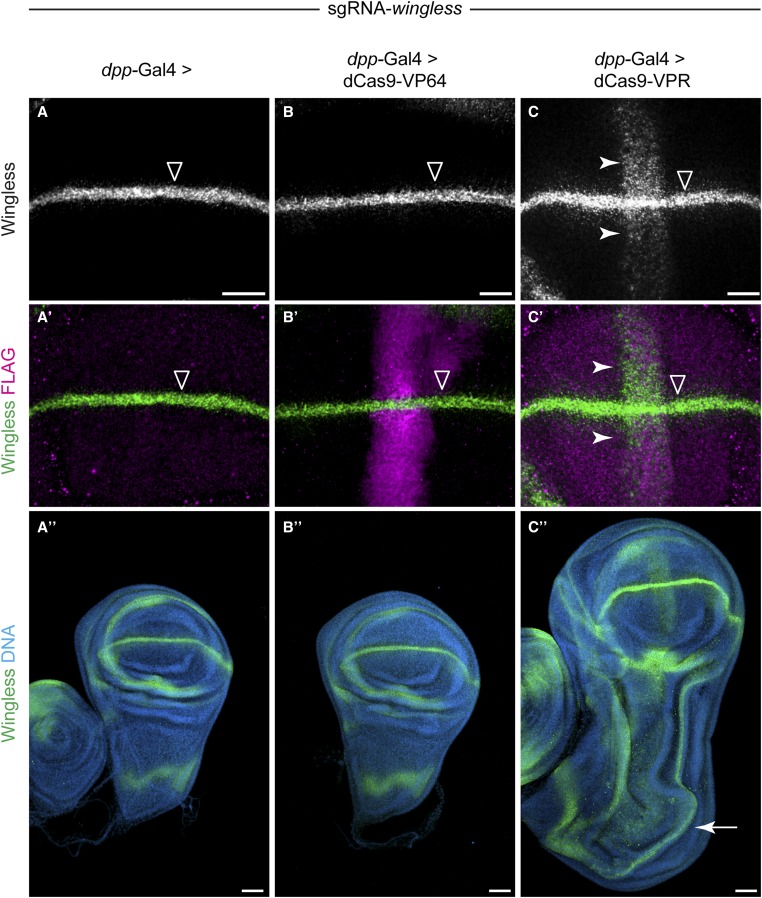
We generated transgenic flies expressing either dCas9-VP64 or dCas9-VPR under UAS control, as well as a line that constitutively expresses two sgRNAs targeting wg. Expression of these transgenes was not toxic, as driving these constructs ubiquitously using Actin-Gal4 was not lethal (data not shown). As a proof of principle, we used dpp-Gal4 to drive expression of the dCas9-VP64 or dCas9-VPR in a stripe of expression along the anterior–posterior margin in the larval wing disc. We crossed dpp-Gal4 > UAS:dCas9-activator flies to sgRNA-wg flies and examined Wg expression using immunostaining. In the wild type, Wg is expressed in a stripe along dorsal–ventral margin, perpendicular to the dpp-Gal4 expression domain (Figure 4, A and A′). Strikingly, the dCas9-VPR construct drove ectopic Wg expression (Figure 4, C and C′), while the dCas9-VP64 did not (Figure 4, B and B′), consistent with our cell culture data. To show that this ectopic Wg expression is physiologically relevant, we examined the morphology of these wing discs and observed a partial duplication of the wing pouch and other patterning abnormalities, consistent with ectopic activation using dpp-Gal4 > UAS:Wg (Figure 4, A′′–C′′) (Ng et al. 1996). These dpp-Gal4 > dCas9-VPR, sgRNA-wg larvae died during early pupal stages, precluding analysis of adult wing morphology. Thus, dCas9-VPR can activate physiologically relevant levels of target gene expression and can generate dominant phenotypes in vivo.
Figure 4.
In vivo activation using dCas9-VPR. Flies homozygous for sgRNA-wg (two sgRNAs) were crossed to flies containing dpp-Gal4 driving expression of UAS-3X-FLAG:dCas9 activators. (A and A′) In the absence of dCas9 activator, Wg is expressed in a stripe along the dorsal–ventral wing margin (open arrowhead). (B and B′) dpp-Gal4 > dCas9-VP64 did not activate ectopic Wg, despite high levels of transgene expression. (C and C′) dpp-Gal4 > dCas9-VPR activates a stripe of ectopic Wg expression (white arrowhead). The dCas9-VPR transgene is expressed at relatively low levels compared to dCas9-VP64 (compare C′ to B′). (C′′) Ectopic activation of Wg using dpp-Gal4 > dCas9-VPR leads to a partial duplication of the wing pouch (white arrow). See Materials and Methods for full genotypes. Bar: 20 µm in A–C′ and 50 µm in A′′–C′′.
Conclusion
In this study, we demonstrate the ease and effectiveness of the dCas9-VPR system for activating target genes both in Drosophila cells and in vivo. Based on our observations that a single sgRNA targeting within ∼400 bp upstream of the TSS can be used to activate target genes, but that sgRNAs differ widely in their efficiency, we propose that a good compromise is to express two sgRNAs per target gene from a single plasmid, using a vector such as pCFD4 (Port et al. 2014). Our results also show that sgRNAs targeting downstream of the TSS are not compatible with dCas9-based activation, consistent with previous studies (Qi et al. 2013). In addition, our results also support previous reports (Chavez et al. 2015; Konermann et al. 2015) that target gene activation levels are inversely proportional to that gene’s basal expression level, which suggests that dCas9-based activation is most effective for genes that are expressed at low levels in a given cell type. Furthermore, we have shown that dCas9-VPR, combined with RNAseq, can be applied to identify targets of transcription factors in multiplex.
Finally, we have provided the first demonstration of Cas9-based activation in vivo, demonstrating that this strategy holds great potential for overexpression studies. For in vivo studies involving stable transgenic organisms, the dCas9-VPR strategy has the benefit that it requires only a single dCas9 component, in contrast to the other existing strategies (Gilbert et al. 2014; Tanenbaum et al. 2014; Zalatan et al. 2014; Chavez et al. 2015; Konermann et al. 2015). The dCas9-VPR strategy that we describe here will make it possible to produce genome-scale transgenic sgRNA lines for overexpression screens, thus complementing other approaches such as random UAS-insertion lines (“EP lines”) (Rørth 1996; Staudt et al. 2005) and UAS-ORF lines (Bischof et al. 2013).
Supplementary Material
Acknowledgments
We thank Alex Chavez for the HS-dCas9-VPR plasmid, Arpan Ghosh for invaluable cloning advice, and Richelle Sopko for comments on the manuscript. Sequencing reactions were carried out with an ABI3730xl DNA analyzer at the DNA Resource Core of Dana-Farber/Harvard Cancer Center (funded in part by National Cancer Institute Cancer Center support grant 2P30CA006516-48). B.E.-C. acknowledges funding from the National Institutes of Health (NIH) under the Ruth L. Kirschstein National Research Service Award F32GM113395 from the NIH General Medical Sciences Division. This work was supported in part by R01GM084947 (NP). NP is an investigator of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: J. Sekelsky
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.181065/-/DC1.
Literature Cited
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Nieto M. A., 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132: 3151–3161. [DOI] [PubMed] [Google Scholar]
- Bischof J., Bjorklund M., Furger E., Schertel C., Taipale J., et al. , 2013. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140: 2434–2442. [DOI] [PubMed] [Google Scholar]
- Boulay J. L., Dennefeld C., Alberga A., 1987. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature 330: 395–398. [DOI] [PubMed] [Google Scholar]
- Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M. et al, 2015. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12: 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Sanjana N. E., Zheng K., Shalem O., Lee K., et al. , 2015. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. W., Wang H., Yang H., Shi L., Katz Y., et al. , 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse N., Hirose S., Hayashi S., 1996. Determination of wing cell fate by the escargot and snail genes in Drosophila. Development 122: 1059–1067. [DOI] [PubMed] [Google Scholar]
- Gibson D. G., 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., et al. , 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., et al. , 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden B. E., Lin S., Perrimon N., 2014. Cas9-based genome editing in Drosophila. Methods Enzymol. 546: 415–439. [DOI] [PubMed] [Google Scholar]
- Housden B. E., Valvezan A. J., Kelley C., Sopko R., Lin S., et al. , 2015. Identification of potential drug targets for Tuberous Sclerosis Complex by synthetic screens combining CRISPR-based knockouts with RNAi. Science Signaling (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O., et al. , 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C., Arslan S., Singh R., Thorpe J., Adli M., 2014. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32: 1–9. [DOI] [PubMed] [Google Scholar]
- Leptin M., 1991. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 5: 1568–1576. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- MacArthur S., Li X.-Y., Li J., Brown J. B., Chu H. C., et al. , 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Linder S. J., Cascio V. M., Fu Y., Ho Q. H., et al. , 2013. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., et al. , 2013. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D., 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56: 777–783. [DOI] [PubMed] [Google Scholar]
- Ng M., Diaz-Benjumea F. J., Vincent J. P., Wu J., Cohen S. M., 1996. Specification of the wing by localized expression of wingless protein. Nature 381: 316–318. [DOI] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M. A., 2002. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 3: 155–166. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P., Kocak D. D., Vockley C. M., Adler A. F., Kabadi A. M., et al. , 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T., Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembold M., Ciglar L., Yanez-Cuna J. O., Zinzen R. P., Girardot C., et al. , 2014. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 28: 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T., Girardot C., Brehme M., Tongprasit W., Stolc V., et al. , 2007. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 21: 436–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E., Higashijima S., Emori Y., Saigo K., 1993. Two FGF-receptor homologues of Drosophila: one is expressed in mesodermal primordium in early embryos. Development 117: 751–761. [DOI] [PubMed] [Google Scholar]
- Staudt N., Molitor A., Somogyi K., Mata J., Curado S., et al. , 2005. Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS Genet. 1: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M. E., Gilbert L. A., Qi L. S., Weissman J. S., Vale R. D., 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Stoetzel C., Gorostiza-Thisse C., Perrin-Schmitt F., 1988. Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J. 7: 2175–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Scott D. A., Kriz A. J., Chiu A. C., Hsu P. D., et al. , 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan J. G., Lee M. E., Almeida R., Gilbert L. A., Whitehead E. H., et al. , 2014. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., et al. , 2007. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.