Abstract
Caenorhabditis elegans postembryonic development consists of four discrete larval stages separated by molts. Typically, the speed of progression through these larval stages is investigated by visual inspection of the molting process. Here, we describe an automated method to monitor the timing of these discrete phases of C. elegans maturation, from the first larval stage through adulthood, using bioluminescence. The method was validated with a lin-42 mutant strain that shows delayed development relative to wild-type animals and with a daf-2 mutant that shows an extended second larval stage. This new method is inherently high-throughput and will finally allow dissecting the molecular machinery governing the speed of the developmental clock, which has so far been hampered by the lack of a method suitable for genetic screens.
Keywords: development, larval molt, bioluminescence, luciferase, Caenorhabditis elegans
CAENORHABDITIS elegans is a key model system in which to study development from genetic regulation and cell biological processes to the precisely programmed body plan. Postembryonic development in this nematode initiates in the presence of food after the egg hatches. The larval developmental program consists of four discrete stages (L1–L4). The formation of a new cuticle, the separation of the old and new cuticles (apolysis), and the escape from the old cuticle (ecdysis) occur during transitions between larval stages called molts (M1–M4). Ecdysis is preceded by lethargus, a period during which the animals show behavioral quiescence and cease pharyngeal pumping (Singh and Sulston 1978). This behavioral quiescence is accompanied by a typical posture (Iwanir et al. 2012; Schwarz et al. 2012) and shows other sleep-like properties such as reversibility and increased arousal threshold (Raizen et al. 2008; Schwarz et al. 2011; Cho and Sternberg 2014).
The rhythmic process of molting is sometimes referred to as a clock despite the lack of rigorous quantitative analyses that would support this. The clock analogy stems in part from the fact that C. elegans has a homolog of the period circadian clock gene, lin-42, which, when mutated, slows developmental timing (Jeon et al. 1999; Monsalve et al. 2011). The expression of the period gene homolog oscillates with the molting cycle (Jeon et al. 1999), similar to daily oscillations in per expression in mammals and flies (Hardin et al. 1990; Tei et al. 1997). Furthermore, the timing of lethargus is programmed by the developmental clock (Raizen et al. 2008), similar to how the endogenous circadian clock programs the timing of the sleep episodes (Aschoff 1965). These features suggest a functional conservation between the developmental and circadian clocks, but the molecular mechanism of the rhythmicity of larval development remains elusive.
Developmental timing is usually studied by means of direct observation of the animals at short time intervals using a dissecting microscope. Lethargus, cessation of pharyngeal pumping, ecdysis (Monsalve et al. 2011), and expression of GFP driven by promoters of genes that oscillate with the molts (Kim et al. 2013) have been used as hallmarks of the developmental process. Although these methods have shed light on C. elegans development, they are exceedingly time-consuming, are ill-suited for independent measurement of high numbers of animals, and are semiquantitative. The full power of the genetic model organism for studying developmental timing is thus lost. In contrast, the study of embryonic development in C. elegans has benefited from the use of automated setups to monitor the process with high time resolution (Schnabel et al. 1997). Here, we describe a highly quantitative and high-throughput method for continuous monitoring of development in C. elegans using bioluminescence. The method is validated using mutants that were previously reported to alter developmental timing.
Materials and Methods
Strains and experimental conditions
We used the reporter strains PE255 (feIs5 [Psur-5::luc+::gfp; rol-6 (su1006)] X) and PE254 (feIs4 [Psur-5::luc+::gfp; rol-6 (su1006)] V) (Lagido et al. 2008, 2009). For experiments with the lin-42 mutant, we used the strain MRS189 [lin-42 (ok2385); feIs4 [Psur-5::luc+::gfp; rol-6 (su1006)] V]. For experiments with the daf-2 mutant, we used the strain MRS210 [daf-2 (e1370); feIs5 [Psur-5::luc+::gfp; rol-6 (su1006)] X]. For stock animals, we cultured the animals according to standard methods (Brenner 1974), routinely maintaining the strains at 18° on nematode growth medium) plates with a lawn of Escherichia coli (OP50). For all experiments, we grew the stocks to the stage of gravid adult and obtained eggs by bleaching. We adjusted the concentration of the egg suspension to 20 eggs per microliter in M9 buffer and incubated them at 18° with gentle agitation, leading to hatching and arrest of development at the L1 stage. For the experiment with the daf-2 mutant, this incubation was performed at 20°. After 2 days of starvation, we initiated the experiments by exposing the arrested L1 animals to food.
Luminometry of single animals
L1-arrested larvae were simultaneously exposed to food by adding the M9 suspension of larvae to S-basal media containing 10 g⋅liter−1 E. coli OP50. Then, single L1 animals were transferred into a well of a white, flat-bottom, 96-well plate by manual pipetting. Each well contained 200 μl of S-basal media with 10 g⋅liter−1 E. coli OP50 (wet weight) and 100 μM D-Luciferin. Transferring of 96 animals to complete one plate took ∼30 min. For the experiment with the mutant daf-2, we manually transferred the animals directly from the M9 suspension of larvae to a well of the 96-well plate containing 100 μl of S-basal with 100 μM D-Luciferin (without food) and then resumed development by adding 100 μl of S-basal with 20 g⋅liter−1 E. coli OP50 (wet weight) and 100 μM D-Luciferin. Plates were sealed with a gas-permeable cover (Breathe Easier, Diversified Biotech). We measured luminescence (Berthold Centro LB960 XS3) for 1 sec, typically at 5-min intervals. We performed all experiments inside temperature-controlled incubators (Panasonic MIR-154). Temperature was monitored using HOBO Data Loggers (Onset).
Data analysis
The raw data from the luminometer were analyzed using ChronoOSX 3 (Roenneberg and Taylor 2000), and the period of the oscillation was calculated by autocorrelation. For visualization of the data as heat maps, we trend-corrected the raw data (Supporting Information, Figure S2A), dividing by a centered moving average (Figure S2B). The moving average was calculated for the duration of one cycle (or period) of the oscillation. We used the trend-corrected data to generate heat maps (R software). To determine the timing of the molts, the trend-corrected data were converted to binary, using 75% of the value of the moving average as a threshold (Figure S2, C and D). We then evaluated the data for onset and offset of molting by detecting the transitions in the binarized data (Figure S2E). Transitions from 1 to 0 correspond to onset of the molt and transitions from 0 to 1 correspond to offset of the molt. The variability of the luminometry signal introduces noise in the binarized data (Figure S2D). To prevent this noise from affecting the detection of the transitions, a simple rule was applied: onsets were followed by “0” for at least 1 hr, and offsets were preceded by “0” for at least 1 hr. We calculated the duration of each larval stage and molt from the beginning and the end of the four molts. Before plotting the data, we removed the outliers detected using the Grubb’s test. To test for differences in the duration of each stage between the N2 and mutant strains (Figure 3), we used the two-tailed unpaired t-test.
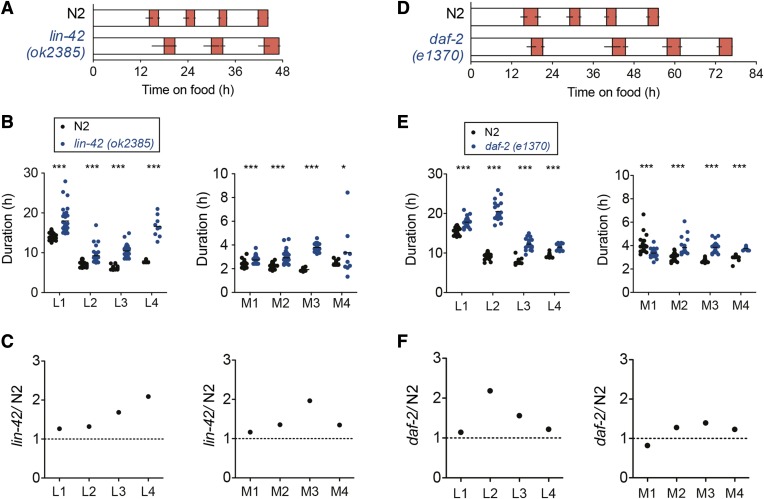
Figure 3.
Developmental timing in lin-42 (ok2385) and daf-2 (e1370) mutants. (A) Average duration of development for N2 (n = 37) and lin-42 (n = 33) at 21.5°. Only three larval stages and molts are represented for lin-42 (see text). Red represents the molts as inferred by low LUC signal. Error bars represent the SD of the duration of each interval. (B) Duration of larval stages (L1–L4) and molts (M1–M4) for N2 and lin-42. Average of each stage is represented by a black horizontal bar. The duration of all stages is significantly different between the N2 and lin-42 backgrounds (***P < 0.001; *P < 0.05). Only nine lin-42 (ok2385) animals progress through a fourth molt. (C) Ratio of the duration of each stage of development comparing lin-42 (ok2385) and N2 backgrounds. (D) Average duration of development for N2 (n = 20) and daf-2 (n = 22) at 19.5°, the permissive temperature for the e1370 mutation. Red represents the molts. Error bars represent the SD of the duration of each interval. (E) Duration of larval stages and molts for N2 and daf-2. Average of each stage is represented by a black horizontal bar. The duration of all stages is significantly different between the N2 and daf-2 backgrounds (***P < 0.001). (F) Ratio between daf-2 (e1370) and N2 of the duration of each stage of development.
Fluorescence and luminescence of populations of animals
To test for fluorescence and luminescence signals around the L1–L2 molt, we added populations of ∼500 arrested L1 larvae to a 96-well plate containing 200 μl of S-basal media with 100 μM D-Luciferin and 10 g⋅liter−1 E. coli OP50 to resume development. Larvae were added to the plate every hour for 10 hr. We incubated the plate at 20.5° and then measured for luminescence and fluorescence (FLUOstar Omega, BMG Labtech) 12 hr after the last inoculation, covering a period of 12–22 hr after resumption of development. For fluorescence, excitation was set at 485 nm and emission at 535 nm.
In vitro ATP measurements
For in vitro measurements of ATP around the L1–L2 molt, we added populations of ∼500 arrested L1 larvae to a 96-well plate containing 200 μl of S-basal media with 100 μM D-Luciferin and 10 g⋅liter−1 E. coli OP50 to resume development. Larvae were added to the plate every hour for 9 hr. We incubated the plate at 20.5° for 13 hr after the last inoculation, covering a period of 13–22 hr after resumption of development. We measured luminescence as a control of developmental stage and then collected animals for ATP measurement. The collected larvae (∼500 per sample) were washed three times with S-basal, frozen in liquid nitrogen in 50 μl of S-basal, and stored at –80°. Samples were then boiled for 15 min and centrifuged to pellet the debris. ATP content in 25 μl of the supernatant was measured using a time-stable ATP determination kit (Biaffin) and normalized to protein content (DC Protein Assay, Bio-Rad).
Results and Discussion
A quantitative, automated method to measure the timing of development in C. elegans
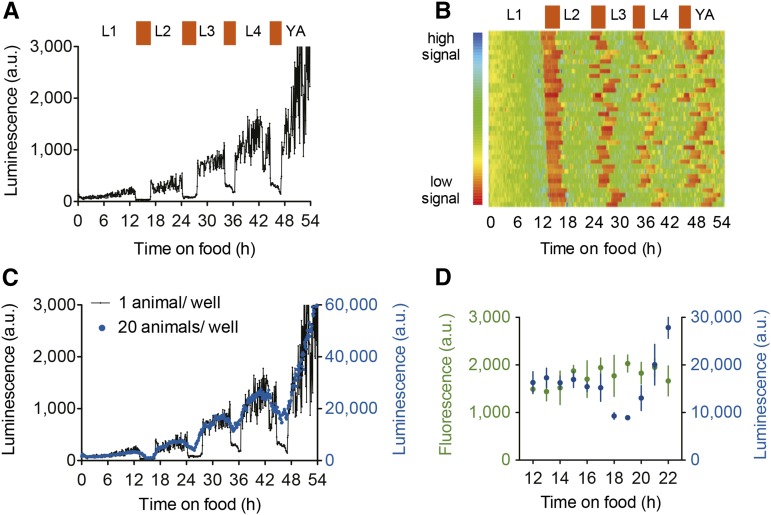
We followed two independent strains throughout development that constitutively and ubiquitously express the luciferase protein fused to GFP (Psur-5::luc+::gfp) (Lagido et al. 2008, 2009). We measured bioluminescence from single animals in 96- or 384-well plates. Experiments were initiated by exposing arrested L1 larvae to food at time = 0, a stimulus that launches the larval developmental program. The luminescence signal emitted by a single animal generally increased throughout development. However, the signal is punctuated and structured by four interruptions (Figure 1A). Using visual inspection, we confirmed that these interruptions are coincident with behavioral quiescence, characteristic of the molting stage. It is these temporal structures that allowed a quantitative assessment of the timing of development. Entry into the first molt occurs at 14.08 ± 1.06 hr at 21.5° when starved L1 are monitored (Figure 1B). The transitions between intermolt and molt are short (5–10 min; Figure S1A), allowing precise determination of the beginning and end of the molt (Figure S2). This method can also report the timing of development for populations, although the precise information about the beginning and the end of the molts is lost due to animal-to-animal variability (Figure 1C).
Figure 1.
Luminescence signal reports larval development of single animals and populations. (A) Bioluminescence from a single animal (LUC::GFP reporter; strain PE255) over 54 h at 21.5° as it develops from arrested L1 to adult. The absence of substrate intake during the molts provokes a decrease in the bioluminescence signal. (B) Heat map showing trend-corrected data for 37 independently monitored animals. (C) Raw data for 20 animals in one well are compared with the signal from a single animal (as in Figure 1A). (D) Fluorescence and luminescence of a population of animals grown at 20.5° during late L1 to early L2. The graph shows data from three experimental replicates, each of which was derived from three to four replicate wells; error bars show SEM.
To determine if the reduction in luminescence is due to a change in the expression level of the LUC::GFP fusion protein, we measured luminescence and fluorescence in parallel in a population of animals before, during, and after the L1–L2 molt. The expression of the LUC::GFP protein in these strains is driven by the regulatory region of the constitutively expressed sur-5 (K03A1.5) gene (Lagido et al. 2008). The fluorescence signal remains stable during the molt, indicating that the LUC::GFP fusion protein is not cycling through development to yield rhythmic light emission (Figure 1D). This is furthermore predicted by work showing that sur-5 expression does not oscillate during larval development (Kim et al. 2013; Hendriks et al. 2014).
Firefly (Photinus pyralis) LUC catalyzes the conversion of the substrate luciferin, supplied in the media, into oxyluciferin. This occurs in a two-step reaction that additionally requires ATP and oxygen and produces AMP, CO2, and light (Wet et al. 1987). Given that oxygen diffuses through the cuticle and luciferin does not (Lagido et al. 2008), the decrease in the signal must be due to depletion of ingested luciferin, endogenous ATP, or a combination of both. In vitro measurements of endogenous ATP levels do not correlate with luminescence during the molt (R2 = 0.0305). ATP levels increased during the last hours of the L1 stage and then decreased to a trough at the end of the molting period, 2 hr after the trough of the luminescence signal (Figure S1B). Upon transfer of an adult to luciferin-free media, luminescence decreases at the same rate as during the entry into a molt (Figure S1C). Thus, luciferin is rapidly depleted in living animals when its uptake from media is blocked. During each molt, C. elegans form a plug of extracellular material in the buccal cavity and they cease pharyngeal pumping (Singh and Sulston 1978; George-Raizen et al. 2014). We conclude that a shortage in the supply of ingested luciferin is sufficient to explain the decrease in luminescence signal during molts.
Quantitative analysis of developmental timing
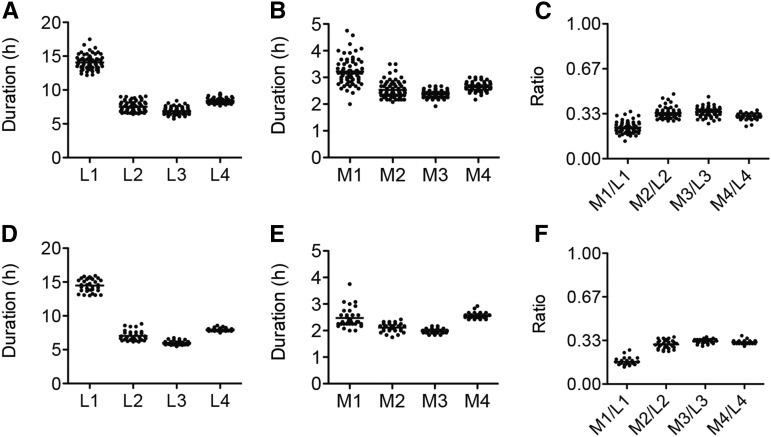
Our method allows for longitudinal measurement of developmental timing of single animals. The assay permits quantitative analysis of the timing of each larval stage (L1, L2, L3, and L4; Figure 2, A and D) as well as that of the molts (M1, M2, M3, and M4; Figure 2, B and E). L1 is longer than L2, L3, and L4. This is in accordance with original observations on the timing of molting (Byerly et al. 1976), although in this case we cannot disregard additional extension of the L1 stage due to a lag in the resumption of development after starvation-induced arrest. Similarly, M1 is longer than M2, M3, and M4. We calculated the ratio of the molt/larval duration for each stage. Except for the first stage (M1/L1), the molts are about one-third as long as the preceding larval stage (Figure 2, C and F).
Figure 2.
Quantitative analysis of the developmental program of C. elegans at 21.5° based on luminescence. Duration of larval stages (L1–L4) (A), molts (M1–M4) (B), and ratio between the duration of each molt and its preceding larval stage (C) for strain PE255. Duration of larval stages (L1–L4) (D), molts (M1–M4) (E), and ratio between the duration of each molt and its preceding larval stage (F) for strain PE254. Average is represented with a black horizontal bar.
Analysis of mutant phenotypes in developmental timing: lin-42 and daf-2
Several lin-42 alleles have been investigated for the progression of larval development. The first description of these mutants indicated a precocious seam cell terminal differentiation that leads to the formation of adult-specific alae during the third molt (Abrahante et al. 1998; Jeon et al. 1999). Many lin-42 animals fail to shed the L4 cuticle and execute the fourth molt (Abrahante et al. 1998). However, a detailed analysis of the timing of molting has been performed with only two lin-42 alleles, ok2385 and n1089, showing reduced speed of development (Monsalve et al. 2011). Consistent with this developmental delay, the cyclic expression of molting defective-10 (mlt-10), encoding a protein necessary for molting, shows an increased period in a lf allele of lin-42 (McCulloch and Rougvie 2014).
We therefore challenged our method using a lin-42 (ok2385) mutant. We crossed this mutant with the LUC::GFP reporter strain and analyzed developmental timing. At 21.5°, the lin-42 animals progress through three larval stages in the time the N2 worms require to complete the developmental program (Figure 3A). The reduction of speed is significant for all steps of development (L1–L4, M1–M4; Figure 3B). The majority of animals (24 of 33) fail to execute the fourth molt—hence the relative dearth of data for L4 and M4 in the mutant animals (n = 9) (Figure 3B). All stages of development are significantly longer in lin-42 mutants, with the third molt (M3) and the fourth larval stage (L4) taking twice as long as in the control strain (Figure 3C).
Environmental cues including food and temperature also control developmental speed and, when unfavorable, can lead to arrested development, namely as an alternative third larval stage called the dauer diapause (Cassada and Russell 1975). Mutations in the gene daf-2 (Insulin/IGF-1 receptor) result in constitutive dauer formation (Daf-c) (Vowels and Thomas 1992). The allele daf-2 (e1370) enters dauer at restrictive temperatures (Vowels and Thomas 1992) and shows a reduced developmental speed at permissive temperature, especially during the second larval stage (Ruaud et al. 2011). We used the daf-2 (e1370) mutant for a second validation experiment. We grew daf-2 animals at 19.5° (permissive temperature for the Daf-c phenotype) and, for the first time, were able to calculate the duration of all larval stages and molts of individual daf-2 animals (Figure 3D). The L2 stage was prolonged (more than twofold) in the mutant compared to the wild-type animals (validation; Figure 3, E and F). Furthermore, we observed a lengthening of all stages of development in daf-2 (P < 0.001), including the molts.
In summary, we describe a quantitative, high-throughput method using bioluminescence to report the fundamental process of development in a genetic model organism. We used the alternation between feeding and fasting in C. elegans larval development to generate a periodic bioluminescence signal, continuously recorded, to quantify the duration of the molts and larval stages in individual animals. The oscillation of the bioluminescence signal is ultimately generated by substrate availability, not by differential gene expression. In adult worms, when the mouth of the animal is not blocked, the luminescence signal could potentially be used to report for food intake. This is a plausible application that would contribute to the study of nutrition and metabolism in C. elegans.
In validating the method with the lin-42 and daf-2 mutants, we were able to describe the defects in developmental timing due to mutations in the circadian clock gene homolog and the C. elegans insulin receptor. We note that the traditional (e.g., visual inspection) methods are obviously sufficient to capture the lin-42 and daf-2 mutant phenotypes (Monsalve et al. 2011; Ruaud et al. 2011), but they are time-consuming and not feasible for screening of large numbers of animals. The assay that we describe lends itself to high-throughput protocols, with individual animals in each well of a 384-well plate giving robust signals (Figure S3). Automated phenotyping of large numbers of worms, as described here, should allow rapid progress in the identification of the developmental clock using genetics methods. The elucidation of the molecular mechanism of this biological clock will allow assessment of the mechanistic conservation with the circadian clock of insects and mammals and may help to tackle the molecular bases for the circadian rhythms described in C. elegans (Kippert et al. 2002; Saigusa et al. 2002; Simonetta et al. 2009; van der Linden et al. 2010; Olmedo et al. 2012). In addition, this method will allow the assessment of the effect of environmental factors, like temperature, diet, or presence of chemicals, in the progression of development.
Supplementary Material
Acknowledgments
Strains PE254 and PE255 were a gift from C. Lagido (University of Aberdeen). We thank the Caenorhabditis Genetics Center and the International C. elegans Gene Knockout Consortium for strains. We thank D. Lenssen for comments on data analysis; A. Fernández-Yáñez, Tanja Popp, and Angela Meckl for technical help; A. van der Linden for comments on an earlier version of the manuscript; and the Conradt and Lambie labs for support. Our work is supported by the Ludwig-Maximilians-University Munich and the Friederich-Bauer-Stiftung (M.M., M.O., and M.G.); the European Research Council (ERC-2011-StG-281691) and the Spanish Ministerio de Economía y Competitividad (BFU2012-35509) (M.A.-S.); and a Marie-Curie Intra-European Fellowship (FP7-PEOPLE-2013-IEF/GA Nr: 627263) (to M.O. and M.A.-S.).
Author contributions: M.O. and M.G. performed the experiments; M.O., M.G. ,and M.M. designed the project; and M.O., M.G., M.A.-S., and M.M interpreted results and wrote the manuscript.
Footnotes
Communicating editor: P. Sengupta
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179242/-/DC1.
Literature Cited
- Abrahante J. E., Miller E. A., Rougvie A. E., 1998. Identification of heterochronic mutants in Caenorhabditis elegans: temporal misexpression of a collagen:green fluorescent protein fusion gene. Genetics 149: 1335–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschoff J., 1965. Circadian rhythms in man. Science 148: 1427–1432. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly L., Cassada R. C., Russell R. L., 1976. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev. Biol. 51: 23–33. [DOI] [PubMed] [Google Scholar]
- Cassada R. C., Russell R. L., 1975. The Dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. [DOI] [PubMed] [Google Scholar]
- Cho J. Y., Sternberg P. W., 2014. Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell 156: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George-Raizen J. B., Shockley K. R., Trojanowski N. F., Lamb A. L., Raizen D. M., 2014. Dynamically-expressed prion-like proteins from a cuticule in the pharynx of Caenorhabditis elegans. Biol. Open 3: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P. E., Hall J. C., Rosbash M., 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540. [DOI] [PubMed] [Google Scholar]
- Hendriks G.-J., Gaidatzis D., Aeschimann F., Groshans H., 2014. Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell 53: 380–392. [DOI] [PubMed] [Google Scholar]
- Iwanir S., Tramm N., Nagy S., Wright C., Ish D., et al. , 2012. The microarchitecture of C. elegans behavior during lethargus: homeostatic bout dynamics, a typical body posture, and regulation by a central neuron. Sleep 36: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon M., Gardner H. F. E. A. Miller, J. Deshler, and A. E. Rougvie, 1999. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286: 1141–1146. [DOI] [PubMed] [Google Scholar]
- Kim D. H., Grün D., van Oudenaarden A., 2013. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat. Genet. 45: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert F., Saunders D. S., Blaxter M. L., 2002. Caenorhabditis elegans has a circadian clock. Curr. Biol. 12: R47–R49. [DOI] [PubMed] [Google Scholar]
- Lagido C., Pettitt J., Flett A., Glover L. A., 2008. Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagido C., McLaggan D., Flett A., Pettitt J., Glover L. A., 2009. Rapid sublethal toxicity assessment using bioluminescent Caenorhabditis elegans, a novel whole-animal metabolic biosensor. Toxicol. Sci. 109: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch K. A., Rougvie A. E., 2014. Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression. Proc. Natl. Acad. Sci. USA 111: 15450–15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve G. C., Van Buskirk C., Frand A. R., 2011. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21: 2033–2045. [DOI] [PubMed] [Google Scholar]
- Olmedo M., O’Neill J. S., Edgar R. S., Valekunja U. K., Reddy A. B., et al. , 2012. Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 109: 20479–20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y.-J., et al. , 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572. [DOI] [PubMed] [Google Scholar]
- Roenneberg T., Taylor W., 2000. Automated recordings of bioluminescence with special reference to the analysis of circadian rhythms. Methods Enzymol. 305: 104–119. [DOI] [PubMed] [Google Scholar]
- Ruaud A. F., Katic I., Bessereau J. L., 2011. Insulin/insulin-like growth factor signaling controls non-Dauer developmental speed in the nematode Caenorhabditis elegans. Genetics 187: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigusa T., Ishizaki S., Watabiki S., Ishii N., Tanakadate A., et al. , 2002. Circadian behavioural rhythm in Caenorhabditis elegans. Curr. Biol. 12: R46–R47. [DOI] [PubMed] [Google Scholar]
- Schnabel R., Hutter H., Moerman D., Schnabel H., 1997. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev. Biol. 184: 234–265. [DOI] [PubMed] [Google Scholar]
- Schwarz J., Lewandrowski I., Bringmann H., 2011. Reduced activity of a sensory neuron during a sleep-like state in Caenorhabditis elegans. Curr. Biol. 21: R983–R984. [DOI] [PubMed] [Google Scholar]
- Schwarz J., Spies J. P., Bringmann H., 2012. Reduced muscle contraction and a relaxed posture during sleep-like Lethargus. Worm 1: 11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta S. H., Migliori M. L., Romanowski A., Golombek D. A., 2009. Timing of locomotor activity circadian rhythms in Caenorhabditis elegans. PLoS ONE 4: e7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. N., Sulston J. E., 1978. Some observations on moulting in Caenorhabditis elegans. Nematologica 24: 63–71. [Google Scholar]
- Tei H., Okamura H., Shigeyoshi Y., Fukuhara C., Ozawa R., et al. , 1997. Circadian oscillation of a mammalian homologue ofthe Drosophila period gene. Nature 389: 512–516. [DOI] [PubMed] [Google Scholar]
- van der Linden A. M., Beverly M., Kadener S., Rodriguez J., Wasserman S., et al. , 2010. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 8: e1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J., Thomas J. H., 1992. Genetic analysis of chemosensory control of Dauer formation in Caenorhabditis elegans. Genetics 130: 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S., 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.