Abstract
Centromeres of the fission yeast Schizosaccharomyces pombe lack the highly repetitive sequences that make most other "regional" centromeres refractory to analysis. To map fission yeast centromeres, we applied H4S47C-anchored cleavage mapping and native and cross-linked chromatin immunoprecipitation with paired-end sequencing. H3 nucleosomes are nearly absent from the central domain, which is occupied by centromere-specific H3 (cenH3 or CENP-A) nucleosomes with two H4s per particle that are mostly unpositioned and are more widely spaced than nucleosomes elsewhere. Inner kinetochore proteins CENP-A, CENP-C, CENP-T, CENP-I, and Scm3 are highly enriched throughout the central domain except at tRNA genes, with no evidence for preferred kinetochore assembly sites. These proteins are weakly enriched and less stably incorporated in H3-rich heterochromatin. CENP-A nucleosomes protect less DNA from nuclease digestion than H3 nucleosomes, while CENP-T protects a range of fragment sizes. Our results suggest that CENP-T particles occupy linkers between CENP-A nucleosomes and that classical regional centromeres differ from other centromeres by the absence of CENP-A nucleosome positioning.
Keywords: Schizosaccharomyces pombe, CENP-A, CENP-C, CENP-T, CENP-I
CENTROMERES are specialized sites on eukaryotic chromosomes that are responsible for chromosome segregation during mitosis and meiosis. Centromeres act as sites for the assembly of a complex multi-protein structure called the kinetochore that attaches to microtubules and orchestrates chromosome movements. Centromere DNA sequences are not conserved between species and vary in size from ∼125 bp in the point centromeres (Pluta et al. 1995) of the budding yeast Saccharomyces cerevisiae to the entire length of chromosomes in holocentromeres in some plants and animals (Melters et al. 2012). Intermediate between these extremes are the regional centromeres (Pluta et al. 1995) of the fission yeast Schizosaccharomyces pombe, which consist of 4–7 kb of unique or low copy DNA (cnt1, cnt 2, and cnt3) surrounded by inverted innermost repeats (imr), which in turn are flanked by tens of kilobases of outer repeats (otr) consisting of the dg and dh repeats (Chikashige et al. 1989). The cnt + imrs form a central domain in each centromere that assembles kinetochore proteins, while the otrs assemble pericentric heterochromatin (Polizzi and Clarke 1991; Takahashi et al. 1992; Saitoh et al. 1997; Takahashi et al. 2000; Cam et al. 2005). Fission yeast centromeres are viewed as an important model for understanding the centromeres and pericentric heterochromatin of plants and animals, both of which are typically composed of megabase-sized arrays of highly tandemly repeated sequences, rendering these centromeres refractory to complete mapping (Plohl et al. 2014). Centromeric tandem repeats are species specific, and monomers come in many sizes, but are most commonly ∼100–200 bp (Melters et al. 2013).
Although centromeric sequences are diverse, the components of the kinetochore are largely conserved among different eukaryotes (Meraldi et al. 2006; Perpelescu and Fukagawa 2011). The kinetochore is often conceptually divided into the outer kinetochore that binds microtubules and the inner kinetochore composed of proteins that bind DNA or centromeric chromatin, also known as the Constitutive Centromere-Associated Network (CCAN) in vertebrates (Hori et al. 2008). How inner kinetochore proteins interact with DNA to form centromeric chromatin is not well understood. The best studied DNA-binding kinetochore protein is the centromere-specific histone H3 variant (cenH3), which replaces canonical H3 in nucleosomes that wrap centromeric DNA, creating a centromere-specific chromatin structure that is thought to epigenetically mark the centromere and to serve as an essential foundation for assembling the kinetochore (Henikoff and Furuyama 2012; Westhorpe and Straight 2013). In many animal and plant centromeres, tandem repeats position both H3 nucleosomes (Musich et al. 1977; Musich et al. 1982; Fischer et al. 1994; Vershinin and Heslop-Harrison 1998) and cenH3 nucleosomes (Hasson et al. 2013; Zhang et al. 2013; Henikoff et al. 2015) into periodic arrays. Arrays of cenH3 nucleosomes are interspersed with arrays of H3 nucleosomes along the chromosome (Blower et al. 2002; Chueh et al. 2005; Wolfgruber et al. 2009; Ribeiro et al. 2010; Wu et al. 2011; Gong et al. 2012; Ishii et al. 2015). Despite this interspersed pattern, inside cells cenH3 and H3 nucleosomes occupy physically distinct regions in space (Blower et al. 2002; Zhang et al. 2005). In vertebrates, cenH3 is known as CENP-A and was discovered together with another conserved inner kinetochore protein, CENP-C (Earnshaw and Rothfield 1985), which also binds DNA (Sugimoto et al. 1994; Yang et al. 1996; Politi et al. 2002; Trazzi et al. 2002; Hori et al. 2008). More recently, inner kinetochore proteins CENP-T, CENP-W, CENP-S, and CENP-X were found to be histone-fold-containing proteins that form a heterotetrameric nucleosome-like complex made up of one CENP-TW dimer and one CENP-SX dimer that together can wrap DNA and induce positive supercoils in vitro (Nishino et al. 2012; Takeuchi et al. 2014). CENP-C and CENP-T are thought to form alternative connections to the outer kinetochore (Hori et al. 2008; Gascoigne et al. 2011; Nishino et al. 2013). Both CENP-C and CENP-T form complexes with CENP-A nucleosomes that are sensitive to disruption by micrococcal nuclease (MNase) digestion (Ando et al. 2002; Politi et al. 2002; Foltz et al. 2006; Hori et al. 2008). However, under high MNase conditions in chicken cell nuclei, neither CENP-T nor CENP-C co-immunoprecipitated with CENP-A, but both co-immunoprecipitated with H3, leading to the proposal that CENP-C and CENP-T associate with H3 nucleosomes (Hori et al. 2008). Subsequently, human CENP-C was found to preferentially bind CENP-A nucleosomes over H3 nucleosomes, suggesting that the co-immunoprecipitation of H3 with CENP-C and CENP-T might have been misleading, due to the much greater abundance of H3 over CENP-A (Carroll et al. 2010). CENP-T, however, is still commonly thought to be associated with H3 nucleosomes (Perpelescu and Fukagawa 2011; Westermann and Schleiffer 2013; Fukagawa and Earnshaw 2014), although this appears to be inconsistent with the physically separate domains of pericentric H3 and centromeric CENP-A nucleosomes (Blower et al. 2002), and with recent suggestions that CENP-T interacts with the N-terminal tail of CENP-A (Folco et al. 2015; Logsdon et al. 2015). The tandem repeats of vertebrate centromeres are an obstacle to mapping the precise DNA locations of inner kinetochore proteins on centromeric DNA and resolving this apparent contradiction.
Many features of tandem repeat centromeres have similarities to the classical regional centromeres of fission yeast, in which cenH3 nucleosomes and other kinetochore proteins are found in the central domain (cnt + imr) of each chromosome (Takahashi et al. 2000; Pidoux et al. 2003; Hayashi et al. 2004; Liu et al. 2005) between blocks of canonical nucleosomes containing H3 methylated on lysine 9 (H3K9me) in the pericentric otrs that are bound by heterochromatin proteins (Partridge et al. 2000; Cam et al. 2005). H3 nucleosomes in the pericentric otrs of fission yeast are well positioned, particularly in the dh repeats (Moyle-Heyrman et al. 2013), but the occupancy and positioning of nucleosomes in the central domain has been unclear, with different methods leading to inconsistent conclusions of discrete, metastable, or "fuzzy" nucleosome positions (Song et al. 2008; Lando et al. 2012; Moyle-Heyrman et al. 2013; Yao et al. 2013). As in plants and animals, the cenH3-containing central domains and H3-containing pericentric heterochromatin are physically separate in 3-D space (Kniola et al. 2001; Appelgren et al. 2003). Unlike repeat-based centromeres, however, the low-copy-number sequences of fission yeast centromeres can be precisely mapped.
In fission yeast, cenH3 is encoded by the cnp1 gene and is usually known as Cnp1 or CENP-A (Takahashi et al. 2000), like its vertebrate homolog. Fission yeast CENP-C and CENP-T are encoded by cnp3 and cnp20, respectively (Tanaka et al. 2009). CENP-I (Mis6) is another inner kinetochore protein (Saitoh et al. 1997) that does not bind DNA directly (Hori et al. 2008), but is part of the Mis6/Sim4/Mal2 complex (Pidoux et al. 2003; Hayashi et al. 2004; Liu et al. 2005), corresponding to the vertebrate CENP-HIK complex (Okada et al. 2006) that physically interacts with CENP-T/W (Basilico et al. 2014). To produce a high-resolution map of the locations of inner kinetochore proteins in fission yeast centromeres, we first determined that H3 nucleosomes are virtually absent in the central domains and then performed H4S47C-anchored chemical cleavage mapping to determine the centromeric locations of histone H4 in CENP-A nucleosomes (Brogaard et al. 2012). We then used chromatin immunoprecipitation (ChIP) of MNased chromatin followed by paired-end sequencing of DNA fragments (ChIP-Seq) to map the locations of CENP-A, CENP-T, CENP-C, CENP-I, and the CENP-A-specific chaperone, Scm3 (Pidoux et al. 2009; Williams et al. 2009). Native and formaldehyde cross-linking ChIP protocols have different limitations (Zentner and Henikoff 2014). To clarify how these alternative immunoprecipitation methods affect the recovery of chromatin in mapping experiments, we used both native (N-ChIP) and cross-linking (X-ChIP) conditions and varied the amounts of MNase and the salt concentration of the extraction buffer. We find that CENP-A nucleosomes contain two H4 molecules and show very little positioning and are more widely and variably spaced in the central domain than are the canonical H3 nucleosomes found elsewhere in the genome. CENP-A, CENP-C, CENP-T, CENP-I, and Scm3 are all highly enriched throughout the central domain, except over transfer RNA (tRNA) genes, and are less enriched over the H3-rich otrs, where they appear to be more sensitive to disruption by MNase, suggesting that contiguous arrays of CENP-A nucleosomes may exhibit enhanced stability. CENP-A nucleosomes on average protect slightly less DNA than H3 nucleosomes from MNase digestion, while CENP-C appears to protect little if any DNA beyond that of the CENP-A nucleosome to which it is cross-linked. CENP-T associates predominantly with CENP-A nucleosomes and protects a broad range of fragment sizes in the central domain, suggesting that it may bind DNA primarily in larger protein complexes. In heterochromatin it appears to interact with H3 nucleosomes in a manner that does not protect additional DNA. The results suggest a model in which CENP-T complexes fill in variable spaces between unpositioned CENP-A nucleosomes.
Materials and Methods
Strains, media, and antibodies
S. pombe cultures were grown in yeast extract supplement at 30°. S. pombe strains used in this study are listed in Supporting Information, Table S1. Antibodies used for the ChIP assays are: Anti-Myc antibody (Santa Cruz no. Sc-40), Anti-FLAG magnetic beads (Sigma anti-FLAG M2, no. 8823), Anti-HA magnetic beads (MBL anti-HA-tag mAB, no. M132-9).
Native chromatin immunoprecipitation (N-ChIP)
Native ChIP in S. pombe cells was performed as described (Krassovsky et al. 2012) with minor modifications. Briefly, nuclei were prepared following a previously published protocol (Polizzi and Clarke 1991) and digested with MNase (Sigma N3755, 4 unit/ml) for 5–20 min. The resulting chromatin solution was needle extracted and soluble chromatin was subjected to N-ChIP. The resulting pellet was further solubilized with extraction buffer (0.1% Triton X-100, 10 mM phosphate buffer, 0.75 mM EDTA, 150 mM NaCl) for 4 hr at 4o. The solubilized chromatin was subjected to N-ChIP.
Cross-linking chromatin immunoprecipitation (X-ChIP)
Cross-linking ChIP assays were performed as previously described (Thakur and Sanyal 2012) with minor modifications. An exponentially growing culture of S. pombe was fixed with 1% formaldehyde for 15 min and the reaction was quenched for 5 min at room temperature using glycine to a final concentration of 125 mM. Cells were resuspended in 0.2 mM Tris-HCl, pH 9.4, 10 mM DTT and incubated at 37° for 15 min on a shaker at 180 rpm. Spheroplasting (to 95%) was performed using Zymolyase-100T (Amsbio, no. 120493-1) in 1.2 M Sorbitol, 20 mM Na-HEPES, pH 7.5 at 37° at low speed (80 rpm). Spheroplasting was terminated by adding ice-cold 1.2 M Sorbitol, 1 mM MgCl2, 20 mM Na-PIPES, pH 6.8. Spheroplasts were pelleted at low speed and subsequently washed with ice-cold 1× PBS, buffer I (0.25% TritonX-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM Na-HEPES, pH 6.5), buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Na-HEPES). To release chromatin, washed spheroplasts were resuspended in 140 mM NaCl, 1 mM EDTA, 50 mM K-HEPES, 0.1% sodium deoxycholate, 1% Triton X-100, pH 7.5 containing a protease inhibitor cocktail. Released chromatin was digested with MNase (Sigma N3755, 7–8 unit/ml) for 5–20 min in the presence of 3 mM CaCl2 at 37°. Chromatin was sheared using a Diagenode Bioruptor, and the soluble fraction was clarified by centrifuging at 13000 rpm for 15 min at 4°. Soluble chromatin was diluted 5-fold with extraction buffer (167 mM NaCl, 1.1 mM EDTA, 1.1% Triton X-100, 167 mM Tris-HCl, pH 8.0). Antibody-conjugated magnetic beads were added and the sample was incubated at 4° for 3 hr. Next, beads were sequentially washed twice with extraction buffer and once each with extraction buffer plus 500 mM NaCl, LiCl wash buffer (10 mM Tris-HCl, pH 8, 250 mM LiCl, 1 mM NP40, 0.5% sodium deoxycholate/1 mM EDTA) and TE buffer. Magnetic beads were resuspended in equal volumes of elution buffer I (50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS) and elution buffer II (10 mM Tris-HCl, pH 8, 1 mM EDTA, 0.67% SDS) and incubated at 65° overnight to reverse the cross-links. Samples were treated with RNase A (Roche no. 1119915 at a final concentration of 40 µg/ml) at 37° for 1 hr and Proteinase K (Roche no. 1000144 at a final concentration of 100 µg/ml) at 55° for 3 hr. The DNA was extracted with an equal volume of phenol/chloroform/isoamyl alcohol and precipitated in 100% ethanol overnight at −20°. Precipitated DNA was resuspended in TE.
Data processing and analysis
We used Novoalign 2.08 (http://www.novocraft.com) to map both paired-end and single-end reads to release 2.20 of the S. pombe genomic sequence obtained from http://www.pombase.org. If a read was mapped to multiple locations, one location was picked at random. For paired-end samples, we extracted properly paired reads and analyzed the resulting fragments. For single-end H3 reads downloaded from the NCBI Sequence Read Archive (SRA, DRR003329) we analyzed the full length of mapped reads. For H4S47C-anchored cleavage data, we counted only the endpoints (left and right) of the mapped paired-end fragments and analyzed the raw counts without normalization. Watson–Crick strand cleavage distances were calculated as previously described (Henikoff et al. 2014), and auto- and cross-correlations were calculated using an R program (https://stat.ethz.ch/R-manual/R-devel/library/stats/html/acf.html). For all noncleavage data, we normalized as follows: For each basepair (i) in the reference sequence, we counted the number of fragments aligned over it (ni) and normalized by dividing by the total number of fragments (N) and multiplying by the annotated S. pombe genome size [=(ni/N) × 12,570,000]. This scale factor would result in the number one at each basepair position if the counts were evenly distributed across the genome, and so facilitates comparing different regions. Correlation matrix values used to construct Figure 5 are for the normalized counts at each basepair position within the heterochromatin and central domains. Except as noted, tracks in the figures were autoscaled together by IGV (Thorvaldsdottir et al. 2013) for display.
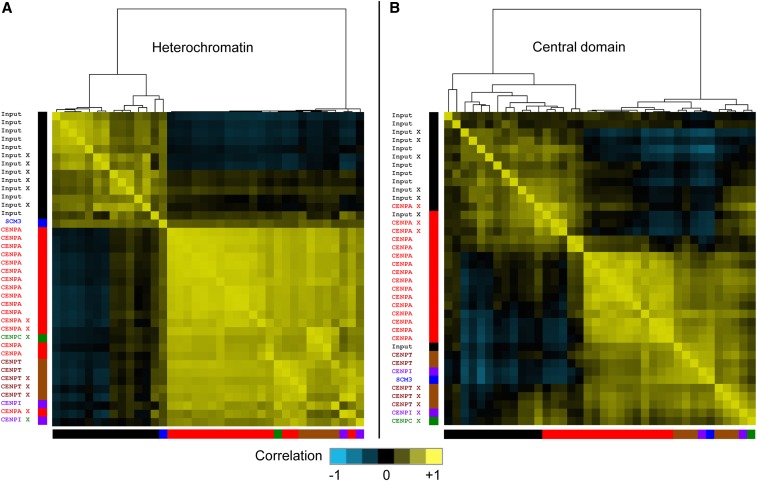
Figure 5.
Heterogeneous correlations between CENP-A ChIP data sets in central domains. Pearson correlation coefficients for all 38 data sets within the combined (A) pericentric heterochromatin and (B) central domains were subjected to hierarchical clustering. Correlation coefficients between normalized counts from cen1, cen2, and cen3 are represented as a heat map. Colors of the bars at the bottom and left side of each heat map indicate the protein targeted by ChIP for: input (black); CENP-A (red); CENP-C (green); CENP-I (purple); CENP-T (brown); and Scm3 (blue). X-ChIP samples are indicated (X). No unique clustering of CENP-A separate from input or other CENPs is seen for central domains, suggesting that the preferences seen in the CENP-A profiles are more likely attributable to technical differences between data sets than to biological differences.
We used the following coordinates to define chromosomal regions based on Pombase release 2.20: central domain (=cnt + imr: I, 3765506–3776767; II,1619257–1629158; III,1093156–1105839), pericentric heterochromatin (=otrs: I, 3753687–3765505 and I, 3776768–3789421; II, 1602264–1619256 and II, 1629159–1644747; III, 1070904–1093155 and III, 1105840–1137003), and euchromatin (distal to otrs: I, 1–3753686 and I, 3789422–end; II, 1–1602263 and II, 1644748–end; III, 1–1070903 and III, 1137004–end). A euchromatic region (ChrI, 3719838–3753687) of the same total length as the three central cores was chosen as a control for calculating Watson–Crick cleavage distances.
Heat maps of base-by-base Pearson correlation coefficients over all 33,849 central domain and 144,316 pericentric heterochromatin base positions were constructed by successive application of Gene Cluster 3.0 (Euclidean distance of correlation coefficients with centroid linkage) and Java TreeView (v. 1.1.6r2, contrast = 1.2) (Eisen et al. 1998). Sliding windows of 1, 11, 101, and 1001 bp were used for triangular smoothing. Correlation analysis with GC content was done similarly, using sliding windows of 11 bp.
Data availability
Strains are listed in Table S1 and are available upon request. The sequence-based data sets generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE64294.
Results
H3 nucleosomes are depleted from the central domain
In previous studies, low levels of H3K4me were observed in the central domains of fission yeast centromeres (Cam et al. 2005), and H3 was found to be reduced in the central domain relative to other select loci (Pidoux et al. 2009; Williams et al. 2009), although these studies did not quantify the amount relative to total H3 or to CENP-A nucleosomes. To determine the relative contributions of CENP-A and H3 nucleosomes to chromatin in the central domain, we compared the occupancy of H3 nucleosomes between the euchromatic arms, the pericentric heterochromatin, and the central domains using published anti-H3 X-ChIP-Seq data sets (Kato et al. 2013). Plotting the log ratio of H3/input, we observed mostly a flat profile in euchromatin and heterochromatin, confirming that canonical H3 nucleosomes comprise almost all of the input in these regions, but we found that H3 is strongly depleted in the central domain (Figure 1A and Figure S1). Occupancy of H3 nucleosomes present in the pericentric heterochromatin was 150 ± 21% of H3 occupancy in the euchromatic arms, similar to the 155 ± 17% occupancy of bulk chromatin (input) in heterochromatin relative to euchromatin (Figure 1B). The occupancy of bulk chromatin in the central domains was 44 ± 7%, but the H3 occupancy was only 4.8 ± 1% relative to euchromatin, indicating the central domains are nearly devoid of H3 and suggesting that these regions are largely occupied by CENP-A nucleosomes. To confirm this, we performed X-ChIP-Seq on a CENP-A-HA strain with wild-type growth (Takahashi et al. 2000) and found that in contrast to H3, CENP-A-HA was strongly enriched in the central domains (Figure 1A). Therefore the vast majority of nucleosomes in the central domain are CENP-A nucleosomes.
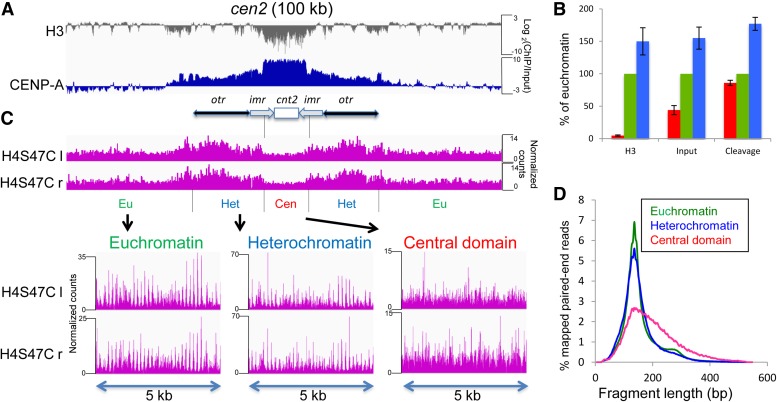
Figure 1.
CENP-A nucleosomes are not positioned in the central domain. (A) A 100-kb region around cen2 is shown. The gray track plots the log-ratio of H3/input from published anti-H3 X-ChIP-Seq data sets (Kato et al. 2013). The blue track shows the log-ratio of CENP-A/input from an X-ChIP-Seq data set for CENP-A-HA. (B) Chromatin occupancy of H3 and Input, or cleavage density are shown in the central domain (red), euchromatin (green), and heterochromatin (blue) as percentages of the average value in euchromatin. (C) The magenta tracks show the frequency of the left (l) and right (r) endpoints of H4S47C-anchored cleavage fragments around cen2, respectively. Expansions of cleavage fragment endpoints in selected 5-kb regions from euchromatin, heterochromatin, and the central domain are shown below. Similar profiles were obtained for cen1 and cen3 (Figure S1). For visual clarity, normalized count occupancies were scaled to the maximum occupancy in each track using the IGV Genome Browser, and the same scaling method was used for the bottom three tracks in the expansions. (D) Distribution of nucleosome-to-nucleosome H4S47C-anchored cleavage fragment lengths in euchromatin, heterochromatin, and the central domain.
CENP-A nucleosomes are not positioned within the central domain
To investigate nucleosome positioning and occupancy within the central domain at single-basepair resolution, we performed H4S47C-anchored cleavage mapping. Chemical cleavage mapping of nucleosomes corresponds well with MNase mapping throughout yeast genomes, but gives more precise positioning and higher resolution and avoids the sequence preferences of MNase mapping (Brogaard et al. 2012; Moyle-Heyrman et al. 2013). Cleavages targeted by H4S47C precisely map histone H4 in both cenH3-containing and canonical H3-containing nucleosomes (Henikoff et al. 2014). As centromeric nucleosomes might be spaced farther apart than other nucleosomes to accommodate the binding of other kinetochore proteins, we performed cleavage mapping without including gel-based size selection for mononucleosomes in the library preparation step. This led to an estimate of the average cleavage density over the three central domains that was 86 ± 4% of that on the euchromatic arms (Figure 1, B and C, and Figure S1). This modest reduction in cleavage density could represent slightly reduced cleavage efficiency in the central domains, but more likely indicates that nucleosome occupancy is somewhat lower in these regions, consistent with the reduced occupancy of bulk chromatin (input) in the central domain that we observed in the H3 data, and with previous reports that the central domain is not saturated for CENP-A (Castillo et al. 2007; Gaither et al. 2014). A similar cleavage density estimate for pericentric heterochromatin is 177 ± 10% of that on euchromatic arms, presumably reflecting high occupancy of nucleosomes in the pericentromere. Regular spikes in the cleavage pattern in euchromatin and heterochromatin reflect consistent nucleosome spacing and positioning (Figure 1C and Figure S1). However, cleavages in the central domains lack any regularity and to a first approximation are similar at most positions, indicating that there is little or no consistent positioning of nucleosomes in these regions.
The length distribution of H4-to-H4 internucleosomal fragments generated by H4S47C cleavage (Figure 1D) peaks at ∼136 bp in euchromatin and in heterochromatin, with an average length of 139 bp, which is shorter than the 147 bp length of a canonical nucleosome. This is because there is very little linker between adjacent nucleosomes (Moyle-Heyrman et al. 2013), and the short intranucleosomal fragments between two H4S47C molecules in the same canonical nucleosome are not recovered (Brogaard et al. 2012; Henikoff et al. 2014). The small secondary peak in euchromatic fragment lengths indicates that dinucleosome-length fragments are relatively rare and the vast majority of nucleosomes are cleaved. In the central domains, the length distribution of fragments has a similar peak but is very strongly skewed toward longer lengths, indicating that the nucleosomes in this region are farther apart overall than nucleosomes elsewhere and are variably spaced. Because there is very little H3 in the central domains, we conclude that these variably spaced nucleosomes are predominantly CENP-A nucleosomes.
The number of CENP-A nucleosomes in fission yeast centromeres has been variously estimated as ∼13/centromere (Yao et al. 2013), ∼113/centromere (680 total CENP-A molecules in an anaphase cluster) (Coffman et al. 2011), and as 21, 19, and 24 nucleosomes for cen1, cen2, and cen3, respectively (Lando et al. 2012). We determined the mean center-to-center fragment length between nucleosomes in the central domain to be 192 bp. From this number we estimated the number of nucleosomes in the central domains of cen1, cen2, and cen3, to be 57, 50, and 64, respectively.
Fission yeast CENP-A nucleosomes contain two H4 molecules
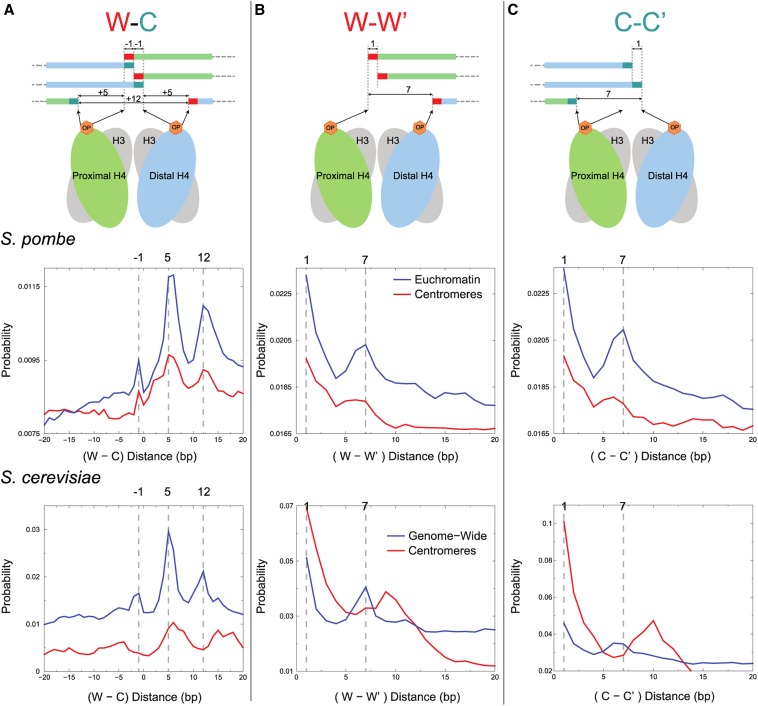
The structure of cenH3 nucleosomes has been controversial, with evidence for several mutually exclusive models in different organisms (Henikoff and Furuyama 2012; Fukagawa and Earnshaw 2014). In budding yeast, H4S47C-anchored cleavage mapping demonstrated precise positioning of cenH3 (Cse4) nucleosomes over the ∼80-bp CDEII sequence in two rotational and two reflectional positions for all 16 centromeres, consistent with heterotypic tetramers (hemisomes) (Henikoff et al. 2014), as previously inferred from high-resolution ChIP (Krassovsky et al. 2012). Such an inference based on cleavage mapping data requires strongly phased nucleosomes, which are lacking for fission yeast CENP-A over the central domain. Nevertheless, distances between fragment ends can be used to infer particle composition and conformation, and this analysis had previously shown that cleavage distances characteristic of octasomes, hexasomes or (H3/H4)2 tetrasomes found in the vast majority of H3 nucleosomes genome-wide were missing from Cse4 nucleosomes at CDEII in budding yeast (Henikoff et al. 2014; Ramachandran et al. 2015). Specifically, the left H4 molecule cleaves mostly at 0 or −1 on the Watson (W) strand and −6 on the Crick (C) strand, and the right H4 cleaves at 0 or +1 on W and +6 on C, such that expected distances between left and right cleavage sites are −1, +5, and +12 (Figure 2A, top). If there is only one H4 per nucleosome, then left–right cleavage distances across the dyad would be missing, and so only +5 would be seen. All three left–right cleavage distances are observed genome-wide in both fission yeast (Figure 2A, middle) and budding yeast (Figure 2A, bottom, data from Henikoff et al. 2014); however, only budding yeast CDEII shows loss of −1 and +12 cleavage distance peaks as expected for hemisomes with only a single H4. The presence of conspicuous −1, +5, and +12 left-right cleavage distance peaks for fission yeast centromeric nucleosomes with a profile that is similar to that seen genome-wide indicates that at least the bulk of CENP-A nucleosomes have two H4 molecules, one on each side of the dyad axis. This conclusion is confirmed by the observation that fission yeast centromeric W–W′ and C–C′ distances peak at +1 and +6 and +7 (Figure 2, B and C, middle), where only +1 would be expected for a single H4 per nucleosome, as observed for budding yeast, where there is a dip at +6 and +7 (Figure 2, B and C, bottom). The nearly complete absence of H3 in fission yeast central domains implies that these cleavage peaks characteristic of H3 nucleosomes genome-wide derive from CENP-A nucleosomes and that in contrast to budding yeast, most or all CENP-A nucleosomes in fission yeast are not hemisomes, but may be tetrasomes, hexasomes, or octasomes.
Figure 2.
CENP-A nucleosomes contain two H4 molecules. (A–C) Top: a model of where H4S47C-anchored cleavage is predicted to occur on the Watson (W) and Crick (C) strands around the dyad of an H3 nucleosome and the predicted distances between cleavage fragment ends. Middle: distribution of distances between cleavage fragment ends for euchromatin (blue line) and for the central domain of fission yeast (red line). Bottom: comparable distributions for budding yeast (Henikoff et al. 2014). Distributions were normalized by the total number of combinations within the 40-bp distance range. (A) W–C distances. (B) W–W′ distances. (C) C–C′ distances.
Budding yeast centromeres show a conspicuous peak at 10 bp in W–W′ and C–C′ cleavage distance plots, indicative of strong rotational phasing of centromeric nucleosomes (Henikoff et al. 2014). In contrast, fission yeast centromeres lack this feature, indicating little if any rotational phasing of CENP-A nucleosomes.
CENP-A is strongly enriched throughout the central domain except at tRNA genes
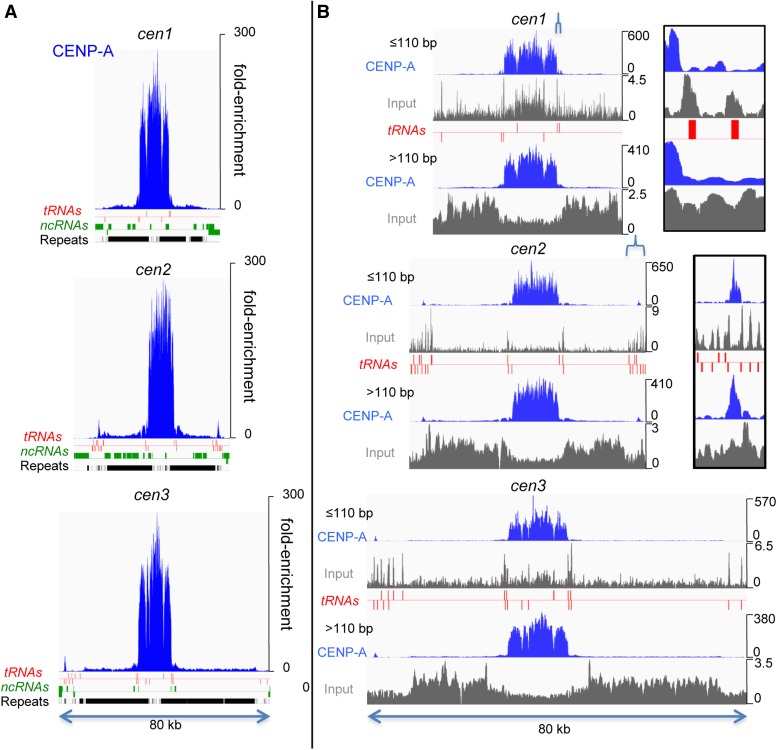
To determine precisely where CENP-A is located within the fission yeast genome, we performed high resolution CENP-A mapping in strains containing either HA- or FLAG-tagged CENP-A expressed under the cnp1 promoter (Takahashi et al. 2000; Shiroiwa et al. 2011) using two different mapping techniques: X-ChIP-Seq and N-ChIP-Seq. N-ChIP typically provides higher dynamic range, better resolution, and avoids cross-linking artifacts, while high-resolution X-ChIP using both MNase and sonication can better solubilize large protein complexes like the kinetochore (Zentner and Henikoff 2014). N-ChIP-Seq revealed that CENP-A-FLAG is highly enriched relative to input, on average ∼200-fold, throughout the central domains and much more weakly over pericentromeric heterochromatin, except at the sites of tRNA genes (Figure 3A). tRNA genes mark the boundaries of the enriched central domain (Partridge et al. 2000; Cam et al. 2005; Scott et al. 2006, 2007), and that of the heterochromatic otr regions, except on the right of cen1. Whereas CENP-A was depleted over the tRNA genes, these genes were enriched in the small subnucleosomal-sized fragments (<110 bp) of the input, where peaks of fragments coincide with each tRNA gene, slightly offset to the upstream side of the gene (Figure 3B). These fragments might be protected from MNase by RNA Polymerase III and its transcription factors.
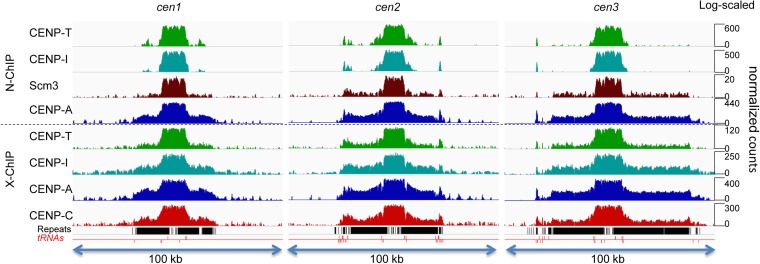
Figure 3.
CENP-A densely occupies central domains, except at tRNA genes. (A) N-ChIP profiles of CENP-A-FLAG are shown for all three centromeric and pericentric regions. Annotations for tRNA genes, noncoding RNAs (ncRNAs), and repeats are indicated on the x-axis, and the y-axis shows fold-enrichment relative to input. (B) CENP-A-FLAG N-ChIP fragments and input fragments were divided into <110-bp and >110-bp size classes. Note that large and small CENP-A ChIP fragments, but not input fragments, show similar profiles. tRNA genes are indicated below the tracks, with those above the red line transcribed left to right and those below transcribed from right to left. Insets show expansions of regions in cen1 and cen2 indicated by brackets.
Although there were apparent peaks of CENP-A-FLAG fragments within the central domain, the most striking feature of the profile is that, except over the tRNA genes, even the valleys between peaks were highly occupied by CENP-A-FLAG, indicating that CENP-A is found on all sequences in the central domain, with possibly minor preferences for some sites over others, forming a ‘fringe’ in the occupancy profile. When fragments were arbitrarily separated into size classes of <110 bp and >110 bp (Figure 3B), we found little difference in the CENP-A-FLAG profiles, indicating that the failure to resolve fixed nucleosome positions is not a result of poor mapping resolution because of long fragment lengths.
No evidence for preferred positions of kinetochore assembly within the central domain
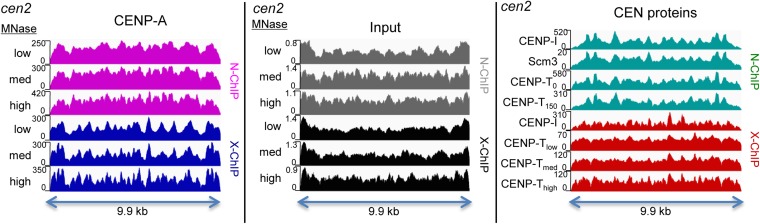
We next tested if other inner kinetochore proteins might form kinetochore structures at preferred sites in the central domains. Using MNase digestion followed by passages through a needle (Jin and Felsenfeld 2007), we were able to solubilize CENP-T-FLAG, CENP-I-HA, CENP-I-GFP, Scm3-FLAG, and myc-CENP-C, and carry out ChIP under native and cross-linking conditions. ChIP-seq mapping revealed that these proteins bind to sequences throughout the central domain, indicating that kinetochore assembly is not restricted to specific sites within the central domain (Figure 4). We observed consistency between profiles for these proteins under native or under cross-linking conditions, which might suggest that peaks in the profiles represent preferred kinetochore positions. However, there was very little correspondence between the N-ChIP and X-ChIP patterns (Figure 4), indicating that these peaks primarily reflect technical variations between different ChIP protocols rather than differences between proteins, strains, or tags. For Cenp-A-FLAG, CENP-A-HA, and CENP-T-FLAG, we found that changing the degree of MNase digestion made little difference in the profile, and we also saw little difference between the profiles of successive chromatin extractions using buffer containing first no salt and next 150 mM salt (Figure 4).
Figure 4.
Kinetochore proteins are strongly enriched throughout the central domain. CENP-A (left) and other kinetochore proteins (right) are distributed throughout the central domain. Sites that appear to be preferred differ between N-ChIP (top) and X-ChIP (bottom). (Left) Effects of increasing MNase on the CENP-A profiles for N-ChIP (CENP-A-FLAG) and X-ChIP (CENP-A-HA) in the central domain of cen2. (Middle) Profiles from inputs to CENP-A ChIP of cen2. (Right) Comparison of N-ChIP and X-ChIP profiles of cen2 for different inner kinetochore proteins. CENP-T0 and CENP-T150 are successive extractions of CENP-T-FLAG fragments with increasing salt.
Correlation analysis fails to detect consistent preferences in CENP-A nucleosome positions within the central domain
If technical differences between otherwise robust methodologies obscure weak underlying positioning signals, then we would expect to see concordance between ChIP data sets by correlation analysis, which provides sensitive detection of weak patterns shared between entire data sets. To systematically test this possibility, we calculated pairwise Pearson correlation coefficients for all 33,849-bp positions within the central domains and all 144,316 positions within heterochromatin for the 38 ChIP and input chromatin samples characterized in this study. To detect shared correspondences between data sets we applied hierarchical clustering to the correlation matrix representing all 38 data sets. As expected for positioning of inner kinetochore proteins and nucleosomes within heterochromatin, all CENP-A, CENP-T, CENP-I, and CENP-C ChIP correlation coefficients clustered together to the exclusion of all input correlation coefficients, which also clustered together (Figure 5A and Figure S2). Within the central domain, all input correlation coefficients but one clustered together and most ChIP correlation coefficients clustered together, including CENP-A, CENP-T, CENP-I, Scm3, and CENP-C (Figure 5B and Figure S2). However, five CENP-A data sets, including those from both X-ChIP and N-ChIP samples, clustered together with input data sets to the exclusion of the other CENP-A data sets. This clustering procedure is robust, as correlation heat maps based on 11-bp, 101-bp, and 1001-bp sliding windows were virtually identical to the 1-bp correlation heat map (Figure S2). We conclude that the peak-and-valley fluctuations that we observed in CENP-A ChIP occupancy (Figure 4) reflect differences in technical details, rather than biologically significant differences in CENP-A occupancy or positioning.
To further investigate the nature of these technical differences, we used 11-bp sliding windows to determine the correlation of each data set with GC content (Figure S3). We found a clear distinction between the X-ChIP data sets in Figure 4, which are all correlated (r = +0.15 to +0.25), and the N-ChIP data sets, which are essentially uncorrelated (r = +0.01 to −0.08). This weak GC-preference in X-ChIP but not N-ChIP might be sufficient to account for the differences between the “fringes” seen in the figure. GC preference in X-ChIP may arise because formaldehyde-induced crosslinks between lysine and deoxyguanosine are more frequent than other deoxynucleoside-to-amino acid crosslinks (Lu et al. 2010), which could result in preferential ChIP of GC rich sequences in cross-linked samples. Because central domains are more AT-rich than the genome as a whole (28% GC vs. 36% GC); (Wood et al. 2002), the data suggest than N-ChIP may sample the central domains more equitably than X-ChIP.
Centromere proteins in the pericentric heterochromatin are less stably incorporated than in the central domain
CENP-A-FLAG was also enriched throughout the pericentric heterochromatin compared with the flanking euchromatic arms (Figure 3), although enrichment in heterochromatin was only 2.5% of the enrichment in the central domains. Both CENP-A-FLAG and CENP-A-HA have been previously reported to be confined to the central domain using X-ChIP followed by endpoint-PCR assays (Takahashi et al. 2000; Shiroiwa et al. 2011). Our ability to detect CENP-A-FLAG in the pericentric heterochromatin is likely to be due to the increased sensitivity of ChIP-Seq over endpoint-PCR assays. We compared this result with profiles of ChIP-Seq using CENP-A-HA expressed from the cnp1 promoter and using five other tagged inner kinetochore proteins expressed in strains with wild-type cnp1 genes: CENP-T-FLAG, CENP-I-HA, CENP-I-GFP, myc-CENP-C, and Scm3-FLAG, using both native and cross-linking conditions. The results were plotted on a log scale to better visualize enrichment in the pericentric heterochromatin (Figure 6). Using X-ChIP, all kinetochore proteins were highly enriched throughout the central domain, and less enriched over the pericentric heterochromatin, at a level of 2.7–6.3% of their enrichment in the central domain. Previously, overexpression of CENP-A has been reported to lead to enrichment of CENP-A over the pericentric heterochromatin (Castillo et al. 2013), but our results indicate that enrichment of CENP-A and other kinetochore proteins in the pericentric heterochromatin occurs even with wild-type cnp1 expression.
Figure 6.
Inner kinetochore proteins are stable over central domains but less stable over heterochromatin. Occupancy profiles of a 100-kb region around each centromere are shown with the y-axis on a log10 scale. Annotations of repeats (black) and tRNA genes (red) are indicated on the x-axis. From top to bottom: CENP-T-FLAG, CENP-I-HA, Scm3-FLAG, CENP-A-FLAG, CENP-T-FLAG, CENP-I-GFP, CENP-A-HA, Myc-CENP-C.
Although myc-CENP-C was coextensive with the other inner kinetochore proteins using X-ChIP, we were unable to detect enrichment of myc-CENP-C using N-ChIP, suggesting that it is easily removed from centromeric chromatin by MNase if not cross-linked. This is reminiscent of the sensitivity of CENP-B/CENP-C complexes to MNase in HeLa cells (Ando et al. 2002) and is consistent with CENP-C ChIP results in nematodes (Steiner and Henikoff 2014). The other inner kinetochore proteins were immunoprecipitated from both the central domains and the pericentric heterochromatin by N-ChIP, although to varying degrees (Figure 6). CENP-T-FLAG and CENP-I-HA were highly enriched over the central domains coextensively with CENP-A-FLAG, but their enrichment over the pericentric heterochromatin is quite limited. This suggests that CENP-T and CENP-I are more easily lost from heterochromatin than CENP-A, and like CENP-C, they can be released from DNA by MNase digestion in the absence of cross-linking.
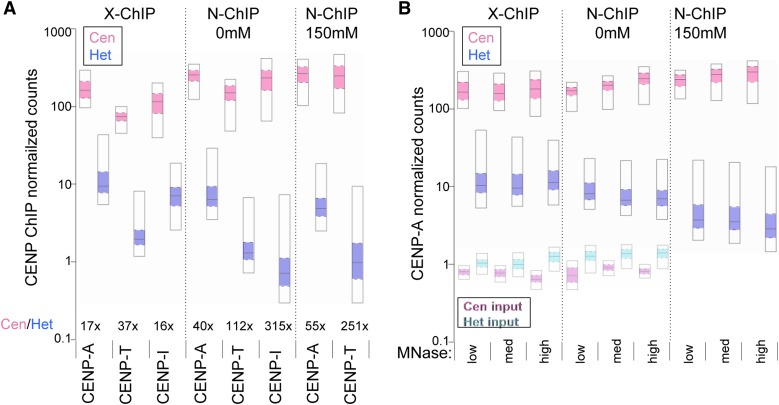
To quantitate this preferential loss of kinetochore proteins from heterochromatin in the absence of cross-linking, we calculated the ratio of the enrichment of each inner kinetochore protein over the central domain to that over the heterochromatin (Cen/Het) for X-ChIP and N-ChIP data. Using X-ChIP, the fold-enrichment of the central domains over heterochromatin ranged from 17X for CENP-A to 37X for CENP-T and 16X for CENP-I (Figure 7A). However, using N-ChIP these ratios grew to 40X for CENP-A, 112X for CENP-T, and 315X for CENP-I, reflecting the limited recovery of fragments from heterochromatin in the absence of crosslinking. The largest increase in fold-enrichment in N-ChIP (least recovery from heterochromatin) is for CENP-I, which is not known to bind DNA directly, while CENP-A and CENP-T, which can wrap DNA, show more modest increases. X-ChIP therefore appears to be better at preserving kinetochore proteins on heterochromatin, especially those not bound directly to DNA.
Figure 7.
Occupancies of inner kinetochore proteins on heterochromatin and central domains. Distributions of the normalized read counts from ChIP-Seq of kinetochore proteins over pericentric heterochromatin (Het) and central domains (Cen) are displayed as percentile plots, in which the median is marked by the solid line, the 25th and 75th percentile by the colored box and the 5th and 95th percentile by the open box. (A) Comparisons between centromere proteins for X-ChIP and N-ChIP. (B) Comparisons between CENP-A samples as a function of MNase digestion. CENP-A-HA was used for X-ChIP and CENP-A-FLAG was used for N-ChIP. For N-ChIP, chromatin was extracted in no-salt buffer (0 mM) and then the pellet was extracted with 150 mM NaCl (150 mM).
We looked quantitatively at how the mean occupancy (normalized counts) of CENP-A-FLAG was affected by increasing MNase digestion. Using N-ChIP, CENP-A-FLAG mean occupancy in heterochromatin decreased slightly with increasing MNase digestion (Figure 7B), reflecting the progressive loss of unstably incorporated proteins in the absence of cross-linking. In contrast, in the central domain, CENP-A-FLAG mean occupancy increased slightly with increasing MNase digestion, probably because of normalization to decreasing total DNA with longer digestion times. Using X-ChIP, mean occupancy of CENP-A-HA showed little change with increasing MNase digestion, both in heterochromatin and the central domain.
For N-ChIP of CENP-A-FLAG and CENP-T-FLAG, we extracted chromatin first in buffer without salt and successively in buffer containing 150 mM salt. For the central domain, slightly more fragments were recovered in the 150 mM buffer than in the no-salt buffer, but more fragments from heterochromatin were extracted in the no-salt buffer than in the 150 mM buffer (Figure 7, A and B), consistent with other indications that CENP-A-FLAG and CENP-T-FLAG are less stable when incorporated in heterochromatin.
Protection of centromeric chromatin from MNase digestion by inner kinetochore proteins
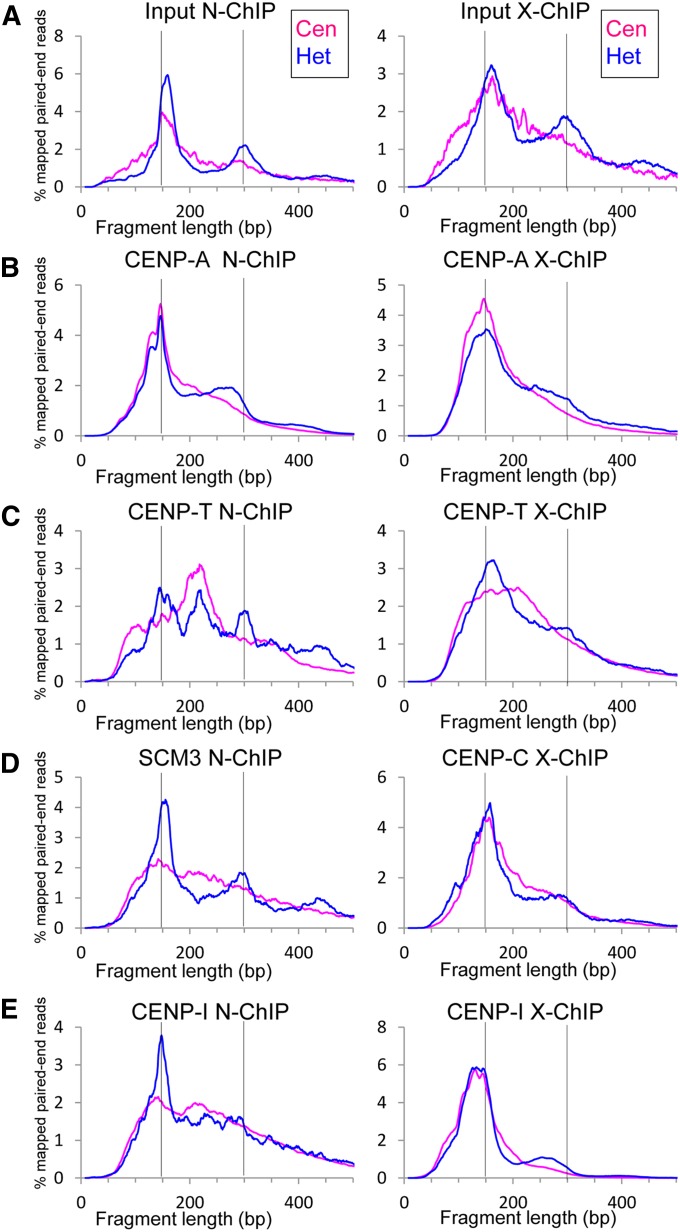
As particle dimensions can be inferred from the length of DNA fragments protected from MNase, we plotted the distribution of fragment sizes in ChIP experiments. Figure 8A shows the fragment length distributions from the heterochromatic otrs and central domains of the inputs to CENP-A X-ChIP and N-ChIP experiments. The fragment length distribution in the heterochromatin, which is dominated by H3 nucleosomes, had a peak at ∼150–160 bp, a secondary peak at ∼300 bp, and a small tertiary peak at ∼450 bp, reflecting the sizes of DNA protected by mono-, di- and trinucleosomes, corresponding to the bottom three bands of a nucleosome ladder. In the central domain, in contrast, fragment lengths formed a broad, skewed peak with no secondary or tertiary peak visible. This corresponds to the chromatin "smear" that has been observed in the central domain (Polizzi and Clarke 1991; Takahashi et al. 1992). The lack of a secondary peak is consistent with the variable internucleosome distances seen in H4S47C-anchored cleavage. The central domain fragments comprise <0.3% of the genome, accounting for the relatively noisy appearance of their distribution in bulk chromatin libraries.
Figure 8.
Length distributions of MNase-protected input and ChIP fragments. Fragment lengths are shown for fragments of N-ChIP and X-ChIP experiments from the otrs (Het) and central domain (Cen) from (A) the bulk chromatin (input to CENP-A ChIP), (B) ChIP pulldown for CENP-A-FLAG (N-ChIP) and CENP-A-HA (X-ChIP), (C) CENP-T-FLAG, (D) Scm3-FLAG (N-ChIP) and Myc-CENP-C (X-ChIP), (E) CENP-I-HA. The horizontal axis represents the fragment length and the vertical axis indicates the relative frequency when smoothed using a 9-bp sliding window. Vertical bars mark 150- and 300-bp positions.
The distributions of protected ChIP fragments containing CENP-A-HA or CENP-A-FLAG from the central domain were smoother, and the peaks were shifted leftward relative to bulk chromatin (Figure 8B), indicating that the most abundant CENP-A-protected fragment lengths on average were somewhat smaller than in bulk chromatin, similar to what has been observed for other cenH3 nucleosomes (Dalal et al. 2007; Krassovsky et al. 2012; Hasson et al. 2013; Zhang et al. 2013; Steiner and Henikoff 2014; Henikoff et al. 2015). The N-ChIP fragments form a double peak, with the smaller peak at ∼125 bp. Multiple peaks of cenH3-protected fragments have been observed in human and rice ChIP (Hasson et al. 2013; Zhang et al. 2013) and reflect preferred MNase cleavage sites either within cenH3 nucleosomes or within cenH3 nucleosome-associated complexes. These peaks are fused in the distribution of X-ChIP fragments. The rightward skew in both distributions suggests that other kinetochore proteins prevent MNase from efficiently trimming the longer linkers between CENP-A nucleosomes to produce a minimal size of CENP-A nucleosomes. In the pericentric heterochromatin, the peak of the distribution of CENP-A-protected fragments was similar in size to that from the central domain. However, the rightward skew of the distribution of fragments was less pronounced in heterochromatin, especially in N-ChIP, and it was replaced by a secondary nucleosome peak located ∼150 bp to the right of the primary peak, but still shifted leftward (<300 bp) from the secondary peak in bulk chromatin. Since this secondary peak is not observed in the central domain, we suggest that it represents dinucleosomes with one CENP-A and one H3 nucleosome and that CENP-A is interspersed with regularly spaced H3 in heterochromatin.
In the distribution of CENP-T fragment lengths from N-ChIP, there was a small peak at ∼100 bp that may reflect putative CENP-TWSX particles (Figure 8C). The main peak in the N-ChIP distribution at ∼220 bp may reflect a CENP-T particle complexed with a CENP-A nucleosome. It is highly unlikely that the peak represents a complex with an H3 nucleosome, since H3 is nearly absent in the central domain. The peak had a similar rightward skew as is seen in CENP-A ChIP and in bulk chromatin from the central domain. In the corresponding distribution of cross-linked CENP-T fragments, a broad peak ranging from ∼100 to ∼250 bp appeared to be a fusion of the peaks seen in N-ChIP. The modest rightward shift of the smaller fragments in X-ChIP relative to N-ChIP indicates that the DNA is not trimmed as effectively on the smaller particles in X-ChIP. The main peak seen in N-ChIP from the central domain appeared also in the N-ChIP fragment distribution from heterochromatin, where 100-fold fewer CENP-T fragments were recovered. This peak was accompanied by peaks absent from the central domain and corresponding in size to H3 mono-, di-, and trinucleosomes, suggesting that CENP-T can associate with canonical H3 nucleosomes in heterochromatin, but in a way that does not protect additional DNA. Similarly, in cross-linked heterochromatin, CENP-T fragment lengths showed primary and weak secondary peaks corresponding to nucleosomal peaks in bulk chromatin, indicating that in heterochromatin cross-linking enriches these fragment sizes over the 220-bp peak. The broad distribution of fragment sizes greater than the ∼100 bp protection expected from CENP-TWSX particles in both N-ChIP and X-ChIP data sets suggests that CENP-T is more likely to protect DNA as part of a larger complex than as an individual CENP-TWSX particle.
Cross-linked CENP-C had a peak at ∼150 bp in the central domain (Figure 8D). Human CENP-C has been shown to bind DNA nonspecifically (Sugimoto et al. 1994; Yang et al. 1996), but it is unlikely that the ∼150-bp fragments were protected directly by CENP-C, as no protection was found in N-ChIP. CENP-C binds to CENP-A preferentially over H3 nucleosomes or DNA (Carroll et al. 2010), so the ∼150-bp peak probably reflects fragments protected by a CENP-A/CENP-C complex, implying that CENP-C protects little additional DNA beyond what is protected by CENP-A. In heterochromatin, the primary peak was shifted slightly rightward and a secondary peak appeared, suggesting a complex with H3 nucleosomes or a mix of CENP-A and H3 nucleosomes.
Scm3 and CENP-I are not known to bind DNA directly, so the fragments they immunoprecipitated in N-ChIP presumably reflect their interactions with DNA-binding proteins (Figure 8, D and E). Scm3 binds to CENP-A and to a complex of CENP-A loading factors including Mis16, Mis18, and Mis19/Eic1/Kis1, which in turn binds members of the Mis6/Sim4/Mal2 complex (Pidoux et al. 2009; Hayashi et al. 2014; Hirai et al. 2014; Subramanian et al. 2014) that corresponds to the vertebrate CENP–HIK complex (Okada et al. 2006). In human cells, the CENP–HIK complex binds to CENP-TW, and these two complexes are interdependent for stable localization at the centromere (Foltz et al. 2006; Basilico et al. 2014). In fission yeast the Mis6/Sim4/Mal2 complex depends for localization on both CENP-T and CENP-C (Tanaka et al. 2009) and is required for localization of Scm3 (Pidoux et al. 2009; Williams et al. 2009), suggesting that the fragments immunoprecipitated by Scm3 and CENP-I reflect DNA protected by CENP-T particles and CENP-A/CENP-C particles. In the central domain, the distributions seemed to reflect primarily the size distribution of bulk chromatin fragments, with very slight peaks corresponding to CENP-A and CENP-T peaks (Figure 8D and Figure 7E). In heterochromatin, the fragment distribution peaks for Scm3 also corresponded to bulk chromatin, with peaks marking H3 mono-, di-, and trinucleosomes (Figure 8D). Scm3 has previously been reported to interact with H3 as well as CENP-A in vitro (Pidoux et al. 2009), which might explain the distribution of peaks in heterochromatin. The correspondence of CENP-I peaks in the heterochromatin, where it is 300-fold less abundant than in the central domain, was less clear and might reflect CENP-A/H3 heterotypic nucleosomes (Arimura et al. 2014) or a mix of CENP-A and H3 nucleosomes. For cross-linked CENP-I, there was a significant enrichment of fragments with the peak size of CENP-A nucleosome fragments, both in the central domain and in heterochromatin where there was a <300-bp secondary peak similar to the one observed in CENP-A ChIP. This suggests that CENP-I is found in close proximity to CENP-A nucleosomes, but is not directly bound to them.
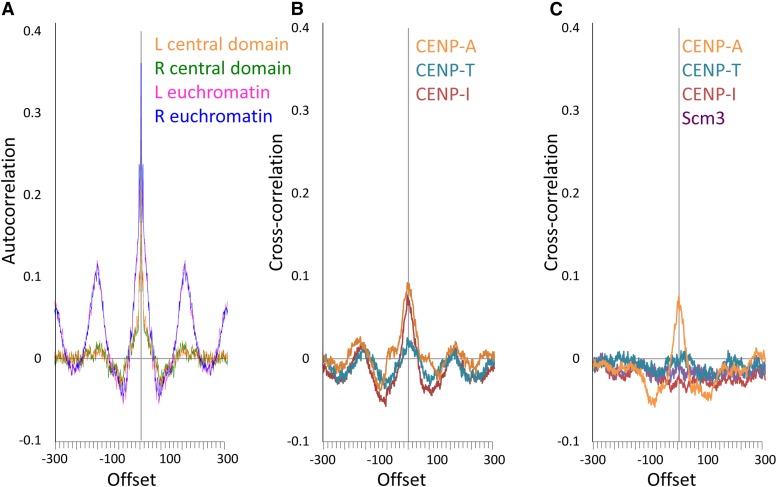
We sought to further investigate whether different kinetochore proteins have particular relationships to each other along the chromosome. We first performed autocorrelation analysis using the ends of H4S47C-anchored cleavage fragments to see if there is any periodicity in centromeric nucleosome positions despite the overall smooth distribution of fragment lengths. The left and right ends of control fragments from euchromatin showed clear periodicity with an autocorrelation of ∼0.13 (Figure 9A) at +150 or −150 bp. Cleavage fragments from the central domain, in contrast, showed a barely detectable autocorrelation of ∼0.02 at these positions. Using the predicted distances between the W and C ends of fragments (Figure 2) to call nucleosome dyad positions, we then used cross-correlation to determine the offsets of the midpoints of ChIP fragments from nucleosome dyads. For X-ChIP fragments all kinetochore proteins showed similar cross-correlations with nucleosome dyads and evidence of weak periodicity in nucleosome positions, with CENP-T showing lower cross-correlation than CENP-A, CENP-I, and CENP-C (Figure 9B). In N-ChIP the correlations of fragment midpoints with the dyad and periodicity essentially vanish except for CENP-A (Figure 9C). The difference between X-ChIP and N-ChIP for CENP-I is particularly striking and appears to confirm that CENP-I becomes cross-linked to CENP-A nucleosomes in X-ChIP but shows no relation to dyad positions in N-ChIP. The weak periodicity observed in X-ChIP cross-correlations may disappear in N-ChIP because of loss of uncrosslinked kinetochore proteins during chromatin extraction. These differences between X-ChIP and N-ChIP underscore the high sensitivity of mapping within the central domain to technical details of methods.
Figure 9.
Correlation of offsets between kinetochore proteins. (A) Autocorrelations of left (L) or right (R) ends of H4S47C-anchored cleavage fragments from euchromatin or from the central domain. (B) Cross-correlations of dyad positions inferred from H4S47C-anchored cleavage fragments with midpoints of X-ChIP fragments for CENP-A-HA, CENP-T-FLAG, and CENP-I-GFP. (C) Cross-correlations of dyad positions inferred from H4S47C-anchored cleavage fragments with midpoints of N-ChIP fragments for CENP-A-FLAG, CENP-T-FLAG, and CENP-I-HA.
Discussion
Dense arrays of mostly unpositioned CENP-A nucleosomes occupy the central domain
We have used H4S47C-anchored cleavage mapping and high-resolution ChIP mapping to determine the locations of several inner kinetochore proteins that occupy fission yeast regional centromeres, and the lengths of fragments they protect from MNase digestion. Using published H3 ChIP-Seq data sets, we observed very low levels of canonical H3-containing nucleosomes in the central domains (∼5% of the occupancy on the euchromatic arms), but our CENP-A ChIP data sets showed strong enrichment there for CENP-A-containing nucleosomes. We conclude that the central domains are almost exclusively occupied by CENP-A nucleosomes.
Two groups have used MNase to map cross-linked nucleosomes in the central domain and reported positioned CENP-A nucleosomes, although one group concluded that there was considerable variation in positioning in wild-type cells (Song et al. 2008; Lando et al. 2012; Yao et al. 2013). A previous H4S47C-anchored cleavage mapping study found fuzzy positions of nucleosomes in the central domain, but with very low cleavage density, leaving the question of nucleosome positioning unresolved (Moyle-Heyrman et al. 2013). Our in vivo H4S47C-anchored cleavage mapping without size selection revealed that cleavage density in the central domain is nearly as high (86%) as in the chromosome arms, but the pattern of cleavages indicates that the central domain nucleosomes are not positioned, in sharp contrast to nucleosomes in much of the rest of the fission yeast genome. We attribute the low cleavage density in the previous report (Moyle-Heyrman et al. 2013) at least in part to the selection of a mononucleosome-sized band of DNA for sequencing. We found that the distances between H4 cleavages in the central domain are skewed toward longer lengths than those from euchromatin or heterochromatin, and so the selection of a gel band would have eliminated longer fragments and thereby reduced cleavage density. Because we found that cleavage density in the central domain is nearly as high as on the euchromatic arms, it seems implausible that the longer internucleosomal distances in the central domain are a result of incomplete cleavage, indicating that central domain nucleosomes are more widely and variably spaced than elsewhere. The near absence of H3 nucleosomes in the central domains leads us to conclude that the wider and more variable nucleosome spacing in these domains is characteristic of CENP-A nucleosomes in the functional centromere. Cleavage mapping also revealed that CENP-A nucleosomes have two H4 molecules and show no evidence of rotational phasing, in sharp contrast to cenH3 nucleosomes from distantly related budding yeast.
Using high-resolution N-ChIP and X-ChIP mapping, we observed strong enrichment of CENP-A and other inner kinetochore proteins throughout the central domain except at the tRNA genes, which have been shown to act as barriers to the spread of the flanking heterochromatin, dependent on binding of PolIII and the transcription factor TFIIIC (Scott et al. 2007). Binding of the PolIII transcription complex or active transcription of tRNAs might prevent CENP-A localization at these sites. Our data argue against significantly preferred positions within the central domain for kinetochore assembly and in favor of the view that each CENP-A nucleosome is capable of acquiring a full set of kinetochore proteins with the potential to capture a microtubule. This hypothesis is supported by the observation that deletion of half or more of a central domain does not reduce centromere function (Yao et al. 2013). The speed of capture of microtubules is predicted to correlate with the size of kinetochores (Wollman et al. 2005), so the equivalence of sites in regional centromeres for kinetochore assembly may serve to increase the size of the kinetochore surface and the efficiency of microtubule capture in mitosis compared with capture only at a set of preferred assembly sites.
We also observed weak enrichment of CENP-A and other inner kinetochore proteins in pericentric heterochromatin. CENP-A incorporated into pericentric heterochromatin is more easily disrupted by increasing MNase in N-ChIP than CENP-A incorporated into the central domain, and comparison of N-ChIP and X-ChIP for other kinetochore proteins also appears to indicate less stable incorporation into heterochromatin. This suggests that the inner kinetochore proteins might be more stable when they are part of a continuous array than when they are present in heterochromatin, perhaps offering insight into why animal and plant centromeres are composed of alternating arrays of CENP-A and H3 nucleosomes. The basis of this apparent greater stability is not known, but might involve interdependent interactions among kinetochore proteins, as seen in the regional centromeres of Candida albicans (Thakur and Sanyal 2012). Similarly, CENP-A arrays in human centromeres have been proposed to permit stabilizing multivalent interactions by CENP-C via its dimerization domain (Carroll et al. 2010).
We doubt that the lower-level incorporation of inner kinetochore proteins into heterochromatin is functionally significant, given the two order-of-magnitude greater enrichment over the central domain. Lower level incorporation of CENP-A and CENP-C into pericentric sequences was also observed in a fission yeast strain in which central domains are not surrounded by heterochromatic repeats (Brown et al. 2014), suggesting that inner kinetochore proteins can incorporate into any pericentric DNA in a process that gives the appearance of one-dimensional diffusion from regions of strong enrichment. This incorporation resembles the low level incorporation of CENP-A over 2 Mb of DNA flanking chicken nonrepetitive centromeres (Shang et al. 2013) and may be related to the arrangement of smaller CENP-A arrays surrounding a large central CENP-A array in a 330-kb human neocentromere (Chueh et al. 2005). The smaller size of the region of low-level incorporation around fission yeast centromeres compared with chicken or human may reflect the smaller size of the source centromere. The pericentric heterochromatin in fission yeast has been proposed to "corral" centromeric chromatin (Sullivan 2002). The higher H3 nucleosome occupancy that we observed for pericentric heterochromatin relative to euchromatin would limit the amount of free DNA on which CENP-A nucleosomes can assemble and may serve to help confine the kinetochore to the central domain.
Association of CENP-T with CENP-A nucleosomes
CENP-C and CENP-T have been thought to associate with H3 nucleosomes (Hori et al. 2008), based on their loss from CENP-A nucleosomes during MNase digestion to mononucleosomes. In fission yeast, we likewise find that CENP-C can be removed from CENP-A chromatin by MNase, but the near absence of H3 nucleosomes in the central domain where CENP-C and CENP-T are highly enriched is strong evidence that they associate predominantly with CENP-A nucleosomes. Consistent with this, recent evidence suggests that CENP-T may interact with the CENP-A N-terminal tail (Folco et al. 2015; Logsdon et al. 2015). We find that CENP-A nucleosomes protect slightly less DNA from MNase than do H3 nucleosomes and that CENP-C protects little if any additional DNA beyond what is protected by CENP-A nucleosomes, suggesting that CENP-C immunoprecipitates DNA primarily through contacting CENP-A nucleosomes. In heterochromatin, both CENP-C and CENP-T immunoprecipitate fragment sizes corresponding to H3 nucleosomes, consistent with observations that they can immunoprecipitate H3 in chicken cells (Hori et al. 2008). However, these interactions with H3 in heterochromatin are minor, and neither protein appears to protect additional DNA beyond what is protected by H3 nucleosomes.
The structure of CENP-T-containing particles in vivo is not known. CENP-T can assemble onto centromeres prior to CENP-W (Krizaic et al. 2015). In both vertebrates and fission yeast, mutations in CENP-T or CENP-W are lethal, while those in CENP-S and CENP-X are viable, although with segregation defects (Hori et al. 2008; Amano et al. 2009; Tanaka et al. 2009; Bhattacharjee et al. 2013). Together these observations suggest that CENP-TW dimers may be able to bind to centromeric DNA and/or other kinetochore proteins to provide an essential function without necessarily forming a CENP-TWSX heterotetramer. Both TW dimers and TWSX tetramers bind to DNA in vitro (Nishino et al. 2012; Takeuchi et al. 2014). The CENP-TWSX particle wraps 60–100 bp of linker DNA between nucleosomes in vitro (Nishino et al. 2012; Takeuchi et al. 2014), but in fission yeast, the most common linker length between H3 nucleosomes is 3 bp (Moyle-Heyrman et al. 2013), leaving no room for such a particle. The broader spacing of CENP-A nucleosomes in the central domain presumably allows room for incorporation of CENP-T-containing particles into central domain DNA.
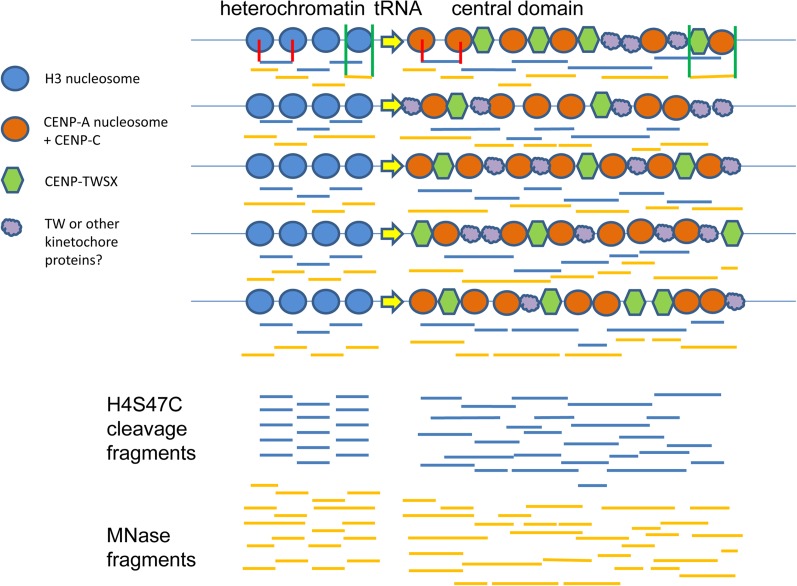
MNase digestion of chicken CENP–TWSX particles assembled on plasmid DNA produces a protected fragment of ∼100 bp, together with a smear of larger fragments, rather than a ladder of discrete particles (Nishino et al. 2012; Takeuchi et al. 2014). CENP-T-W dimers assembled on short DNA fragments can form DNA complexes in vitro that protect any available DNA but also lack discrete sizes (Nishino et al. 2012; Takeuchi et al. 2014). The mode of CENP–TW or CENP–TWSX DNA binding that causes the apparent continuous protection of DNA and smearing of fragments on MNase digestion is not understood, but an attractive hypothesis is that similar CENP-T-containing particles in fission yeast are the basis of the broad, skewed distribution of CENP-T-FLAG fragment lengths and their lack of a discrete ladder of sizes and that they also are responsible for the classical smeared digestion pattern of central domain chromatin and the skewed distribution of ChIP fragments from CENP-A and other inner kinetochore proteins that do not appear to be trimmed to minimum sizes. We propose a model of fission yeast inner kinetochore structure in which the variable linkers between irregularly placed CENP-A nucleosomes are occupied by CENP-T-containing particles, at least some of which are CENP–TWSX particles in close proximity to CENP-A nucleosomes bound by CENP-C (Figure 10). Together with CENP-C these particles are necessary to maintain the inner kinetochore proteins of the Mis6/Sim4/Mal2 complex (Tanaka et al. 2009), which regulates kinetochore microtubule polymerization and metaphase plate organization (Amaro et al. 2010).
Figure 10.
Model of centromeric chromatin. H3- and CENP-A-containing nucleosomes are shown at the tRNA boundary of the central domain for five different cells. H4S47C-mediated cleavage nucleosome-to-nucleosome fragments are shown as horizontal blue lines. Red vertical lines show how the fragments correspond to nucleosomes for one nucleosome pair in heterochromatin and one in the central domain. MNase-generated fragments are depicted with amber horizontal lines. Green vertical lines indicate their correspondence to chromatin proteins for one nucleosome in heterochromatin and for a CENP-A nucleosome complexed with a CENP-TWSX tetramer.
Nonrepetitive centromeres and repeat-rich centromeres
Although fission yeast centromeres and the repeat-rich centromeres of plants and animals have both been described as “regional,” our data reveal profound differences in how they are organized at the nucleosome level as well as in size and sequence. Plant and animal centromeric tandem repeat arrays precisely position cenH3 nucleosomes (Hasson et al. 2013; Zhang et al. 2013; Henikoff et al. 2015). Precise positioning is found for sequence-dependent single-wrap particles, such as hemisomes in budding yeast and 100-bp CENP-A protected particles at homogeneous human α-satellite dimers (Henikoff et al. 2015), whereas diffuse positioning is found for sequence-independent octasomes, such as those likely to be present at pericentric human HORs and neocentromeres (Hasson et al. 2013; Henikoff et al. 2015). Our detection of two H4 molecules per nucleosome in the fission yeast central domain fits this paradigm. Tandem repeat arrays have been proposed to evolve through competition for transmission in asymmetric female meiosis (Malik and Henikoff 2001) and to stabilize nucleosomes through translational and rotational phasing (Zhang et al. 2013), which may give them a competitive transmission advantage in asymmetric meiosis. In contrast, the classical regional centromeres of fission yeast are characterized by a lack of positioning and rotational phasing of CENP-A nucleosomes on low-copy-number sequences. Like fission yeast, the yeast C. albicans also has centromeres on gene-free single-copy sequences and has a smeared electrophoresis pattern of centromeric chromatin (Baum et al. 2006), suggesting that the wider and irregular spacing of CENP-A nucleosomes may be a conserved feature of fungal regional centromeres. Similarly, cenH3 nucleosomes in the 4.1- to 4.6-kb gene-free centromeres of Plasmodium falciparum also appear to lack positioning (Hoeijmakers et al. 2012), suggesting that this may be a common feature of regional centromeres in organisms with symmetric meiosis, where centromere variants do not compete. For these classical regional centromeres, it may suffice to have a less precisely organized region if it is of sufficient size to assure timely microtubule capture.
In sequence and organization, fission yeast central domains appear to have more in common with nonrepetitive centromeres and neocentromeres in multicellular organisms than with repeat-rich centromeres. In rice Cen8, which has both satellite and unique sequence DNA, internucleosomal distances on unique sequences are irregular, and positioning of cenH3 nucleosomes is less well defined than that on centromeric tandemly repetitive sequences in the same centromere, consistent with the notion that centromeric repeats confer consistent cenH3 nucleosome positioning, while low-copy-number sequences tend to position nucleosomes with less regularity (Zhang et al. 2013). Positioned CENP-A nucleosomes were reported in human neocentromeres on nonrepetitive sequences in cell lines derived from three individuals (Hasson et al. 2013), indicating that nucleosome positions can be stably inherited in these centromeres. However, a statistical model of one of these neocentromeres found that while occupancy at some sites was 80%, the average occupancy at identified CENP-A sites was only 6%, indicating both some strongly preferred positions and also considerable variation in occupancy at different positions in different cells (Bodor et al. 2014).
Interestingly, in the centromere of horse chromosome 11, which also lacks tandem repeats, large (80–160 kb) continuous domains of CENP-A nucleosomes can occupy different positions within a 500-kb region (Purgato et al. 2014) in cell lines derived from different individuals, suggesting that CENP-A domains can “slide” or migrate along a chromosome. Although the domains are positionally consistent within a cell line, the high number of positional epialleles indicates significant plasticity in positions over time. Despite this, the occurrence of a single large CENP-A domain in all epialleles suggests that continuous arrays of CENP-A nucleosomes may have a transmission advantage. This resembles fission yeast centromeres in the metastable positions of CENP-A nucleosomes, their occurrence in continuous arrays, and apparent one-dimensional diffusion of CENP-A into adjacent sequences.
Taken together, these observations suggest that vertebrate centromeres and neocentromeres lacking centromeric tandem repeats may form a continuum with the much smaller fission yeast central domains. In plants, the total size of cenH3 domains correlates with genome size (Zhang and Dawe 2012), so genome size could be the primary difference between classical regional centromeres of micro-organisms and nonrepetitive centromeres in plants and animals. In the latter organisms, however, competition between centromeres is expected to lead eventually to the replacement of low-copy centromere sequences with repeat-rich centromeres of well-positioned cenH3 nucleosomes.
Supplementary Material
Acknowledgments
We thank Jorja Henikoff for data processing and analysis, Srinivas Ramachandran for Watson/Crick cleavage distance analysis, Christine Codomo for Illumina library preparation, the Fred Hutch Shared Genomic Resource for performing DNA sequencing, and Gerry Smith and Takehito Furuyama for critical comments on the manuscript. We thank the National BioResource Project of Japan, Ji-Ping Wang, Takeshi Sakuno, and Songtao Jia for strains. The authors declare no competing interests.
Footnotes
Communicating editor: M. P. Colaiácovo
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179788/-/DC1.
Sequence data from this article have been deposited with the National Center for Biotechnology Information Gene Expression Omnibus under access no. GSE64294.
Literature Cited
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., et al. , 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro A. C., Samora C. P., Holtackers R., Wang E., Kingston I. J., et al. , 2010. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12: 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S., Yang H., Nozaki N., Okazaki T., Yoda K., 2002. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22: 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelgren H., Kniola B., Ekwall K., 2003. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 116: 4035–4042. [DOI] [PubMed] [Google Scholar]
- Arimura Y., Shirayama K., Horikoshi N., Fujita R., Taguchi H., et al. , 2014. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 4: 7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J. R., Prumbaum D., Rojas A. M., et al. , 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife 3: e02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M., Sanyal K., Mishra P. K., Thaler N., Carbon J., 2006. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl. Acad. Sci. USA 103: 14877–14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Osman F., Feeney L., Lorenz A., Bryer C., et al. , 2013. MHF1-2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination. Open Biol. 3: 130102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blower M. D., Sullivan B. A., Karpen G. H., 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D. L., Mata J. F., Sergeev M., David A. F., Salimian K. J., et al. , 2014. The quantitative architecture of centromeric chromatin. eLife 3: e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K., Xi L., Wang J. P., Widom J., 2012. A map of nucleosome positions in yeast at base-pair resolution. Nature 486: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. R., Thomas G., Lee N. C., Blythe M., Liti G., et al. , 2014. Kinetochore assembly and heterochromatin formation occur autonomously in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 111: 1903–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H. P., Sugiyama T., Chen E. S., Chen X., FitzGerald P. C., et al. , 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37: 809–819. [DOI] [PubMed] [Google Scholar]
- Carroll C. W., Milks K. J., Straight A. F., 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A. G., Mellone B. G., Partridge J. F., Richardson W., Hamilton G. L., et al. , 2007. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 3: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A. G., Pidoux A. L., Catania S., Durand-Dubief M., Choi E. S., et al. , 2013. Telomeric repeats facilitate CENP-A(Cnp1) incorporation via telomere binding proteins. PLoS One 8: e69673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., et al. , 1989. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell 57: 739–751. [DOI] [PubMed] [Google Scholar]
- Chueh A. C., Wong L. H., Wong N., Choo K. H., 2005. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14: 85–93. [DOI] [PubMed] [Google Scholar]
- Coffman V. C., Wu P., Parthun M. R., Wu J. Q., 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y., Wang H., Lindsay S., Henikoff S., 2007. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 5: e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N., 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321. [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D., 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T. C., Groner S., Zentgraf U., Hemleben V., 1994. Evidence for nucleosomal phasing and a novel protein specifically binding to cucumber satellite DNA. Z. Nat. C. J. Biosci. 49: 79–86. [DOI] [PubMed] [Google Scholar]
- Folco H. D., Campbell C. S., May K. M., Espinoza C. A., Oegema K., et al. , 2015. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 25: 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, et al. , 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8: 458–469. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Earnshaw W. C., 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaither T. L., Merrett S. L., Pun M. J., Scott K. C., 2014. Centromeric barrier disruption leads to mitotic defects in Schizosaccharomyces pombe. G3 Genes Genomes Genet. 4: 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K. E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., et al. , 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Wu Y., Koblizkova A., Torres G. A., Wang K., et al. , 2012. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24: 3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D., Panchenko T., Salimian K. J., Salman M. U., Sekulic N., et al. , 2013. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20: 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., et al. , 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Ebe M., Nagao K., Kokubu A., Sajiki K., et al. , 2014. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes Cells 19: 541–554. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Furuyama T., 2012. The unconventional structure of centromeric nucleosomes. Chromosoma 121: 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ramachandran S., Krassovsky K., Bryson T. D., Codomo C. A., et al. , 2014. The budding yeast centromere DNA element II wraps a stable Cse4 hemisome in either orientation in vivo. eLife 3: e01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, J. G., J. Thakur, S. Kasinathan and S. Henikoff, 2015 A unique chromatin complex occupies young α-satellite arrays of human centromeres. Sci. Adv. 1: e1400234. [DOI] [PMC free article] [PubMed]
- Hirai H., Arai K., Kariyazono R., Yamamoto M., Sato M., 2014. The kinetochore protein Kis1/Eic1/Mis19 ensures the integrity of mitotic spindles through maintenance of kinetochore factors Mis6/CENP-I and CENP-A. PLoS One 9: e111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers W. A., Flueck C., Francoijs K. J., Smits A. H., Wetzel J., et al. , 2012. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell. Microbiol. 14: 1391–1401. [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C. B., Welburn J. P., et al. , 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052. [DOI] [PubMed] [Google Scholar]
- Ishii T., Karimi-Ashtiyani R., Banaei-Moghaddam A. M., Schubert V., Fuchs J., et al. , 2015. The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res. 23: 277–284. [DOI] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G., 2007. Nucleosome stability mediated by histone variants H3.3 and H2A. Z. Genes Dev 21: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Okazaki K., Iida T., Nakayama J., Murakami Y., et al. , 2013. Spt6 prevents transcription-coupled loss of posttranslationally modified histone H3. Sci. Rep. 3: 2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B., O’Toole E., McIntosh J. R., Mellone B., Allshire R., et al. , 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 12: 2767–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassovsky K., Henikoff J. G., Henikoff S., 2012. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 109: 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizaic I., Williams S. J., Sanchez P., Rodriguez-Corsino M., Stukenberg P. T., et al. , 2015. The distinct functions of CENP-C and CENP-T/W in centromere propagation and function in Xenopus egg extracts. Nucleus 6: 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Endesfelder U., Berger H., Subramanian L., Dunne P. D., et al. , 2012. Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast. Open Biol. 2: 120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J. R., 3rd, He X., 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24: 2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon G. A., Barrey E. J., Bassett E. A., DeNizio J. E., Guo L. Y., et al. , 2015. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 208: 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Ye W., Zhou L., Collins L. B., Chen X., et al. , 2010. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132: 3388–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H. S., Henikoff S., 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157: 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters, D. P., L. V. Paliulis, I. F. Korf and S. W. Chan, 2012 Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chrom. Res. 20: 579–593. [DOI] [PubMed]
- Melters D. P., Bradnam K. R., Young H. A., Telis N., May M. R., et al. , 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A. D., Rheinbay E., Sorger P. K., 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle-Heyrman G., Zaichuk T., Xi L., Zhang Q., Uhlenbeck O. C., et al. , 2013. Chemical map of Schizosaccharomyces pombe reveals species-specific features in nucleosome positioning. Proc. Natl. Acad. Sci. USA 110: 20158–20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musich P. R., Brown F. L., Maio J. J., 1977. Subunit structure of chromatin and the organization of eukaryotic highly repetitive DNA: nucleosomal proteins associated with a highly repetitive mammalian DNA. Proc. Natl. Acad. Sci. USA 74: 3297–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musich P. R., Brown F. L., Maio J. J., 1982. Nucleosome phasing and micrococcal nuclease cleavage of African green monkey component alpha DNA. Proc. Natl. Acad. Sci. USA 79: 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K. E., Suzuki A., Hori T., et al. , 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148: 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I. M., et al. , 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., et al. , 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8: 446–457. [DOI] [PubMed] [Google Scholar]
- Partridge J. F., Borgstrom B., Allshire R. C., 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 14: 783–791. [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T., 2011. The ABCs of CENPs. Chromosoma 120: 425–446. [DOI] [PubMed] [Google Scholar]
- Pidoux A. L., Richardson W., Allshire R. C., 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A. L., Choi E. S., Abbott J. K., Liu X., Kagansky A., et al. , 2009. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M., Mestrovic N., Mravinac B., 2014. Centromere identity from the DNA point of view. Chromosoma 123: 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A. F., Mackay A. M., Ainsztein A. M., Goldberg I. G., Earnshaw W. C., 1995. The centromere: hub of chromosomal activities. Science 270: 1591–1594. [DOI] [PubMed] [Google Scholar]
- Politi V., Perini G., Trazzi S., Pliss A., Raska I., et al. , 2002. CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 115: 2317–2327. [DOI] [PubMed] [Google Scholar]
- Polizzi C., Clarke L., 1991. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J. Cell Biol. 112: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purgato S., Belloni E., Piras F. M., Zoli M., Badiale C., et al. , 2014. Centromere sliding on a mammalian chromosome. Chromosoma 124: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Zentner G. E., Henikoff S., 2015. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res. 25: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S. A., Vagnarelli P., Dong Y., Hori T., McEwen B. F., et al. , 2010. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA 107: 10484–10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Takahashi K., Yanagida M., 1997. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90: 131–143. [DOI] [PubMed] [Google Scholar]
- Scott K. C., Merrett S. L., Willard H. F., 2006. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16: 119–129. [DOI] [PubMed] [Google Scholar]
- Scott K. C., White C. V., Willard H. F., 2007. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS One 2: e1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W. H., Hori T., Martins N. M., Toyoda A., Misu S., et al. , 2013. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell 24: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroiwa Y., Hayashi T., Fujita Y., Villar-Briones A., Ikai N., et al. , 2011. Mis17 is a regulatory module of the Mis6-Mal2-Sim4 centromere complex that is required for the recruitment of CenH3/CENP-A in fission yeast. PLoS One 6: e17761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. S., Liu X., Liu X. S., He X., 2008. A high-resolution map of nucleosome positioning on a fission yeast centromere. Genome Res. 18: 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner F. A., Henikoff S., 2014. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife 3: e02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L., Toda N. R., Rappsilber J., Allshire R. C., 2014. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 4: 140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Yata H., Muro Y., Himeno M., 1994. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J. Biochem. 116: 877–881. [DOI] [PubMed] [Google Scholar]
- Sullivan B. A., 2002. Centromere round-up at the heterochromatin corral. Trends Biotechnol. 20: 89–92. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., et al. , 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 3: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E. S., Yanagida M., 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219. [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Nishino T., Mayanagi K., Horikoshi N., Osakabe A., et al. , 2014. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 42: 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H. L., Kagami A., Watanabe Y., 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17: 334–343. [DOI] [PubMed] [Google Scholar]
- Thakur J., Sanyal K., 2012. A coordinated interdependent protein circuitry stabilizes the kinetochore ensemble to protect CENP-A in the human pathogenic yeast Candida albicans. PLoS Genet. 8: e1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Bernardoni R., Diolaiti D., Politi V., Earnshaw W. C., et al. , 2002. In vivo functional dissection of human inner kinetochore protein CENP-C. J. Struct. Biol. 140: 39–48. [DOI] [PubMed] [Google Scholar]
- Vershinin A. V., Heslop-Harrison J. S., 1998. Comparative analysis of the nucleosomal structure of rye, wheat and their relatives. Plant Mol. Biol. 36: 149–161. [DOI] [PubMed] [Google Scholar]
- Westermann S., Schleiffer A., 2013. Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol. 23: 260–269. [DOI] [PubMed] [Google Scholar]
- Westhorpe F. G., Straight A. F., 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 25: 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. S., Hayashi T., Yanagida M., Russell P., 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgruber T. K., Sharma A., Schneider K. L., Albert P. S., Koo D. H., et al. , 2009. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet. 5: e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R., Cytrynbaum E. N., Jones J. T., Meyer T., Scholey J. M., et al. , 2005. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol. 15: 828–832. [DOI] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M. A., Lyne M., Lyne R., et al. , 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Wu Y., Kikuchi S., Yan H., Zhang W., Rosenbaum H., et al. , 2011. Euchromatic subdomains in rice centromeres are associated with genes and transcription. Plant Cell 23: 4054–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Tomkiel J., Saitoh H., Johnson D. H., Earnshaw W. C., 1996. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 16: 3576–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Liu X., Sakuno T., Li W., Xi Y., et al. , 2013. Plasticity and epigenetic inheritance of centromere-specific histone H3 (CENP-A)-containing nucleosome positioning in the fission yeast. J. Biol. Chem. 288: 19184–19196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G. E., Henikoff S., 2014. High-resolution digital profiling of the epigenome. Nat. Rev. Genet. 15: 814–827. [DOI] [PubMed] [Google Scholar]
- Zhang H., Dawe R. K., 2012. Total centromere size and genome size are strongly correlated in ten grass species. Chromosome Res. 20: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Talbert P. B., Zhang W., Wu Y., Yang Z., et al. , 2013. The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc. Natl. Acad. Sci. USA 110: E4875–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li X., Marshall J. B., Zhong C. X., Dawe R. K., 2005. Phosphoserines on maize CENTROMERIC HISTONE H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell 17: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are listed in Table S1 and are available upon request. The sequence-based data sets generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE64294.