Abstract
We designed a system to determine whether dicentric chromosomes in Drosophila melanogaster break at random or at preferred sites. Sister chromatid exchange in a Ring-X chromosome produced dicentric chromosomes with two bridging arms connecting segregating centromeres as cells divide. This double bridge can break in mitosis. A genetic screen recovered chromosomes that were linearized by breakage in the male germline. Because the screen required viability of males with this X chromosome, the breakpoints in each arm of the double bridge must be closely matched to produce a nearly euploid chromosome. We expected that most linear chromosomes would be broken in heterochromatin because there are no vital genes in heterochromatin, and breakpoint distribution would be relatively unconstrained. Surprisingly, approximately half the breakpoints are found in euchromatin, and the breakpoints are clustered in just a few regions of the chromosome that closely match regions identified as intercalary heterochromatin. The results support the Laird hypothesis that intercalary heterochromatin can explain fragile sites in mitotic chromosomes, including fragile X. Opened rings also were recovered after male larvae were exposed to X-rays. This method was much less efficient and produced chromosomes with a strikingly different array of breakpoints, with almost all located in heterochromatin. A series of circularly permuted linear X chromosomes was generated that may be useful for investigating aspects of chromosome behavior, such as crossover distribution and interference in meiosis, or questions of nuclear organization and function.
Keywords: dicentric chromosome, intercalary heterochromatin, fragile X, FLP
DROSOPHILA is noted for a combination of facile genetics and high-resolution cytology that allows the recovery and characterization of a variety of chromosome rearrangements. Simple structural changes such as duplications, deficiencies, inversions, and translocations are readily produced through a variety of classical and modern techniques. More complex rearrangements such as balancers, compound chromosomes, and ring chromosomes also have been generated. Ring chromosomes historically have been of interest for a number of unique properties, such as a propensity for loss during early embryonic mitoses and dominant zygotic lethality (Leigh 1976; Ashburner et al. 2005). Despite their “instability,” ring chromosome stocks can be remarkably stable, with very few reported instances of spontaneous opening into linear chromosomes.
In this work, we produced a number of linearized ring chromosomes. One goal was to determine whether there are preferred sites of breakage for dicentric chromosomes. Although we have previously shown that dicentric Y, 3, and 4 chromosomes can break and heal in the male germline and be recovered in offspring, these chromosomes allowed little opportunity to determine whether there were preferred sites of breakage (Ahmad and Golic 1998; Titen and Golic 2008, 2010; Titen et al. 2014). In the case of the Y chromosome, low resolution of mitotic cytology and the required role of the Y in male fertility limit the chromosomes that can be recovered. On chromosome 3, aneuploidy limits the range of broken chromosomes that can be recovered, and because chromosome 4 is so small, there is no opportunity to recover a wide range of breakpoints.
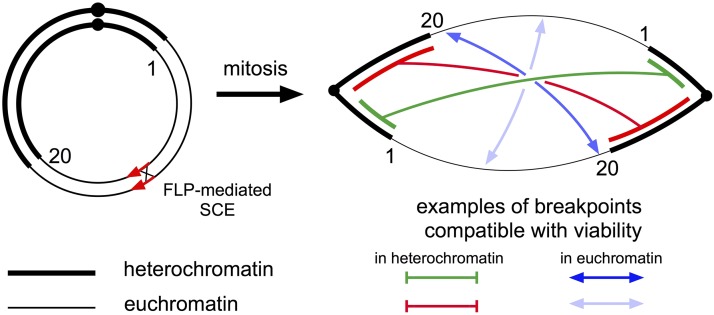
We designed a scheme that would allow us to determine whether dicentric chromosomes break at preferred sites. Dicentric chromosomes with two bridging arms were produced by FLP recombinase mediated sister chromatid exchange in a Ring-X chromosome (Figure 1). If the two arms break at similar sites and then heal, a nearly euploid linear X chromosome may be recovered. We expected the vast majority of linear X chromosomes to be broken in heterochromatin because the breakpoints simply need to be in the same block of heterochromatin on each arm but, in molecular terms, may be megabases apart. We expected euchromatic breaks to be rare because the sites of breakage on each arm would need to be at nearly the same site. However, half the recovered linear chromosomes were broken in euchromatin, and the breakpoints were clustered at a limited number of euchromatic sites. This result is most easily explained by a limited number of preferred regions for breakage.
Figure 1.
Dicentric chromosomes produced by FLP-mediated sister chromatid exchange. The form of a dicentric chromosome produced by sister chromatid exchange is shown. Numbers 1 and 20 indicate the polytene chromosome coordinates of euchromatin at the normal X tip and base, respectively. Examples of combinations of breaks that are compatible with viability are indicated at the right. Breaks in heterochromatin need not be precisely matched for viability, but breakpoints in euchromatin must be very nearly at the same site to produce a chromosome that is viable in males (two of many possible examples indicated). The lengths of each segment of heterochromatin are not known precisely.
Although the X chromosome is a frequent target of experimental structural manipulation, one form of the X that has not been recovered is a metacentric single X. The Drosophila melanogaster X is normally acrocentric, with all euchromatin and most heterochromatin lying to the left of the centromere and only a very short heterochromatic arm located to the right. A number of X chromosomes with inversions that encompass varying portions of the left arm have been generated, but to our knowledge, a single X with its centromere in a medial position has not been generated. Among the linear chromosomes that we recovered are many with the centromere located approximately in the middle of the chromosome.
Materials and Methods
The ring chromosomes we used for these experiments were generated as described previously (Golic and Golic 2010). In addition to an entire X chromosome, they also carry a portion of the Y. They are maintained in stock as R(1;Y)/Y males by C(1)DX/Y females. To look for evidence of somatic breakage of the dicentric, FLP was expressed by heat-shock induction of the 70FLP transgene in females with a normal X and a ring chromosome that carried an insertion of the FRT-bearing element P{>whs>} (Golic and Lindquist 1989; Golic et al. 1997).
The ring chromosomes that we generated tended to be quite filicidal. To produce ring chromosomes that were not filicidal, we treated males with 2000 rad of X-rays and crossed individual males to C(1)DX/Y females. We then propagated progeny of these crosses by simple transfer to fresh food for several generations to allow selection of a healthy derivative. The chromosome used for the germline experiments reported here is called R(1;Y)6AX2. Its improved viability is likely a consequence of deletion or rearrangement of a portion(s) of its heterochromatin (Hinton 1955; Stone 1982; Ferree et al. 2014). The relative arrangement of centromere and heterochromatic segments in this derivative has not been determined, and in the figures, the centromere and heterochromatic segments are indicated as they were in the original ring. A single male from this stock was crossed to FM7/y w females to produce a R(1;Y)/FM7 stock. Genetic and cytologic tests confirmed that this stock carried a ring chromosome. Females from this stock were used in crosses to generate an open ring.
Crosses to recover opened-ring chromosomes
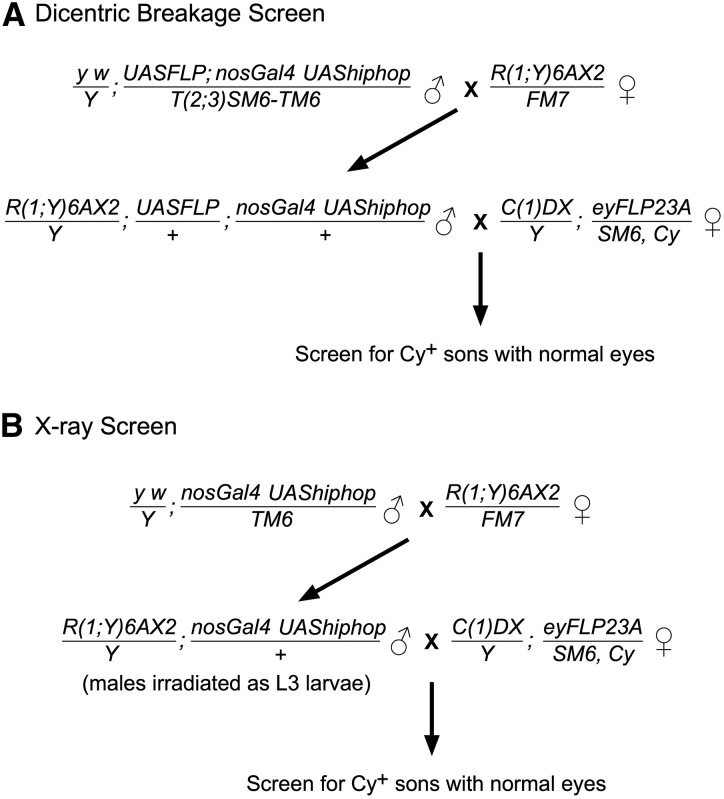
Figure 2 shows the crossing schemes used to recover linear chromosomes by opening of the R(1;Y)6AX2 chromosome. For generation of dicentric chromosomes, nosGal4 (Van Doren et al. 1998) drove UASFLP (Duffy et al. 1998) expression in the male germline. In many crosses, the UAShiphop construct HRH008H (Gao et al. 2010) was included because in other experiments we have seen that it can increase the recovery of broken-and-healed chromosomes. Males were mated singly to two to three C(1)DX/Y; eyFLP23A/SM6, Cy females or as four males by five females. In Table 1 and Table 2, the number 95 specifies a particular combination of nosGal4 and UASFLP on chromosome 3, and 1C and 3A refer to highly expressing insertions of nosGal4 on chromosome 3. These are our laboratory’s stock designations.
Figure 2.
The crosses used in screens to recover opened-ring chromosomes. See Materials and Methods for a full description.
Table 1. Recovery of linear chromosomes after FLP-mediated dicentric induction.
| Single-male crosses | Multiple-male crosses | |||||
|---|---|---|---|---|---|---|
| Paternal genotype | Total vials | Fertile vials | Linear X | Total vials | Fertile vials | Linear X |
| R(1;Y)6AX2/Y; (nosGal4 UASFLP)95/+ | 12 | 12 | 2 | — | — | — |
| R(1;Y)6AX2/Y; nosGal41C UASFLP/+ | 22 | 2 | 2 | — | — | — |
| R(1;Y)6AX2/Y; UASFLP/+; nosGal41C HRH008H/+ | 70 | 16 | 8 | 6 | 4 | 4 |
| R(1;Y)6AX2/Y; UASFLP/+; nosGal43A HRH008H/+ | 79 | 10 | 3 | 19 | 10 | 6 |
| Total | 183 | 40 | 15 | 25 | 14 | 10 |
Table 2. Recovery of linear chromosomes after irradiation.
| Treatment | Paternal genotype | Single-male crosses | Fertile | Linear X |
|---|---|---|---|---|
| 100 rad | R(1;Y)6AX2/Y; nosGal4(1C or 3A) HRH008H/+ | 80 | 72 | 0 |
| R(1;Y)6AX2/Y; TM6/+ | 21 | 21 | 1 | |
| 50 rad | R(1;Y)6AX2/Y; nosGal4(1C or 3A) HRH008H/+ | 195 | 183 | 6 |
| R(1;Y)6AX2/Y; TM6/+ | 59 | 56 | 2 | |
| Total | 355 | 332 | 9 |
For the X-ray experiment, third instar larvae were collected and placed on the surface of a small petri dish containing a thin layer of fly food, exposed to 50 or 100 rad of X-rays, and returned to a vial of food to complete development. The males that eclosed were individually mated with three to five C(1)DX; eyFLP23A/SM6, Cy females each.
In some cases, linear X chromosomes were recovered that were clearly normal X chromosomes and not opened rings. In the X-ray experiment, males with a normal X were easily recognized as yellow-bodied, white-eyed males. The R(1;Y)6AX2 chromosome is marked with recessive y and w alleles but also carries the P{>whs>} element, which confers red pigment to the eyes of males that carry it. We expected virtually all males with an opened ring to have red eyes in the X-ray experiment, but a few white-eyed males were recovered in this experiment. They most likely emanate from R/FM7/Y (XXY) females in the initial cross, which are present at low frequency in the R(1;Y)6AX2 stock. If these females produce a Y-bearing egg, which is then fertilized by an X, y w sperm that also carries nosGal4 HR008H (both of which are marked with w+), the flies will be red-eyed and indistinguishable from sons that carry the R(1;Y)6AX2 chromosome. In the next generation, they will produce many sons with a linear X, which is simply the X, y w chromosome they inherited from their father. If the chromosome were actually an opened ring, it should still carry the P{>whs>} element and always give red-eyed sons. Cytologic examination of one such chromosome confirmed that it appeared to be a normal X, with none of the Y chromatin that is carried by R(1;Y)6AX2. In the experiments with FLP, it was not possible to immediately identify these chromosomes because FLP expression in the germline excises the whs gene, and even legitimate opened rings produce white-eyed males. However, mitotic chromosome cytology revealed that four of the potential opened rings looked like normal X chromosomes. They were not considered further.
Cytology
Mitotic chromosomes were prepared as described by Gatti and Pimpinelli (1983), stained with DAPI, and examined under UV fluorescence on an Olympus IX-81 microscope with a 100× objective. Images were captured with an Orca-ER camera. Polytene chromosomes were prepared according to Lefevre (1976) and examined on a Zeiss Axioskop with phase-contrast 40× and 100× objectives. Photographs were taken with an Olympus EVOLT E-330 camera.
Data availability
The Drosophila strains described or generated in this work are available on request from the authors.
Results
Ring opening by dicentric breakage
When recombination is induced between sister chromatids of a Ring-X chromosome, a dicentric chromosome with a double anaphase bridge is produced (Figure 1). If both bridges break and the ends heal, linear X chromosomes may be recovered. In previous work, we found that single-bridge dicentric chromosomes typically break during mitosis, and in the male germline, the broken end can be healed by addition of a new telomere (Ahmad and Golic 1998; Titen and Golic 2008, 2010; Titen et al. 2014). We did not know whether a double bridge would break, so we first looked for evidence of breakage in somatic cells. We used a ring chromosome that carries an insertion of the P element P{>whs>} with a w+ gene flanked by directly repeated copies of the FRT (FLP Recombination Target). This chromosome also carries some portion of the Y chromosome that has not been fully characterized. Exchange between FRTs on the same chromosome will excise the white gene, while exchange between FRTs on sister chromatids will produce a dicentric chromosome. We heat shocked larvae carrying this chromosome and a heat-inducible FLP transgene for 1 hr at 38° and returned them to normal growth temperature of 25°. Several hours later, we examined metaphase spreads from brain squashes to see whether we could identify linear X chromosomes resulting from breakage. We saw several such examples (Figure 3). We concluded that it was feasible to try to generate linear X chromosomes by germline breakage and healing of a ring chromosome.
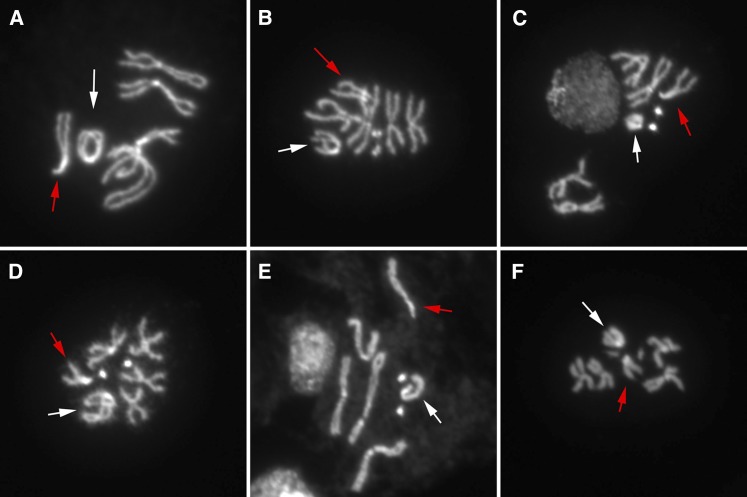
Figure 3.
Breakage of a ring chromosome in somatic cells after induction of FLP-mediated sister chromatid exchange. In all panels, the normal X is indicated with a red arrow, and the ring or derivatives are indicated with a white arrow. (A) The original R(1;Y) and a normal linear X. (B) A broken ring. (C) A ring that appears to have broken to generate a very short chromosome and then recircularized by end joining. (D) A ring that broke and replicated before fusing ends of sister chromatids. (E and F) Broken rings.
To generate dicentric chromosomes in the germline, we used nosGal4 and UASFLP to express FLP recombinase. In some crosses, we also included a UAShiphop (Gao et al. 2010) construct because we have seen that it can increase the recovery of healed chromosomes (unpublished observations). To detect linear X chromosomes, we crossed FLP-expressing males with the ring [R(1;Y)6AX2] to attached-X females carrying an eyFLP transgene [C(1)DX/Y; eyFLP/SM6, Cy], which drives FLP expression in the developing eye disk. If flies with eyFLP also carry a ring chromosome with an FRT, the dicentric chromosomes that are produced in the developing eye make adults with small, rough eyes (Golic and Golic 2010). However, FLP expression has no deleterious effect in flies that carry an FRT on a linear X chromosome. Therefore, the occurrence of eyFLP sons with normal eyes indicates that the male carries a linear X. When such males were identified, they were crossed to C(1)DX/Y females to maintain the stock. These males often arose in clusters and likely represent the mitotic multiplication of a single germline break-and-heal event. To ensure that all stocks represent independent events, though, a single son was chosen as the progenitor of each stock.
A total of 289 males carrying R(1;Y)6AX2 and nosGal4 UASFLP (and in some cases UAShiphop) were crossed to C(1)DX/Y; eyFLP/SM6, Cy females. Fifty-four of these crosses were fertile, and from these fertile crosses we recovered 25 independent linear X chromosomes (Table 1). The ring chromosome that we used in this screen is slightly filicidal, and in addition, males with the ring and eyFLP survive poorly. Together these factors generate a strong selection for recovery of linearized chromosomes.
Approximately half the males that were fertile produced daughters but no sons. We suspect that these may represent cases where a linear chromosome was produced that allowed male germ cell viability but was not compatible with viability of the whole animal. This suggests that a wider variety of linear chromosomes might be found if the screen were designed to recover them in daughters, where they would be heterozygous with a complete X.
Our initial tests used single-male crosses: of a total of 183 crosses, 40 were fertile, and 15 yielded linear X chromosomes. Based on the high rate of male sterility, we realized that we could cross several males per vial, and despite picking only a single linear X son per vial, we would lose very few independent ring-opening events. In 25 vials with 4 males each, 14 were fertile, and 10 produced offspring with a linear X.
The false-positive rate of this screen is very low. In addition to 25 verified linear X chromosomes, we recovered only a single X chromosome that passed the initial screen yet was still a ring chromosome. Four more linear X chromosomes appeared to be normal X chromosomes and almost certainly did not represent ring opening but arose by nondisjunction (see Materials and Methods for discussion).
We first characterized the recovered chromosomes by mitotic and polytene chromosome cytology. Eleven of the opened-ring chromosomes had heterochromatin at both ends, 11 had euchromatin at both ends, and 3 had heterochromatin at one end and euchromatin at the other (Figure 4 and Figure 5). In male polytene chromosome spreads, the chromosomes with heterochromatin at each end most often appeared as a loop that emanated from and returned to the chromocenter. Occasionally, one end of a chromosome was pulled out of the chromocenter, and in such cases, it often showed a thin thread of chromatin trailing from the end of the chromosome. In some cases, the entire X chromosome was pulled away from the remaining chromosomes and appeared as a solitary circular structure. These polytene circles simply reflect the fact that all heterochromatin typically associates to form the chromocenter. The mitotic karyotypes clearly show that these chromosomes are linear.
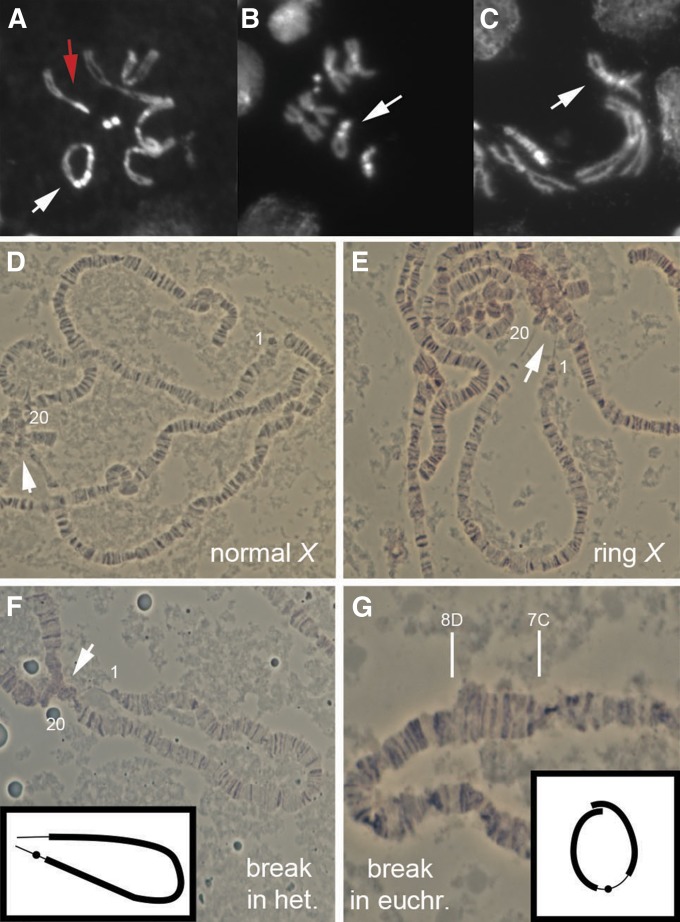
Figure 4.
Representative examples of linearized ring chromosomes. (A–C) Mitotic chromosome cytology, with ring chromosome or linear derivatives (white arrow): (A) R(1;Y)6AX2 in female with normal X (red arrow); (B) chromosome #17F, with heterochromatin at each end, recognizable by lack of sister chromatid separation in regions of heterochromatin at metaphase; (C) chromosome #23F, with euchromatin at each end. (D–G) Polytene chromosome cytology, with chromocenter indicated by arrow and normal tip and base indicated as 1 and 20, respectively: (D) a normal linear X; (E) the ring X; (E) chromosome #4F, with heterochromatin at each end, and interpretive diagram showing heterochromatin (thin lines) looping together at chromocenter; (G) chromosome #29F, with euchromatin at each end, showing duplication that extends from 7C–8D, with interpretive diagram showing euchromatin (thick lines) overlapping at the duplicated region.
Figure 5.
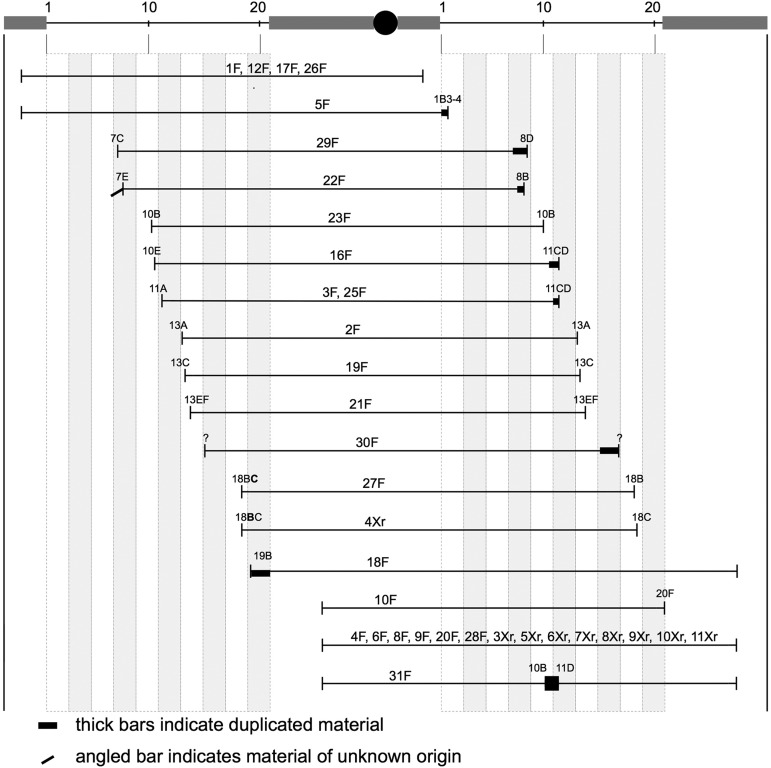
The recovered linear chromosomes. The span of each chromosome is indicated underneath a diagram that shows the centromere (circle), heterochromatin (thick gray line), and euchromatin (thin black line), with polytene chromosome coordinates indicated. The termini in heterochromatin are arbitrarily indicated as ending at the same point, though it is clear that there are differences (see text). The name of each chromosome is indicated above each line. Chromosomes with names ending in F were produced by dicentric breakage; names ending in Xr were produced after irradiation. The termini of #30F represent an educated guess based on suppression of B.
In the chromosomes with heterochromatin at each end, the euchromatin might be in normal or inverted orientation with respect to the centromere. To distinguish these possibilities, we generated females that were heterozygous for the opened ring, which is marked by y and f+, and a normal X, marked by y+ and f, and scored sons for recombination. The standard distance between y and f is 56.7 cM. If the opened ring is in normal orientation with respect to the centromere, recombination between these genes should be frequent, but if the opened ring is inverted with respect to the centromere, recombination should be rare. We found that 4 of 11 were in normal orientation (#1F, #12F, #17F, and #26F) and 7 of 11 were inverted (#4F, #6F, #8F, #9F, #20F, #28F, and #31F) (Table 3 and Figure 5). To avoid confusion with polytene chromosome cytologic locations, the # symbol is used to signify a recovered linear chromosome.
Table 3. Recombination tests of linear chromosomes.
| Progeny | |||||
|---|---|---|---|---|---|
| Parentals | Recombinants | Percent recombinants | |||
| Chromosome | y f+ | y+ f | y f | y+ f+ | |
| 1F | 46 | 99 | 62 | 74 | 48.40 |
| 2F | 75 | 54 | 0 | 0 | 0.00 |
| 3F | 74 | 179 | 0 | 0 | 0.00 |
| 4F | 148 | 168 | 0 | 4 | 1.25 |
| 5F | 112 | 121 | 78 | 77 | 39.95 |
| 6F | 51 | 75 | 2 | 4 | 4.55 |
| 8F | 85 | 118 | 1 | 1 | 0.98 |
| 9F | 44 | 74 | 0 | 0 | 0.00 |
| 10F | 53 | 87 | 2 | 4 | 4.11 |
| 12F | 43 | 46 | 28 | 28 | 38.62 |
| 16F | 94 | 159 | 2 | 0 | 0.78 |
| 17F | 28 | 57 | 33 | 26 | 40.97 |
| 18F | 55 | 77 | 0 | 3 | 2.22 |
| 19F | 166 | 189 | 1 | 0 | 0.28 |
| 20F | 58 | 78 | 1 | 4 | 3.55 |
| 21F | 174 | 182 | 0 | 0 | 0.00 |
| 22F | 99 | 118 | 2 | 2 | 1.81 |
| 23F | 148 | 115 | 1 | 1 | 0.75 |
| 25F | 156 | 201 | 0 | 0 | 0.00 |
| 26F | 110 | 130 | 91 | 97 | 43.93 |
| 27F | 158 | 187 | 1 | 0 | 0.29 |
| 28F | 199 | 203 | 5 | 11 | 3.83 |
| 29F | 18 | 77 | 0 | 0 | 0.00 |
| 31F | 56 | 181 | 0 | 0 | 0.00 |
| 3xr | 178 | 193 | 2 | 5 | 1.85 |
| 4xr | 104 | 155 | 0 | 1 | 0.38 |
| 5xr | 128 | 147 | 2 | 1 | 1.08 |
| 6xr | 162 | 197 | 2 | 3 | 1.37 |
| 7xr | 113 | 109 | 2 | 0 | 0.89 |
| 8xr | 188 | 193 | 7 | 6 | 3.30 |
| 9xr | 170 | 200 | 1 | 7 | 2.12 |
| 10xr | 162 | 182 | 2 | 2 | 1.15 |
| 11xr | 175 | 176 | 2 | 3 | 1.40 |
We identified at least five different sets of breakpoints in heterochromatin within these chromosomes. First, the fact that there are two orientations of euchromatin indicates that there were breaks on each side of the centromere. Additionally, we examined inverted chromosomes more closely and observed at least four different morphologies (Figure 6). Thus, the linear chromosomes produced by dicentric breakage may terminate at several different heterochromatic sites.
Figure 6.
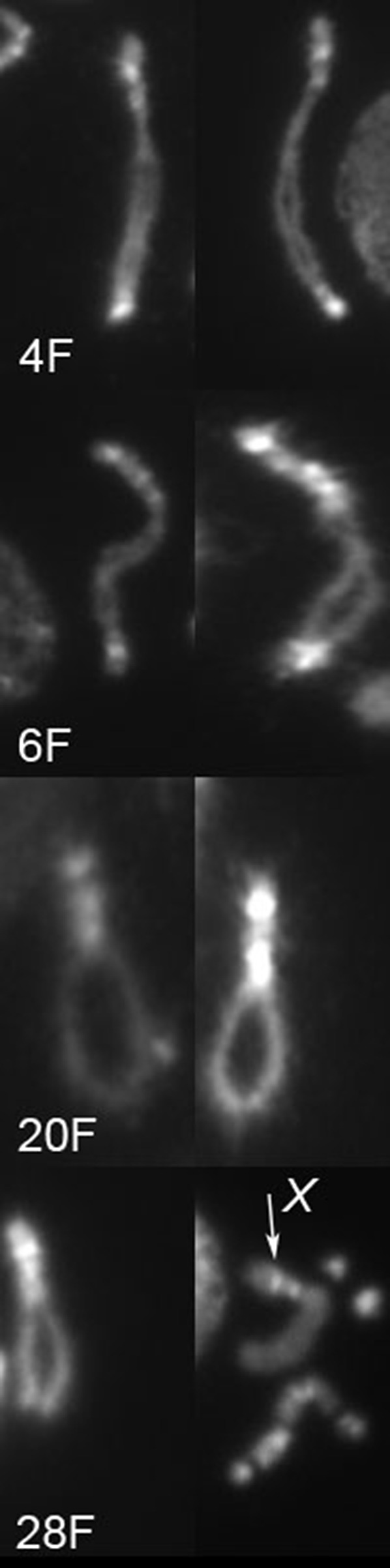
Four different heterochromatin morphologies found in chromosomes with inverted orientation. Two examples of each of four chromosomes (as labeled) are shown. In the last panel of #28F, the X chromosome is indicated with an arrow; there are also two 4 chromosomes and a Y chromosome present in this picture.
One of the chromosomes with both ends terminating in heterochromatin is worth further mention. In male polytene spreads, the #31F chromosome had a large tangle in the middle of its euchromatin that stretched from ∼10B–11D. This section of the chromosome appeared to be at least doubled and more likely tripled or quadrupled. We were unable to obtain a completely clear view of this segment because it appeared to adopt multiple pairing configurations that disturbed the morphology. This chromosome may have broken more than once, perhaps repeatedly breaking in the 10D region on one arm and in the 11D region on the other and rejoining to produce a ring with duplication(s) and then finally breaking in heterochromatin and healing to produce a linear chromosome.
Three chromosomes terminated in heterochromatin at one end and euchromatin at the other (#5F, #18F, and #10F). Two of these chromosomes carried the entire euchromatic portion of the X on one side of the centromere and a small duplication of euchromatin on the other side. One was in normal orientation with respect to the centromere, and two were inverted. Although chromosome #10F appears to terminate at cytologic region 20F, we cannot be certain that there is not a small piece of heterochromatin present at the tip.
Finally, 11 of the recovered linear chromosomes terminated in euchromatin on both ends. As expected, all exhibited low rates of recombination between y and f. The higher-resolution view of euchromatin afforded by polytene chromosomes allowed us to map the left and right termini with a relatively high degree of precision. Five of these 11 appeared to be perfect openings with no duplication or deficiency at either end. There were five clear examples of chromosomes with small duplications, where the left and right ends overlapped slightly (Figure 4). Additionally, one of the duplicated chromosomes (#22F) carried what seemed to be a small piece of nonhomologous chromatin appended to the end of one arm. The origin of this segment was not determined, but the lack of pairing with the opposite arm indicates that it is likely derived from elsewhere in the genome. The eleventh chromosome (#30F) showed very poor viability in males and could be maintained in stock-only heterozygotes with an FM7, y B f balancer chromosome. We suspect that this chromosome carries a duplication of the 16A region, which includes B+, because the Bar phenotype of FM7 is strongly suppressed in the FM7/#30F heterozygous females. A large duplication also could easily account for the reduced viability of males with this chromosome. However, we were unable to obtain polytene chromosomes to verify this supposition.
Ring opening after irradiation
In a separate series of experiments, we treated third instar male larvae with low doses of X-rays to see whether linear chromosomes might be recovered after X-ray-induced double-strand breaks. The results were quite different from those of the previous experiments. First, the males in this experiment were much more fertile than males expressing FLP in their germline. A total of 355 single-ring-bearing males were irradiated and crossed to C(1)DX; eyFLP/Cy females, and 332 were fertile (Table 2). Second, although linear chromosomes were recovered, they were much less frequent, with only nine such chromosomes recovered. There also was one false positive that was still a ring and three males with normal linear X chromosomes that arose by nondisjunction. Third, the recovered chromosomes had a distinctly different array of breakpoints, with only one broken in euchromatin and the remaining eight broken within heterochromatin on the same side of the centromere to produce X chromosomes with euchromatin that was inverted with respect to the centromere (Figure 5 and Table 3). None of these was distinguishable by mitotic cytology. Although linear chromosomes were recovered from this experiment, it was much less efficient than using FLP to generate dicentrics, and the array of chromosomes recovered was much more restricted.
Discussion
There have been a handful of reported cases of ring chromosomes that spontaneously opened into linear chromosomes (Morgan 1933; Bender 1958; Leigh 1976). All except one had opened in heterochromatin. The exception was a compound ring, i.e., incorporating two X chromosomes into a single chromosome, that opened in euchromatin to generate a linear compound X chromosome (Traverse and Pardue 1988). None of these generated the novel X chromosomes that were produced by our experiments, namely, single X chromosomes with the centromere in the middle of the chromosome. Because the chromosomes we recovered opened at several different sites, the result was a collection of X chromosomes that are circularly permuted with respect to gene and centromere location. These chromosomes may be useful for investigating chromosome behavior in meiosis or for questions of nuclear organization and function. If desired, it should be possible to convert the metacentric chromosomes back into ring chromosomes by two-strand double-crossover events with a normal X chromosome when one crossover occurs in each arm.
For a ring chromosome to be converted to a recoverable linear chromosome, it must break to produce a linear molecule with a single centromere, both ends must heal, and germ cells and whole organisms with the chromosome must be viable. Naively, we expected a predominance of breakpoints in heterochromatin in the chromosomes recovered after dicentric breakage. Heterochromatin constitutes approximately half the R(1;Y) used in these experiments. Because this heterochromatin carries no vital genes (apart from rDNA, which is also supplied by the Y), the breakpoints in heterochromatin could be offset from each other by a substantial distance and still be tolerated. However, when breaks occur in euchromatin, they must be at nearly the same site on each arm to be recovered in a viable male. If the breakpoints are offset, only very small deficiencies and somewhat larger duplications will be tolerated (Ashburner et al. 2005; Cook et al. 2010). We imagined that the occurrence of breakpoints at nearly the same site on both arms would be rare, thus favoring the recovery of chromosomes with two breakpoints in heterochromatin. However, if linear chromosomes could be recovered after irradiation, we expected a broader distribution of breakpoints than linear chromosomes generated by dicentric breakage. A ring that broke in heterochromatin and then healed almost certainly would be viable as the only X in males. Additionally, because many genes are not required for viability, it seemed likely that many of the random breaks in euchromatin would be tolerated. In light of these expectations, it was a surprise to find precisely the opposite results from the dicentric breakage and X-ray experiments. The 25 linear chromosomes recovered after dicentric breakage have 25 new termini in euchromatin and 25 in heterochromatin. In contrast, the 9 linear chromosomes recovered after exposure to X-rays have 16 of 18 new termini in heterochromatin (2 × 2 contingency test, P = 0.0045).
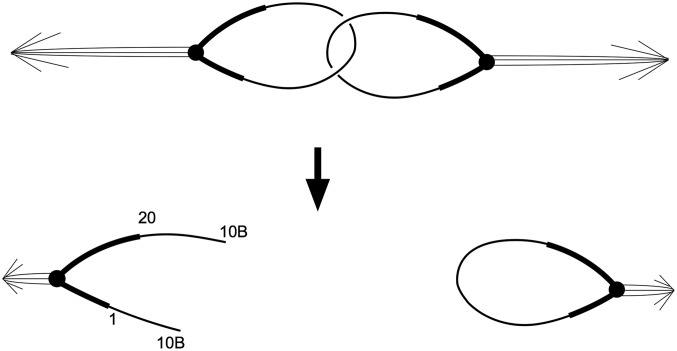
One explanation that might account for the recovery of so many ring openings in euchromatin in the first set of experiments is that the linear chromosomes were derived by a mechanism other than dicentric breakage. For instance, if catenated rings were to segregate and topoisomerase failed to resolve the interlocked rings, one of the rings might break (Figure 7). If the ends healed, many of these breaks, either in euchromatin or in heterochromatin, might be compatible with viability. Any other form of spontaneous breakage of a single ring followed by healing could give similar results. However, if the opened rings were primarily the result of spontaneous events rather than dicentric breakage, we would expect the X-ray experiment to produce at least as many opened rings, and it did not. Furthermore, the occurrence of short duplicated segments at the termini of many of the opened rings is a predicted outcome of dicentric breakage but not of spontaneous ring opening followed by healing.
Figure 7.
Breakage of catenated rings. At the top, interlocked rings are shown segregating to opposite poles in anaphase. This situation could be resolved by breakage of one ring, allowing completion of mitosis. As a result of this break, both arms terminate at the same site (indicated arbitrarily as 10B in the example at the bottom left).
In addition to generating far fewer opened rings, the X-ray experiment gave a very different distribution of ring-opening breakpoints. It is possible that this result may have a trivial explanation. The eight linear chromosomes with similar or identical appearance could be replicates of one spontaneous opening event that occurred within the stock of the R(1;Y)6AX2 chromosome. Many of the rings we constructed are filicidal (Golic and Golic 2010), and for those rings, there is a strong selection for spontaneous opening. However, this does not appear to be the case with R(1;Y)6AX2, which was chosen for these experiments precisely because it is only slightly filicidal, and the stock is healthy. Even so, a spontaneously opened ring might have a slight advantage that would allow it to increase in frequency within the stock, leading to its recovery eight times in the X-ray experiment. Arguing against this is the fact that ∼80% of the FLP-mediated openings were generated several months subsequent to the X-ray experiments, and if there were a healthier linear X within the stock, we would expect it to account for many more of those opened rings. We also recently generated 15 iso-X lines from the R(1;Y)6AX2 stock, and cytologic analysis shows them all to be rings. Either there was at one time a spontaneously opened ring segregating within the stock that has since been lost from the stock, or the eight seemingly identical linear chromosomes derived from the X-ray experiment resulted from eight independent events. We cannot at this time definitively conclude which explanation is correct, though we favor the latter.
Even if only a single newly opened ring were generated by X-rays, it would be more than seen previously. Catcheside and Lea (1945) examined 789 daughters carrying the X of irradiated Ring-X chromosome fathers—no linear derivatives were observed. Our experiments using eyFLP to detect linearized rings are more efficient, allowing us to screen many more offspring for opened rings, perhaps accounting for the difference. The experiments of Catcheside and Lea (1945) also involved irradiation of mature sperm. We have argued that healing of broken dicentric chromosomes in the male germline most likely occurs before meiosis (Titen et al. 2014). After meiosis, it may be too late for healing to occur, and instead, breaks are repaired by joining broken ends in the zygote. We also deliberately chose to use very low doses of X-rays relative to those typically used for mutagenesis because we wished to generate only one or a few breaks per cell. Finally, overexpression of hiphop, a vital component of the telomere cap, may have played a role in allowing us to recover linearized chromosomes after irradiation.
A striking feature of the new chromosome termini produced by dicentric breakage, beyond the fact that half are found in euchromatin, is clustering of the euchromatic breakpoints. By placing the locations of new telomeres on a circular map, it becomes clear that the breaks are found primarily in regions 10, 11, and 13 (Figure 8). A closer look at those breakpoints (Figure 5) shows that although similar, there were only a few that might be identical. Chromosomes #3F and #25F could not be distinguished by polytene cytology, and the right breakpoint of #16F could not be distinguished from either of these. Although the internal rearrangement breakpoints of #31F could not be determined precisely, they were similar to breakpoints found in #23F, #16F, #3F, and #25F. The left breakpoints of #27F and #4Xr could not be distinguished, but all other breakpoints in euchromatin were clearly distinct. The clustering shown in Figure 8 represents regional clustering rather than specific common sites. We have not overlooked the fact that half the termini of the linear chromosomes produced by dicentric breakage are in heterochromatin, but the cytologic resolution is insufficient to say whether there might also be preferred regions of breakage within heterochromatin.
Figure 8.
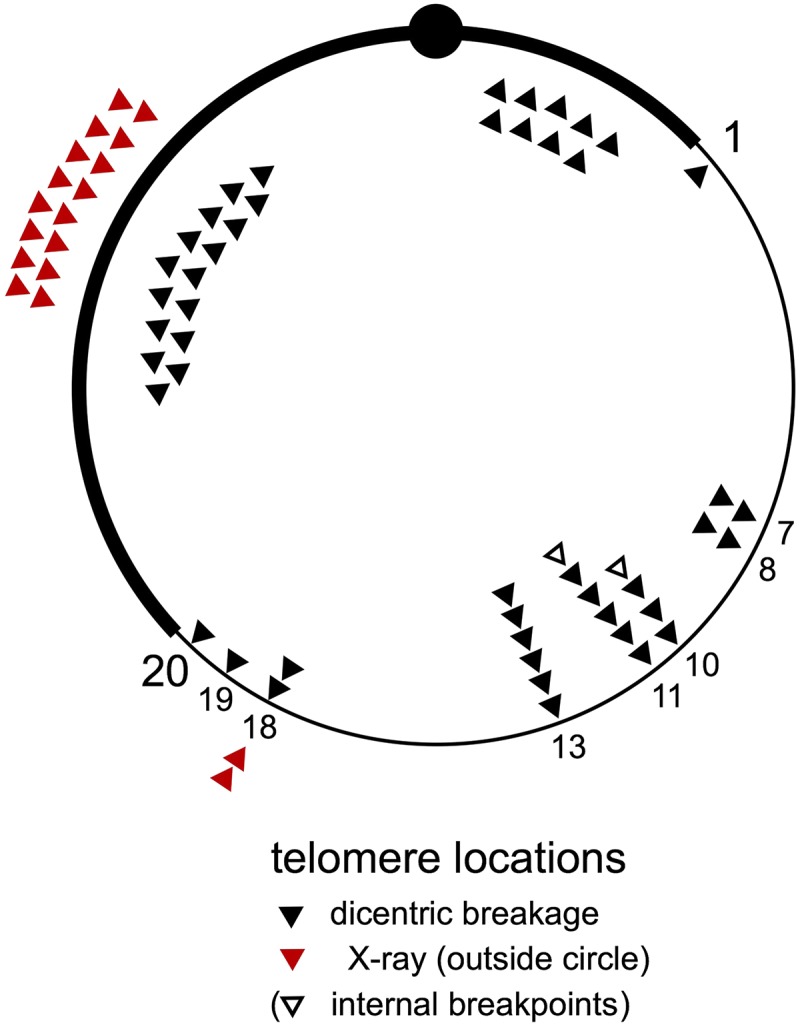
Telomere locations of linearized chromosomes. The location of each of the new telomeres on the opened rings is indicated by a solid triangle on this diagram of the R(1;Y)6AX2 chromosome and are grouped according to numbered divisions of polytene chromosomes. The telomeres of linear chromosomes generated by dicentric breakage are mapped inside the ring; those generated after irradiation are indicated outside the ring (in red). The internal rearrangement breakpoints of chromosome #31F are indicated with open triangles. Chromosome #30F is not included in this diagram. Breakpoints in heterochromatin are correctly located to the left or right of the centromere, but no further specification is intended.
A number of explanations for this breakpoint clustering can be imagined. For instance, if chromosome healing was very inefficient except at a few sites, it might account for these results. However, previous results show that healing can occur at many sites in heterochromatin and euchromatin (Mason et al. 1984; Levis 1989; Biessmann et al. 1990; Tower et al. 1993; Ahmad and Golic 1998; Mikhailovsky et al. 1999; Titen and Golic 2010; Beaucher et al. 2012; Titen et al. 2014).
We suggest that nonrandom breakage of the dicentric chromosome, as has been seen in other organisms (Shimizu et al. 2005; Pobiega and Marcand 2010; Song et al. 2013; Lopez et al. 2015), is the most likely explanation for the clustered locations of new telomeres. If breakage in euchromatin tends to occur at a few preferred sites, then viable two-break combinations should be much more frequent than if breakage were random. In salivary gland polytene chromosomes, there are a few dozen sites throughout the genome that possess a unique set of properties, which include “weakness” in squashed chromosome preparations, under-replication, and late replication. These sites have been termed intercalary heterochromatin (IH) (Bridges 1935; Kaufmann 1939; Zhimulev et al. 1982; Hammond and Laird 1985; Lamb and Laird 1987). The clustered breakpoints in polytene regions 10 and 11 surround a notable “weak point” and site of under-replication in 11A. Similarly, the breakpoint cluster in region 13 is near sites of IH in 12E and 13B. As mentioned earlier, breakpoint clustering is regional and not limited to a specific site. Similarly, late replication and under-replication are regional properties of chromosomes, extending over hundreds of kilobases (Belyaeva et al. 2012; Yarosh and Spradling 2014).
The correlation between sites of IH and breakpoint clusters suggests the possibility of a cause-and-effect relationship. However, salivary glands are composed of nondividing somatic cells, and the relevance to chromosome structure in mitotically dividing germline cells is not immediately obvious. Still, there is a connection: the timing of IH replication in salivary glands is highly correlated with the timing of IH replication in mitotically dividing cells (Belyaeva et al. 2012). Thus, the structure of polytene chromosomes, insofar as it is based on replication timing, is likely to provide clues to the structure of mitotic chromosomes, including possible weak points.
Laird and colleagues proposed that the properties of intercalary heterochromatin can explain fragile sites in human chromosomes (Laird et al. 1987; Laird and Lamb 1988). The fragile-X syndrome in humans is associated with late replication of hundreds of kilobases in the region of the fragile site (Hansen et al. 1993, 1997; Subramanian et al. 1996), just as late replication is associated with weak points in polytene chromosomes. Our results, coupled with the high correlation of replication timing in mitotic cells and salivary gland polytene cells, support the hypothesis that a common mechanism underlies the occurrence of weak points in polytene and mitotic chromosomes.
Acknowledgments
This work was funded by grant R01-GM065604 from the National Institutes of Health to K.G.G. H.H. was partially funded by the University of Utah Undergraduate Research Opportunities Program.
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Ahmad K., Golic K. G., 1998. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148: 775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Golic K. G., Hawley R. S., 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Beaucher M., Zheng X.-F., Amariei F., Rong Y. S., 2012. Multiple pathways suppress telomere addition to DNA breaks in the Drosophila germline. Genetics 191: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva E. S., Goncharov F. P., Demakova O. V., Kolesnikova T. D., Boldyreva L. V., et al. , 2012. Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS ONE 7: e30035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M. A., 1958. A comparison of crossover relationships in a Drosophila ring X chromosome and a rod X derivative. Genetics 43: 48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H., Carter S. B., Mason J. M., 1990. Chromosome ends in Drosophila without telomeric DNA sequences. Proc. Natl. Acad. Sci. USA 87: 1758–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C. B., 1935. Salivary chromosome maps. J. Hered. 26: 60–64. [Google Scholar]
- Catcheside D. G., Lea D. E., 1945. Dominant lethals and chromosome breaks in ring X-chromosomes of Drosophila melanogaster. J. Genet. 7: 25–40. [Google Scholar]
- Cook R. K., Deal M. E., Deal J. A., Garton R. D., Brown C. A., et al. , 2010. A new resource for characterizing X-linked genes in Drosophila melanogaster: systematic coverage and subdivision of the X chromosome with nested, Y-linked duplications. Genetics 186: 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B., Harrison D. A., Perrimon N., 1998. Identifying loci required for follicular patterning using directed mosaics. Development 125: 2263–2271. [DOI] [PubMed] [Google Scholar]
- Ferree P. M., Gomez K., Rominger P., Howard D., Kornfeld H., et al. , 2014. Heterochromatin position effects on circularized sex chromosomes cause filicidal embryonic lethality in Drosophila melanogaster. Genetics 196: 1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Walser J.-C., Beaucher M. L., Morciano P., Wesolowska N., et al. , 2010. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 29: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Pimpinelli S., 1983. Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster. Chromosoma 88: 349–373. [Google Scholar]
- Golic K. G., Lindquist S., 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- Golic M. M., Golic K. G., 2010. A simple and rapid method for constructing ring-X chromosomes in Drosophila melanogaster. Chromosoma 120: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic M. M., Rong Y. S., Petersen R. B., Lindquist S. L., Golic K. G., 1997. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 25: 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond M. P., Laird C. D., 1985. Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma 91: 279–286. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Canfield T. K., Fjeld A. D., Mumm S., Laird C. D., et al. , 1997. A variable domain of delayed replication in FRAXA fragile X chromosomes: X inactivation-like spread of late replication. Proc. Natl. Acad. Sci. USA 94: 4587–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R. S., Canfield T. K., Lamb M. M., Gartler S. M., Laird C. D., 1993. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell 73: 1403–1409. [DOI] [PubMed] [Google Scholar]
- Hinton C. W., 1955. The behavior of an unstable ring chromosome of Drosophila melanogaster. Genetics 40: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B. P., 1939. Distribution of induced breaks along the X-chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 25: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird C., Jaffe E., Karpen G., Lamb M., Nelson R., 1987. Fragile sites in human chromosomes as regions of late-replicating DNA. Trends Genet. 3: 274–281. [Google Scholar]
- Laird C. D., Lamb M. M., 1988. Intercalary heterochromatin of Drosophila as a potential model for human fragile sites. Am. J. Med. Genet. 30: 689–691. [DOI] [PubMed] [Google Scholar]
- Lamb M. M., Laird C. D., 1987. Three euchromatic DNA sequences under-replicated in polytene chromosomes of Drosophila are localized in constrictions and ectopic fibers. Chromosoma 95: 227–235. [DOI] [PubMed] [Google Scholar]
- Lefevre G., 1976. A photographic representation and interpretation of the polytene chromosomes of Drosophila melanogaster salivary glands, pp. 32–66 in The Genetics and Biology of Drosophila, Vol. 1a, edited by Ashburner M., Novitski E. Academic Press, London. [Google Scholar]
- Leigh B., 1976. Ring chromosomes and radiation induced chromosome loss, pp. 505–528 in The Genetics and Biology of Drosophila, Vol. 1b, edited by Ashburner M., Novitski E. Academic Press, London. [Google Scholar]
- Levis R. W., 1989. Viable deletions of a telomere from a Drosophila chromosome. Cell 58: 791–801. [DOI] [PubMed] [Google Scholar]
- Lopez V., Barinova N., Onishi M., Pobiega S., Pringle J. R., et al. , 2015. Cytokinesis breaks dicentric chromosomes preferentially at pericentromeric regions and telomere fusions. Genes Dev. 29: 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Strobel E., Green M. M., 1984. mu-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc. Natl. Acad. Sci. USA 81: 6090–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailovsky S., Belenkaya T., Georgiev P., 1999. Broken chromosomal ends can be elongated by conversion in Drosophila melanogaster. Chromosoma 108: 114–120. [DOI] [PubMed] [Google Scholar]
- Morgan L. V., 1933. A closed X chromosome in Drosophila melanogaster. Genetics 18: 250–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobiega S., Marcand S., 2010. Dicentric breakage at telomere fusions. Genes Dev. 24: 720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N., Shingaki K., Kaneko-Sasaguri Y., Hashizume T., Kanda T., 2005. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp. Cell Res. 302: 233–243. [DOI] [PubMed] [Google Scholar]
- Song W., Gawel M., Dominska M., Greenwell P. W., Hazkani-Covo E., et al. , 2013. Nonrandom distribution of interhomolog recombination events induced by breakage of a dicentric chromosome in Saccharomyces cerevisiae. Genetics 194: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. C., 1982. Filicidal ring-Y chromosomes in D. melanogaster. Genetics 102: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian P. S., Nelson D. L., Chinault A. C., 1996. Large domains of apparent delayed replication timing associated with triplet repeat expansion at FRAXA and FRAXE. Am. J. Hum. Genet. 59: 407–416. [PMC free article] [PubMed] [Google Scholar]
- Titen S. W. A., Golic K. G., 2008. Telomere loss provokes multiple pathways to apoptosis and produces genomic instability in Drosophila melanogaster. Genetics 180: 1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W. A., Golic K. G., 2010. Healing of euchromatic chromosome breaks by efficient de novo telomere addition in Drosophila melanogaster. Genetics 184: 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W. A., Lin H.-C., Bhandari J., Golic K. G., 2014. Chk2 and P53 regulate the transmission of healed chromosomes in the Drosophila male germline. PLoS Genet. 10: e1004130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Karpen G. H., Craig N., Spradling A. C., 1993. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse K. L., Pardue M. L., 1988. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres. Proc. Natl. Acad. Sci. USA 85: 8116–8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L., Lehmann R., 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8: 243–246. [DOI] [PubMed] [Google Scholar]
- Yarosh W., Spradling A. C., 2014. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev. 28: 1840–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev I. F., Semeshin V. F., Kulichkov V. A., Belyaeva E. S., 1982. Intercalary heterochromatin in Drosophila. I. Localization and general characteristics. Chromosoma 878: 197–228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Drosophila strains described or generated in this work are available on request from the authors.