Abstract
The rate at which new mutations arise in the genome is a key factor in the evolution and adaptation of species. Here we describe the rate and spectrum of spontaneous mutations for the fission yeast Schizosaccharomyces pombe, a key model organism with many similarities to higher eukaryotes. We undertook an ∼1700-generation mutation accumulation (MA) experiment with a haploid S. pombe, generating 422 single-base substitutions and 119 insertion-deletion mutations (indels) across the 96 replicates. This equates to a base-substitution mutation rate of 2.00 × 10−10 mutations per site per generation, similar to that reported for the distantly related budding yeast Saccharomyces cerevisiae. However, these two yeast species differ dramatically in their spectrum of base substitutions, the types of indels (S. pombe is more prone to insertions), and the pattern of selection required to counteract a strong AT-biased mutation rate. Overall, our results indicate that GC-biased gene conversion does not play a major role in shaping the nucleotide composition of the S. pombe genome and suggest that the mechanisms of DNA maintenance may have diverged significantly between fission and budding yeasts. Unexpectedly, CpG sites appear to be excessively liable to mutation in both species despite the likely absence of DNA methylation.
Keywords: mutation accumulation, effective population size, biased gene conversion, fission yeast
MUTATION is the ultimate source of all genetic differences within and among species. Mutation may alter the sequence of the genome via single-base substitutions; change the length of the genome by means of insertions, deletions, and duplications; or alter genome architecture via changes in chromosomal ploidy. The rate at which each of these events occurs in nature is largely obscured in a comparison of individuals or species owing to the action of natural selection and drift. An unbiased estimate of the rate and spectrum of spontaneous mutations can be obtained by passing replicate individuals through thousands of generations of mutation accumulation (MA). Under this protocol, which imposes a very small effective population size during passage, selection will have little influence on the fate of new mutations.
The fission yeast Schizosaccharomyces pombe is a member of the largest and most diverse fungal phylum, Ascomycota, and is a key model system in cell biology. In the wild, S. pombe occurs in small, incompletely isolated populations with only weak population structure resulting from recent global dispersal (Brown et al. 2011; Jeffares et al. 2015). Beyond this, little is known of its natural ecology. It is, however, often recovered from natural fermentations and was isolated originally from East African millet beer.
Notwithstanding the uncertainty regarding its ecology, the life cycle of S. pombe in the wild is likely to be predominantly haploid. In a collection of 81 natural isolates, all but one were haploid, with the sole diploid most likely resulting from biparental inbreeding (Brown et al. 2011). Under laboratory conditions, transient diploids do occur after mating but are generally unstable.
By measuring the rate and types of mutations that occur during a long-term MA experiment with S. pombe, we estimate the effective population size, the rate of outcrossing in the wild, and the strength of selection acting on the GC composition of the genome. Comparison to budding yeast indicates that while the overall base-substitution mutation rates are similar, the spectrum of both insertion-deletion mutations (indels) and point mutations differs significantly between these two yeast species.
Materials and Methods
Mutation accumulation
The 96 sequenced lines were established from a single progenitor cell of wild-type haploid S. pombe (mating type h+; ade6-M216 kanMX4 ura4-D18 leu1-32, strain id = ED668,BG_0000H8; obtained from Bioneer, Republic of Korea) and passed through single-cell bottlenecks on a 3- to 4-day cycle on YES medium plates at 32°.
At the conclusion of the experiment, isolates from 10 random lines from months 1, 3, and 6 were used to estimate the S. pombe generation time under the culture conditions used. Total cell counts per colony on days 2, 3, and 4 were obtained using a hemocytometer (four technical replicates per colony, between two and four colonies per line per day) after colonies were excised with a scalpel and resuspended in PBS by vortexing for 10 sec. The number of generations per passage n for each line was calculated as n = log2N, where N is the average cell count on days 3 and 4. The variance in mutations per line, generations per line, and sites analyzed per line were combined according to equation A1.19b from Lynch and Walsh (1998, p. 818). No significant differences were observed in the generation time between lines or over the course of the experiment (Supporting Information, Figure S1).
Sequencing, mutation identification, and validation
Genomic DNA was phenol-chloroform extracted. DNA libraries for Illumina sequencing were constructed using the Nextera DNA Sample Prep Kit with an insert size of 300 bp. Then 150-bp paired-end sequencing on an Illumina HiSeq 2500 Sequencing System was carried out at the Hubbard Center for Genome Studies, University of New Hampshire.
After trimming adaptor sequences, paired-end reads were mapped with the Burrows-Wheeler Aligner (BWA) 0.7.10 (Li and Durbin 2009) to the PomBase Data Version History data set version ASM294v2.25 of the reference genome (pombebase.org). We also independently mapped reads using NOVOCRAFT 2.08.1 to detect potential program-specific false-positive results in the BWA pipeline. Only uniquely mapped reads with quality scores > 20 were retained, and only sites in the reference genome with a minimum coverage of three forward and three reverse reads were considered.
To account for differences between the progenitor strain and the reference genome, the major allele across all samples was defined as the consensus, and mutations were called when both mappers indicated that at least 80% of reads in an individual supported an alternate state. Likewise, an indel or structural variant call required at least 30% read support from mpileup files, and indel breakpoints were also confirmed after realignment with either BreakDancer 1.1.2 (Chen et al. 2009) or Pindel 0.2.4w (Ye et al. 2009). These ad hoc filtering steps have proven to be robust in previous MA experiments (Ossowski et al. 2010; Sung et al. 2012b; Long et al. 2015); however, we undertook PCR and Sanger sequencing of the progenitor and mutant lines for 96 randomly selected substitutions and indels and a further 42 events of particular interest.
To detect large duplications, we parsed out the depth-of-coverage information for each site from the mpileup files in the BWA pipeline and constructed a coverage matrix for each chromosome of the 96 MA lines (96 × chromosome size matrix) and plotted the coverage distribution of each chromosome using mean coverages of 1-kb sliding windows with 500-bp steps [R script: rollapply(coverage.matrix,1000,mean,by = 500)]. Large duplications were then visually identified and confirmed by mean coverage of the candidate region being approximately two times that of 20-kb flanking regions. PCR/optical duplicate reads might potentially create artifact duplications, but the two large duplications were still observed once duplicate reads were removed using picard-tools-1.119.
Effective population size and A+T content
We calculated the average pairwise genetic distance at intergenic sites θπ from a worldwide collection of 32 natural isolates of S. pombe (Fawcett et al. 2014) using SNPs called with the approach discussed earlier (Table S2). The effective population size Ne of S. pombe was estimated by assuming that θπ = 2Neµ (considering a predominantly haploid life history), where µ is the mutation rate determined in this study. After excluding seven near-identical lines, the same SNP calls were used to calculate the folded spectrum of polymorphism present in this population.
The expected equilibrium A+T proportion of the genome is given by
where u is the A|T → G|C mutation rate (where | denotes “or”), and v is the G|C → A|T mutation rate (Lynch 2010). The population-scaled selection coefficient S favoring G|C over A|T is given by
where p is the observed A+T proportion of the genome, and .
Data availability
Paired-end reads of the 96 MA lines and the progenitor line have been deposited in NCBI SRA, with project accession number PRJNA295384 and study number SRP063607.
Results
Base-substitution mutation rate in S. pombe
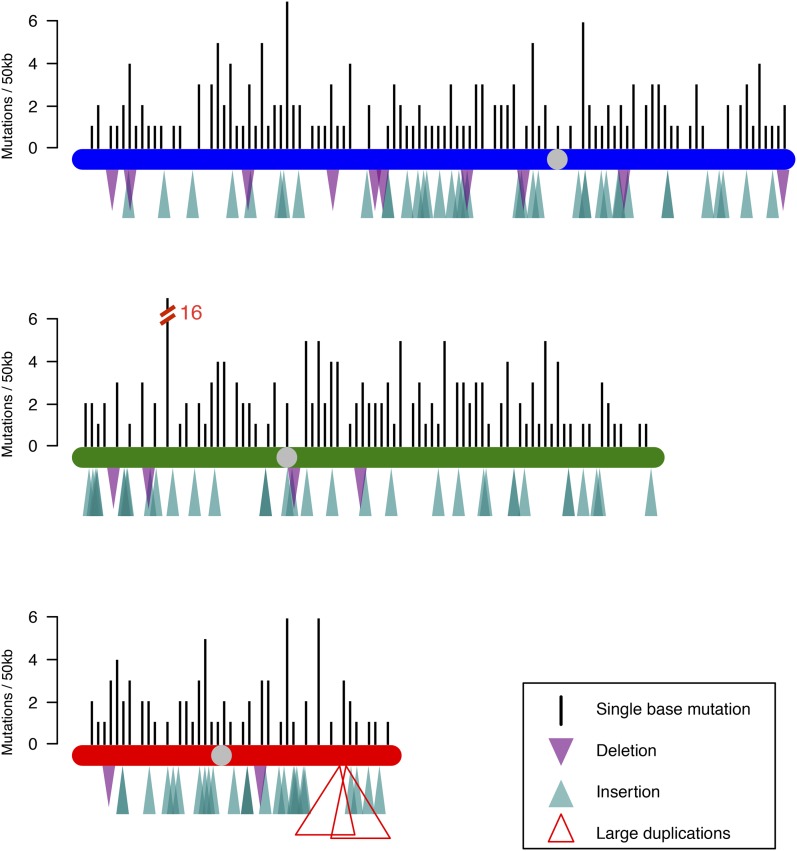
To observe the full spectrum of spontaneous mutations in the S. pombe genome, we sequenced 96 replicate haploid lines after ∼1716 ± 5.9 generations of MA (Figure S1). Lines were sequenced to an average coverage of 62× (ranging from 15 to 137 times, excluding mitochondrial coverage), and high-quality base calls were obtained for 83–96% of the reference genome (average 94.9%, or 12.0 Mb). In total, 422 single-base substitutions were identified (Figure 1), including three double mutations at consecutive bases and no mutations in the mitochondria (Table S1 and Table S2). This equates to a rate of 2.13 ± 0.11 × 10−10 base-substitution mutations per site per generation (however, consider the revised estimate presented later).
Figure 1.
The distribution of mutations across the three S. pombe chromosomes. Insertion-deletion mutations (indels) are depicted as triangles below the chromosomes. Gray circles indicate centromere positions.
Sanger sequencing confirmed 53 of 53 substitutions (including four mutations in flocculation genes, discussed later) and 41 of 41 indel events, suggesting a very low false-positive rate. Does this low false-positive rate come at the expense of a high false-negative rate? Following the approach of Keightley et al. (2014a), the false-negative rate was assessed by introducing 1000 random “synthetic” mutations into the SAM files of 10 randomly chosen MA lines and attempting to recover these events via the approach described earlier. In total, 900 of 1000 of these synthetic mutations were identified, with the remaining 100 events all being located at sites that failed our accepted filtering criteria for analyzed sites (either the site had a quality score < 20, fewer than three forward and three reverse reads spanned the site, or the synthetic mutation was called in fewer than 80% of the reads), implying that the false-negative rate at analyzed sites (∼94.9% of the genome) in our study is close to zero. We did not attempt to estimate the mutation rate or spectrum in the remaining 5.1% of the genome, which may or may not differ from the rest of the genome because of an excess of low-complexity and repetitive elements.
Over the course of the experiment, lines accrued an average of 4.4 single-base substitutions (range 0–10). Consistent with the expectation that selection had a minimal impact on the fate of new mutations during the experiment, the ratio of synonymous to nonsynonymous mutations in coding regions did not differ significantly from the expectation of randomly placed mutations (0.317 vs. 0.272; χ2 = 0.77, P = 0.38). Furthermore, there was no significant excess of events in any line or on any chromosome (χ2 = 3.6, P = 0.16) or in any functional context (relative to the lengths of sequence in each category; CDS: χ2 = 0.61, P = 0.43; nonsynonymous: χ2 = 2.9, P = 0.088; intronic: χ2 = 0.0, P = 0.94), with the exception of a slight deficiency of mutations in the UTRs of mRNAs (χ2 = 16.4, P = 5.1 × 10−5) (Table S2). Also, once three mutational hotspots were removed (discussed later), there was no excess of mutations in nonessential over essential genes (χ2 = 2.99, P = 0.083) (i.e., genes that show a viable vegetative cell population on deletion compared to those that fail to grow; www.pombase.org). Taken together, these observations suggest that the base-substitution mutation rate and spectrum measured are not grossly biased by selection purging particular mutations during the experiment.
Does DNA replication timing or RNA production influence the mutation rate in S. pombe? In this study, regions of the genome previously shown to undergo DNA replication late in the cell cycle did not suffer higher mutation rates than regions of early or average replication timing (two-sided t-test, P = 0.70) (Eshaghi et al. 2007). Nor did we observe any relationship between mutation rate and the mRNA expression level of genes (Marguerat et al. 2012).
Inadvertent selection on the flocculation pathway?
On average, across all lines we observed one mutation every 25 kb. One gene, however, gained 16 mutations across 11 lines (Figure 1). This represents a highly significant excess of mutations (bootstrapping, P < 0.00001), suggesting either a mutational hotspot or that mutations in this gene conferred a selective advantage during our experiment. This gene, SPBPJ4664.02, is an uncharacterized cell surface glycoprotein, unique to S. pombe, that is upregulated during flocculation (Kwon et al. 2012). Two other genes also showed a highly significant excess of mutations (four each; bootstrapping, P = 0.0001): Galactose-Specific Flocculation 2 (gsf2, SPCC1742.01) and Pombe Flocculin 2 (pfl2, SPAPB15E9.01c), both characterized members of the flocculation pathway (Matsuzawa et al. 2011; Kwon et al. 2012). In total, 20 of the 96 lines acquired mutations in flocculation-associated genes. Interestingly, however, not a single mutation was observed in gsf1, the major repressor of flocculation.
Flocculation is a phenomenon in which yeast cells aggregate in liquid culture for protection from environmental stresses. In S. pombe, the pathway plays a role in colony morphology and hyphal growth on solid medium. Considering the excess of mutations in this pathway, either this family of cell surface glycoproteins has an above-average mutation rate, or our protocol of picking isolated colonies inadvertently applied a strong selective pressure for mutations that reduce the propensity for cell adhesion.
Interestingly, the gene to undergo the most mutations, SPBPJ4664.02, has been duplicated in a single natural isolate (JB912, CBS2777). This line shows the maximum cell length and calcofluor staining (a marker of incomplete cell division and filamentous growth) among 160 lines that were recently phenotyped (Jeffares et al. 2015). However, no overt morphologic differences were observed between MA lines with and without mutations in this gene.
In comparison, only 1 of 867 mutations observed in Saccharomyces cerevisiae MA lines carried a mutation in the flocculation pathway (Zhu et al. 2014). We note, however, that the progenitor strain used in that study carried two mutations in the key flocculation gene flo9, suggesting that some aspects of this pathway may have already been altered prior to the experiment.
Considering the possibility that selection may have been acting on the flocculation pathway under our experimental regime, we excluded the 24 base substitutions in the three flocculation-related genes mentioned earlier from the estimated mutation rate and all subsequent analyses. The remaining 398 events equate to a revised base-substitution mutation rate of 2.00 ± 0.10 × 10−10 mutations per site per generation, or ∼1 mutation per genome every 400 generations.
Similarities and differences between fission and budding yeasts
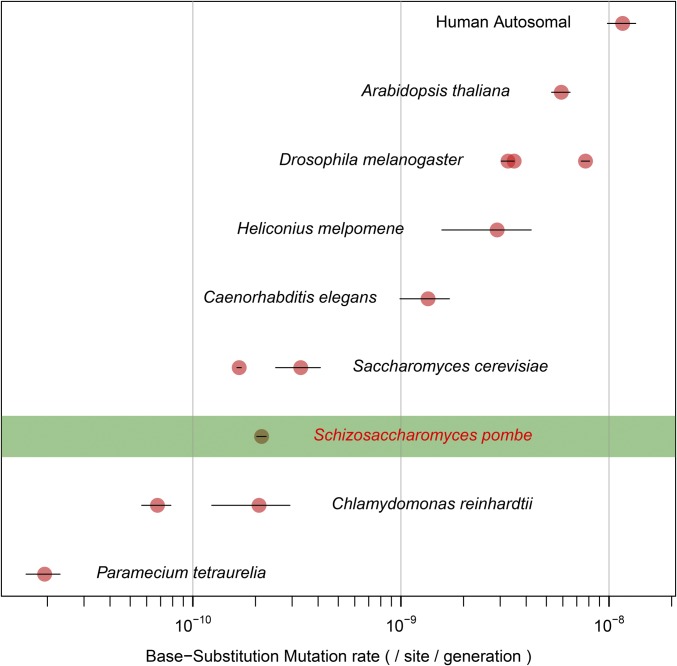
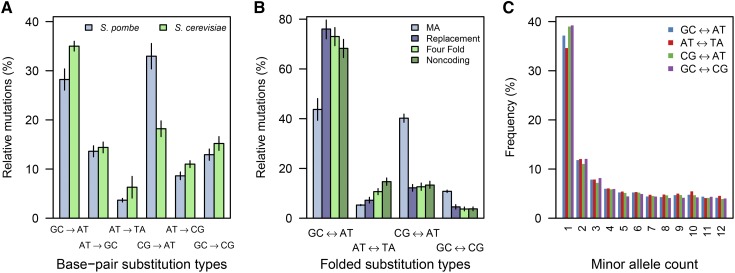
Our mutation-rate estimate for S. pombe falls between the two previous estimates for S. cerevisiae of 3.3 ± 0.8 × 10−10 (Lynch et al. 2008) and 1.67 ± 0.04 × 10−10 (Zhu et al. 2014) (Figure 2), suggesting a similar mutation rate in these two yeast species. We do, however, see a highly significant difference in the spectrum of mutations (χ2 = 27.0, P = 5.8 × 10−5) (Figure 3A). Whereas G:C → A:T transitions (where “:” denotes Watson-Crick base pairing) are the dominant mutation class in S. cerevisiae (Zhu et al. 2014), G:C → T:A transversions are more common in S. pombe. As a result, the transition:transversion ratio of 0.78 in S. pombe is smaller than the ratio of 0.95 observed in S. cerevisiae.
Figure 2.
The spontaneous mutation rate in S. pombe and eight other eukaryotes. Error bars represent SE (Lynch et al. 2008, 2010; Keightley et al. 2009; Ossowski et al. 2010; Denver et al. 2012; Ness et al. 2012; Sung et al. 2012b; Schrider et al. 2013; Keightley et al. 2014b; Zhu et al. 2014).
Figure 3.
The spectrum of base-substitution mutations differs between fission and budding yeasts. (A) The proportion of mutations during MA for each of the six possible transitions and transversions based on 398 mutations in S. pombe and 867 mutations in S. cerevisiae (Zhu et al. 2014). (B) The spectrum of mutations seen during S. pombe MA folded into the four possible unpolarized classes compared to the proportion of SNPs seen in the S. pombe population from 25 natural isolates (Fawcett et al. 2014). (C) The minor-allele count for SNPs in each class at all sites in the genome (see Figure S2 for replacement, fourfold, and intergenic sites). Error bars represent SE.
The S. cerevisiae and S. pombe genomes do not encode a characterized DNA methyltransferase (Becker et al. 2012), and neither shows evidence of DNA methylation (Capuano et al. 2014) [however, see Tang et al. (2012)]. It is therefore interesting that C → T transitions are significantly more common at CpG sites than in the non-CpG context in both S. pombe (χ2 = 70.12, P < 2 × 10−16) and S. cerevisiae (Zhu et al. 2014). This suggests that cytosines remain more labile to deamination when in a CpG context, regardless of the methylation status (Sung et al. 2015).
Under a number of assumptions, the level of polymorphism present in a population and the mutation rate allow one to estimate the effective population size Ne for a species. Here, using the polymorphism observed in a worldwide collection of S. pombe (Table S3) (Fawcett et al. 2014; Jeffares et al. 2015) and assuming a predominantly haploid life history, we estimate the Ne of S. pombe to be 12 million. In comparison, less polymorphism is observed at noncoding sites in S. cerevisiae (Schacherer et al. 2009), leading to an Ne estimate of 3.4 million.
Fission and budding yeasts display broadly similar levels of recombination, polymorphism, and mutation. Linkage disequilibrium (LD), however, is dramatically more extensive in S. pombe. Whereas LD decays to half the maximum value in fewer than 3 kb in S. cerevisiae (Liti et al. 2009), the same drop in S. pombe requires between 20 kb (Jeffares et al. 2015) and 50 kb (data not shown). A possible explanation is that S. pombe undergoes less frequent outcrossing in the wild. This does in fact appear to be the case. Using the approach of Tsai et al. (2008), which compares the observed level of LD in a population with that expected from experimentally observed recombination rates, we estimate that S. pombe undergoes mating with a genetically dissimilar individual on average every ∼800,000 generations, which is at least an order of magnitude less often than outcrossing estimates of once every 50,000 generation for S. cerevisiae (Ruderfer et al. 2006).
Mutation elevates the genomic A|T composition
Consistent with observations in other species, there is a strong bias in S. pombe for G|C sites (where the “|” denotes “or”) to mutate into A|T. After accounting for the incidence of each base in the genome, G|C sites mutate 2.7 times more frequently into A|T sites than the reverse (2.96 × 10−10 vs. 1.12 × 10−10 mutations per site per generation). This bias toward A|T is thought to be a universal property of the mutational process [however, see Long et al. (2015)], arguably driven largely by the deamination of cytosine and 5-methylcytosine and the oxidative conversion of guanine into 8-oxo-guanine (Lynch 2007, 2010; Hershberg and Petrov 2010).
Under a situation where this mutation bias is the sole determinant of the base composition of the genome, we would expect the S. pombe genome to be 73.3 ± 6.7% (SE) A+T, far higher than the observed 64%. This difference is highly significant at replacement sites (positions where any mutation will alter the amino acid sequence), which are on average only 56% A+T, and nonsignificant at fourfold degenerate, intronic, and intergenic sites, all of which are ∼69% A|T. This pattern is consistent with selective constraint on protein-coding sequence and less likely to be due to GC-biased gene conversion, which only operates on heterozygous sites, regardless of their functional annotation, that occur rarely in a predominantly haploid, non-outcrossing species such as S. pombe.
GC-biased gene conversion may play a larger role in shaping the AT percent of the genome in other species, including S. cerevisiae, that do show a strong departure from their expected mutation-driven A+T percent equilibrium at silent sites (sites that do not alter the amino acid sequence) (Lynch 2010). Assuming that the genome is at mutation-selection-drift equilibrium in S. pombe, we estimate that the population-scaled selection coefficient 2Nes favoring G|C variants over A|T equals 0.76 at replacement sites.
The spectrum of mutation does not reflect the spectrum of polymorphism
Mutation is the ultimate source of all genetic variation. Therefore, in the absence of selection and biased gene conversion, the spectrum of polymorphism should mirror the underlying mutational process. A discrepancy between these distributions might imply selection. The high level of sequence divergence between S. pombe and its closest relatives (Rhind et al. 2011) means that in general the ancestral state of a polymorphism is unknown. One is therefore unable to determine if a G:A polymorphism is the result of G → A or an A → G mutation. The six classes of substitutions thus become four.
The levels of polymorphism in each of these four classes differ dramatically from the expectation under the absence of selection (Figure 3B). Most notably, there are substantially more G:C ↔ A:T and fewer C:G ↔ A:T SNPs than expected at both coding and noncoding sites. The two classes of substitutions that occur less than expected are slightly skewed toward rare alleles (Figure 3C and Figure S2), suggesting that selection is playing a role.
Insertion bias among insertion-deletion mutations (indels)
The 119 short indels identified (Table S4) equate to an indel rate of 6.0 ± 0.6 × 10−11. This is 3.5 times less than the single-base substitution rate and 10 times greater than the indel rate reported in S. cerevisiae (Zhu et al. 2014), although differences in sequencing depth and mutation filtering criteria make this comparison difficult.
Unlike the deletion bias observed in S. cerevisiae (Zhu et al. 2014), indels in S. pombe are strongly biased toward insertions (102 events, +153 bp) over deletions (17 events, −34 bp) such that, on average, each line gained 1.2 bp after ∼1700 generations. Nearly all indels occurred in homopolymer runs, consistent with strand slippage during DNA replication (Table S4). However, most events between 4 and 10 nucleotides in length resulted in short duplications or the deletion of one copy of a short repeat, which is also consistent with inappropriate pairing of single-stranded overhangs during nonhomologous end joining of DNA double-strand breaks (Lynch 2007).
Two lines also experienced large duplications on chromosome III (46 kb and 104 kb) resulting in the duplication of 17 and 45 genes (Figure S3). The left and right breakpoints of the shorter event are within the highly similar repeat elements wtf19 and wtf21 (Bowen et al. 2003), and the breakpoints of the longer event are within highly similar LTR Tf2 elements. Despite being haploid, S. pombe undergoes an extended G2 phase of the cell cycle, presenting an opportunity for unequal crossover between sister chromatids, a likely mechanism for such large duplications. We note that large duplications are commonly observed segregating in the S. pombe population (Brown et al. 2011; Fawcett et al. 2014).
Discussion
Here we estimate that the single-base mutation rate in fission yeast is similar to that reported for budding yeast. These two species have a similar genome size (∼12.5 Mb), gene number (∼5,000), and effective population size (3.4 million vs. 12 million for S. cerevisiae and S. pombe, respectively), with the distinction of a predominantly haploid life history for S. pombe vs. diploid for S. cerevisiae. Combined with a number of studies in other species, our observations are in general agreement with the drift-barrier hypothesis of mutation-rate evolution. This postulates that selection operates primarily to increase replication fidelity, with the efficiency of selection eventually being thwarted when the mutation rate has been reduced to such a low level that further improvement is compromised by the power of random genetic drift (Sung et al. 2012a). However, the significant difference observed in the spectrum of base substitutions and indel lengths implies that there need not be strict maintenance of the underlying components of DNA replication fidelity and repair, provided that the summed consequences are the same. In other words, while the overall mutation rate is a central target of selection, the molecular spectrum provides some degrees of freedom by which the individual underlying components responsible for determining DNA fidelity may diverge among species. At some level, such mutational divergence should come as no surprise given the many other well-established differences in cell biology between S. pombe and S. cerevisiae (Frost et al. 2012).
A major motivation for estimating the empirical mutation spectrum in an MA setting is that it should reflect the spectrum of polymorphism at truly neutral sites in the genome. Our results suggest that no general class of sites in the S. pombe genome is completely free of selection. A similar difference between the spectrum of mutations and polymorphism also has been observed in Caenorhabditis (Denver et al. 2012) and Arabidopsis (Hagmann et al. 2015). This is consistent with either pervasive selection and/or a fluctuating mutation spectrum in the wild. We note that environmental stress has been shown to influence the mutation rate in Arabidopsis (Hagmann et al. 2015), and the presence of rare variants that alter the effectiveness of DNA replication or repair (mutator alleles) is likely to cause heterogeneity in the mutation rate within a population.
Summary
DNA replication and repair are molecular traits under the influence of selection. It has been proposed that selection will drive the mutation rate down to reduce the burden of deleterious mutations and that genetic drift will present a barrier below which selection will be ineffective. Here we observe that two yeast species with a similar effective population size, and hence influence of genetic drift, do in fact have very similar mutation rates. However, they differ dramatically in the frequency of each underlying mutation class, indicating that while selection has arrived at a similar overall mutation rate, the underlying mechanisms that maintain the DNA sequence have diverged.
Supplementary Material
Acknowledgments
We thank Jacki Heraud, Weiyi Li, and Matt Ackerman for helpful discussions and Peter Keightley for providing scripts to generate synthetic mutations. This work is supported by the Multidisciplinary University Research Initiative (award W911NF-09-1-0444 to M.L.) from the U.S. Army Research Office and the National Institutes of Health (grant R01GM101672 to M.L.).
Author contributions: A.F. and M.L. designed the research. A.F., H.L., and S.A. performed the research. W.S. and T.G.D. contributed new analytic tools and reagents. A.F., H.L., and M.N. analyzed the data. A.F. wrote the paper.
Footnotes
Communicating editor: D. M. Weinreich
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177329/-/DC1
Literature Cited
- Becker M., Müller S., Nellen W., Jurkowski T. P., Jeltsch A., et al. , 2012. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 40: 11648–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen N. J., Jordan I. K., Epstein J. a., Wood V., Levin H. L., 2003. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 13: 1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. R. A., Liti G., Rosa C., James S., Roberts I., et al. , 2011. A geographically diverse collection of Schizosaccharomyces pombe isolates shows limited phenotypic variation but extensive karyotypic diversity. G3 1: 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano F., Mülleder M., Kok R., Blom H. J., Ralser M., 2014. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal. Chem. 86: 3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wallis J. W., McLellan M. D., Larson D. E., Kalicki J. M., et al. , 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Wilhelm L. J., Howe D. K., Gafner K., Dolan P. C., et al. , 2012. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol. Evol. 4: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi M., Karuturi R. K. M., Li J., Chu Z., Liu E. T., et al. , 2007. Global profiling of DNA replication timing and efficiency reveals that efficient replication/firing occurs late during S-phase in S. pombe. PLoS One 2: e722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J. A., Iida T., Takuno S., Sugino R. P., Kado T., et al. , 2014. Population genomics of the fission yeast Schizosaccharomyces pombe. PLoS One 9: e104241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A., Elgort M. G., Brandman O., Ives C., Collins S. R., et al. , 2012. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell 149: 1339–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann J., Becker C., Müller J., Stegle O., Meyer R. C., et al. , 2015. Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLoS Genet. 11: e1004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A., 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6: e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares D. C., Rallis C., Rieux A., Speed D., Převorovský M., et al. , 2015. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nat. Genet. 47: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Ness R. W., Halligan D. L., Haddrill P. R., 2014a Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics 196: 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Pinharanda A., Ness R. W., Simpson F., Dasmahapatra K. K., et al. , 2014b Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol. Biol. Evol. 32: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon E.-J. G., Laderoute A., Chatfield-Reed K., Vachon L., Karagiannis J., et al. , 2012. Deciphering the transcriptional-regulatory network of flocculation in Schizosaccharomyces pombe. PLoS Genet. 8: e1003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14): 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Kucukyildirim S., Sung W., Williams E., Lee H., et al. , 2015. Background mutational features of the radiation-resistant bacterium Deinococcus radiodurans. Mol. Biol. Evol. 32(9): 2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2007. The Origins of Genome Architecture. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Lynch M., 2010. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 107: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., et al. , 2012. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa T., Morita T., Tanaka N., Tohda H., Takegawa K., 2011. Identification of a galactose-specific flocculin essential for non-sexual flocculation and filamentous growth in Schizosaccharomyces pombe. Mol. Microbiol. 82: 1531–1544. [DOI] [PubMed] [Google Scholar]
- Ness R. W., Morgan A. D., Colegrave N., Keightley P. D., 2012. Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Chen Z., Yassour M., Thompson D. a., Haas B. J., et al. , 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer D. M., Pratt S. C., Seidel H. S., Kruglyak L., 2006. Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38: 1077–1081. [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. a., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Houle D., Lynch M., Hahn M. W., 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194: 937–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Gout J.-F., Miller S. F., Williams E., et al. , 2015. Asymmetric context-dependent mutation patterns revealed through mutation-accumulation experiments. Mol. Biol. Evol. 32(7): 1672–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., Lynch M., 2012a Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Tucker A. E., Doak T. G., Choi E., Thomas W. K., et al. , 2012b Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc. Natl. Acad. Sci. USA 109: 19339–19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Gao X.-D., Wang Y., Yuan B.-F., Feng Y.-Q., 2012. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal. Chem. 84: 7249–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai I. J., Bensasson D., Burt A., Koufopanou V., 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl. Acad. Sci. USA 105: 4957–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. O., Siegal M. L., Hall D. W., Petrov D. A., 2014. Precise estimates of mutation rate and spectrum in yeast. Proc. Natl. Acad. Sci. USA 111: E2310–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Paired-end reads of the 96 MA lines and the progenitor line have been deposited in NCBI SRA, with project accession number PRJNA295384 and study number SRP063607.